Abstract
The maturation of immature chondrocytes to hypertrophic chondrocytes is regulated by parathyroid hormone-related peptide (PTHrP). We demonstrate that PTHrP or forskolin administration can block induction of collagen X-luciferase by exogenous Runx2, MEF2, and Smad1 in transfected chondrocytes. We have found that PTHrP/forskolin administration represses the transcriptional activity of MEF2 and that forced expression of MEF2-VP16 can restore expression of the collagen X reporter in chondrocytes treated with these agents. PTHrP/forskolin induces dephosphorylation of histone deacetylase 4 (HDAC4) phospho-S246, which decreases interaction of HDAC4 with cytoplasmic 14-3-3 proteins and promotes nuclear translocation of HDAC4 and repression of MEF2 transcriptional activity. We have found that forskolin increases the activity of an HDAC4 phospho-S246 phosphatase and that forskolin-induced nuclear translocation of HDAC4 was reversed by the protein phosphatase 2A (PP2A) antagonist, okadaic acid. Finally, we demonstrate that knockdown of PP2A inhibits forskolin-induced nuclear translocation of HDAC4 and attenuates the ability of this signaling molecule to repress collagen X expression in chondrocytes, indicating that PP2A is critical for PTHrP-mediated regulation of chondrocyte hypertrophy.
Chondrocyte maturation in the growth plate is regulated by parathyroid hormone (PTH)-related peptide (PTHrP) signals (14, 16, 29, 33). PTHrP signals are thought to be mediated via the PTH/PTHrP receptor, a G protein-coupled receptor which can signal via both Gs, which activates adenylyl cyclase (AC)/protein kinase A (PKA), and the Gq/G11 family, which activates phospholipase C/PKC (10). Several lines of evidence indicate that signaling via the AC/PKA pathway is sufficient for this receptor to slow the rate of chondrocyte maturation (10). Runx2/3 (34) and MEF2C/D transcription factors (2) also play a critical role in modulating chondrocyte hypertrophy. MEF2 function is repressed by class II histone deacetylases (HDACs), one of which (HDAC4) is known to block both precocious and ectopic chondrocyte hypertrophy (30). HDAC4 is known to be phosphorylated at three conserved serines, whose phosphorylation promotes the association of these proteins with 14-3-3 proteins in the cytoplasm (9, 20), which is thought to block both the nuclear localization of these HDACs and consequent repression of MEF2 transcriptional activity. In this work, we demonstrate that PTHrP signals block chondrocyte hypertrophy by promoting dephosphorylation of HDAC4 phospho-S246 by protein phosphatase 2A (PP2A), thereby inducing nuclear translocation of this HDAC and consequent repression of MEF2 activity.
MATERIALS AND METHODS
Plasmids and antibodies.
The following plasmids were used: −4kb ColX-luciferase (31); 6x(Runx2)-luciferase (8); 30x(SBE)-luciferase (12); CMV-Runx2 (17); CMV-Smad1 and CMV-Smad4 (12, 36); pcDNA-MEF2C-Flag, 3XMEF2-luciferase, Gal4-HDAC4(2-740), Gal4-HDAC4(2-740) S246A, Gal4-HDAC4(2-740) 3SA, 14-3-3-VP16, MEF2C-VP16, GFP-HDAC4, HDAC4-Flag, HDAC4-S246-Flag, and HDAC4-3SA-Flag (3); 14-3-3 epsilon-HA (Addgene; deposited by Michael Yaffe); SIK1-CA (5); and CAMKI-CA (20). MEF2C-HA was generated by PCR-cloning mouse Mef2C into pcDNA3.1+; a hemagglutinin (HA) tag was inserted in the C terminus, in front of the Mef2C stop codon. The following antibodies were used: anti-Flag (Sigma; F3165); anti-HDAC4 (Abcam; ab12171); anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) (Chemicon; MAB374); anti-β-actin (Abcam; ab6276); anti-phospho-S246, -S467, and -S632 HDAC4 (6); anti-HA (Santa Cruz; sc-805), anti-PP2A (R&D Systems; AF1653); and antitubulin (Sigma; T9822). All secondary antibodies were from Jackson Immunoresearch. Flag agarose beads used for immunoprecipitation (IP) were purchased from Sigma (A2220), and HA beads were purchased from Covance (AFC-101P).
Cell culture.
All cells were maintained at 37°C in the presence of 5% CO2. Upper sternal chondrocytes (USCs) were isolated from the cephalic core region of day-18 chicken embryo sterna as previously described (15). Cells were cultured for 7 to 10 days in Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (HyClone), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) and plated for transfections. Cells were treated with 25 μM forskolin (Calbiochem), PTHrP [(Tyr36)-pTH-related protein 1 to 36; Bachem], and/or okadaic acid (VWR) at concentrations specified. Proliferating mouse limb bud-derived cells, MLB13MYC clone 14 (MLB14) (28), were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). To induce differentiation, cells were plated at high density and switched to DMEM supplemented with 1% heat-inactivated serum (Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin (Invitrogen), and 100 ng/ml BMP2 (a generous gift from Walter Sebald, Universität Würzburg). Metatarsals were isolated from 15.5-day postcoitum (dpc) MEF2-lacZ reporter mice (24) and cultured as described in reference 11. Beta-galactosidase staining was performed using a beta-galactosidase detection kit from Millipore. All animal studies were approved by the Harvard Medical Area Standing Committee on Animals.
Luciferase reporter assay and HDAC4 localization assay.
USCs were plated at 1.5 × 105 cells/well into six-well plates and transfected with the indicated expression plasmids using Superfect transfection reagent (Qiagen) according to the manufacturer's protocol. To control for transfection efficiency for renilla luciferase reporters, cells were cotransfected with a simian virus 40 (SV40)-driven firefly luciferase, and conversely, for firefly luciferase reporters, cells were cotransfected with SV40-driven renilla luciferase. Cells were treated with 25 μM forskolin or 3 × 10−7 M PTHrP for 48 h before harvesting, with media being changed daily. A total of 50 nM okadaic acid was added to the cultures 24 h before harvesting, where indicated. Cells were lysed 48 h after transfection, and luciferase reporter activity was determined using the dual-luciferase reporter assay system (Promega). For HDAC4 localization experiments, USCs were seeded at 2 × 105 cells/well into two-well glass slides and transfected with green fluorescent protein (GFP)-HDAC4 or HDAC4-Flag plasmids using either Superfect (Qiagen) or Fugene 6 (Roche) transfection reagent. Cells were treated with 25 μM forskolin or 10−6 M PTHrP for 4 h before fixing or pretreated with 50 nM okadaic acid for 24 h, where indicated. USCs were fixed 48 h after transfection in 4% paraformaldehyde for 15 min at room temperature and counterstained with DAPI (4′,6-diamidino-2-phenylindole). HDAC4-Flag localization was visualized using standard immunofluorescence techniques. The number of cells with predominantly nuclearly localized GFP-HDAC4 or HDAC4-Flag was counted.
IP, co-IP, and Western blotting.
For HDAC4-Flag co-IP experiments, 2 μg of each plasmid was transfected into USCs using Superfect reagent. Cells were treated with 25 μM forskolin or 10−6 M PTHrP for 1 h or 4 h before harvesting, as indicated. Whole-cell extracts were prepared from USCs 48 h after transfection using IP buffer (50 mM Tris [pH 7.8], 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 0.5% NP-40, and 1 mM phenylmethylsulfonyl fluoride, supplemented with protease and phosphatase inhibitor tablets [Roche]). The extracts were briefly sonicated and centrifuged for 10 min at 10,000 × g at 4°C. Supernatants were incubated with either Flag agarose beads or HA agarose beads overnight at 4°C on a rotating wheel. Finally, the beads were washed three times in the IP buffer, and proteins were denatured for 5 min at 95°C and separated by gel electophoresis. Western blot analysis was carried out under standard conditions with antibodies listed above.
siRNA experiments, RNA isolation, RT-PCR, and qPCR.
All small interfering RNA (siRNA) oligonucleotides used in these studies were purchased from Ambion and are as follows: HDAC4 (s101930), PP2CA (s72066), PP2CB (s72070), PP1CA (s72053), PP1CB (s72055), PP1CC (s72058), GAPDH (4390849), and control (4390843). siRNA oligonucleotides were transfected into MLB14 cells using X-tremeGENE transfection reagent (Roche), according to the manufacturer's protocol. Cells were treated with 25 μM forskolin (in dimethyl sulfoxide [DMSO]) or DMSO control, with the media being changed daily. Cellular lysates were collected 48 h or 96 h after transfection, and total cellular RNA was purified using RNeasy Plus mini-RNA isolation kit (Qiagen). Reverse transcription (RT)-PCR was performed using standard conditions (35). Quantitative PCR (qPCR) analysis was conducted on a 7900HT real-time PCR machine (Applied Biosystems) using SYBR premix Ex Taq reagent (Takara). Primers used in qPCR include the following: Col10a1 (TGCTGAACGGTACCAAACG [forward] and TGCCTTGTTCTCCTCTTACTGG [reverse]), Hdac4 (CAACAGACAGAAACTGGACAGTAAG [forward] and ACCTCATTCCATATGGTGTCG [reverse]), Tbp (GGGAGCTGTGATGTGAAGTTC [forward] and TAAGGAGAACAATTCTGGGTTTG [reverse]), Mef2c (CTCCACCTCGGCTCTGTAAC [forward] and CAGCTGCTCAAGCTGTCAAC [reverse]), Runx2 (CCCAGCCACCTTTACCTACA [forward] and TATGGAGTGCTGCTGGTCTG [reverse]), Mef2d (TGGCAACACCAAGTTTACTCAG [forward] and AGGTGAACTGAAGGCTGGTAAG [reverse]), and Ihh (CATCTTCAAGGACGAGGAGAAC [forward] and AGTGATGGCCATCTTCATCC [reverse]). Cotransfection of the GFP-HDAC4 expression plasmid and siRNA oligonucleotides was performed with X-tremeGENE transfection reagent according to the manufacturer's protocol. Cells were treated with 25 μM forskolin (in DMSO) or DMSO control for 48 h, fixed with 4% paraformaldehyde for 15 min at room temperature and counterstained with DAPI. The number of cells with predominantly nuclearly localized GFP-HDAC4 was counted, and the percentage of cells with nuclear GFP-HDAC4 was calculated.
RESULTS
PTHrP and forskolin block the ability of ectopic Runx2/Smad1/MEF2C to induce expression of a collagen X luciferase reporter in mature chondrocytes.
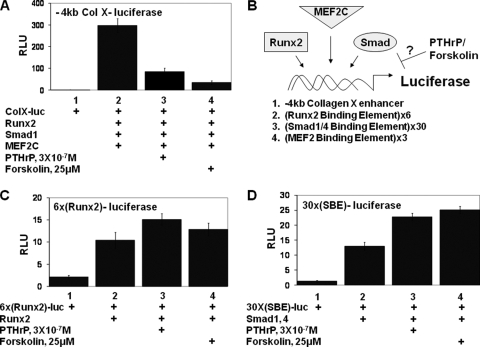
We have previously found that PTHrP signals can block the ability of retrovirally encoded Runx2 to induce collagen X expression in explants of presomitic mesoderm (27). To further explore how PTHrP signals repress expression of chondrocyte hypertrophy markers, we evaluated whether PTHrP could similarly repress expression of a collagen X reporter in transfected chicken embryo USCs. We cotransfected USCs with a luciferase reporter driven by 4 kb of sequences upstream of the chicken collagen X promoter (−4kb ColX-luciferase) (31) together with Runx2, MEF2C, and Smad1 expression vectors. Cotransfected Runx2, MEF2C, and Smad1 robustly activated expression of the −4kb ColX-luciferase reporter in USCs (Fig. 1A, compare lanes 1 and 2). We evaluated whether administration of PTHrP to such cultures would blunt the induction of the collagen X reporter in response to cotransfected Runx2, MEF2C, and Smad1. Addition of 3 × 10−7 M PTHrP led to an approximately 70% decrease in the expression of the −4kb ColX reporter (Fig. 1A, lane 3). To address whether elevation of cAMP levels would similarly block the ability of cotransfected Runx2/MEF2C/Smad1 to induce expression of the −4kb ColX reporter, we treated USCs with 25 μM forskolin. Forskolin treatment led to a precipitous decline in the expression of the −4kb ColX reporter (Fig. 1A, lane 4), suggesting that elevated cyclic AMP (cAMP) levels mimic the effects of PTHrP in this culture system.
FIG. 1.
PTHrP signaling/forskolin blocks expression of a collagen X reporter in chondrocytes without repressing the transcriptional activity of either Runx2 or Smad1/4. (A) USCs were cotransfected with the −4kb ColX-renilla luciferase reporter and an SV40-firefly luciferase reporter plus Runx2, MEF2C, and Smad1 expression vehicles in either the absence or presence of PTHrP or forskolin as indicated. As shown in this and subsequent figures, renilla luciferase units were normalized to the expression of cotransfected SV40-firefly luciferase to obtain relative luciferase units (RLU). (B) The PTHrP/forskolin transcriptional target in the collagen X enhancer was determined by evaluating whether these signals block the expression of luciferase reporters driven by either the intact collagen X enhancer, 6 copies of a Runx2 binding element, 30 copies of a Smad1/4 binding element, or 3 copies of a MEF2 binding element. (C) USCs were transfected with a reporter driven by six copies of a Runx2 binding element [6x(Runx2)-firefly luciferase] and an SV40-renilla luciferase reporter plus a Runx2 expression vehicle in either the absence or presence of PTHrP or forskolin as indicated. (D) USCs were transfected with a reporter driven by 30 copies of a Smad1/4 binding element [30X(SBE)-renilla luciferase] and an SV40-firefly luciferase reporter plus Smad1/4 expression vehicles in either the absence or presence of PTHrP or forskolin as indicated.
PTHrP signaling does not repress either Runx2 or Smad1/4 transcriptional activity in chondrocytes.
Because induction of the −4kb ColX reporter by cotransfected Runx2, Smad1, and MEF2C was efficiently blocked by either PTHrP or forskolin in USCs (Fig. 1A), it seems plausible that these signals may somehow inhibit the activity of one of these transcription factors. To examine this issue, we evaluated whether these signals could block the ability of these transcription factors to induce expression of simplified reporters containing reiterated binding sites for a single transcription factor (diagrammed schematically in Fig. 1B). Runx2 transcriptional activity was assayed by employing an artificial reporter containing six Runx2 binding sites driving expression of the luciferase gene [6x(Runx2)-luciferase] (8). In striking contrast to the −4kb ColX reporter, induction of 6x(Runx2)-luciferase by cotransfected Runx2 was slightly augmented by the presence of either PTHrP or forskolin (Fig. 1C). We evaluated the effect of PTHrP/forskolin on Smad1/4 transcriptional activity by assaying the expression of a reporter containing 30 copies of a GC-rich Smad1/4 binding element [30x(SBE)-luciferase] (12). Induction of 30x(SBE)-luciferase by cotransfected Smad1/4 was also augmented in the presence of either PTHrP or forskolin (Fig. 1D). Thus, neither PTHrP nor forskolin represses the transcriptional activity of either Runx2 or Smad1/4 to induce expression of luciferase reporters driven by their cognate binding sites.
PTHrP signaling represses MEF2 activity in chondrocytes.
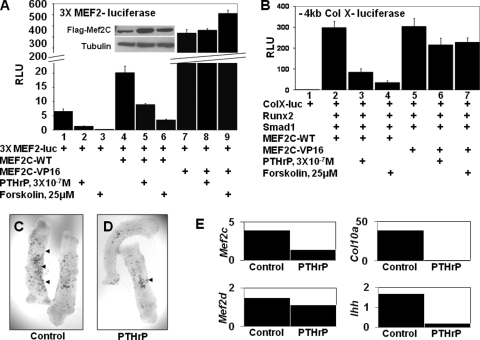
To evaluate whether MEF2 function is modulated by PTHrP/forskolin in chondrocytes, we transfected USCs with a luciferase reporter driven by three reiterated MEF2 binding sites (3xMEF2-luciferase) (18). We observed that administration of either PTHrP or forskolin led to a precipitous drop in the expression of this reporter (Fig. 2A, lanes 1 to 3), suggesting that these signals inhibit either the expression or activity of endogenous MEF2 in the cell. As PTHrP and forskolin could potentially be affecting the expression of endogenous MEF2C and/or the interaction of this protein with repressors of MEF2 function, such as class II HDACs, we assayed whether PTHrP/forskolin would affect the activity of either cotransfected wild-type MEF2C or an activated form of MEF2C (MEF2C-VP16) to drive expression of the 3×MEF2-luciferase reporter. Both PTHrP and forskolin blocked induction of the 3×MEF2-luciferase reporter by cotransfected wild-type MEF2C (Fig. 2A, lanes 4 to 6), without affecting the levels of this transcription factor (inset in Fig. 2A), indicating that these agents block the activity of exogenous MEF2C. MEF2C-VP16 is a fusion protein containing MEF2C fused to the strong transcriptional activation domain of the herpesvirus protein VP16 (23). We found that cotransfected MEF2C-VP16 very strongly activates expression of the 3×MEF2-luciferase reporter in USCs and that administration of PTHrP or forskolin did not blunt this induction (Fig. 2A, lanes 7 to 9). Thus, while PTHrP/forskolin administration can block the transcriptional activity of MEF2C-WT, these agents do not significantly attenuate the activity of MEF2C-VP16.
FIG. 2.
PTHrP inhibits expression of a MEF2-lacZ transgene in developing cartilage and the transcriptional activity of both endogenous and exogenous MEF2C, but not that of MEF2-VP16. (A) USCs were cotransfected with a 3X(MEF2)-firefly luciferase reporter and an SV40-renilla luciferase reporter plus MEF2C-WT or MEF2C-VP16 expression vehicles in either the absence or presence of PTHrP or forskolin as indicated. The insert shows a Western blot of extracts from USCs transfected with wild-type MEF2C-Flag and cultured in either the absence or presence of PTHrP or forskolin as indicated. RLU, relative luciferase units. (B) USCs were cotransfected with a −4kb ColX-renilla luciferase reporter and an SV40-firefly luciferase reporter plus Runx2, Smad1, MEF2C-WT, or MEF2C-VP16 expression vehicles in either the absence or presence of PTHrP or forskolin as indicated. (C and D) Metatarsals were isolated from 15.5-dpc mouse embryos containing a lacZ transgene driven by three MEF2 binding sites (24) cultured in either the absence or presence of 10−6 M PTHrP for 3 days and stained for beta-galactosidase expression. Arrowheads, regions of the explants showing beta-galactosidase activity. (E) Gene expression from three pooled metatarsal explants treated as described in the legends for panels C and D was measured by RT-qPCR and normalized to Tbp expression.
An activated form of MEF2 can reverse the ability of PTHrP and forskolin to repress expression of a collagen X reporter.
Because MEF2 activity is repressed by PTHrP signaling in USCs, we examined whether expression of an activated form of MEF2C (MEF2C-VP16), whose activity is not blunted by these signals, would rescue the expression of the collagen X reporter in the presence of PTHrP. Cotransfections of Runx2/Smad1 plus either MEF2C-WT or MEF2C-VP16 equally activated expression of the −4kb ColX-luciferase reporter (Fig. 2B, compare lanes 2 and 5). As previously observed (as shown in Fig. 1A), induction of this reporter in response to Runx2/Smad1/MEF2C-WT was repressed by either PTHrP or forskolin (by 70% and 90%, respectively; Fig. 2B, compare lanes 2 to 4). In contrast, induction of this reporter by cotransfected Runx2/Smad1/MEF2C-VP16 was significantly less attenuated by either PTHrP or forskolin (by 30% and 25%, respectively; Fig. 2B, compare lanes 5 to 7). Thus, inclusion of MEF2C-VP16, whose transcriptional activity is not attenuated by either PTHrP or forskolin, blunts the ability of either of these signaling molecules to repress expression of the collagen X reporter. These findings suggest that forced expression of an activated form of MEF2C renders expression of chondrocyte hypertrophy markers relatively resistant to the effects of PTHrP signaling and are consistent with prior findings from the laboratory of Olson and colleagues that ectopic expression of MEF2C-VP16 in developing cartilage induces precocious chondrocyte hypertrophy in transgenic mice (2).
PTHrP reduces expression of a MEF2-lacZ reporter in metatarsal explants.
Application of PTH/PTHrP to explants of embryonic metatarsals blocks chondrocyte hypertrophy and expression of collagen X (11). We examined whether inhibition of chondrocyte maturation by PTHrP correlated with a reduction of either MEF2 activity and/or expression. Metatarsals were isolated from 15.5-dpc mouse embryos containing a lacZ transgene driven by three MEF2 binding sites (24), cultured in either the absence or presence of 10−6 M PTHrP for 3 days, and stained for beta-galactosidase expression. Staining with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) revealed punctate expression of beta-galactosidase in untreated metatarsals (Fig. 2C). Interestingly, the level of beta-galactosidase expression decreased in explants cultured in the presence of PTHrP (Fig. 2D), suggesting that PTHrP represses either the activity and/or expression of MEF2 in such cultures. Loss of MEF2 reporter gene expression in PTHrP-treated metatarsals correlated with a precipitous decline in collagen X and Ihh expression in the explants and a notable decrease in the expression of both MEF2C and MEF2D (Fig. 2E). Together, these findings indicate that PTHrP represses the activity of exogenous MEF2 in transfected chondrocytes and the expression of both a MEF2-lacZ reporter and endogenous MEF2 family members in cultured metatarsal explants.
PTHrP and forskolin treatment of chondrocytes induces the nuclear retention of GFP-HDAC4.
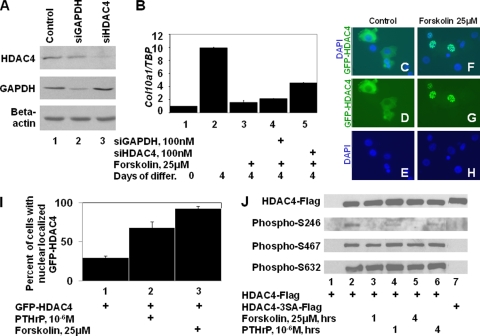
The transcriptional activity of MEF2 family members in chondrocytes is modulated by HDAC4 (30). To examine if HDAC4 function is necessary for forskolin to repress expression of endogenous Col10a1, we employed a line of chondrogenic mouse limb bud cells (MLB14) which express COL10A1 in a BMP-dependent manner (28) (Fig. 3B, lanes 1 to 2). siRNA targeting either Gapdh or Hdac4 significantly decreased expression of the cognate protein 2 to 4 days following transfection into MLB14 cells (Fig. 3A). Treatment of MLB14 cells with forskolin resulted in a significant decrease in Col10a1 expression (Fig. 3B, compare lanes 2 and 3). While knockdown of HDAC4 in forskolin-treated MLB14 cells led to the partial restoration of Col10a1 expression (Fig. 3B, compare lanes 3 and 5), knockdown of GAPDH in such cells did not significantly alter expression of Col10a1 (Fig. 3B, lane 4), indicating that HDAC4 function is necessary for AC/PKA signaling to maximally inhibit Col10a1 expression. The incomplete restoration of Col10a1 expression following knockdown of HDAC4 may be due to either the incomplete knockdown of HDAC4 or the presence of other class II HDACs in MLB14 cells (data not shown).
FIG. 3.
PTHrP and forskolin induce the nuclear localization of HDAC4 and dephosphorylation of HDAC4 phospho-S246. (A) siRNAs targeting either Gapdh or Hdac4 knockdown expression of their respective proteins in chondrogenic MLB14 cells. (B) Knockdown of HDAC4 rescues Col10a1 expression in forskolin-treated MLB14 cells. MLB14 cells, induced to differentiate for 4 days in BMP2-containing medium, were treated as indicated; the ratio of Col10a1 to Tbp was determined by RT-qPCR. (C to H) USCs were transfected with an expression vehicle encoding GFP fused to HDAC4 (GFP-HDAC4) and cultured in either the absence or presence of forskolin. Localization of GFP-HDAC4- and DAPI-stained nuclei is displayed. (I) The percentage of chondrocytes displaying nuclearly localized GFP-HDAC4 when cultured in either the absence or presence of PTHrP or forskolin. (J) USCs were transfected with expression vehicles encoding either Flag-tagged wild-type HDAC4 (HDAC4-Flag) or a Flag-tagged mutant form of HDAC4 containing alanine in place of S246, S467, and S632 (HDAC4-3SA-Flag) and cultured in control medium or in medium containing either PTHrP or forskolin for an additional 1 or 4 h prior to cell lysis. Immunoprecipitated HDAC4-Flag was Western blotted and probed with antibodies to HDAC4-Flag, HDAC4 phospho-S246, HDAC4 phospho-S467, and HDAC4 phospho-S632.
Because HDAC4 function is required for forskolin to efficiently block Col10a1 expression in MLB14 chondrocytes, we examined whether PTHrP/forskolin might indirectly be affecting the activity of MEF2 by altering the cellular localization of HDAC4. Chicken USCs were transfected with a GFP-HDAC4 expression vehicle (22). While chondrocytes cultured in control medium localize GFP-HDAC4 predominantly in the cytoplasm (Fig. 3, panels C, D, and E), those cultured for 4 h in either PTHrP or forskolin localize GFP-HDAC4 predominantly in the nucleus (Fig. 3, panels F, G, and H). Quantification of these results indicated that GFP-HDAC4 was localized to the nucleus in only 30% of control-transfected USCs, in 68% of PTHrP-treated cells, and in 93% of forskolin-treated cells (Fig. 3I). Similar results were observed with MLB14 cells transfected with GFP-HDAC4 upon forskolin treatment (data not shown). Together, these findings indicate that either PTHrP or forskolin can increase the nuclear localization of GFP-HDAC4.
PTHrP signaling or forskolin administration specifically induces the dephosphorylation of HDAC4 phospho-S246.
Because phosphorylation of HDAC4 on residues S246, S467, and S632 promotes the retention of this protein in the cytoplasm (9, 32), we investigated whether phosphorylation of these serines is altered by either PTHrP or forskolin. USCs were transfected with expression vehicles encoding either Flag-tagged wild-type HDAC4 (HDAC4-Flag) or a Flag-tagged mutant form of HDAC4 containing alanine in place of S246, S467, and S632 (HDAC4-3SA-Flag) (21). Forty-eight h posttransfection, the USCs were treated with either PTHrP or forskolin for 1 or 4 h prior to cell lysis. HDAC4-Flag was immunoprecipitated with anti-Flag antibody and Western blotted to detect HDAC4-Flag, phospho-S246, phospho-S467, or phospho-S632. HDAC4 residues S246, S467, and S632 are apparently phosphorylated in chondrocytes, as anti-phospho-S246/S467/S632 antisera recognized their respective phosphorylated epitopes in wild-type HDAC4 but not in the mutant form of HDAC4 (Fig. 3J, compare lanes 2 and 7). While phosphorylation of S467 or S632 was not significantly altered by treatment of chondrocytes with either forskolin or PTHrP, phosphorylation of S246 was markedly decreased following treatment of cells with either forskolin or PTHrP (Fig. 3J, compare lane 2 with lanes 3 to 6). These results suggest that phosphorylation of HDAC4 S246 (but not S467 or S632) is specifically decreased by either forskolin or PTHrP in chondrocytes.
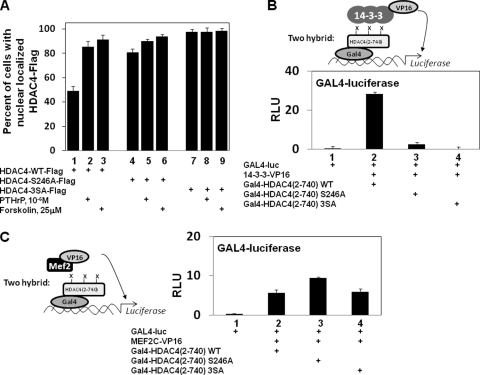
PTHrP/forskolin administration blocks the association of HDAC4 and 14-3-3 and increases the association between HDAC4 and MEF2C as assayed by both two-hybrid and co-IP assays.
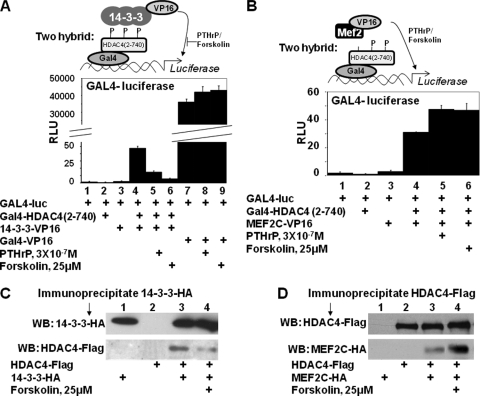
Prior mutational analysis of HDAC4 has indicated that a loss of S246 phosphorylation decreases interaction of HDAC4 with 14-3-3 proteins (3). To investigate whether PTHrP or forskolin administration similarly decreases the interaction between HDAC4 and 14-3-3 proteins, we employed a vertebrate two-hybrid assay. USCs were transfected with the Gal4 DNA binding domain fused to HDAC4 [residues 2 to 740 and lacking the HDAC catalytic domain; Gal4-HDAC4(2-740)] (3) in either the absence or presence of 14-3-3 fused to the VP16 activation domain (14-3-3-VP16) (3). While transfection of chondrocytes with either Gal4-HDAC4(2-740) or 14-3-3-VP16 alone failed to induce the activity of a luciferase reporter driven by reiterated Gal4 binding sites (GAL4-luciferase; Fig. 4A, lanes 1 to 3), transfection of cells with both these constructs led to robust expression of this reporter (Fig. 4A, lane 4) (3). Administration of either PTHrP or forskolin dramatically reduced the induction of GAL4-luciferase activity in response to cotransfected Gal4-HDAC4(2-740) and 14-3-3-VP16 (Fig. 4A, compare lane 4 with lanes 5 and 6), suggesting that administration of either PTHrP or forskolin reduces the interaction between Gal4-HDAC4(2-740) and 14-3-3-VP16. Importantly, neither PTHrP nor forskolin affected the activity of a Gal4-VP16 fusion protein (Fig. 4A, lanes 7 to 9), suggesting that the transcriptional effects of PTHrP and forskolin are due to modulation of HDAC4/14-3-3 interaction.
FIG. 4.
PTHrP/forskolin blocks the association of HDAC4 and 14-3-3 proteins and increases the association between HDAC4 and MEF2C. (A) USCs were transfected with a GAL4-firefly luciferase reporter and an SV40-renilla luciferase reporter plus expression vehicles encoding Gal4-HDAC4(2-740), 14-3-3-VP16, or Gal4-VP16 in either the absence or presence of PTHrP or forskolin as indicated. RLU, relative luciferase units. P, phosphate group. (B) USCs were transfected with a GAL4-firefly luciferase reporter and an SV40-renilla luciferase reporter plus expression vehicles encoding either Gal4-HDAC4(2-740) or MEF2C-VP16 in either the absence or presence of PTHrP or forskolin. (C) USCs were transfected with expression vehicles encoding HDAC4-Flag plus 14-3-3-HA in either the absence or presence of forskolin as indicated. 14-3-3-HA and associated proteins were immunoprecipitated, and Western analysis (WB) was employed to detect co-IP 14-3-3-HA and HDAC4-Flag. (D) USCs were transfected with expression vehicles encoding HDAC4-Flag plus MEF2C-HA in either the absence or presence of forskolin as indicated. HDAC4-Flag and associated proteins were immunoprecipitated and Western analysis was employed to detect co-IP HDAC4-Flag and MEF2C-HA.
To investigate whether PTHrP/forskolin induces an increased association between HDAC4 and MEF2, we again utilized a vertebrate two-hybrid assay, this time employing Gal4-HDAC4(2-740) and MEF2C-VP16. Importantly, the MEF2 interaction domain located in the N terminus of HDAC4 is present in Gal4-HDAC4(2-740). While Gal4-HDAC4(2-740) and MEF2C-VP16 failed individually to activate expression of GAL4-luciferase (Fig. 4B, lanes 1 to 3), the combination of Gal4-HDAC4(2-740) and MEF2C-VP16 led to robust activation of this reporter (Fig. 4B, lane 4). Administration of PTHrP or forskolin resulted in an increase in GAL4-luciferase activity (Fig. 4B, compare lanes 4 to 6), suggesting that the interaction between HDAC4 and MEF2C is augmented by PTHrP/forskolin.
To further investigate whether elevation of cAMP levels by forskolin alters either HDAC4-14-3-3 or HDAC4-MEF2 interaction, we examined whether co-IP of these proteins from transfected chondrocytes would be affected by forskolin treatment. USCs were cotransfected with expression vehicles encoding HDAC4-Flag plus either 14-3-3-HA or MEF2C-HA in either the absence of presence of forskolin. IP of transfected 14-3-3-HA led to the co-IP of HDAC4-Flag (Fig. 4C, lane 3), and addition of forskolin to the cells markedly decreased this interaction (Fig. 4C, lane 4). IP of transfected HDAC4-Flag led to the co-IP of MEF2C-HA (Fig. 4D, lane 3), and in this case addition of forskolin to the cells increased this interaction (Fig. 4D, lane 4). Thus, both the vertebrate two-hybrid assay and co-IP analyses indicated that forskolin treatment leads to a relative decrease in the interaction between HDAC4 and 14-3-3 proteins and a corresponding increase in the interaction between HDAC4 and MEF2C.
Phosphorylation of HDAC4 S246 is required for robust interaction of HDAC4 with 14-3-3 proteins and retention of HDAC4 in the cytoplasm.
Because our analysis indicated that PTHrP/forskolin-induced dephosphorylation of HDAC4 S246 (but not S467 or S632) correlates with PTHrP/forskolin-induced nuclear localization of HDAC4 in chondrocytes, we examined whether an alanine substitution at this position would affect either cellular localization or interaction of HDAC4 with 14-3-3 proteins. While HDAC4-WT-Flag was nuclearly localized in 49% of transfected chondrocytes, HDAC4-S246A-Flag was nuclearly localized in 81% of such cells (Fig. 5A, lanes 1 and 4). In comparison, HDAC4-3SA-Flag, containing alanine substitutions for S246, S467, and S632, was nuclearly localized in 98% of transfected chondrocytes (Fig. 5A, lane 7). Thus, while loss of S246 phosphorylation substantially increases nuclear localization of HDAC4-Flag, nuclear localization of this mutant HDAC4 can be further accentuated by the loss of S467 and S632 phosphorylation. Consistent with these findings, we observed with vertebrate two-hybrid analyses that substitution of HDAC4 S246 with alanine profoundly decreased the interaction of 14-3-3-VP16 with Gal4-HDAC4(2-740) by over 91% (Fig. 5B), while enhancing the interaction between MEF2C-VP16 and Gal4-HDAC4(2-740) (Fig. 5C). These results indicate that loss of HDAC4 S246 phosphorylation alone can significantly weaken interaction between HDAC4 and 14-3-3 proteins, resulting in redistribution of HDAC4 into the nucleus. Interestingly, PTHrP/forskolin treatment induced a substantial increase in nuclear localization of HDAC4-WT-Flag, (Fig. 5A, lanes 1 to 3) and a small but reproducible increase in the nuclear localization of HDAC4-S246A-Flag (Fig. 5A, lanes 4 to 6) and did not affect the cellular localization of HDAC4-3SA-Flag (Fig. 5A, lanes 7 to 9). These findings suggest that PTHrP/forskolin treatment may also decrease phosphorylation of S467 and S632 at a level that is undetectable by Western analysis (Fig. 3J).
FIG. 5.
Phosphorylation of HDAC4 S246 is required for robust interaction of HDAC4 with 14-3-3 proteins and retention of HDAC4 in the cytoplasm. (A) USCs were transfected with an expression vehicle encoding HDAC4-WT-Flag, HDAC4-S246A-Flag, or HDAC4-3SA-Flag and cultured in either the absence or presence of PTHrP or forskolin as indicated. The percentage of chondrocytes displaying nuclearly localized HDAC4-Flag when cultured in either the absence or presence of PTHrP or forskolin is displayed. (B and C) USCs were transfected with a GAL4-firefly luciferase reporter and an SV40-renilla luciferase reporter plus expression vehicles encoding 14-3-3-VP16; MEF2-VP16; and Gal4-HDAC4(2-740)WT, Gal4-HDAC4(2-740)S246A, or Gal4-HDAC4(2-740)3SA as indicated. Relative luciferase units (RLU) are displayed. X, either a phosphate group or hydrogen.
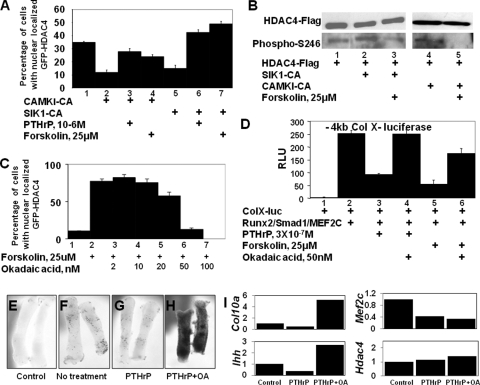
Forskolin decreases both the cytoplasmic localization and phosphorylation of HDAC4 S246 in the presence of activated HDAC4 kinases.
A decrease in the phosphorylation of HDAC4 S246 by either PTHrP or forskolin treatment suggests that elevation of cAMP levels either (i) inhibits the activity/expression of an HDAC4 S246 kinase or (ii) induces the activity/expression of an HDAC4 S246 phosphatase. To distinguish between these two possibilities, we examined whether forced expression of an activated HDAC4 kinase can counter the effects of forskolin treatment. If forskolin administration blocks only the activity or expression of an essential HDAC4 kinase in chondrocytes and does not activate an HDAC4 phosphatase, then forced expression of constitutively activated forms of either CAMKI or SIK1, which phosphorylate HDAC4 and induce both its nuclear exclusion and association with 14-3-3 proteins (3, 5, 18), should promote increased phosphorylation of HDAC4 S246 in both the absence and presence of forskolin. On the other hand, if forskolin administration induces the activity or expression of an essential HDAC4 phosphatase, then the ability of constitutively activated forms of CAMKI or SIK1 to promote both nuclear exclusion and increased phosphorylation of HDAC4 S246 should be blunted by forskolin administration.
Cotransfection of USCs with activated forms of either CAMKI or SIK1 decreased the nuclear localization of GFP-HDAC4 from 35% to 12% or 15%, respectively (Fig. 6A, lanes 1, 2, and 5). Treatment of USCs cotransfected with activated CAMKI with either PTHrP or forskolin increased the nuclear retention of GFP-HDAC4 from 12% to 28% or 24%, respectively (Fig. 6A, lanes 2 to 4). Similarly, PTHrP or forskolin treatment of USCs cotransfected with activated SIK1 increased the nuclear retention of GFP-HDAC4 from 15% to 43% or 49%, respectively (Fig. 6A, lanes 5 to 7). Together these findings suggest that PTHrP or forskolin counters the activity of constitutively activated HDAC4 kinases. Consistent with these results, we found that cotransfection of HDAC4-Flag with an activated form of SIK1 increased the phosphorylation of HDAC4 S246 (Fig. 6B, compare lanes 1 and 2), which was reversed by administration of forskolin (Fig. 6B, compare lanes 2 and 3). Similarly, we found that cotransfection of an activated form of CAMKI also induced increased phosphorylation of HDAC4 S246 (data not shown), which was reversed upon exposure of cells to forskolin (Fig. 6B, compare lanes 4 and 5). Together these data suggest that elevation of cAMP levels by forskolin counters the ability of activated forms of either SIK1 or CAMKI to affect S246 phosphorylation by inducing the expression and/or activity of an HDAC4 phospho-S246 phosphatase.
FIG. 6.
PTHrP/forskolin decreases phosphorylation of HDAC4 phospho-S246, increases nuclear localization of HDAC4, and represses expression of a collagen X reporter via an okadaic acid-sensitive phosphatase. (A) USCs were cotransfected with GFP-HDAC4 plus activated forms of CAMK (CAMKI-CA) or SIK (SIK1-CA) in either the absence or presence of PTHrP or forskolin (4-h treatment), as indicated. The percentage of cells with nuclear localized GFP-HDAC4 is displayed. (B) USCs were transfected with expression vehicles encoding HDAC4-Flag plus expression vehicles encoding either SIK1-CA or CAMKI-CA in either the absence or presence of forskolin as indicated. HDAC4-Flag was immunoprecipitated, and Western analysis was employed to detect immunoprecipitated HDAC4-Flag and HDAC4 phospho-S246. (C) USCs were transfected with a GFP-HDAC4 expression vehicle and cultured for 4 h in either the absence or presence of forskolin and increasing amounts of okadaic acid, as indicated. The percentage of cells with nuclearly localized GFP-HDAC4 is displayed. (D) USCs were cotransfected with a −4kb ColX-renilla luciferase reporter and an SV40-firefly luciferase reporter plus Runx2/Smad1/MEF2C-WT expression vehicles in either the absence or presence of forskolin and okadaic acid as indicated. RLU, relative luciferase units. (E to H) Metatarsals were isolated from 15.5-dpc mouse embryos either lacking (control) or containing a lacZ transgene driven by three MEF2 binding sites (24), cultured in either the absence or presence of 10−6 M PTHrP and 50 nM okadaic acid (OA) as indicated for 3 days, and stained for beta-galactosidase expression. (I) Gene expression in three pooled metatarsal explants treated as described in the legend for panels E to H was measured by RT-qPCR and normalized to Gapdh expression.
Okadaic acid blocks the ability of forskolin to induce nuclear localization of HDAC4 in chondrocytes and to repress induction of collagen X luciferase in chondrocytes.
Recently, it was demonstrated that PP1 and PP2A can both dephosphorylate and regulate the cellular localization of class II HDACs (19, 25, 26). Because forskolin induces the dephosphorylation of S246 by increasing the expression/activity of an HDAC4 phosphatase, we investigated if the PP2A/PP1 inhibitor, okadaic acid, could reverse the effects of forskolin. We found that pretreatment of chicken USCs or chondrogenic MLB14 cells with as little as 50 nM okadaic acid could completely block a forskolin-induced increase in nuclearly localized HDAC4 (Fig. 6C and data not shown), suggesting that cAMP/PKA signaling induces the expression/activity of an okadaic acid-sensitive phosphatase that dephosphorylates phospho-S246 in HDAC4 and thereby induces its nuclear localization.
We examined whether this phosphatase inhibitor would similarly reverse the ability of forskolin to inhibit expression of the −4kb ColX-luciferase reporter. While administration of either PTHrP or forskolin markedly blocks the induction of this reporter by cotransfected Runx2, MEF2C, and Smad1 (Fig. 6D, compare lane 2 with lanes 3 and 5), this inhibition is either completely (in the case of PTHrP) or substantially (in the case of forskolin) reversed by inclusion of okadaic acid (Fig. 6D, compare lanes 3 and 4 or lanes 5 and 6, respectively). These results indicate that the activity of an okadaic acid-sensitive phosphatase is necessary for forskolin to block MEF2 activity. We also examined whether addition of okadaic acid would affect the ability of PTHrP signals to repress expression of the MEF2-lacZ reporter in metatarsals isolated from 15.5-dpc mouse embryos containing a lacZ transgene driven by three MEF2 binding sites (24). While PTHrP administration decreased expression of both the lacZ transgene (Fig. 6F and G) and the chondrocyte hypertrophy markers collagen X and Ihh in these cultures (Fig. 6I), administration of both PTHrP and okadaic acid strikingly increased expression of the MEF2 reporter transgene (Fig. 6H) and similarly increased expression of collagen X and Ihh (Fig. 6I). Interestingly, the increased expression of chondrocyte hypertrophy markers induced by okadaic acid was observed in the absence of increased expression of MEF2C (Fig. 6I). Taken together, our findings suggest that elevation of cAMP levels by PTHrP/forskolin induces the expression/activity of an okadaic acid-sensitive phosphatase that dephosphorylates HDAC4 phospho-S246, thereby promoting the nuclear localization of this class II HDAC and consequent suppression of MEF2 function.
PP2A is the okadaic acid-sensitive phosphatase that is required for PTHrP/forskolin to induce HDAC4 nuclear localization.
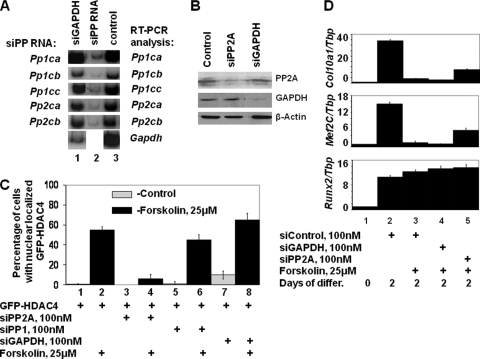
Because okadaic acid is known to block the activity of PP1 and PP2A, we evaluated whether either PP1 or PP2A function is necessary for PTHrP/forskolin to induce nuclear localization of HDAC4. We employed siRNAs to knock down the expression of the catalytic subunit of these phosphatases in both NIH 3T3 cells and the MLB14 chondrogenic cell line. There are three isoforms of the catalytic subunit of PP1 (PP1CA, PP1CB, and PP1CC) and two isoforms of the catalytic subunit of PP2A (PP2CA and PP2CB). We transfected NIH 3T3 cells with siRNAs directed against these various catalytic subunits and monitored the effect of these siRNAs on the steady-state levels of Pp1ca, Pp1cb, Pp1cc, Pp2ca, and Pp2cb catalytic subunit transcripts by RT-PCR analysis. siRNAs directed against the various PP1 or PP2A catalytic subunits specifically decreased transcript levels of the targeted phosphatase subunit (Fig. 7A) and did not alter transcript levels of the other PP1 or PP2A catalytic subunits (data not shown). In addition, we found that transfection of either NIH 3T3 cells or chondrogenic MLB14 cells with the combination of siRNAs targeting both isoforms of the catalytic subunit of PP2A significantly lowered steady-state levels of the PP2A-catalytic subunit proteins, while siRNA directed against Gapdh lowered its protein levels (data not shown and Fig. 7B).
FIG. 7.
PP2A is the okadaic acid-sensitive phosphatase that is required for PTHrP/forskolin to induce HDAC4 nuclear localization. (A) NIH 3T3 cells were transfected with siRNAs directed against Gapdh (lane 1), siRNAs directed against isoforms of the catalytic subunit of either PP1 (Pp1ca, Pp1cb, and Pp1cc) or PP2A (Pp2ca and Pp2cb) (lane 2), or no siRNA (lane 3). Transcript levels of the various genes were assayed by RT-qPCR. (B) MLB14 cells were transfected with no siRNA (control), siRNAs directed against PP2A, or siRNA directed against GAPDH, as indicated. Protein levels were assayed by Western analysis. (C) MLB14 cells were transfected with GFP-HDAC4 plus either no siRNA (control, lanes 1 and 2) or siRNA directed against the PP2A catalytic subunit (lanes 2 and 3), the PP1 catalytic subunit (lanes 5 and 6), or GAPDH (lanes 7 and 8). Cells were cultured for 44 h and treated for an additional 4 h either in control medium (odd lanes) or in medium containing forskolin (even lanes), as indicated. The percentage of cells containing GFP-HDAC4 solely in the nucleus is displayed. (D) Knockdown of PP2A boosts Col10a1 and Mef2C expression without affecting Runx2 expression in chondrocytes cultured in forskolin. MLB14 chondrogenic cells were treated with siRNA directed against either Gapdh transcript or the isoforms of the catalytic subunit of PP2A just prior to plating in BMP2 for 2 days. Cells were cultured in either the absence or presence of forskolin as indicated. Relative amounts of the indicated transcripts were determined by RT-qPCR and normalized to that of the TATA-binding protein (Tbp).
Because we could specifically knock down the expression of the various catalytic isoforms of either PP1 or PP2A with siRNAs, we explored whether forskolin-induced nuclear translocation of HDAC4 would be affected by knocking down either PP1 or PP2A catalytic subunits. The GFP-HDAC4 expression vector was cotransfected with various siRNAs into MLB14 cells, and the percentage of cells containing GFP-HDAC4 solely in the nucleus was quantified. In control-transfected MLB14 cells, forskolin administration increased the percentage of cells containing GFP-HDAC4 localized only in the nucleus from 0% (in nontreated cells) to 55% (in forskolin-treated cells) (Fig. 7C, lanes 1 and 2). Transfection of MLB14 cells with siRNAs directed against both isoforms of the catalytic subunit of PP2A (siPP2A) markedly diminished the ability of forskolin to induce nuclear translocation of GFP-HDAC4 from 55% in non-siRNA-treated cells to only 6% in siPP2A-treated cells (Fig. 7C, lanes 2 and 4, respectively). In contrast, siRNAs directed against either all three isoforms of the catalytic subunit of PP1 (siPP1) or siRNA directed against GAPDH did not significantly affect the ability of forskolin to induce nuclear translocation of GFP-HDAC4 (Fig. 7C, lanes 6 and 8). These findings indicate that PP2A, not PP1, is necessary for forskolin to induce the nuclear translocation of HDAC4.
Knockdown of PP2A boosts Col10a1 expression in chondrocytes cultured in forskolin.
Because loss of PP2A blocks the ability of forskolin to induce nuclear translocation of HDAC4, we wondered whether knockdown of PP2A would similarly block the ability of forskolin to inhibit Col10a1 expression. We treated MLB14 cells with siRNA directed against either Gapdh or Pp2a transcripts prior to plating in BMP2 for 2 days. Treatment of the MLB14 cells with forskolin decreased expression of Col10a1 (Fig. 7D, compare lanes 2 and 3). While knockdown of PP2A enhanced the expression of Col10a1 in such forskolin-treated cultures (Fig. 7D, lane 5), knockdown of GAPDH did not boost Col10a1 expression (Fig. 7D, lane 4), indicating that knockdown of PP2A specifically attenuated the ability of forskolin to repress Col10a1 expression. Interestingly, like Col10a1, expression of Mef2C was repressed by forskolin (Fig. 7D, lanes 2 and 3) and was reactivated by knockdown of PP2A in forskolin-treated cells (Fig. 7D, lane 5). In contrast, these various treatments did not significantly affect Runx2 expression (Fig. 7D).
DISCUSSION
PTHrP signaling inhibits both MEF2 expression and transcriptional activity in chondrocytes.
Prior work has indicated that PTHrP signaling plays a critical role in attenuating the rate of chondrocyte maturation in the growth plate (14, 16, 29, 33). While recent studies have documented an essential role for Runx2/3, MEF2C/D, and Smad1/4 in this process (2, 31, 34), it has not been clear whether PTHrP signals directly modulate the activity of any of these transcription factors. In this work we document that PTHrP or forskolin administration represses the ability of Runx2/Smad1/MEF2C to activate expression of a collagen X reporter in USCs isolated from chicken embryos. To determine how these signals repress induction of the collagen X reporter by these transcription factors, we investigated how PTHrP/forskolin affected the activity of these transcription factors to activate expression of simplified reporter constructs. Like the collagen X reporter, we found that PTHrP/forskolin inhibits the transcriptional activity of either endogenous or exogenous MEF2C to activate a luciferase reporter driven by reiterated MEF2 binding sites. In striking contrast, these same signals augment the transcriptional activity of Runx2 or Smad1/4 to activate expression of luciferase reporters driven by their cognate binding sites. Moreover, we found that cotransfection of an activated form of MEF2C, whose activity is not blunted by either PTHrP or forskolin, could restore expression of the collagen X reporter in chondrocytes treated with these agents. These findings strongly suggest that MEF2 transcriptional activity is a target for PTHrP signals in chondrocytes. In addition, we found that PTHrP treatment of embryonic metatarsal cultures decreased expression of a MEF2-lacZ reporter transgene and repressed expression of both endogenous Col10a1 and Mef2C, while increasing the expression of Hdac4 (data not shown). Taken together, these findings indicate that PTHrP/AC/PKA signal transduction blocks both the expression and activity of MEF2 transcription factors. PTHrP may, in addition, control either the expression or transcriptional activity of other factors necessary for collagen X expression, as cotransfected MEF2-VP16, whose activity is not attenuated by either PTHrP or forskolin, was unable to completely restore expression of a collagen X luciferase reporter in USCs treated with either PTHrP or forskolin. Interestingly, forskolin treatment of skeletal muscle cells has similarly been demonstrated to inhibit the activity of MEF2D by inducing both PKA-mediated phosphorylation of MEF2D and nuclear translocation of HDAC4 (7).
PTHrP inhibits MEF2 by promoting the nuclear translocation of HDAC4.
Because HDAC4 is known to block both precocious and ectopic chondrocyte hypertrophy (30), we investigated whether PTHrP signals modulate the interaction of this HDAC with MEF2 family members. Indeed we found that PTHrP or forskolin administration to chondrocytes induced a striking relocalization of a GFP-HDAC4 fusion protein from the cytoplasm into the nucleus. This nuclear relocalization of HDAC4 correlated with a decreased association of this HDAC with 14-3-3 proteins and an increased association with MEF2C as assayed by both vertebrate two-hybrid and co-IP analyses. As HDAC4 is known to be phosphorylated at specific conserved residues (S246, S467, and S632), whose phosphorylation promotes the association of these proteins with 14-3-3 proteins in the cytoplasm (9, 20), we assayed the effect of PTHrP and forskolin on the phosphorylation of these residues. We observed that these signals specifically induced the dephosphorylation of HDAC4 phospho-S246 but did not affect the apparent level of either phospho-S467 or phospho-S632 as detected by Western analysis. In addition, we noted that substitution of alanine for HDAC4 S246 profoundly disrupted interaction of HDAC4 with 14-3-3 proteins and substantially increased the nuclear localization of GFP-HDAC4 in chondrocytes. Consistent with our findings, HDAC4 phospho-S246 has previously been shown to be the most important of the three conserved HDAC4 phosphorylation sites necessary to promote high-affinity interaction between HDAC4 and 14-3-3 proteins (3). In addition, it is possible that PTHrP signaling may also induce dephosphorylation of other residues on HDAC4, such as S298, whose phosphorylation is also thought to tether this protein in the cytoplasm (25). As forskolin treatment has similarly been noted to drive nuclear localization of both HDAC4 in myoblasts (7) and HDAC5 in neurons (4), it seems likely that PKA-mediated dephosphorylation of class II HDACs may be employed generally in other cell types to regulate the activity of MEF2 family members.
PTHrP/forskolin requires PP2A activity to both induce nuclear translocation of HDAC4 and maximally inhibit expression of a collagen X reporter.
We found that PTHrP/forskolin reversed the ability of activated HDAC4 kinases to induce cytoplasmic localization of GFP-HDAC4 and could induce the dephosphorylation of HDAC4 phospho-S246 in the presence of activated forms of either SIK1 or CAMKI, suggesting that elevation of cAMP levels induces the expression and/or activity of an HDAC4 phospho-S246 phosphatase in chondrocytes. Consistent with this notion, we observed that a low concentration (50 nM) of the phosphatase inhibitor okadaic acid blocked the ability of PTHrP to promote nuclear localization of GFP-HDAC4; inhibit the induction of a −4kb ColX-luciferase reporter by cotransfected Runx2, MEF2C, and Smad1; or repress expression of MEF2-lacZ, collagen X, and Ihh in embryonic metatarsal explants. Together, these findings suggest that PTHrP signals block chondrocyte hypertrophy by inducing the activity of an okadaic acid-sensitive phosphatase, which in turn dephosphorylates HDAC4 phospho-S246, thereby inducing nuclear translocation of this HDAC and consequent repression of MEF2 activity. While our findings indicate that PTHrP/forskolin induces the expression/activity of an HDAC4 phospho-S246 phosphatase, these signals may also drive nuclear entry of HDAC4 by blocking the activity of an HDAC4 phospho-S246 kinase, which has yet to be identified in chondrocytes.
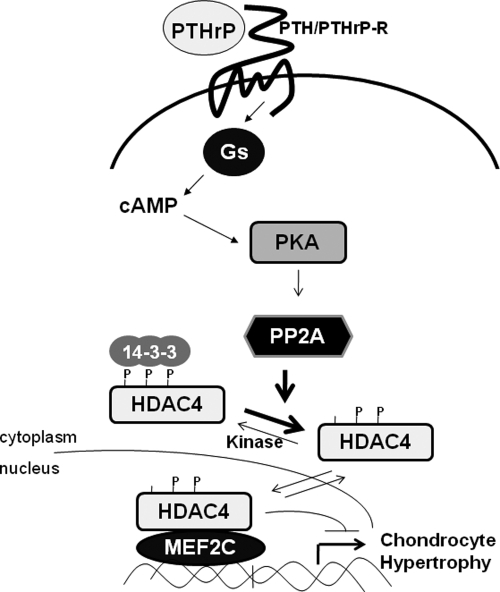
We have found that siRNA-mediated knockdown of the okadaic acid-sensitive phosphatase PP2A both blocks the ability of forskolin to induce nuclear translocation of HDAC4 and boosts Col10a1 expression in forskolin-treated cultures of chondrogenic MLB14 cells. Interestingly, HDAC4 can be isolated in a complex with the PP2A holoenzyme (25), which is a heterotrimeric enzyme that is composed of a scaffolding A subunit, a catalytic C subunit, and one of multiple regulatory B subunits. The regulatory B subunits are thought to influence enzyme activity, substrate specificity, and subcellular localization of the PP2A holoenzyme (reviewed in reference 13). PTHrP/forskolin-induced nuclear translocation of HDAC4 correlates with dephosphorylation of S246 and decreased interaction of HDAC4 with 14-3-3 proteins. Because PP2A activity is necessary for forskolin to induce nuclear translocation of HDAC4, it seems likely that elevation of cAMP levels and consequent PKA activation may be indirectly inducing dephosphorylation of HDAC4 phospho-S246 by altering either the expression or activity of PP2A or the interaction of this phosphatase with HDAC4. Indeed, it was recently demonstrated that PKA-mediated phosphorylation of a B-subunit isoform of PP2A (termed B56 delta) is critical to target this phosphatase to certain substrates in neurons (1). Thus, PTHrP signaling may similarly induce PKA-mediated phosphorylation of a B subunit of PP2A, which in turn targets this phosphatase to HDAC4, resulting in the dephosphorylation of HDAC4 phospho-S246, nuclear translocation of HDAC4, and consequent inhibition of both MEF2 activity and chondrocyte hypertrophy (outlined in Fig. 8). Future studies are necessary to both determine the identity of the relevant PP2A B subunit(s) in chondrocytes which targets this phosphatase to HDAC4 and determine whether interaction between HDAC4 and PP2A is indeed regulated by PKA phosphorylation.
FIG. 8.
PTHrP signals repress chondrocyte hypertrophy via a PKA-activated dephosphorylation of HDAC4 phospho-S246 by PP2A, which enhances the nuclear localization of HDAC4 and thereby inhibits MEF2 function.
Acknowledgments
This work was supported by grants to A.B.L. (AR048524 from NIAMS/NIH) and T.-P.Y. (AR055613 from NIAMS/NIH).
We thank Johannes Backs, Rebecca Berdeaux, Gerard Karsenty, Marc Montminy, Eric Olson, Stuart Schreiber, and Alex Toker for generously providing us with plasmids; Eric Olson, Tetsuo Konno, and Jon Seidman for supplying us with MEF2-lacZ mice; and Walter Sebald for supplying us with BMP2.
Footnotes
Published ahead of print on 24 August 2009.
REFERENCES
- 1.Ahn, J. H., T. McAvoy, S. V. Rakhilin, A. Nishi, P. Greengard, and A. C. Nairn. 2007. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc. Natl. Acad. Sci. USA 104:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, M. A., Y. Kim, M. P. Czubryt, D. Phan, J. McAnally, X. Qi, J. M. Shelton, J. A. Richardson, R. Bassel-Duby, and E. N. Olson. 2007. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell 12:377-389. [DOI] [PubMed] [Google Scholar]
- 3.Backs, J., K. Song, S. Bezprozvannaya, S. Chang, and E. N. Olson. 2006. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Investig. 116:1853-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belfield, J. L., C. Whittaker, M. Z. Cader, and S. Chawla. 2006. Differential effects of Ca2+ and cAMP on transcription mediated by MEF2D and cAMP-response element-binding protein in hippocampal neurons. J. Biol. Chem. 281:27724-27732. [DOI] [PubMed] [Google Scholar]
- 5.Berdeaux, R., N. Goebel, L. Banaszynski, H. Takemori, T. Wandless, G. D. Shelton, and M. Montminy. 2007. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13:597-603. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, T. J., D. S. Waddell, T. Barrientos, Z. Lu, G. Feng, G. A. Cox, S. C. Bodine, and T. P. Yao. 2007. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 282:33752-33759. [DOI] [PubMed] [Google Scholar]
- 7.Du, M., R. L. Perry, N. B. Nowacki, J. W. Gordon, J. Salma, J. Zhao, A. Aziz, J. Chan, K. W. Siu, and J. C. McDermott. 2008. Protein kinase A represses skeletal myogenesis by targeting myocyte enhancer factor 2D. Mol. Cell. Biol. 28:2952-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducy, P., and G. Karsenty. 1995. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15:1858-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, J., U. I. Chung, H. Kondo, F. R. Bringhurst, and H. M. Kronenberg. 2002. The PTH/PTHrP receptor can delay chondrocyte hypertrophy in vivo without activating phospholipase C. Dev. Cell 3:183-194. [DOI] [PubMed] [Google Scholar]
- 11.Guo, J., U. I. Chung, D. Yang, G. Karsenty, F. R. Bringhurst, and H. M. Kronenberg. 2006. PTH/PTHrP receptor delays chondrocyte hypertrophy via both Runx2-dependent and -independent pathways. Dev. Biol. 292:116-128. [DOI] [PubMed] [Google Scholar]
- 12.Ionescu, A. M., H. Drissi, E. M. Schwarz, M. Kato, J. E. Puzas, D. J. McCance, R. N. Rosier, M. J. Zuscik, and R. J. O'Keefe. 2004. CREB cooperates with BMP-stimulated Smad signaling to enhance transcription of the Smad6 promoter. J. Cell. Physiol. 198:428-440. [DOI] [PubMed] [Google Scholar]
- 13.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karaplis, A. C., A. Luz, J. Glowacki, R. T. Bronson, V. L. Tybulewicz, H. M. Kronenberg, and R. C. Mulligan. 1994. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 8:277-289. [DOI] [PubMed] [Google Scholar]
- 15.Koyama, E., E. B. Golden, T. Kirsch, S. L. Adams, R. A. Chandraratna, J. J. Michaille, and M. Pacifici. 1999. Retinoid signaling is required for chondrocyte maturation and endochondral bone formation during limb skeletogenesis. Dev. Biol. 208:375-391. [DOI] [PubMed] [Google Scholar]
- 16.Lanske, B., A. C. Karaplis, K. Lee, A. Luz, A. Vortkamp, A. Pirro, M. Karperien, L. H. K. Defize, C. Ho, R. C. Mulligan, A. B. Abou-Samra, H. Juppner, G. V. Segre, and H. M. Kronenberg. 1996. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science 273:663-666. [DOI] [PubMed] [Google Scholar]
- 17.Li, T. F., Y. Dong, A. M. Ionescu, R. N. Rosier, M. J. Zuscik, E. M. Schwarz, R. J. O'Keefe, and H. Drissi. 2004. Parathyroid hormone-related peptide (PTHrP) inhibits Runx2 expression through the PKA signaling pathway. Exp. Cell Res. 299:128-136. [DOI] [PubMed] [Google Scholar]
- 18.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin, M., M. Potente, V. Janssens, D. Vertommen, J. C. Twizere, M. H. Rider, J. Goris, S. Dimmeler, R. Kettmann, and F. Dequiedt. 2008. Protein phosphatase 2A controls the activity of histone deacetylase 7 during T cell apoptosis and angiogenesis. Proc. Natl. Acad. Sci. USA 105:4727-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1996. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16:2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naya, F. J., C. Wu, J. A. Richardson, P. Overbeek, and E. N. Olson. 1999. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development 126:2045-2052. [DOI] [PubMed] [Google Scholar]
- 25.Paroni, G., N. Cernotta, C. Dello Russo, P. Gallinari, M. Pallaoro, C. Foti, F. Talamo, L. Orsatti, C. Steinkuhler, and C. Brancolini. 2008. PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell 19:655-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parra, M., T. Mahmoudi, and E. Verdin. 2007. Myosin phosphatase dephosphorylates HDAC7, controls its nucleocytoplasmic shuttling, and inhibits apoptosis in thymocytes. Genes Dev. 21:638-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provot, S., H. Kempf, L. C. Murtaugh, U. I. Chung, D. W. Kim, J. Chyung, H. M. Kronenberg, and A. B. Lassar. 2006. Nkx3.2/Bapx1 acts as a negative regulator of chondrocyte maturation. Development 133:651-662. [DOI] [PubMed] [Google Scholar]
- 28.Rosen, V., J. Nove, J. J. Song, R. S. Thies, K. Cox, and J. M. Wozney. 1994. Responsiveness of clonal limb bud cell lines to bone morphogenetic protein 2 reveals a sequential relationship between cartilage and bone cell phenotypes. J. Bone Miner. Res. 9:1759-1768. [DOI] [PubMed] [Google Scholar]
- 29.Schipani, E., B. Lanske, J. Hunzelman, A. Luz, C. S. Kovacs, K. Lee, A. Pirro, H. M. Kronenberg, and H. Juppner. 1997. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc. Natl. Acad. Sci. USA 94:13689-13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555-566. [DOI] [PubMed] [Google Scholar]
- 31.Volk, S. W., P. Luvalle, T. Leask, and P. S. Leboy. 1998. A BMP responsive transcriptional region in the chicken type X collagen gene. J. Bone Miner. Res. 13:1521-1529. [DOI] [PubMed] [Google Scholar]
- 32.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weir, E. C., W. M. Philbrick, M. Amling, L. A. Neff, R. Baron, and A. E. Broadus. 1996. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc. Natl. Acad. Sci. USA 93:10240-10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, C. A., H. Yamamoto, T. Fujita, T. Furuichi, K. Ito, K. Inoue, K. Yamana, A. Zanma, K. Takada, Y. Ito, and T. Komori. 2004. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18:952-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng, L., H. Kempf, L. C. Murtaugh, M. E. Sato, and A. B. Lassar. 2002. Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes Dev. 16:1990-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou, H., R. Wieser, J. Massague, and L. Niswander. 1997. Distinct roles of type I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes Dev. 11:2191-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]