Abstract
Analysis of methicillin-resistant Staphylococcus aureus (MRSA) characterized as USA300 by pulsed-field gel electrophoresis identified two distinct clones. One was similar to community-associated USA300 MRSA (ST8-IVa, t008, and Panton-Valentine leukocidin positive). The second (ST8-IVa, t024, and PVL negative) had different molecular characteristics and epidemiology, suggesting independent evolution. We recommend spa typing and/or PCR to discriminate between the two clones.
The methicillin-resistant Staphylococcus aureus (MRSA) clone USA300, having multilocus sequence type (MLST) ST8 and staphylococcal protein A (spa) type 008 and carrying staphylococcal cassette chromosome (SCCmec) IVa, has disseminated in the United States, as well as to other parts of the world (16, 26, 27). USA300 carries the luk-PV genes encoding Panton-Valentine leukocidin (PVL), has been identified in a variety of community populations, and has been associated with skin and soft tissue infections (SSTI), as well as more severe infections, such as sepsis, pneumonia, and necrotizing fasciitis (7, 22, 28).
The identification of USA300 isolates is primarily based on pulsed-field gel electrophoresis (PFGE) (10). Other genetic markers have also been suggested for identification of USA300 isolates, including (i) the arcA gene of the arginine catabolic mobile element (ACME) (3, 8, 26, 29), (ii) sequencing of the direct repeat unit (dru) region (9), and (iii) different USA300-specific multiplex PCRs targeting luk-PV and a “signature” six-AT-repeat sequence within the conserved hypothetical gene SACOL0058 (2).
In Denmark, MRSA isolates have been consecutively typed by PFGE since 1999, with the addition of sequence-based methods, such as MLST and spa typing, on selected isolates since 2001 (6). This process identified some of the first USA300 isolates in Europe but, surprisingly, also identified isolates with USA300 PFGE banding patterns but a different spa type (1, 15, 16). In this study, we investigated the epidemiology and genetic diversity of these isolates and USA300 and USA500 reference strains (20) using PFGE (23), spa typing (11), MLST (5), SCCmec typing (21, 24), dru typing (9), ACME (26), the six-AT signature sequence (2), detection of luk-PV (4), and antimicrobial susceptibility testing (Neo-Sensitabs), as well as microarray analysis (18, 19).
Clinical and epidemiological information was obtained consecutively (17). Infections were categorized into four different groups: import, hospital associated, community associated, and health care associated with community onset (14).
Where appropriate, statistical significance (P < 0.05) was assessed using the Mann-Whitney test or Fischer's exact test.
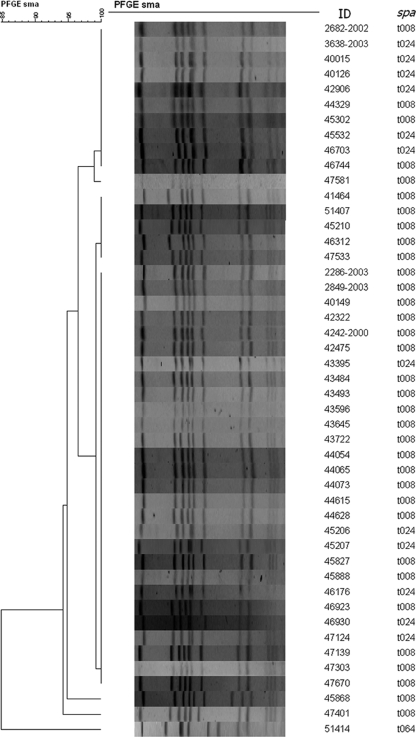
Between 1999 and 2006, 80 MRSA isolates from Denmark had USA300 PFGE profiles (50 representative PFGE profiles are shown in Fig. 1). However, by spa typing, two different spa types, t008 (11-19-12-21-17-34-24-34-22-25 [n = 38]) and the single repeat variant t024 (11-12-21-17-34-24-34-22-25 [n = 42]), both belonging to ST8, were identified (the extra repeat [19] is shown in boldface). All isolates typed as SCCmec IVa. However, significant genetic, epidemiological, and clinical differences were found, as shown in Tables 1 and 2. Patients infected with t008 isolates were significantly younger than patients infected with t024 isolates (P < 0.01). Patients acquired t008 MRSA in the community (50%) or through travel abroad (21%), and infections were predominantly SSTIs (94%). In contrast, the majority of patients with t024 MRSA were either hospitalized (26%) or had health care-associated risk factors (38%), and they presented with SSTIs (64%) but, also, a larger variety of infections, including fatal respiratory tract infections (11%) and operation- and procedure-related infections (11%). Remarkably, no t024 MRSA cases were imported.
FIG. 1.
Unweighted pair group method with arithmetic mean (UPGMA) dendrogram of SmaI PFGE profiles from Danish MRSA isolates (2000 to 2005) constructed by the use of Dice determinations (optimization, 1.0%; tolerance, 2.1 to 3.1%). The reference strains are USA300 and USA500. Note that t008 and t024 isolates do not cluster separately by PFGE.
TABLE 1.
Bacteriological, epidemiological, and clinical data obtained for patients infected or colonized with either t008 or t024 in Denmark (1999 to 2005)
| Characteristicb | t008 (n = 38) | t024 (n = 42) |
|---|---|---|
| Typing | ||
| ST | 8 | 8 |
| SCCmec | IVa | IVa |
| luk-PV | + | − |
| ACME | + | +/− |
| dru type | 7d, 7e, 9g,a 9i, 10a | 10a |
| Six-AT repeats | + | +/− |
| Resistance (%) | ||
| Kanamycin | 74 | 12 |
| Tetracycline | 8 | 5 |
| Erythromycin | 82 | 88 |
| Clindamycin | 13 | 86 |
| Norfloxacin | 42 | 55 |
| Fusidic acid | 5 | 12 |
| Median ages of patients (yrs)* | 31 | 72 |
| Sources of isolates (%) | ||
| Community associated# | 50 | 2 |
| Health care associated with community onset | 18 | 38 |
| Hospital associated# | 5 | 26 |
| Import# | 21 | 0 |
| Active surveillance culture# | 5 | 33 |
| Locations of infections (%) | n = 36 | n = 28 |
| Skin and soft tissue# | 94 | 64 |
| Respiratory tract | 3 | 11 |
| Deep seated | 3 | 7 |
| Operation and procedure related | 11 | |
| Urinary tract | 3.5 | |
| Other | 3.5 |
Boldface indicates the predominant dru type.
*, #, statistically significant difference (P < 0.05) between t008 and t024 isolates with either Mann-Whitney U or Fischer's exact test, respectively.
TABLE 2.
Summary comparison of MGEs between isolates as detected by multistrain microarray
| Strain | PFGE pattern | spa type | Variant; relevant gene(s) carried by indicated MGEa |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteriophage |
SaPI1 (COL) | SCCmec IV (MW2) | Plasmid class |
Tn554 | Transposon | ||||||
| φSa1 | φSa2 (MW2) | φSa3 | I (COL) | II (MW2) | |||||||
| 2849-2003 | USA300 | t008 | v1; luk-PV | (MRSA252) chp, scn, sak | v1; sek, seq | v1 | v1; bla, cad | ||||
| 44073 | USA300 | t008 | v1; luk-PV | (MRSA252) chp, scn, sak | v1; sek, seq | v1 | v1; bla, cad | ||||
| 45532 | USA300 | t024 | v2 | v2 | v2 | v2; bla, cad | ermA, spc | ||||
| 46703 | USA300 | t024 | v2 | v2 | v2 | v2; bla, cad | ermA, spc | ||||
| USA300 | USA300 reference | t008 | v1; luk-PV | (MRSA252) chp, scn, sak | v1; sek, seq | v1 | tet | v1; bla, cad | |||
| USA500 | USA500 reference | t064 | (Mu50) | v3 | (MW2) scn, sak | v3; sek, seq, seb, ear | v3 | v3; bla, cad | (Mu50) aacA | ||
MGEs have been clustered into families according to the method of Lindsay and Holden (18), and the sequenced strain with the most closely matched MGE is given in parentheses (i.e., MRSA252, Mu50, COL, and MW2) (17). Note that the closest match is never identical, and there is substantial variation in MGEs between most S. aureus isolates. “v” indicates a variant compared to the other strains; strains with the same variant have the same v number. aacA, aminoglycoside resistance; bla, β-lactamase resistance; chp, chemotaxis inhibitory protein; cad, cadmium resistance; ear, putative β-lacatamase like protein; ermA, erythromycin resistance; scn, staphylococcal complement inhibitor; sak, staphylokinase; seb, enterotoxin B; sek, enterotoxin K; seq, enterotoxin Q; spc, spectinomycin resistance; tet, tetracycline resistance.
In contrast to most t008 isolates, t024 isolates were often constitutively resistant to clindamycin and susceptible to kanamycin (Tables 1 and 2). Furthermore, t024 isolates did not carry the luk-PV genes, exhibited different dru types, and inconsistently carried the ACME-related arcA gene and the conserved hypothetical gene SACOL0058 containing the six-AT-repeat sequence characteristic of typical t008 USA300 isolates (Table 1). Whole-genome microarray analysis of the CDC USA300 and USA500 reference strains and four clinical isolates, including two t008 (2849-2003 and 44073) and two t024 (45532 and 46703) isolates, revealed additional genetic differences. All isolates, including the USA500 reference strain, had a typical ST8 gene distribution pattern, including the genomic islands GIα and GIβ. However, a marked difference in the carriage of mobile genetic elements (MGEs) was observed, including resistance and putative virulence genes (Table 2). The t008 clinical isolates were very closely related by bacteriophage, Staphylococcus aureus pathogenicity island (SaPI), plasmid, transposon, and SCCmec element content and were virtually identical to the USA300 reference strain, except that the latter carried an additional tetracycline resistance cassette. In contrast, the t024 isolates, while very closely related in their core genomes, were distinct from the t008 isolates due to dissimilar bacteriophages, SaPI's, SCCmec elements, plasmids, and transposons (Table 2). These differences represent multiple horizontal gene transfer and/or rearrangement events, suggesting a substantial difference between t024 and t008 isolates. The USA500 reference isolate was distinct from the t008 and t024 isolates by PFGE profile (Fig. 1), spa type (t064), and MGE profile, especially regarding phage, SaPI, and transposon carriage.
These results suggest that two different clones with a typical USA300 PFGE pattern and SCCmec IVa are common in Denmark. Half of the isolates seem to be “classic” USA300 both epidemiologically and genetically (spa type t008, luk-PV positive, ACME positive, and with six or more AT repeats within SACOL0058), whereas the other half belong to a genetically and epidemiologically different clone principally characterized as spa type t024. This latter clone has recently been shown to be inadequately detected by the BD GeneOhm MRSA kit compared to the detection of t008 isolates, emphasizing sequence differences in the primer binding sites of the SCCmec right extremity junction (1). At present, t024 comprises 1.01% of the spa sequences deposited in the Ridom database (www.Ridom.de), originating from most of Europe, as well as the United States and Canada. However, not all t024 isolates are identical either, as a Dutch isolate with luk-PV has been identified (12).
The observations in this study suggest that reports of USA300 could include isolates with important genetic variations if PFGE, MLST, or SCCmec typing is the method used, as supported by findings of ACME- and Panton-Valentine leukocidin-negative USA300 isolates (10, 13).
Already, the need for more discriminatory methods has been debated in areas of high USA300 prevalence (25). The results of this study suggest that spa type t008 may identify the ST8 lineage related to community-associated MRSA infections more accurately than PFGE. A marker such as luk-PV is generally enough to identify “classic” USA300 isolates. Microarray analysis has revealed a range of other genes that could also be considered to discriminate isolates (Table 2). The results of this study clearly suggest that USA300 MRSA identified solely by PFGE should be confirmed by at least one PCR analysis, which could be for luk-PV, or a sequence-based typing method, such as spa typing.
Microarray data accession numbers.
Fully annotated microarray data have been deposited in BμG@Sbase (accession number E-BUGS-77; http://bugs.sgul.ac.uk/E-BUGS-77) and also ArrayExpress (accession number E-BUGS-77).
Acknowledgments
The Wellcome Trust supports the Bacterial Microarray Group at St. George's (www.bugs.sgul.ac.uk).
We thank Denise Waldron, Adam Witney, and Phil Butcher for additional assistance with microarrays.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Bartels, M. D., K. Boye, A. R. Larsen, R. L. Skov, and H. Westh. 2007. Rapid increase of genetically diverse methicillin-resistant Staphylococcus aureus, Copenhagen, Denmark. Emerg. Infect. Dis. 13:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnstetter, K. K., D. J. Wolter, F. C. Tenover, L. K. McDougal, and R. V. Goering. 2007. Rapid multiplex PCR assay for identification of USA300 community-associated methicillin-resistant Staphylococcus aureus isolates. J. Clin. Microbiol. 45:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 4.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 5.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 43:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis, J. S., M. C. Doherty, U. Lopatin, C. P. Johnston, G. Sinha, T. Ross, M. Cai, N. N. Hansel, T. Perl, J. R. Ticehurst, K. Carroll, D. L. Thomas, E. Nuermberger, and J. G. Bartlett. 2005. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin. Infect. Dis. 40:100-107. [DOI] [PubMed] [Google Scholar]
- 8.Goering, R. V., L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, D. J. Wolter, and F. C. Tenover. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 45:1981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goering, R. V., D. Morrison, Z. Al Doori, G. F. S. Edwards, and C. G. Gemmell. 2008. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 14:964-969. [DOI] [PubMed] [Google Scholar]
- 10.Goering, R. V., R. M. Shawar, N. E. Scangarella, F. P. O'Hara, H. Amrine-Madsen, J. M. West, M. Dalessandro, J. A. Becker, S. L. Walsh, L. A. Miller, S. F. van Horn, E. S. Thomas, and M. E. Twynholm. 2008. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J. Clin. Microbiol. 46:2842-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huijsdens, X. W., M. Janssen, N. H. Renders, A. Leenders, P. van Wijk, M. G. Santen Verheuvel, J. K. van Driel, and G. Morroy. 2008. Methicillin-resistant Staphylococcus aureus in a beauty salon, the Netherlands. Emerg. Infect. Dis. 14:1797-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy, A. D., M. Otto, K. R. Braughton, A. R. Whitney, L. Chen, B. Mathema, J. R. Mediavilla, K. A. Byrne, L. D. Parkins, F. C. Tenover, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. USA 29. 105:1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klevens, R. M., M. A. Morrison, S. K. Fridkin, A. Reingold, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, G. Fosheim, L. K. McDougal, and F. C. Tenover. 2006. Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg. Infect. Diseas. 12:1991-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen, A., R. Goering, M. Stegger, K. Boye, M. Sorum, H. Westh, and R. Skov. 2007. Characterisation of an MRSA clone in Denmark similar to USA300 in PFGE pattern but with different properties: a potential pitfall when identifying USA300 isolates. Int. J. Antimicrob. Agents 29:S441. [Google Scholar]
- 16.Larsen, A., M. Stegger, R. Goering, M. Sorum, and R. Skov. 2007. Emergence and dissemination of the methicillin resistant Staphylococcus aureus USA300 clone in Denmark (2000-2005). Euro. Surveill. 12:22-24. [Google Scholar]
- 17.Larsen, A. R., M. Stegger, S. Bocher, M. Sorum, D. L. Monnet, and R. L. Skov. 2009. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999 to 2006. J. Clin. Microbiol. 47:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindsay, J. A., and M. T. Holden. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6:186-201. [DOI] [PubMed] [Google Scholar]
- 19.Lindsay, J. A., C. E. Moore, N. P. Day, S. J. Peacock, A. A. Witney, R. A. Stabler, S. E. Husain, P. D. Butcher, and J. Hinds. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, L. G., F. Perdreau-Remington, G. Rieg, S. Mehdi, J. Perlroth, A. S. Bayer, A. W. Tang, T. O. Phung, and B. Spellberg. 2005. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 352:1445-1453. [DOI] [PubMed] [Google Scholar]
- 23.Murchan, S., M. E. Kaufmann, A. Deplano, R. De Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel, M., K. B. Waites, C. J. Hoesley, A. M. Stamm, K. C. Canupp, and S. A. Moser. 2008. Emergence of USA300 MRSA in a tertiary medical centre: implications for epidemiological studies. J. Hosp. Infect. 68:208-213. [DOI] [PubMed] [Google Scholar]
- 26.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tietz, A., R. Frei, and A. F. Widmer. 2005. Transatlantic spread of the USA300 clone of MRSA. N. Engl. J. Med. 353:532-533. [DOI] [PubMed] [Google Scholar]
- 28.Young, L. M., and C. S. Price. 2008. Community-acquired methicillin-resistant Staphylococcus aureus emerging as an important cause of necrotizing fasciitis. Surg. Infect. 9:469-474. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2008. Novel multiplex PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and Panton-Valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 46:1118-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]