Abstract
Bacillus anthracis, the causative agent of anthrax disease, could be used as a biothreat reagent. It is vital to develop a rapid, convenient method to detect B. anthracis. In the current study, three high affinity and specificity monoclonal antibodies (mAbs, designated 8G3, 10C6 and 12F6) have been obtained using fully washed B. anthracis spores as an immunogen. These mAbs, confirmed to direct against EA1 protein, can recognize the surface of B. anthracis spores and intact vegetative cells with high affinity and species-specificity. EA1 has been well known as a major S-layer component of B. anthracis vegetative cells, and it also persistently exists in the spore preparations and bind tightly to the spore surfaces even after rigorous washing. Therefore, these mAbs can be used to build a new and rapid immunoassay for detection of both life forms of B. anthracis, either vegetative cells or spores.
Introduction
Bacillus anthracis, the causative agent of anthrax disease, is a Gram-positive spore-forming bacterium. Fully virulent bacilli carry two plasmids, pXO1 and pXO2, which contain genes to produce the lethal factor and edema factor toxins, and a poly-γ-D-glutamic acid capsule, respectively [1], [2]. In response to nutrient deprivation, B. anthracis will produce spores that can withstand harsh conditions, including temperature, radiation, chemical assault, time and even the vacuum of outer space [3]. These remarkable characteristics allow B. anthracis to be used as a biological threat agent.
Recent studies have indicated that B. anthracis is genetically similar to other members of the Bacillus genus. The 16S rRNA, 23S rRNA and 16S–23S internal transcribed spacer sequences of B. anthracis share a high degree of similarity with those in B. cereus, B. subtilis, B. megaterium, B. mycoides, and B. thuringiensis [4], [5]. These sequence similarities make identification of B. anthracis challenging, and substantial effort has been devoted to developing identification methods. The conventional method is the bacteriological assay, which is reliable but time consuming. Other advanced approaches have been proposed for B. anthracis detection, including immunological assays [6], [7], PCR-based methods [8], [9], [10], [11], [12], [13], molecular fingerprinting [14], [15], [16] and mass spectrometric (MS) analyses [17], [18]. The detection targets have mainly focused on the pXO1 and pXO2, gene polymorphisms, specific gene sequences (Ba813, rpoB, bclA) and small acid soluble proteins (SASPs) in the spores. However, these methods fail to eliminate the need for complicated protocols such as cell disruption, nucleic acid extraction and protein purification, and cannot support convenient, rapid and real-time detection of B. anthracis. Therefore, direct detection of B. anthracis is attractive for on-site application. So far, several immunoreagents, directed against the surface of B. anthracis, have been developed for the direct detection of B. anthracis vegetative cells or B. anthracis spores [19], [20], [21], [22], [23].
In the current study, attempts were also made to generate mAbs using fully washed B. anthracis spores as an immunogen. Unexpectedly, we identified three high affinity mAbs (8G3, 10C6 and 12F6) capable of direct and species-specific recognition of both B. anthracis spores and vegetative cells. Furthermore, these mAbs were all directed against the EA1 protein, which is well known as a major S-Layer protein in B. anthracis vegetative cells [24], [25]. Some recent studies have revealed that EA1 is also retained in the proteomic profiling of rigorous washed spores and even salt/detergent washed exosporium [26], [27]. Although Williams and Turnbough suggested EA1 was not a true spore surface protein, they stated this protein persistently existed in the spore surface [28]. Therefore, it is valid to use EA1 as a detection target of B. anthracis spores herein, as EA1 is highly associated with the spore surface. In this study, we conclude that the mAbs we prepared, directed against EA1, can recognize the surface of B. anthracis spores as well as vegetative cells, and we also suggest EA1 protein can serve as a potential marker for the detection of B. anthracis. This study is significant because until now, there have been no monoclonal antibodies reported for direct and species-specific detection of both life forms of B. anthracis.
Results
Atomic-force microscopy (AFM) analysis of B. anthracis spores
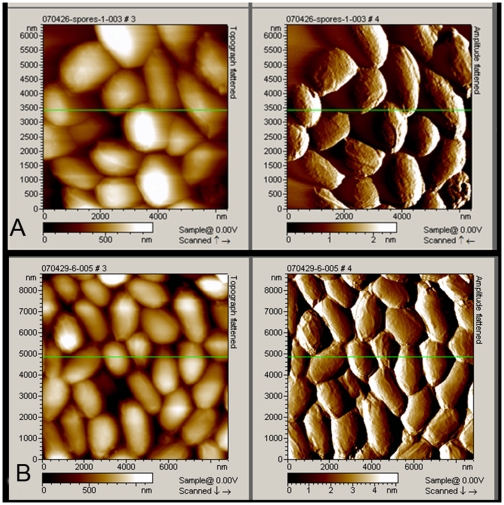
To guarantee the purity of spores that we prepared, both unwashed and fully washed spores were analysed by AFM. As is shown in Figure 1, a larger amount of free spores appeared. Even the unwashed spores were surprisingly clean, as no intact vegetative cells and little vegetative cell debris were present (Fig. 1A). However, because it is likely that some vegetative cell proteins will bind accidentally to the spore surface, the rigorous washing method, as described above, was still employed. There were few differences between the unwashed spores and fully washed spores, but the latter seemed to have a much smoother surface than the former, indicating that our spore purification protocol had removed some unknown material (Fig. 1B). Therefore, we used these fully washed B. anthracis spores as our immunogen and employed them in the subsequent experiments.
Figure 1. Images captured by AFM analysis of B. anthracis spores.
Topography images (left) and amplitude images (right) of the spores were collected in tapping mode. A: Unwashed spores. B: Extensively washed spores.
Preparation and screening of mAbs against B. anthracis spores
By the fusion protocol [29], approximately 600 hybridoma cultures were screened with the indirect ELISA method. Fifteen hybridomas produced high affinity antibodies and were cloned successfully. Following isotyping, five mAbs were IgMs, while the other 10 were IgGs (data not shown). These mAbs were then examined for their reactivity against a range of Bacillus spores using ELISAs. Three mAbs, designated 8G3 (IgG3), 10C6 (IgG1) and 12F6 (IgG1), specifically recognized B. anthracis spores (Fig. 2A), and showed no cross-reaction with high concentrations of B. cereus, B. subtilis, B. megaterium, B. mycoides and other Bacillus spores. Additionally, compared to the other mAbs, these three mAbs all had higher affinity against B. anthracis spores. Even with a range of low concentrations, these mAbs, especially mAb 8G3, always showed strong signals when reacting against B. anthracis spores (Fig. 2B). Therefore, these three mAbs were examined further.
Figure 2. The specificity and reactivity of mAbs with B. anthracis spores.
A: Reaction of the mAbs with a series of Bacillus spores, as detected by indirect ELISA. B: Reactions of different concentrations of mAbs with B. anthracis spores by indirect ELISA. The experiment was repeated three times.
Antigen identification
Exosporium, the outermost structure of spores, was isolated from B. anthracis A16 spores using mild sonication in order to avoid the loss of some exosporium proteins and the damage of the remaining spore protein denaturation. Using electron microscopy, it was observed that the exosporium fragments were successfully extracted (Fig. 3A), and there was no obvious disruption to the spores after sonication (Fig. 3B). The exosporium fragments were boiled in sample buffer for 30–40 min to release more soluble proteins. The soluble exosporium proteins were separated by SDS-PAGE, and the mAbs (8G3, 10C6 and 12F6) recognized a 91–93 kDa protein by Western blotting (Fig. 4A). The band was excised and subjected to DE-MALDI-TOF-MS analysis; the protein was identified as EA1 (Fig. 4B). To determine whether the mAbs were all directed against EA1, the coding region of the gene for EA1 was cloned into the pQE-30 plasmid and expressed in E. coli M15, and the expressed proteins were purified with affinity chromatography. By the method reported previously [30], the affinity constants (Kaff) of the mAbs (8G3, 10C6 and 12F6) for the recombinant EA1 protein were measured to be 1 to 3×109 M−1 Thus, the results not only established that EA1 was the target protein of the three mAbs, but also indicated that the mAbs had a high affinity for EA1.The reaction curves for the mAbs with 4 µg ml−1 recombinant EA1 are shown in Figure 5.
Figure 3. Transmission electron micrographs.
A: B. anthracis exosporium fragments and the B. anthracis spores after sonication. B: Scale bars: 500 nm.
Figure 4. Identification of the mAbs' target proteins.
A: The protein profiles of the fully washed B. anthracis exosporium following SDS-PAGE (Lane 1) and immunoblotting with mAb 8G3 (Lane 2), 10C6 (Lane 3), 12F6 (Lane 4) and mouse IgG (Lane 5). B: Mass spectrometric analysis of the target protein, which was determined to be EA1.
Figure 5. The reactivity of the mAbs with purified EA1 protein.
Reactions of a range of concentrations of the mAbs with the 4 µg/ml EA1 protein using indirect ELISA. The experiment was repeated three times.
The mAbs could also react with B. anthracis vegetative cells
Since EA1 is a major S-Layer protein in B. anthracis vegetative cells [24], [31], the mAbs against EA1 were tested for their reactivity and specificity to B. anthracis vegetative cells. The sandwich ELISA method was employed, rather than indirect ELISA, because the intact vegetative cells were so large that were not adequately coated to the microwells in our indirect ELISA assays (data not shown).
As shown in Figure 6, the mAbs specifically recognized B. anthracis vegetative cells with no significant cross-reaction with other strains at high concentrations (107 CFU/ml). Of these mAbs, 12F6 seemed the best candidate, because it reacted strongly with different kinds of B. anthracis vegetative cells. However, the other mAbs, especially 8G3, did not strongly recognize B. anthracis strains lacking the pXO1 or pXO2 plasmids. Therefore, these results indicated that the mAbs had different characteristics, despite having been raised against the same protein.
Figure 6. The specificity and reactivity of the mAbs with B. anthracis vegetative cells.
Reactions of the mAbs with a series of vegetative cells, as detected by sandwich ELISA.
The mAbs bound to surfaces of both B. anthracis spores and vegetative cells
Binding of mAbs (8G3, 10C6 and 12F6) to the surface of B. anthracis A16 spores and vegetative cells was investigated with immunofluorescence. Normal mouse IgG was used as a negative control (Figure S1), and photographs from the 8G3 experiments as examples are shown in Figure 7. In the immunofluorescence assays, the fully washed spores incubated with 8G3 were extensively stained with R-phycoerythrin-conjugated goat anti-mouse IgG (red). The vegetative cells were incubated with the mAb, and were visualized with FITC-conjugated goat anti-mouse IgG (green). The pattern of fluorescence indicated that these mAbs not only recognized the surfaces of spores, but also bound to the intact vegetative cells.
Figure 7. Representative confocal microscopy images of 8G3 binding to fully washed B. anthracis spores (top) and vegetative cells (bottom).
The secondary antibodies were R-phycoerythrin-conjugated goat anti-mouse IgG. (red) or FITC-conjugated goat anti-mouse IgG (green). The corresponding phase contrast images (A and D) and fluorescence images (B and E) were merged to show overlap (C and F). Scale bars: spores, 5 µm; vegetative cells, 10 µm.
EA1 was persistently present in B. anthracis spore extracts
During preparation, the B. anthracis spores were washed extensively with water and pelleted through 20% and 50% Renografin to remove vegetative cell debris [10]. The spores were monitored and collected at four stages of the standard washing procedure: unwashed spores from the culture plate (Preparation 1), spores washed with MilliQ water for 24 h, three times (Preparation 2), Renografin purified spores (Preparation 3), and spores subjected to three additional washes (Preparation 4). In addition, the supernatants from sedimentation during Preparation 3 were diluted 1∶10 with water and centrifuged at 10,000×g for 30 min to collect the debris. Equal volumes of all preparations and the debris were subjected to SDS-PAGE analysis.
As a result, the B. anthracis A16 SDS-PAGE profiles showed no obvious differences between Preparations 1 and 2, except that a negligible amount of protein was removed by the washing steps (Figure 8). The same results were observed when Preparations 3 and 4 were compared. In contrast, most proteins, including EA1, decreased after sedimentation through Renografin. These removed proteins were present in the “debris” of the supernatant, which contained a similar amount of protein to Preparations 2 and 4. These results suggest that the Renografin step washed out some of the EA1 along with many other spore proteins, which were then detected by SDS-PAGE. Despite the presence of EA1 in the debris, EA1 was also persistently present in the B. anthracis A16 spore extracts of each preparation. Furthermore, B. anthracis strains lacking pXO1 or pXO2 produced similar results to those presented above (data not shown). Therefore, EA1 was persistently present in B. anthracis spore extracts.
Figure 8. Comparison of the protein profiles of spores and debris.
Proteins were separated on a 10% Tricine gel and visualized with silver staining. Lane 1, unwashed spores (Preparation 1); Lane 2, spores washed three times with ultrapure water (Preparation 2); Lane 3, spores prepared by Renografin purification (Preparation 3); Lane 4, fully washed spores (Preparation 4); Lane 5, debris in the supernatant of centrifuged spores after Renografin purification. (The staining time of Preparation 3, 4 and 5 was longer than that of Preparation 1 and 2, in order that more bands of each sample could be visualized clearly).
Discussion
The primary goal of this study was to generate mAbs with high affinity and specificity that could be applied to rapid detection of B. anthracis spores. The mAbs were produced against formaldehyde-inactivated B. anthracis A16 spores and reacted with a range of live Bacillus spores, including B. anthracis. Most of the mAbs we produced were highly specific for B. anthracis spores. For each screening of the hybridoma cultures, spores from B. cereus and B. thuringiensis, the two closest relatives of B. anthracis, were used as negative controls. To identify mAbs with high affinity and specificity, hybridomas were selected if the mAbs reacted strongly with B. anthracis but did not recognize either negative antigen. As a result, the three mAbs (8G3, 10C6 and 12F6) we prepared have no cross reaction with many B. thuringienesis subspecies and B. cereus isolates (Table S1).
The three mAbs recognized not only the surface of B. anthracis spores but could also detect intact B. anthracis vegetative cells (Fig. 7). Furthermore, these mAbs were capable of reaction with live B. anthracis as well as dead B. anthracis (inactivated by 1.5% formaldehyde), which is critical for the detection of biological warfare agents in unknown “white powders”, since it has been suggested that Bacillus inactivation would affect antibody detection assays [32]. Although these three mAbs were directed toward the same target protein, EA1, they had different characteristics. The mAb 12F6 was superior at reacting with different kinds of B. anthracis vegetative cells, while 8G3 had a higher affinity for B. anthracis spores and the target protein EA1 (Fig. 2, Fig. 5 and Fig. 6). Besides this, in the epitope mapping, the epitopes of mAb 8G3 and 10C6 were concluded to be located from the amino acid 275 to 435 on the EA1 protein, and the epitope of mAb 12F6 was exactly located from the amino acid 465 to 554 (data not shown). We suggested that the different positions of the mAb epitopes caused the mAbs to exhibit different behavior the detection of B. anthracis.
As to whether EA1, a major S-layer component of B. anthracis vegetative cells, is also a spore protein, much research has indicated that this protein is retained in the proteomic profiling of spores and salt/detergent washed exosporium [26], [27]. However, Williams and Turnbough stated that this protein was merely a persistent contaminant in spore preparations [28]. Whichever is correct, it does not matter for the detection of B. anthracis spores, because this protein does persistently exist in each of the spore preparations (Fig. 8), and the B. anthracis spores, even after full washing, can be detected with our anti-EA1 mAbs (Fig. 2 and Fig. 7).
Although EA1 could be partially washed out during the rigorous washing step in our study, the other proteins detected by SDS-PAGE also apparently decreased and were present as debris in the supernatant of centrifuged purified samples (Fig. 8). The supernatant debris contained significant amounts of true spore proteins with similar protein profiles to fully washed spores. Therefore, rigorous washing methods, such as Renografin purification, can cause serious loss of spore surface associated proteins and are not suggested for proteomic analysis [26], [27]. This suggests that EA1 is certainly at least a highly spore-associated protein: it might be a true spore protein, or may anchor onto particular spore surface components.
In conclusion, this study reports three mAbs (8G3, 10C6 and 12F6) that can bind to B. anthracis spores and intact vegetative cells with high species-specificity and affinity. It also indicates that EA1, the target protein of our mAbs, could serve as a potential detection target of B. anthracis, establishing a new immunoassay protocol that realizes sensitive, rapid, on-site and simultaneous detection of both life forms of B. anthracis.
Materials and Methods
Bacterial strains
The bacterial strains used in this study are listed in Table 1. All assays involving live B. anthracis spores and vegetative cells were carried out in a P3 biosafe laboratory. The experimenters were equipped with masks, gloves and exposure suits.
Table 1. Bacterial strains examined in this study.
| Strains | Source | Plasmid | |
| pXO1 | pXO2 | ||
| B. anthracis A16 | Institute of Microbiology and Epidemiology, AMMS | + | + |
| B. anthracis 170044 | Institute of Microbiology and Epidemiology, AMMS | + | + |
| B. anthracis CMCC(B) 63002 | National Center for Medical Culture Collections, China | + | − |
| B. anthracis CMCC(B) 63005 | National Center for Medical Culture Collections, China | − | + |
| B. cereus CCTCCAB 93038+ | Institute of Microbiology and Epidemiology, AMMS | ||
| B. cereus ATCC 33018 | Huazhong Agricultural University, China | ||
| B. thuringienesis subsp. alesti | Huazhong Agricultural University, China | ||
| B. thuringienesis subsp. sotto | Huazhong Agricultural University, China | ||
| B.licheniformis | Institute of Microbiology and Epidemiology, AMMS | ||
| B. subtilis | Institute of Microbiology and Epidemiology, AMMS | ||
| B. megaterium | Institute of Microbiology and Epidemiology, AMMS | ||
| B. mycoides | Wuhan Institute of Virology, Chinese Academy of Sciences | ||
| B. pumilus | Wuhan Institute of Virology, Chinese Academy of Sciences | ||
| Proteus vulgaris | Wuhan Institute of Virology, Chinese Academy of Sciences | ||
| E. coli BL21 | Wuhan Institute of Virology, Chinese Academy of Sciences | ||
| E. coli M15 | Wuhan Institute of Virology, Chinese Academy of Sciences | ||
AMSS: the Academy of Military Medical Sciences, China.
Preparation of spores
Spores were prepared by growing bacteria at 37°C on modified Difco sporulation medium (DSM) [33], containing 6 g tryptone, 3 g yeast extract, 10 g NaCl, 1 g KCl, 0.25 g Mg2SO4·7H2O, 0.23 g Ca(NO3)2, 0.197 g MnCl2·4H2O, 0.0002 g FeSO4, and 15 g agar per liter. When more than 95% free spores appeared, the spores were collected with cold, sterile, ultrapure water, centrifuged and washed extensively three times, before sedimentation through 20% and 50% Renografin (Bracco Sine, China) [10] and another three washes. The washed spores were then resuspended in sterile saline and stored at 4°C. The spores were quantified by serially diluting the spore stock, and plating 100 µl aliquots on Luria broth (LB) plates, in triplicate. The number of spores (CFU/ml) was counted following overnight culture at 37°C.
Atomic-force microscopy (AFM) analysis of spores
An aqueous spore suspension of 10 µl, containing 108–109 spores, was spread on a 1 cm×1 cm silicon wafer and air-dried. The sample was not placed in the AFM chamber for imaging until the spores settled on the substrate. Picoscan™ 2500 AFM (Molecular Imaging, US) and commercial single MAClever type II cantilevers (Molecular Imaging, US) were used to obtain images in tapping mode.
Exosporium extraction
Approximately 0.5 g of purified B. anthracis spores (wet weight) were collected by centrifugation at 9,000×g for 5 min at 4°C and resuspended in 15 ml cold sterile TE buffer (50 mmol l−1 Tris-HCl, 0.5 mmol l−1 EDTA, pH 7.2), containing 1 µl ml−1 protease inhibitor cocktail (Sigma, US). All subsequent centrifugations were performed at 4°C. To extract larger exosporium fragments, the spore suspension was divided into three partitions and each was treated using a Vibra-Cell Ultrasonic Processor (Sonics & Materials, USA) with 120 pulse cycles on ice (5 s pulse - 9 s interval - 5 s pulse). The three partitions were sonicated, pooled and centrifuged for 45 min at 1,200×g. Then the pellet was washed five times with cold TE buffer and centrifuged as above. All the supernatant samples were saved and centrifuged at 184,000×g for 1 h. The sediments, which were composed of exosporium fragments, were then resuspended in 500 µl sterile PBS for further study.
Production of monoclonal antibodies and polyclonal antibodies
Preparations containing 106 spores of B. anthracis strain A16 and inactivated by 1.5% formaldehyde were injected subcutaneously into six-week-old BALB/c mice. The immunization was repeated three times at two-week intervals before boosting by intraperitoneal injection. The spleen cells were removed 3 d later and fused with SP2/0 myeloma cells, according to the procedures of Kohler and Milstein (1975)[29]. The hybridomas were cloned by limit dilution, screened using ELISA (described below) and then injected intraperitoneally into BALB/c mice. The mAbs were isotyped using an isotyping kit (Sigma, US), according to the manufacturer's directions, and were purified by caprylic acid-ammonium sulfate precipitation of ascites [34].
For the polyclonal antibodies against anthracis vegetative cells, the rabbit was injected subcutaneously with 107 cells (inactivated by 1.5% formaldehyde) for three times at two week before boosting by double injection dose. The polyclonal antibodies were purified from the antiserum by caprylic acid-ammonium sulfate precipitation of ascites.
ELISA
For indirect ELISA detection, 96 well microtiter plates were coated with 100 µl per well of carbonate-bicarbonate (CB) buffer (pH 9.6) containing either 107–108/ml spores or recombinant EA1 (2, 4, 8, 16 µg/ml) and incubated overnight at 4°C. The wells were blocked with 200 µl of blocking buffer (5% skim milk in phosphate buffered saline (PBS) for 2 h at 37°C. Hybridoma culture supernatants or a concentration series of purified mAbs were added in 100 µl aliquots to individual wells and incubated for 30 min at 37°C. Horseradish peroxidase (HRP)-conjugated goat anti-mouse antibodies were added at a dilution of 1/900 and incubated for 20 min.
For sandwich ELISA, the microtiter plates were coated with purified mAbs (10 µg/ml) and blocked as described above. The plates were incubated with a range of Bacillus vegetative cells at 37°C for 1 h and then reacted with 10 µg/ml polyclonal antibodies against anthrax at 37°C for 40 min. HRP-conjugated goat anti-rabbit antibodies were added to the plate at a dilution of 1/3000 (Boster, China) and reacted for 30 min at 37°C.
For both indirect ELISA and sandwich ELISA, five washes with PBS with 0.05% Tween 20 (PBST) were carried out between each step. Normal mouse IgG was used as a negative control, and all antibody dilutions were prepared in PBS, containing 1% skim milk. A tetramethylbenzidine substrate (0.1 mg/ml 100 µl per well) was added for approximately 10–15 min at 37°C to start the reaction, and 50 µl of 2 M H2SO4 was added to stop the reaction. Then the absorbance was measured at 450 nm, and each assay was performed in quadruplicate.
Analysis by SDS-PAGE and immunoblotting
Samples were mixed with an equal volume of sample buffer (50 mM Tris-HCl at pH 6.8, 0.5 mol l−1 β-mercaptoethanol, 20% v/v glycerol, 10% w/v SDS, 0.2% w/v bromophenol blue) and boiled for 10–40 min. The soluble proteins were analyzed by SDS-PAGE using a Bio-Rad gel apparatus, according to the manufacturer's instructions. For immunological detection, the proteins were transferred from the gels onto polyvinylidene difluoride (PVDF) membranes (Millipore, US). The membranes were blocked in Tris-buffered saline (TBS) containing 5% skim milk at 4°C overnight, and were washed three times for 5 min with Tris-buffered saline Tween-20 (TBST, with 0.05% Tween 20). Then the membranes were incubated in 5 µg ml−1 mAbs, which were diluted in TBS containing 1% skim milk, for 1.5 h at room temperature, followed by three additional 10 min washes. The membranes were incubated for 1 h with a 1/100 dilution of HRP-conjugated goat anti-mouse IgG. Four thorough 10 min washes were completed before the substrate buffer (6 mg 3,3′-Diaminobenzidine (DAB) and 10 µl of 37% H2O2 in 10 ml TBS) was added and allowed to react for 3–5 min. Images were acquired using a Canon digital camera and analyzed using Adobe Photoshop software.
MS protein analysis
The protein band identified on the SDS-PAGE gel was excised, washed, and subjected to tryptic digestion [26]. The peptides were collected and analyzed by delayed extraction-matrix assisted laser desorption ionization-time of flight mass spectrometry (DE-MALDI-TOF-MS). The MS spectra were obtained using an Axima-CFR Plus Mass Spectrometer (Shimadzu Biotech, Japan). All MS analyses were performed at the Institute of Biophysics, Chinese Academy of Sciences, China.
Gene cloning and expression
Sequence data for the B. anthracis genome were obtained from the National Center for Biotechnology Information (NCBI) web site at http://www.ncbi.nlm.nih.gov. Based on the sequence information, DNA primers were designed to amplify the entire eag open reading frame of the B. anthracis A16 strain using polymerase chain reaction (PCR). The chromosomal DNA of A16 was used as the template, and the primer sequences were 5′-TATTGGATCCATGGCAAA GACTAACT-3′ and 5′-CTATAGAGCTCGTATAGATTTGGGTT A -3′. The PCR conditions were as follows: 30 cycles of 94°C for 5 min, 94°C for 45 s, 40°C for 45 s, and 72°C for 2 min for 30 cycles; and lastly 72°C for 5 min. The PCR products were digested with BamHI and SacI and were inserted into plasmid pQE-30 (Qiagen, Germany). The recombinant plasmids were transformed into E. coli M15 to express the cloned gene in vitro. Firstly, the E. coli M15, carrying recombinant plasmids, were incubated at 37°C at 160 rpm to an average OD600 of 0.5 followed by addition of IPTG to 1 mM final concentration, and were incubated for another 4 h at 30°C. Secondly, the cells were harvested by centrifugation at 3000 g for 5 min, and were sonicated by a Vibra-Cell Ultrasonic Processor (Sonics & Materials, USA) as described above. Lastly, after centrifugation at 12,000 g for 20 min, the supernatant containing recombinant EA1 protein with His tag was purified using Ni-nitrilotriacetic acid agarose (Qiagen, Germany) affinity chromatography, according to the manufacturer's instructions.
Determination of the mAbs affinities
As the antibody affinities reported previously [30], different concentrations of recombinant EA1 (2, 4, 8, 16 µg/ml) were coated on the microwells and incubated with serial dilutions of mAbs for indirect ELISAs. The affinity constants (Kaff) of the mAbs (8G3, 10C6 and 12F6) for the recombinant EA1 protein were measured using the equation: Kaff = (n−1)/2(n[Ab′]t−[Ab]t), where n = [Ag]/[Ag′]. Briefly, [Ag] and [Ag′] are antigen (EA1) concentrations. [Ab′]t and [Ab]t are the measurable total antibody concentrations at the half maximum OD (OD-50) for plates coated with [Ag′] and [Ag], respectively.
Indirect immunofluorescence
For the indirect immunofluorescence assays, 200 µl aliquots of spore or vegetative cell suspension (109 CFU ml−1) were pelleted by centrifugation at 10,000×g for 1 min at room temperature. The spores and vegetative cells were resuspended by adding 500 µl blocking buffer and incubated for 2 h at 37°C with gentle shaking (70–100 rpm). Aliquots of mAbs (500 µl; 1 µg ml−1) were added and the mixture was incubated for 1 h at 37°C with shaking. R-phycoerythrin-conjugated goat anti-mouse IgG (Sigma, US) or FITC-conjugated goat anti-mouse IgG (Boster, China) was used as the secondary antibody (100 µl, 1/50 dilution), and was added to the spores or vegetative cell solution respectively. The mixture was incubated for 40 min at room temperature in the dark. Each step was followed by three 10 min washes in PBST, followed by centrifugation at 10,000×g for 1 min to remove the supernatant. The antibodies were collected in PBS containing 1% skim milk, and images were acquired using a Leica confocal microscope.
Supporting Information
Confocal microscopy images of negative control antibody binding to B. anthracis. The secondary antibodies were R-phycoerythrin-conjugated goat anti-mouse IgG. (B. anthracis spores, top) and FITC-conjugated goat anti-mouse IgG (vegetative cells, bottom). Scale bars: spores, 5 µm; vegetative cells, 10 µm
(0.53 MB DOC)
Additional B. thuringienesis subspecies and B. cereus isolates than the strains shown in the manuscript were reacted with our mAbs to further determine the mAbs species specificity. The B. cereus strains were from Wuhan Institute of Virology, Chinese Academy of Sciences. The B. thuringiensis strains were from Huazhong Agricultural University, China.
(0.07 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Natural Science Foundation of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikesell P, Ivins BE, Ristroph JD, Dreier TM. Evidence for plasmid-mediated toxin production in Bacillus anthracis. Infect Immun. 1983;39:371–376. doi: 10.1128/iai.39.1.371-376.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ash C, Collins MD. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol Lett. 1992;73:75–80. doi: 10.1016/0378-1097(92)90586-d. [DOI] [PubMed] [Google Scholar]
- 5.Daffonchio D, Cherif A, Borin S. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S–23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group”. Appl Environ Microbiol. 2000;66:5460–5468. doi: 10.1128/aem.66.12.5460-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SH, Zhang JB, Zhang ZP, Zhou YF, Yang RF, et al. Construction of single chain variable fragment (ScFv) and BiscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal Chem. 2006;78:997–1004. doi: 10.1021/ac0512352. [DOI] [PubMed] [Google Scholar]
- 7.Kleine-Albers C, Bohm R. The detection of Bacillus anthracis protective antigens by enzyme immunoassay (EIA) using polyclonal and monoclonal antibodies. Zentralbl Veterinarmed B. 1989;36:226–230. [PubMed] [Google Scholar]
- 8.Henderson I, Duggleby CJ, Turnbull PC. Differentiation of Bacillus anthracis from other Bacillus cereus group bacteria with the PCR. Int J Syst Bacteriol. 1994;44:99–105. doi: 10.1099/00207713-44-1-99. [DOI] [PubMed] [Google Scholar]
- 9.Beyer W, Glockner P, Otto J, Bohm R. A nested PCR method for the detection of Bacillus anthracis in environmental samples collected from former tannery sites. Microbiol Res. 1995;150:179–186. doi: 10.1016/S0944-5013(11)80054-6. [DOI] [PubMed] [Google Scholar]
- 10.Henriques AO, Beall BW, Roland K, Moran CP., Jr Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J Bacteriol. 1995;177:3394–3406. doi: 10.1128/jb.177.12.3394-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MA, Brightwell G, Leslie D, Bird H, Hamilton A. Fluorescent detection techniques for real-time multiplex strand specific detection of Bacillus anthracis using rapid PCR. J Appl Microbiol. 1999;87:218–223. doi: 10.1046/j.1365-2672.1999.00908.x. [DOI] [PubMed] [Google Scholar]
- 12.Qi Y, Patra G, Liang X, Williams LE, Rose S, et al. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl Environ Microbiol. 2001;67:3720–3727. doi: 10.1128/AEM.67.8.3720-3727.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skottman T, Piiparinen H, Hyytiainen H, Myllys V, Skurnik M, et al. Simultaneous real-time PCR detection of Bacillus anthracis, Francisella tularensis and Yersinia pestis. Eur J Clin Microbiol Infect Dis. 2007 doi: 10.1007/s10096-007-0262-z. [DOI] [PubMed] [Google Scholar]
- 14.Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, et al. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl Environ Microbiol. 2004;70:1068–1080. doi: 10.1128/AEM.70.2.1068-1080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolsto AB. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl Environ Microbiol. 2004;70:191–201. doi: 10.1128/AEM.70.1.191-201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, et al. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castanha ER, Fox A, Fox KF. Rapid discrimination of Bacillus anthracis from other members of the B. cereus group by mass and sequence of “intact” small acid soluble proteins (SASPs) using mass spectrometry. J Microbiol Methods. 2006;67:230–240. doi: 10.1016/j.mimet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 18.Elhanany E, Barak R, Fisher M, Kobiler D, Altboum Z. Detection of specific Bacillus anthracis spore biomarkers by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:2110–2116. doi: 10.1002/rcm.491. [DOI] [PubMed] [Google Scholar]
- 19.Williams DD, Benedek O, Turnbough CL., Jr Species-specific peptide ligands for the detection of Bacillus anthracis spores. Appl Environ Microbiol. 2003;69:6288–6293. doi: 10.1128/AEM.69.10.6288-6293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozel TR, Murphy WJ, Brandt S, Blazar BR, Lovchik JA, et al. mAbs to Bacillus anthracis capsular antigen for immunoprotection in anthrax and detection of antigenemia. Proc Natl Acad Sci U S A. 2004;101:5042–5047. doi: 10.1073/pnas.0401351101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swiecki MK, Lisanby MW, Shu F, Turnbough CL, Jr, Kearney JF. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J Immunol. 2006;176:6076–6084. doi: 10.4049/jimmunol.176.10.6076. [DOI] [PubMed] [Google Scholar]
- 22.Mechaly A, Zahavy E, Fisher M. Development and implementation of a single-chain Fv antibody for specific detection of Bacillus anthracis spores. Appl Environ Microbiol. 2008;74:818–822. doi: 10.1128/AEM.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love TE, Redmond C, Mayers CN. Real time detection of anthrax spores using highly specific anti-EA1 recombinant antibodies produced by competitive panning. J Immunol Methods. 2008;334:1–10. doi: 10.1016/j.jim.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol Microbiol. 1997;23:1147–1155. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- 25.Fritz DL, Jaax NK, Lawrence WB, Davis KJ, Pitt ML, et al. Pathology of experimental inhalation anthrax in the rhesus monkey. Lab Invest. 1995;73:691–702. [PubMed] [Google Scholar]
- 26.Delvecchio VG, Connolly JP, Alefantis TG, Walz A, Quan MA, et al. Proteomic profiling and identification of immunodominant spore antigens of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Appl Environ Microbiol. 2006;72:6355–6363. doi: 10.1128/AEM.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai EM, Phadke ND, Kachman MT, Giorno R, Vazquez S, et al. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J Bacteriol. 2003;185:1443–1454. doi: 10.1128/JB.185.4.1443-1454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams DD, Turnbough CL., Jr Surface layer protein EA1 is not a component of Bacillus anthracis spores but is a persistent contaminant in spore preparations. J Bacteriol. 2004;186:566–569. doi: 10.1128/JB.186.2.566-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 30.Beatty JD, Beatty BG, Vlahos WG. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J Immunol Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- 31.Mesnage S, Tosi-Couture E, Gounon P, Mock M, Fouet A. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J Bacteriol. 1998;180:52–58. doi: 10.1128/jb.180.1.52-58.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang JL, Heroux K, Kearney J, Arasteh A, Gostomski M, et al. Bacillus spore inactivation methods affect detection assays. Appl Environ Microbiol. 2001;67:3665–3670. doi: 10.1128/AEM.67.8.3665-3670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonenshein AL, Cami B, Brevet J, Cote R. Isolation and characterization of rifampin-resistant and streptolydigin-resistant mutants of Bacillus subtilis with altered sporulation properties. J Bacteriol. 1974;120:253–265. doi: 10.1128/jb.120.1.253-265.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perosa F, Carbone R, Ferrone S, Dammacco F. Purification of human immunoglobulins by sequential precipitation with caprylic acid and ammonium sulphate. J Immunol Methods. 1990;128:9–16. doi: 10.1016/0022-1759(90)90458-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confocal microscopy images of negative control antibody binding to B. anthracis. The secondary antibodies were R-phycoerythrin-conjugated goat anti-mouse IgG. (B. anthracis spores, top) and FITC-conjugated goat anti-mouse IgG (vegetative cells, bottom). Scale bars: spores, 5 µm; vegetative cells, 10 µm
(0.53 MB DOC)
Additional B. thuringienesis subspecies and B. cereus isolates than the strains shown in the manuscript were reacted with our mAbs to further determine the mAbs species specificity. The B. cereus strains were from Wuhan Institute of Virology, Chinese Academy of Sciences. The B. thuringiensis strains were from Huazhong Agricultural University, China.
(0.07 MB DOC)