Abstract
Acetylcholinesterases (AChEs) and their genes from susceptible and resistant insects have been extensively studied to understand the molecular basis of target site insensitivity. Due to the existence of other resistance mechanisms, however, it can be problematic to correlate directly a mutation with the resistant phenotype. An alternative approach involves recombinant expression and characterization of highly purified wild-type and mutant AChEs, which serves as a reliable platform for studying structure-function relationships. We expressed the catalytic domain of Anopheles gambiae AChE1 (r-AgAChE1) using the baculovirus system and purified it 26,000-fold from the conditioned medium to near homogeneity. While KM's of r-AgAChE1 were comparable for ATC, AβMTC, PTC, and BTC, Vmax's were substantially different. The IC50's for eserine, carbaryl, paraoxon, BW284C51, malaoxon, and ethopropazine were 8.3, 72.5, 83.6, 199, 328, and 6.59×104 nM, respectively. We determined kinetic constants for inhibition of r-AgAChE1 by four of these compounds. The enzyme bound eserine or paraoxon stronger than carbaryl or malaoxon. Because the covalent modification of r-AgAChE1 by eserine occurred faster than that by the other compounds, eserine is more potent than paraoxon, carbaryl, and malaoxon. Furthermore, we found that choline inhibited r-AgAChE1, a phenomenon related to the enzyme activity decrease at high concentrations of acetylcholine.
Keywords: insecticide resistance, mosquito, malaria, organophosphate, carbamate, cholinergic synapse
1. Introduction
Acetylcholinesterases (AChEs) play an essential role in neurotransmission at cholinergic synapses by rapidly hydrolyzing acetylcholine in insects and other animals including humans (Massoulié et al., 1993). Organophosphorous and carbamate pesticides have been developed to inhibit AChEs, leading to acetylcholine accumulation and continuous stimulation of the insect nervous system (Fournier and Mutero, 1994). Because vertebrate AChEs are similar in structure and function to the insect enzymes, application of these chemical pesticides is strictly regulated to prevent accidental exposure of people and livestock to these toxicants. On the other hand, severe resistance has developed in many arthropod species, rendering some existing insecticides ineffective against crop pests and disease vectors (Oakeshott et al., 2005). This situation calls for the development of a new generation of chemical compounds highly selective against the target enzymes in arthropods (Pang, 2006). A Cys residue, located near the active site of AChEs from insects but not vertebrates, was selectively modified by a series of irreversible inhibitors (Pang et al., 2009). To further develop these and other compounds, in-depth understanding of the structure, function, and catalytic mechanism of insect AChEs is needed.
Insect AChEs have been purified to various degrees from at least twenty insect species for biochemical and toxicological analyses (Zhu and Zhang, 2005). These enzymes are more susceptible to inhibition by eserine (physostigmine) than ethopropazine, a relatively selective butyrylcholinesterase inhibitor. The hydrolytic activity of insect AChEs is also inhibited by high concentrations of acetylcholine (Marcel et al., 1998), and this phenomenon is known as “substrate inhibition”. Typically, specific activities of insect AChEs are much lower than those of vertebrate AChEs. Molecular cloning of AChEs has been achieved in flies, mosquitoes, aphids, moths, beetles, and cockroaches (Hall and Spierer, 1986; Weill et al., 2002; Gao et al., 2002; Shang et al., 2007; Kozaki et al., 2008; Kim et al., 2006). Sequence comparison and phylogenetic analysis indicate that most insects contain two AChE genes (ace-1 and ace-2), which arose from an ancient gene duplication before the radiation of arthropod species. Insecticide-insensitive AChEs mainly associate with mutations in ace-1 but not ace-2 (Cassanelli et al., 2006). In Drosophila and other true flies whose ace-1 is lost, AChE2 plays an indispensable role at cholinergic synapses (Weill et al., 2004). The physiological roles of AChE1 and AChE2 are largely unclear in other insect species, although one or both have to function at cholinergic synapses.
Anopheles gambiae is a principal vector of malarial parasites that cause over one million human deaths each year (Seidlein et al., 1998). This species carries ace-1 and ace-2 but none of their gene products have been characterized in sufficient detail. One A. gambiae strain carrying AChE1 (G119S) is insensitive to ten insecticides with resistance ratios similar to those of Culex pipiens expressing AChE1 (G119S) (Alout et al., 2008). In an effort to develop new strategies of vector control, we expressed the catalytic domain of wild-type (i.e. susceptible) A. gambiae acetylcholinesterase-1 (r-AgAChE1) in a baculovirus-insect cell system and characterized the purified enzyme. Kinetic analysis demonstrated that the active anticholinesterase metabolite of the insecticide malathion (i.e. malaoxon) strongly inhibited r-AgAChE1 by tight binding and rapid modification of the active site serine residue. Additionally, choline at concentrations higher than 2 mM inhibited r-AgAChE1.
2. Materials and methods
2.1. Chemicals
acetylthiocholine iodide (ATC), acetyl-(β-methyl)thiocholine iodide (AβMTC), 1,5-bis(4-allyldimethylammoniumphenyl)-pentan-3-one dibromide (BW284C51) (Olivera-Bravo et al., 2005), propionylthiocholine iodide (PTC), S-butyrylthiocholine iodide (BTC), 5,5-dithio-bis(2-nitrobenzoic acid) (DTNB), choline chloride, eserine hemisulfate, ethopropazine hydrochloride, and carbaryl were purchased from Sigma-Aldrich (St. Louis, MO). Paraoxon (O,O'-diethyl p-nitrophenyl phosphate, >99%, the active metabolite of parathion) and malaoxon (O,O'-dimethyl S-(1,2-dicarbethoxy)ethyl phosphorothioate, >99%) were both purchased from Chem Service (West Chester, PA).
2.2. Construction of AgAChE1/pMFH6 and the recombinant baculovirus
The AgAChE1 cDNA clone (BM629847), provided by MR4/ATCC, was completely sequenced and confirmed to encode a polypeptide identical in sequence to AgAChE1 from the susceptible strain (Weill et al., 2002). The cDNA sequence was also compared with its gene to detect sequence variations. The catalytic domain-coding region was amplified from the cDNA template in a polymerase chain reaction using Advantage cDNA Polymerase Mix (BD Biosciences) and two oligonucleotide primers (j910: 5′-GGAATTCACGACAACGATCCGCTG, nucleotides 702-725 and j911: 5′-ACTCGAGGCTGCTTTCGCACG, reverse complement of nucleotides 2353-2373). The thermal cycling conditions were 35 cycles of 94°C for 10 s, 45°C for 5 s and 60°C for 4 min. After electrophoresis, the 1.67 kb product was recovered from the agarose gel and inserted to pGem-T (Promega). Plasmid DNA was prepared from transformants and verified by complete sequence analysis. The cDNA fragment, retrieved by EcoRI-XhoI digestion, was cloned into the same sites in pMFH6 to generate AgAChE1/pMFH6. This cloning strategy allows in-frame fusion with the amino-terminal honeybee mellitin signal peptide and the carboxyl-terminal hexahistidine tag, both encoded by the vector (Lu and Jiang, 2007). In vivo transposition of the expression cassette, selection of bacterial colonies carrying the recombinant bacmid, and isolation of bacmid DNA were performed as previously described (Ji et al., 2004). The initial viral stock (V0) was obtained by transfecting Spodoptera frugiperda Sf21 cells with the bacmid DNA-CellFECTIN mixture, and its titer was improved through serial infection. The V5 viral stock, containing the highest level of baculovirus (1∼2×108 pfu/ml) was stored at -70°C for further experiments.
2.3. Expression and purification of r-AgAChE1
Sf21 cells (at 2.4×106 cells/ml) in 600 ml of Ultimate Insect Serum-Free Medium (Invitrogen Life Technologies) were infected with the viral stock at a multiplicity of infection of 5∼10 and grown at 27°C for 75 h with agitation at 100 rpm. After the cells were removed by centrifugation at 5,000g for 10 min, the culture supernatant was diluted with equal volume of water and the solution pH was adjusted to 6.3. Seventy-five ml of dextran sulfate-Sepharose CL-6B (Nakamura et al., 1985), equilibrated in buffer A (0.01% Tween-20, 10 mM potassium phosphate, pH 6.3), was gently mixed with the protein sample on ice for 1 h. The suspension was loaded into a column (2.5 cm i.d.×20 cm), washed with 200 ml buffer A, and eluted with 300 ml 1 M NaCl in buffer A. Active fractions were pooled and supplemented with 2 mM MgCl2, and its pH was adjusted to 7.5. After incubating with 10 ml Concanavalin A-Sepharose (GE Healthcare Life Sciences) on ice for 1 h, the suspension was loaded into a column, washed with 50 ml buffer B (0.01% Tween-20, 0.5 M NaCl, 20 mM sodium phosphate, pH 7.5), and eluted with 250 ml 0.4 M methyl-α-D-mannopyranoside in buffer B. The combined active fractions were mixed with 3 ml nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) on ice for 1 h and loaded into a column. After washing with 15 ml buffer C (0.01% Tween-20, 10 mM imidazole, 0.3 M NaCl, 20 mM Tris-HCl, pH 8.0), the bound proteins were eluted with 80, 100 and 250 mM imidazole in buffer C (6.0 ml in each step). The purified enzyme was stored at -20°C in the presence of 5% glycerol.
2.4. Measurement of protein concentration and enzyme activity
Protein concentrations were determined by a modified Bradford method using a commercial kit (Pierce) and bovine serum albumin as a standard. AChE activity was measured based on the modified Ellman method (Zhu and Gao, 1999) using ATC and DTNB in a total volume of 100 μl. Absorbance at 405 nm was monitored immediately for 2 min on a VERSAmax microplate reader (Molecular Devices). One unit of AChE activity is defined as the amount of enzyme hydrolyzing one μmole of ATC in 1 min.
2.5. Characterization of the AgAChE1 catalytic domain
Optimal pH for the enzymatic reaction was determined by mixing diluted r-AgAChE1 (3 μl, 3.55 ng/μl) with amphoteric buffer (17 μl, 1:5 diluted Polybuffer 96, GE Healthcare Life Sciences) adjusted to various pH and 80 μl substrate solution. For control, r-AgAChE1 was replaced by buffer C (3 μl). After activity measurement, the final pH readings of the test and control mixtures were taken using a microelectrode (Sentron pH-System). Enzyme-catalyzed ATC hydrolysis was plotted against pH to reveal the optimal pH of r-AgAChE1. Apparent molecular mass of r-AgAChE1 was determined by gel filtration chromatography on an HPLC column and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing or nondenaturing condition. The protein was visualized by Coomassie blue or silver staining and by immunoblot analysis using anti-(His)5 monoclonal antibodies (Qiagen). N-linked glycosylation was detected by treating the enzyme with 1×glycoprotein denaturing buffer (Sigma) at 100°C for 10 min. After adding one-tenth volume each of 10×G7 buffer (50 mM sodium phosphate, pH 7.5) and 10% Nonidet P-40, the protein was incubated with 2.5 μl PNGase F (Sigma) at 37°C for 1 h. The reaction mixture, as well as untreated control, was heated at 95°C for 5 min in the presence of 1×SDS sample buffer containing dithiothreitol (DTT), separated by SDS-PAGE, and stained with Coomassie blue.
2.6. Kinetics of substrate hydrolysis
The purified r-AgAChE1 (36.6 μg/μl) was diluted 1:500 (for ATC, AβMTC, and PTC) and 1:100 (for BTC), and aliquots of the diluted enzyme (20 μl) were separately incubated with the substrates at various concentrations. Substrate hydrolysis was monitored at room temperature for 2.5 min in the microplate assay described above. The activity data (mOD/min) were converted to specific activity (μmol/min/mg protein) as follows: (mOD/min×5×enzyme dilution factor)/(1.36×104M-1cm-1×0.30cm×[Et]), where [Et] = 36.6 μg/ml. KM and Vmax for each substrate were calculated by fitting the velocity (v) and substrate concentration ([S]) data to v = Vmax[S]/(KM + [S]) using Prism 3.0 (GraphPad Software Inc).
2.7. Determination of IC50's and inhibitory kinetic constants
The purified r-AgAChE1 (0.244 ng/μl, 10 μl) was separately incubated with BW284C51, carbaryl, eserine hemisulfate, ethopropazine, malaoxon, or paraoxon (10 μl) at various concentrations for 10 min at room temperature. The residual activity (v) was determined by the microplate reader assay for 5 min and plotted against log10[I]. IC50 for each inhibitor was obtained by nonlinear regression analysis of v and log10[I] data using the sigmoidal dose-response equation (Prism 3.0). Dissociation equilibrium constant (Kd) (i.e. affinity constant Ka), unimolecular rate constant (k2), and bimolecular reaction constant (ki) for SgAChE1 inhibition were determined according to Hart and O'Brien (1973). Aliquots of the diluted enzyme (0.157 ng/μl, 10 μl) were individually added to 80 μl ATC-DTNB premixed with 10 μl carbaryl, eserine, malaoxon, or paraoxon at different concentrations. Absorbance at 405 nm was monitored immediately for 5 min on the microplate reader at fifteen-second intervals. k was calculated by fitting the A405nm and t data to one-phase exponential association equation: A = A∞(1-e−kt). Then, 1/k and 1/[I] values were plotted and analyzed by linear regression (Gray and Duggleby, 1989): 1/k2 is the intercept on 1/k axis whereas Kd is calculated from the intercept on 1/[I] axis, i.e. −KM/Kd(KM+[S]), where [S] (600 μM) and KM are for ATC. ki is calculated from ki = k2/Kd.
2.8. Effect of acetylthiocholine and choline on r-AgAChE1 activity
After the purified enzyme (1.12 ng/μl, 10 μl) was mixed with DTNB (480 μM, 10 μl) and ATC at various concentrations (18.75-625 mM, 80 μl), absorbance at 405 nm was monitored using the microplate reader and enzyme-catalyzed substrate hydrolysis was plotted against ATC concentrations. To test whether the reduction of ATC hydrolysis was due to substrate and/or product inhibition, this experiment was repeated by measuring AChE activities at 0 and 30 min in the presence of 15, 30, 60, or 120 mM ATC. The reaction mixtures (20 μl) were then transferred into new wells containing 200 μl buffer (control) or ATC at the same concentration. Rates of absorbance change were directly plotted in a bar graph along with the activity at 0 and 30 min.
To prove that product inhibition does occur, acetate, choline, or both (10 μl) at different concentrations, as well as a buffer, was mixed with the same amount of r-AgAChE1 (1.12 ng/μl, 10 μl) for 5 min at room temperature. After the substrate solution (80 μl, 60 μM DTNB and 750 μM ATC) was added, the enzyme activities were determined immediately using the microplate assay and plotted against product concentrations.
3. Results
3.1. Features of A. gambiae AChE1 cDNA and gene
The full-length cDNA, 3,574 bp long, contains an open reading frame ranging from nucleotides 276-2441 (Fig. S1). The 5′ untranslated region corresponds to exon 1, exon 2, and 5′ end of exon 3 of the gene. The sizes of introns 1-3 (954, 3925 and 1938 bp) are significantly longer than those of introns 4-8 (86, 79, 86, 66, 107 bp). The rest of exon 3 encodes a 24-residue signal peptide for secretion and, along with exon 4, encodes a Ser/Ala-rich pro-region. Since its counterpart was absent in the purified Schizaphis graminum AChE1 (Gao and Zhu, 2001), we suspect proteolytic processing also occurs in the maturation of A. gambiae pro-AChE1. Exons 5-8 encode the entire catalytic domain, followed by a carboxyl-terminal tail critical for self-association and membrane anchorage. Exon 9 encodes the tail and 3′ untranslated region (1118 bp). The AATAAA motif near the 3′ end may act as the signal for polyadenylation. A comparison of the cDNA and gene sequences revealed eight synonymous substitutions.
3.2. Production of the catalytic domain of A. gambiae AChE1 (r-AgAChE1)
To express the enzyme for functional analysis, we amplified the region coding for the catalytic domain which is defined as D122NDP…CESS673. Based on our sequence comparison (data not shown), D122 corresponded to the first residue of mature D. melanogaster AChE2 and the third residue of processed S. graminum AChE1 (Gao et al., 2002). While C670 was kept for forming a possible disulfide bond, we deleted the hydrophobic tail starting at A674 to prevent the recombinant enzyme from associating with cell membrane. The PCR product, after digestion with EcoRI and XhoI, was inserted into the same sites to yield AgAChE1/pMFH6. Complete sequence analysis showed one synonymous change caused by Taq DNA polymerase. The recombinant plasmid was used to generate a viral stock through transposition, transfection and serial amplification. Under the optimal conditions, r-AgAChE1 was secreted by the baculovirus-infected Sf21 cells at a final concentration of 1.54 μg/ml and 80.6 U/ml. Following an ion exchange step, the captured protein was eluted from the polycationic resin in an enriched form free of medium components or nucleic acids that interfere with affinity chromatography. A fifteen-fold increase in specific activity was achieved using Concanavalin A-Sepharose (Table 1). Similar to the ion exchange step, A. gambiae AChE1 strongly associated with the resin and came off the column in a large volume. The recombinant protein in pooled fractions tightly bound to the Ni2+-NTA agarose and while 80-100 mM imidazole efficiently removed loosely associated proteins, most r-AgAChE1 remained attached to the column until 250 mM imidazole was applied. The eluted enzyme (0.48 mg) was essentially pure and recognized by the monoclonal antibodies against the hexahistidine tag (Fig. 1). The isolated A. gambiae AChE1 had a high specific activity of 523 U/mg, while the overall purification factor and yield were 26,000-fold and 51.8%, respectively (Table 1).
Table 1. Purification of A. gambiae AChE1.
| sample | volume (ml) |
protein | activity | yield (%) |
specific activity (U/mg) |
purification (fold) |
||
|---|---|---|---|---|---|---|---|---|
| (μg/ml) | (mg) | (U/ml) | (U) | |||||
| medium | 600 | 3820 | 2292 | 0.81 | 4.8×102 | 100 | 0.21 | 1 |
| dextran sulfate | 300 | 978 | 293 | 1.18 | 3.5×102 | 73.1 | 1.21 | 6 |
| Concanavalin A | 210 | 81 | 17.0 | 1.47 | 3.1×102 | 64.0 | 18.2 | 86 |
| Ni-NTA | 8.5 | 56 | 0.48 | 29.5 | 2.5×102 | 51.8 | 523 | 2491 |
Fig. 1. Electrophoretic separation of A. gambiae AChE1 samples from different purification steps on 10% SDS-polyacrylamide gels followed by silver staining.
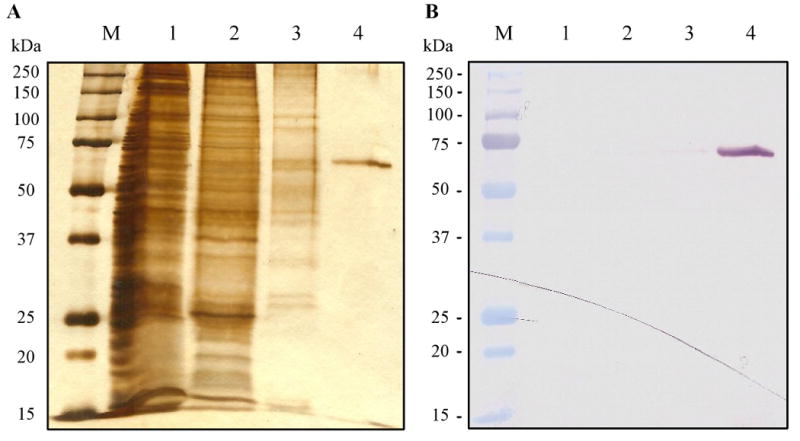
(A) and immunoblot detection (B) The pre-stained molecular weight markers (lane M), the culture supernatant (lane 1), and proteins eluted from dextran sulfate-Sepharose (lane 2), Concanavalin A-Sepharose (lane 3), and Ni2+-NTA agarose (lane 4) columns were separated on the gel under reducing condition. Recombinant A. gambiae AChE1 was detected using monoclonal antibodies against its carboxyl-terminal hexahistidine tag.
3.3. Biochemical properties of r-AgAChE1
The calculated Mr and isoelectric point of r-AgAChE1 are 62,921 Da and 5.97. Deduced from the inserted cDNA sequence, mature r-AgAChE1 has the following amino acid sequence: G1IHDNDPL…PCESSLEHHHHHH563, in which the underlined portion is identical to residues D122 through S673 of A. gambiae AChE1 (Fig. S1). There are three predicted N-linked glycosylation sites (N62, N455, and N512) in the sequence but no O-linked glycosylation site. The enzyme contains the catalytic triad comprising S202, E328 and H442, as well as the hydrophobic residues (W87, W117, Y124, Y133, W235, F291, F293, F332, Y335, and W434) lining the active site gorge. Six absolutely conserved Cys residues may form three disulfide bonds (C70-C97, C156-C269, and C404-C525). While C289 is located at the entry point of the active site in insect AChE1s (Pang, 2006), Cys552 of the adjacent subunits may form an interchain disulfide linkage.
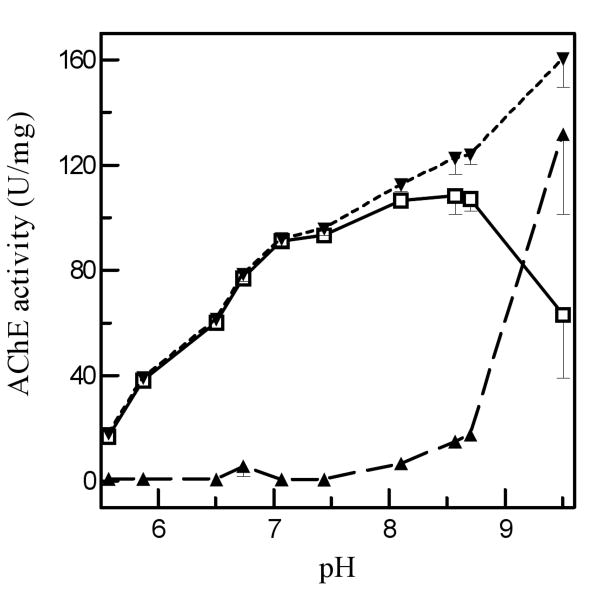
The AChE activity increased from pH 5.5 to 7.0 and remained at a high level from pH 7.0 to 8.5 (Fig. 2). While the activity peaked at pH 8.5, spontaneous substrate hydrolysis occurred at pH 8.1 and became severe as pH increased. The optimal pH for the enzymatic reaction is 7.0-7.5.
Fig. 2. Effect of pH on acetylthiocholine hydrolysis by purified A. gambiae AChE1.
As described in Materials and Methods, 3.0 μl aliquots of the purified enzyme or buffer were mixed with amphoteric buffers (17 μl, adjusted to various pH) and 80 μl substrate solution. After activity and pH measurements, enzyme-catalyzed ATC hydrolysis (○—○), which is equal to the activity difference between test (▼---▼) and control (▲- - -▲), is plotted against pH and vertical bars indicates standard errors of the means (n = 3).
The purified enzyme had an apparent molecular mass of 65 and 130 kDa on SDS-PAGE gels under reducing and nonreducing conditions, respectively (Fig. 3A). This suggested that the recombinant protein existed as a homodimer stabilized by an interchain disulfide bridge. The native protein eluted from the HPLC gel filtration column at 12.2 min (Fig. 3B), corresponding to an apparent Mr of 60 kDa. While this is significantly lower than the estimate (130 kDa) from the nonreducing SDS-PAGE analysis, we suspect that the native enzyme had interacted with the column and the direct interpretation based on retention time was erroroneous. The association status of r-AgAChE1 remained unclear since the homodimer may associate with each other non-covalently, as demonstrated in mammals (Massoulié et al., 2005).
Fig. 3. Determination of the association status of A. gambiae AChE1 by 7.5% SDS-PAGE and HPLC gel filtration chromatography.
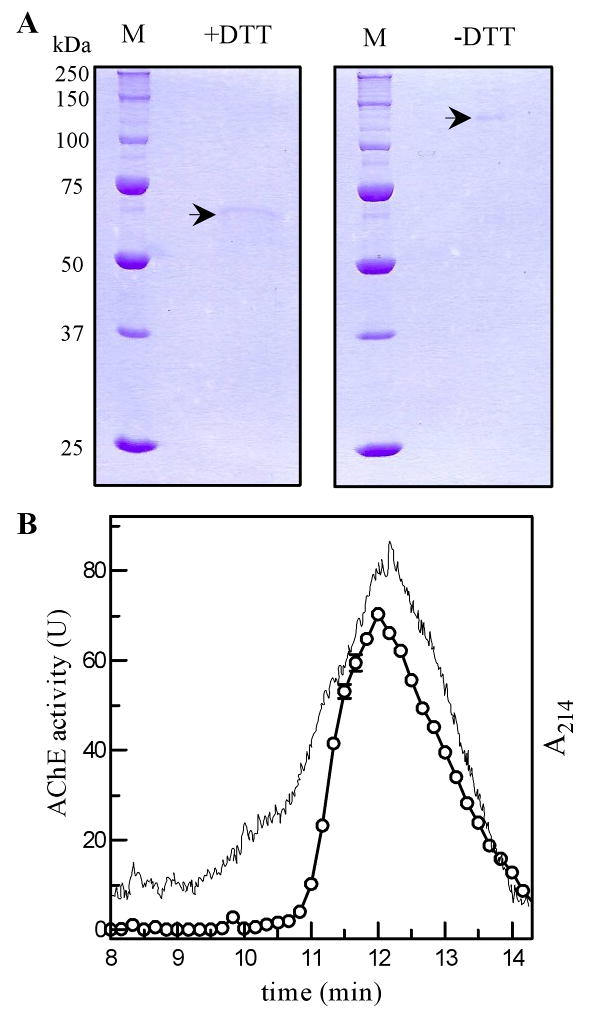
(A) The purified protein was treated with SDS sample buffer with (left panel) or without (right panel) dithiothreitol, separated on SDS gel, and visualized by Coomassie blue staining. (B) The purified A. gambiae AChE1 was separated on the HPLC gel filtration column calibrated with molecular weight standards. The elution times for 670 kDa thyroglobulin, 158 kDa bovine γ-globulin, 44 kDa chicken ovalbumin, 17 kDa equine myoglobin, and 1.35 kDa vitamin B12 were 7.50, 9.97, 11.28, 12.98, and 15.13 min, respectively. The activity in the fractions (○—○) were measured and shown as mean ± S.E. (n = 3), along with the absorbance at 214 nm (—).
In order to examine whether or not A. gambiae AChE1 is glycosylated, the purified enzyme was treated with N-glycosidase and analyzed by SDS-PAGE under reducing conditions (Fig. 4). The mobility increase indicated that the recombinant protein was posttranslationally modified. In contrast, after the enzyme had been treated with O-glycosidase, there was no such change (data not shown). These data are consistent with predictions based on the amino acid sequence.
Fig. 4. Deglycosylation of A. gambiae AChE1 by N-glycosidase F.
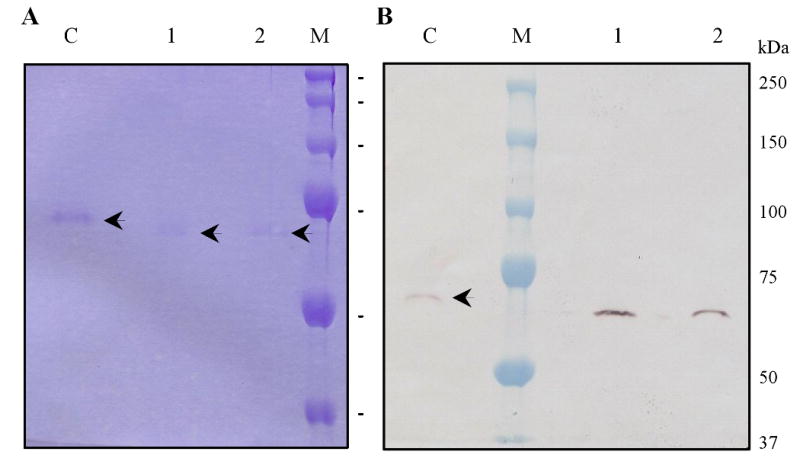
The purified enzyme was treated with buffer (lane “C”) or PNGase F (lanes 1 and 2), separated by 10% (A) or 7.5% (B) SDS-PAGE under reducing condition, and visualized by Coomassie Blue staining (A) and monoclonal antibodies against the hexahistidine tag (B). Sizes and positions of the molecular weight markers are indicated on the right.
3.4. Substrate and inhibitor specificities of A. gambiae AChE1
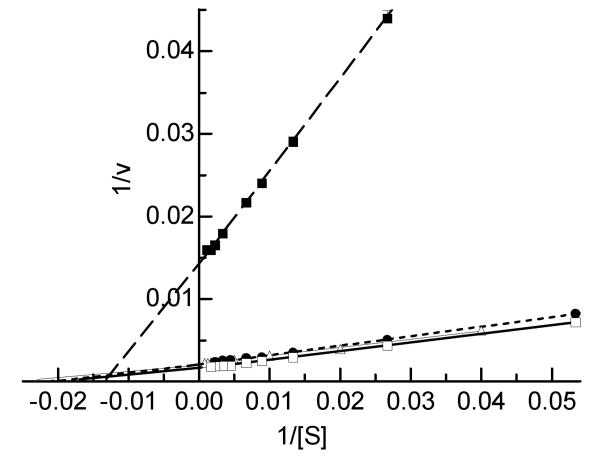
We determined the kinetic parameters of A. gambiae AChE1 using a panel of four substrates at various concentrations and found that the enzyme-catalyzed hydrolysis generally followed the Michaelis-Menten kinetics (Fig. 5). As revealed by the comparable KM values (Table 2), A. gambiae AChE1 seemed to bind these artificial substrates with similar affinity. On the other hand, the Vmax's for these reactions were significantly different: 622.0±12.9, 467.8±7.1, 460.8±3.4 and 70.1±0.6 U/mg for ATC, AβMTC, PTC and BTC, respectively. The relative catalytic efficiencies of ATC, AβMTC and PTC, as indicated by their Vmax/KM ratios (9.34, 8.95 and 10.34, respectively), are similar to each other but are 9.9- to 11.5-fold as high as that of BTC (0.90). Acetylcholine (rather than butyrylcholine) is likely the natural substrate of A. gambiae AChE1.
Fig. 5. Determination of the enzymatic properties of A. gambiae AChE1 using four different substrates.
Hydrolysis of ATC (□—□), AβMTC (●---●), PTC (Δ—Δ) and BTC (■- - -■) by the purified enzyme was measured as described in Materials and Methods. Each point on the double reciprocal plot represents mean ± S.E. (n = 4). KM and Vmax values for each substrate were derived from v versus [S] plot (data not shown) by the nonlinear regression analysis.
Table 2. Kinetic parameters of recombinant A. gambiae AChE1*.
| substrate | KM (μM) | Vmax (U/mg) | Vmax/KM (L·mg-1·min-1) | kcat ×10-4 (min-1) |
|---|---|---|---|---|
| ATC | 63.85 ± 3.23 | 622.0 ± 12.9 | 9.74 | 3.91±0.08 |
| AβMTC | 52.30±1.70 | 467.8±7.1 | 8.95 | 2.94±0.04 |
| PTC | 44.49 ± 1.10 | 460.8 ± 3.4 | 10.34 | 2.90±0.02 |
| BTC | 77.96 ± 1.43 | 70.1 ± 0.6 | 0.90 | 0.44±0.00 |
Results are presented as the mean ± S.D. (n=4). Correlation coefficients (r2) for the four substrates are 0.98-1.00 on the double-reciprocal plot. Turnover rates (kcat) are calculated based on the molecular mass (62,921 Da) and maximum velocity (Vmax) of r-AgAChE1.
3.5. Inhibition kinetics
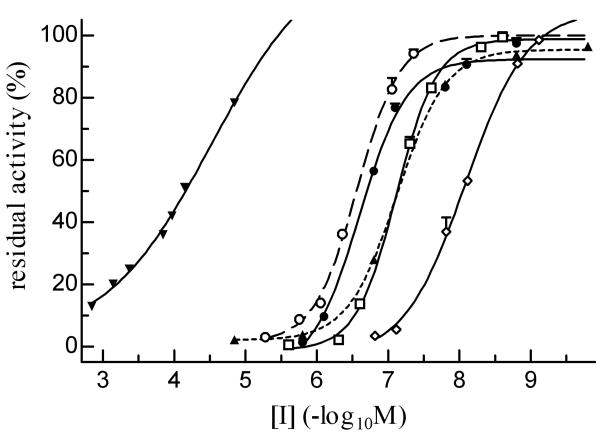
AChE was strongly inhibited by eserine, carbaryl and paraoxon, and less strongly by BW284C51 and malaoxon (Fig. 6). The IC50 of ethopropazine (65.9 μM) was approximately 200, 330, 790, and 910 times as high as those of malaoxon (0.328 μM), BW284C51 (0.199 μM), paraoxon (83.6 nM) and carbaryl (72.5 nM), respectively (Table 3). The other carbamate, eserine blocked 50% of A. gambiae AChE1 activity at a low concentration of 8.27 nM.
Fig. 6. Inhibition of r-AgAChE1 by organophosphates, carbamates and other compounds at various concentrations.
After incubation with its inhibitors for 10 min at 25°C, the purified enzyme was reacted with ATC-DTNB and monitored by a microplate reader at 405 nm. The residual activities, shown as mean ± S.E. (n = 3), are plotted against the inhibitor concentrations. Ethopropazine (▼―▼), BW284C51 (○- - -○), eserine (◊―◊), carbaryl (▲---▲), paraoxon (□―□), and malaoxon (●―●).
Table 3. IC50 of carbamates, organophosphates, and other compounds towards recombinant A. gambiae AChE1*.
| carbamate | IC50 (M) | r2 |
|---|---|---|
| carbaryl | (7.25 ±0.47)×10-8 | 1.00 |
| eserine | (8.27 ±0.75)×10-9 | 0.99 |
| BW284C51 | (1.99 ±0.26)×10-7 | 0.99 |
| ethopropazine | (6.59 ±0.48)×10-5 | 0.99 |
| malaoxon | (3.28 ±0.38)×10-7 | 0.99 |
| paraoxon | (8.36 ±0.87)×10-8 | 0.99 |
Results are presented as the mean ± S.D. (n=3). r2: average correlation coefficient.
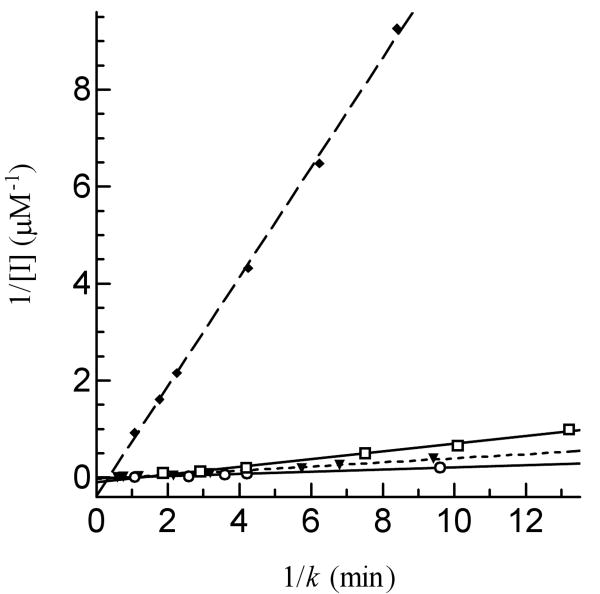
To characterize further the sensitivity of r-AgAChE1 to inhibition by different cholinesterase inhibitors, we determined kinetic parameters for eserine, carbaryl, paraoxon and malaoxon (Fig. 7 and Table 4). The dissociation constants (Kd's) indicated that eserine (0.26 ± 0.02 μM) or paraoxon (1.00 ± 0.14 μM) bound r-AgAChE1 tighter than carbaryl (4.70 ± 1.63 μM) or malaoxon (5.85 ± 0.79 μM). The Michaelis complex of r-AgAChE1 and eserine converted to an acyl-enzyme intermediate at a rate (k2: 3.02 ± 0.21 min-1) higher than that of carbaryl (1.84 ± 0.37 min-1), malaoxon (1.37 ± 0.14 min-1) or paraoxon (0.79 ± 0.07 min-1), eserine has the highest ki value (ki: 11.74 μM-1min-1) and is more potent than the other three inhibitors (ki: 0.24-0.83 μM-1min-1). The ki values of these four compounds have a similar order as their IC50's do.
Fig. 7. Determination of Kd and k2 values of four inhibitors against r-AgAChE1.
Aliquots of the diluted enzyme (10 μl, 1.12 ng/μl) were individually added to 80 μl ATC-DTNB premixed with 10 μl carbaryl (▼---▼), eserine (◆- - -◆), malaoxon (○―○), or paraoxon (□―□) at different concentrations. Absorbance at 405 nm was monitored immediately on the microplate reader at fifteen-second intervals for 5 min and the readings were used to derive k's by curve fitting [A = A∞(1-e−kt)]. Then, 1/k and 1/[I] values were plotted and analyzed by linear regression as described in Materials and Methods.
Table 4. Inhibition constants of carbaryl, eserine, paraoxon, and malaoxon.
| Ka (μM) | ki (μM-1min-1) | k2 (min-1) | r2 | |
|---|---|---|---|---|
| carbaryl | 4.70 ± 1.63 | 0.440 ± 0.023 | 1.84 ± 0.37 | 0.982 |
| eserine | 0.26 ± 0.02 | 11.740 ± 0.223 | 3.02 ± 0.21 | 0.999 |
| malaoxon | 5.85 ± 0.79 | 0.242 ± 0.008 | 1.37 ± 0.14 | 0.996 |
| paraoxon | 1.00 ± 0.14 | 0.830 ± 0.045 | 0.79 ± 0.07 | 0.988 |
r2: correlation coefficient of the linear regression (p-value < 0.0001)
3.6. Inhibitory effect of ATC and choline at high concentrations
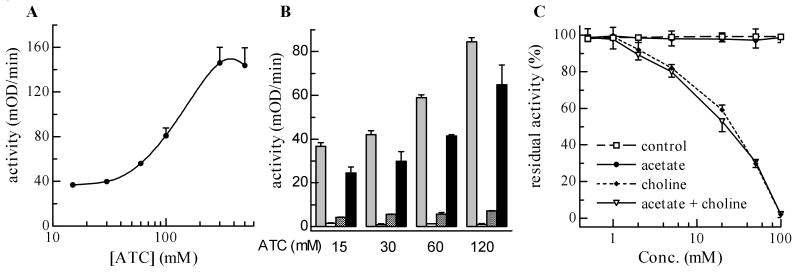
Since substrate inhibition is a common feature of insect AChEs, we tested if the enzyme activity of r-AgAChE1 is affected by ATC at high concentrations. Fig. 8A confirmed the phenomenon by showing an activity decrease in the presence of ATC (≥ 0.4 M). Since we do not know the acetylcholine concentration at synaptic gaps of any insects, the physiological relevance of such inhibition is unknown at this time. As a matter of fact, neither is it clear whether substrate itself (rather than product or both substrate and product) causes the activity loss. To confirm the inhibition and address the latter question, we repeated the experiment by measuring substrate hydrolysis at 1 and 30 min. At four different concentrations of ATC, there was always a significant activity reduction at 30 min and a large increase of substrate hydrolysis after the reaction mixtures were diluted in fresh buffer (Fig. 8B). In other words, the enzyme (not active in the presence of ATC and products at 30 min) was disinhibited by dilution of one or more of the reaction constituents. To distinguish between substrate and product inhibition, we also diluted the reaction mixtures in substrate solutions at the same concentrations and observed greater activity recovery. Since the substrate concentration did not change (substrate consumption in the first 30 min was negligible), the disinhibition appeared due to reaction component(s) rather than ATC, possibly acetate or choline. Therefore, we directly examined whether or not acetate and/or choline negatively impacted r-AgAChE1. Neither acetate (0.5-100 mM) nor buffer caused any significant decrease in activity, whereas choline at 2, 5, 20 and 100 mM led to approximately 10, 20, 50 and 100% activity loss, respectively (Fig. 8C). Inclusion of acetate at the same concentration had no additive effect.
Fig. 8. Activity of r-AgAChE1 activity influenced by acetylthiocholine and choline.
A Effect of substrate concentrations was investigated using ATC as described in Materials and Methods. A cubic spline curve is used to fit the data (mean ± SEM, n=3). B) Effects of reaction time, ATC concentration, and dilution solution on the enzyme activity were studied according to the experimental design (section 2.8). Grey bar, 0 min; white bar, 30 min; shaded bar, diluted with buffer; black bar, diluted with substrate at the same concentration. Whiskers indicate standard errors of the means (n = 3). C) Effect of buffer control, acetate, choline, or a mixture of acetate and choline on r-AgAChE1 was tested as described in Materials and Methods. Residual activity data (mean ± S.E., n=3) were plotted against concentrations of acetate or choline and data points in individual groups are connected by straight lines.
4. Discussion
Since insecticide-insensitive AChEs mainly associate with mutations in ace-1 (Cassanelli et al., 2006; Alout et al., 2007), we expressed the catalytic domain of AChE1 from A. gambiae, a major vector of malarial parasites. The purification procedure, taking approximately 8 h to finish, had an overall yield of 52% and was successfully used for isolating r-AChEs from other insect species (Zhao et al., unpublished data). Baculovirus-insect cell systems have been developed to produce AChEs, in most cases for mutation analysis (Anthony et al., 1995; Harel et al., 2000; Walsh et al., 2001; Chen et al., 2001; Kim et al., 2007; Shang et al., 2007; Carlier et al., 2008). Our system, which takes advantage of an efficient signal peptide and strong affinity binding of r-AgAChE1 to Ni-NTA agarose, provides highly purified r-AgAChE1 in sufficient amount for structural analysis and potential development of a new generation of selective insecticides that target the sulfhydryl group of in arthropod AChEs (Pang, 2006; Pang et al., 2009).
We selected to express the catalytic domain of AgAChE1 because of the following reasons. AChEs from D. melanogaster, Nasonia vitripennis, and Apis mellifera contain no or only a few residues between signal peptide and catalytic domain (data not shown). BLASTP search using the putative pro-region of AgAChE1 as query has not revealed significant sequence similarity beyond mosquitoes (data not shown). However, this region shares a feature with the processed part of S. graminum AChE1: high abundance in hydrophilic and charged residues (Fig. S1 and Gao et al., 2002). While function of the pro-segment (longer than 200 residues in some cases) is unclear at this time, structural flexibility caused by this hydrophilic segment can be detrimental to the structural analysis.
The recombinant AgAChE1 had an apparent molecular mass of 65 kDa and formed a dimer. Its specific activity towards ATC is 523 U/mg, significantly higher than AChEs from other orders of insects. This is consistent with the notion that AChEs from dipteran species are more efficient in hydrolyzing acetylcholine and its surrogate (Toutant, 1989). At its optimal pH of 7.0-7.5, r-AgAChE1 hydrolyzes ATC much faster than BTC. The ratio of Vmax's for BTC and ATC was 0.09, lower than that of D. melanogaster (0.56) (Gnagey et al., 1987) or Diabrotica virgifera (0.1) (Gao at el., 1998) but higher than that of Schizaphis graminum (0.03) (Gao and Zhu, 2001) and Rhyzopertha dominica (0.032) (Guedes et al., 1998). Therefore, AChEs from different insect species differ in their relative specificity toward ATC and BTC even though they all prefer ATC.
Since KM values of an enzyme partly reflect its binding affinity toward different substrates, it is surprising that r-AgAChE1 has similar KM (44.5 ∼ 78.0 μM) for ATC, AβMTC, PTC, and BTC. Perhaps future studies will reveal structural features of the substrate binding site which accommodates the substrates with different acyl groups. An alternative explanation could be that the subdomain flexibility of r-AgAChE1 is unusually high (Stojan et al., 2008). As a result of the small KM variations, differences in catalytic efficiency (Vmax/KM) of r-AgAChE1 to certain extent represent variations in Vmax (Table 2): while hydrolysis of AβMTC (Vmax/KM: 8.95) and PTC (10.34) was equally efficient, r-AgAChE1 catalyzed ATC breakdown (9.74) twelve times as efficiently as with BTC (0.90).
Additional evidence for A. gambiae AChE1 being an esterase of acetylcholine rather than butyrylcholine was obtained from our inhibition analyses. Ethopropazine, a relatively selective inhibitor of butyrylcholinesterases, had an IC50 of 6.59×104 nM, much higher than that of malaoxon (328 nM) or BW284C51 (199 nM), relatively specific inhibitors of AChEs (Table 3). BW284C51, however, was not as potent as paraoxon (83.6 nM), carbaryl (72.5 nM) or eserine (8.3 nM), inhibitors that covalently modify the reactive site Ser residue in r-AgAChE1.
The ki values for eserine, paraoxon and malaoxon (Table 4) are larger than but in the same order as those reported previously (Alout et al., 2008) (4.201, 0.266, 0.210 μM-1min-1). The difference is probably due to systematic errors: we used highly purified AgAChE1 catalytic domain whereas mosquito extracts contain components (e.g. AChE2 and other esterases) that may preferentially sequester the inhibitors and interfere with their binding to AgAChE1.
A reduction in enzyme activity at high substrate concentrations, known as substrate inhibition, is a feature of many insect AChEs (Toutant, 1989). Depending on the species source of the enzyme, substrate inhibition can be seen at concentrations ranging from 50 μM to 10 mM (Zhu and Clark, 1994; Gao and Zhu, 2001). In our studies with r-AgAChE1 substrate inhibition occurred at around 0.4 M (Fig. 8A), thus we questioned its physiological relevance. Nonetheless, this surprising observation led us to evaluate it at lower concentrations of substrate (15-120 mM ATC) and determine whether disinhibition of r-AgAChE1 was influenced by reagent dilution (Fig. 8B). Dilution while maintaining ATC at the same concentration clearly demonstrated a role of reaction constituent(s) other than the substrate in the suppression of AChE1 activity and led to the discovery that choline, one of the hydrolysis products of acetylcholine, is at least partly responsible for the activity loss (Fig. 8C). Similar inhibition by choline was observed when we studied r-AChE1 from the greenbug Schizaphis graminum (Zhao at al., unpublished results).
Supplementary Material
The last nucleotide of each exon is shaded to indicate the splicing junctions. Single nucleotide differences between the cDNA and gene are underlined. The polyadenylation signal (AATAAA) is double underlined. Amino acid residues, shown in one-letter abbreviations, are aligned with the second nucleotide of each codon. The predicted signal peptide (-24 ∼ -1) is double underlined, putative N-linked glycosylation sites are marked “×”, and the stop codon is indicated by “*”. Positions of the cysteine residues conserved in all AChEs are marked “& – &, $ – $, and ● – ●” to show disulfide bond connectivity. The unique Cys residue present in insect AChE1s (Pang, 2006) is double underlined, whereas the one possibly involved in interchain disulfide linkage is labeled “#”. The catalytic triad (Ser, His and Glu) is in bold and shaded, and the hydrophobic residues (F, W, and Y) lining the active site gorge are labeled with “+”. The primer binding sites for recombinant expression in the baculovirus-insect cell system are underlined.
Acknowledgments
We thank Dr. Kun-Yan Zhu for critical comments on the manuscript. Dr. Zhiqiang Lu has provided assistance in the HPLC gel filtration experiment. This work was supported by National Institutes of Health Grant GM58634 (to H.J.). This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450.
Abbreviations
- AChE
acetylcholinesterase
- ATC
acetylthiocholine iodide
- AβMTC
acetyl-(β-methyl)thiocholine iodide
- PTC
propionylthiocholine iodide
- BTC
butyrylthiocholine iodide
- DTNB
5,5-dithio-bis(2-nitrobenzoic acid)
- OP
organophosphorous compounds
- BW284C51
1,5-bis(4-allyldimethylammoniumphenyl)-pentan-3-one dibromide
- HPLC
high performance liquid chromatography
- Ni-NTA
nickel-nitrilotriacetic acid
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- r-AgAChE1
recombinant A. gambiae acetylcholinesterase-1 catalytic domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alout H, Berthomieu A, Hadjivassilis A, Weill M. A new amino-acid substitution in acetylcholinesterase 1 confers insecticide resistance to Culex pipiens mosquitoes from Cyprus. Insect Biochem Mol Biol. 2007;37:41–47. doi: 10.1016/j.ibmb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Anthony N, Rocheleau T, Mocelin G, Lee HJ, ffrench-Constant R. Cloning, sequencing and functional expression of an acetylcholinesterase gene from the yellow fever mosquito Aedes aegypti. FEBS Lett. 1995;368:461–465. doi: 10.1016/0014-5793(95)00711-h. [DOI] [PubMed] [Google Scholar]
- Carlier PR, Anderson TD, Wong DM, Hsu DC, Hartsel J, Ma M, Wong EA, Choudhury R, Lam PC, Totrov MM, Bloomquist JR. Towards a species-selective acetylcholinesterase inhibitor to control the mosquito vector of malaria, Anopheles gambiae. Chem Biol Interact. 2008;175:368–375. doi: 10.1016/j.cbi.2008.04.037. [DOI] [PubMed] [Google Scholar]
- Cassanelli S, Reyes M, Rault M, Carlo Manicardi G, Sauphanor B. Acetylcholinesterase mutation in an insecticide-resistant population of the codling moth Cydia pomonella (L.) Insect Biochem Mol Biol. 2006;36:642–653. doi: 10.1016/j.ibmb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chen Z, Newcomb R, Forbes E, McKenzie J, Batterham P. The acetylcholinesterase gene and organophosphorus resistance in the Australian sheep blowfly, Lucilia cuprina. Insect Biochem Mol Biol. 2001;31:805–816. doi: 10.1016/s0965-1748(00)00186-7. [DOI] [PubMed] [Google Scholar]
- Fournier D, Mutero A. Modification of acetylcholinesterase as a mechanism of resistance to insecticides. Comp Biochem Physiol. 1994;108C:19–31. [Google Scholar]
- Gao JR, Rao JV, Wilde GE, Zhu KY. Purification and kinetic analysis of acetylcholinesterase from western corn rootworm, Diabrotica virgifera (Coleoptera: Chrysomelidae) Arch Insect Biochem Phys. 1998;39:118–125. doi: 10.1002/(SICI)1520-6327(1998)39:3<118::AID-ARCH4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Gao JR, Zhu KY. An acetylcholinesterase purified from the greenbug (Schizaphis graminum) with some unique enzymological and pharmacological characteristics. Insect Biochem Mol Biol. 2001;31:1095–1104. doi: 10.1016/s0965-1748(01)00057-1. [DOI] [PubMed] [Google Scholar]
- Gao JR, Kambhampati S, Zhu KY. Molecular cloning and characterization of a greenbug (Schizaphis graminum) cDNA encoding acetylcholinesterase possibly evolved from a duplicate gene lineage. Insect Biochem Mol Biol. 2002;32:765–775. doi: 10.1016/s0965-1748(01)00159-x. [DOI] [PubMed] [Google Scholar]
- Gnagey AL, Forte M, Rosenberry TL. Isolation and characterization of acetylcholine esterase from Drosophila. J Biol Chem. 1987;262:13290–13298. [PubMed] [Google Scholar]
- Gray PJ, Duggleby RG. Analysis of kinetic data for irreversible enzyme inhibition. Biochem J. 1989;257:419–424. doi: 10.1042/bj2570419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes RNC, Zhu KY, Kambhampti S, Dover BA. Characterization of acetylcholineesterase purified from the lesser grain borer, Rhyzopertha dominica. Comparative Biochem Physiol. 1998;119C:205–210. doi: 10.1016/s0742-8413(97)00208-9. [DOI] [PubMed] [Google Scholar]
- Hall LMC, Spierer P. The Ace locus of Drosophila melanogaster: structural gene for acetylcholinesterase with an unusual 5′ leader. EMBO J. 1986;5:2949–2954. doi: 10.1002/j.1460-2075.1986.tb04591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel M, Kryger G, Rosenberry TL, Mallender WD, Lewis T, Fletcher RJ, Guss JM, Silman I, Sussman JL. Three-dimensional structure of Drosophila melanogaster acetylcholine esterase and of its complex with putative insecticides. Protein Sci. 2000;9:1063–1072. doi: 10.1110/ps.9.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GJ, O'Brien RD. Recording spectrophotometric method for determination of dissociation and phosphorylation constants for the inhibition of acetylcholinesterase by organophosphates in the presence of substrate. Biochemistry. 1973;12:2940–2945. doi: 10.1021/bi00739a026. [DOI] [PubMed] [Google Scholar]
- Ji C, Wang Y, Ross J, Jiang H. Expression and in vitro activation of Manduca sexta prophenoloxidase-activating proteinase-2 precursor (proPAP-2) from baculovirus-infected insect cells. Protein Exp Purif. 2003;29:235–243. doi: 10.1016/s1046-5928(03)00020-2. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Yoon KS, Clark JM. Functional analysis of mutations in expressed acetylcholinesterase that result in azinphosmethyl and carbofuran resistance in Colorado potato beetle. Pestic Biochem Physiol. 2007;88:181–190. [Google Scholar]
- Kim JI, Jung CS, Koh YH, Lee SH. Molecular, biochemical and histochemical characterization of two acetylcholinesterase cDNAs from the German cockroach Blattella germanica. Insect Mol Biol. 2006;15:513–522. doi: 10.1111/j.1365-2583.2006.00666.x. [DOI] [PubMed] [Google Scholar]
- Kozaki T, Kimmelblatt BA, Hamm RL, Scott JG. Comparison of two acetylcholinesterase gene cDNAs of the lesser mealworm, Alphitobius diaperinus, in insecticide susceptible and resistant strains. Arch Insect Biochem Physiol. 2008;67:130–138. doi: 10.1002/arch.20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Jiang H. Expression of Manduca sexta serine proteinase homolog precursors in insect cells and their proteolytic activation. Insect Biochem Mol Biol. 2008;38:89–98. doi: 10.1016/j.ibmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Palacios LG, Pertuy C, Masson P, Fournier D. Two invertebrate acetylcholinesterases show activation followed by inhibition with substrate concentration. Biochem J. 1998;329:329–334. doi: 10.1042/bj3290329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoulié J, Sussman J, Bon S, Silman I. Structure and functions of acetylcholinesterase and butyrylcholinesterase. Prog Brain Res. 1993;98:139–146. doi: 10.1016/s0079-6123(08)62391-2. [DOI] [PubMed] [Google Scholar]
- Massoulié J, Bon S, Perrier N, Falasca C. The C-terminal peptides of acetylcholinesterase: cellular trafficking, oligomerization and functional anchoring. Chem Biol Interact. 2005;157-158:3–14. doi: 10.1016/j.cbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Morita T, Iwanaga S. Intracellular proclotting enzyme in limulus (Tachypleus tridentatus) hemocytes: its purification and properties. J Biochem. 1985;97:1561–1571. doi: 10.1093/oxfordjournals.jbchem.a135213. [DOI] [PubMed] [Google Scholar]
- Oakeshott JG, Devonshire AL, Claudianos C, Sutherland TD, Horne I, Campbell PM, Ollis DL, Russell RJ. Comparing the organophosphorus and carbamate resistance mutations in cholin- and carboxyl-esterases. Chem Biol Interact. 2005;157:269–275. doi: 10.1016/j.cbi.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Olivera-Bravo S, Ivorra I, Morales A. The acetylcholinesterase inhibitor BW284c51 is a potent blocker of Torpedo nicotinic AchRs incorporated into the Xenopus oocyte membrane. Br J Pharmacol. 2005;144:88–97. doi: 10.1038/sj.bjp.0705965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YP. Novel acetylcholinesterase target site for malaria mosquito control. PLoS ONE. 2006;1:e58. doi: 10.1371/journal.pone.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YP, Singh SK, Gao Y, Lassiter TL, Mishra RK, Zhu KY, Brimijoin S. Selective and irreversible inhibitors of aphid acetylcholinesterases: steps toward human-safe insecticides. PLoS ONE. 2009;4:e4349. doi: 10.1371/journal.pone.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang JY, Shao YM, Lang GJ, Yuan G, Tang ZH, Zhang CX. Expression of two types of acetylcholinesterase gene from the silkworm, Bombyx mori, in insect cells. Insect Sci. 2007;14:443–449. [Google Scholar]
- Seidlein L, Bojang K, Jones P, Jaffar S, Pinder M, Obaro S, Doherty T, Haywood M, Snounou G, Gemperli B, Gathmann I, Royce C, McAdam K, Greenwood B. A randomized controlled trial of artemether/benflumetol, a new antimalarial and pyrimethamine/sulfadoxine in the treatment of uncomplicated falciparum malaria in African children. Amer J Trop Med Hyg. 1998;58:638–644. doi: 10.4269/ajtmh.1998.58.638. [DOI] [PubMed] [Google Scholar]
- Stojan J, Ladurantie C, Siadat OR, Paquereau L, Fournier D. Evidence for subdomain flexibility in Drosophila melanogaster acetylcholinesterase. Biochemistry. 2008;47:5599–5607. doi: 10.1021/bi7025479. [DOI] [PubMed] [Google Scholar]
- Toutant JP. Insect acetylcholinesterase: catalytic properties, tissue distribution and molecular forms. Prog Neurobiol. 1989;32:423–446. doi: 10.1016/0301-0082(89)90031-2. [DOI] [PubMed] [Google Scholar]
- Walsh SB, Dolden TA, Moores GD, Kristensen M, Lewis T, Devonshire AL, Williamson MS. Identification and characterization of mutations in housefly (Musca domestica) acetylcholinesterase involved in insecticide resistance. Biochem J. 2001;359:175–181. doi: 10.1042/0264-6021:3590175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M, Fort P, Berthomieu A, Dubois MP, Pasteur N, Raymond M. A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila. Proc R Soc Lond, Ser B: Biol Sci. 2002;269:2007–2016. doi: 10.1098/rspb.2002.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M, Malcolm C, Chandre F, Mogensen K, Berthomieu A, Marquine M, Raymond M. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol. 2004;13:1–7. doi: 10.1111/j.1365-2583.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- Zhu KY, Clark JM. Purification and characterization of acetylcholinesterase from the Colorado potato beetle, Leptinotarsa decemlineata (Say) Insect Biochem Mol Biol. 1994;24:453–461. doi: 10.1016/0965-1748(95)00055-0. [DOI] [PubMed] [Google Scholar]
- Zhu KY, Gao JR. Increased activity associated with reduced sensitivity of acetylcholine esterase in organophosphate-resistant greenbug, Schizaphis graminum (Homoptera: Aphididae) Pesticide Sci. 1999;55:11J17. [Google Scholar]
- Zhu KY, Zhang JZ. Insect acetylcholinesterase and its roles in insecticide resistance. In: Liu TX, Le G, editors. Entomological research: progress and prospect. Beijing: Science Press; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The last nucleotide of each exon is shaded to indicate the splicing junctions. Single nucleotide differences between the cDNA and gene are underlined. The polyadenylation signal (AATAAA) is double underlined. Amino acid residues, shown in one-letter abbreviations, are aligned with the second nucleotide of each codon. The predicted signal peptide (-24 ∼ -1) is double underlined, putative N-linked glycosylation sites are marked “×”, and the stop codon is indicated by “*”. Positions of the cysteine residues conserved in all AChEs are marked “& – &, $ – $, and ● – ●” to show disulfide bond connectivity. The unique Cys residue present in insect AChE1s (Pang, 2006) is double underlined, whereas the one possibly involved in interchain disulfide linkage is labeled “#”. The catalytic triad (Ser, His and Glu) is in bold and shaded, and the hydrophobic residues (F, W, and Y) lining the active site gorge are labeled with “+”. The primer binding sites for recombinant expression in the baculovirus-insect cell system are underlined.