Abstract
Plant cystatins are inhibitors of cysteine-proteases of the papain C1A and legumain C13 families. Cystatin data from multiple plant species have suggested that these inhibitors act as defense proteins against pests and pathogens and as regulators of protein turnover. In this study, we characterize the entire cystatin gene family from barley (Hordeum vulgare), which contain 13 nonredundant genes, and identify and characterize their target enzymes, the barley cathepsin L-like proteases. Cystatins and proteases were expressed and purified from Escherichia coli cultures. Each cystatin was found to have different inhibitory capability against barley cysteine-proteases in in vitro inhibitory assays using specific substrates. Real-time reverse transcription-polymerase chain reaction revealed that inhibitors and enzymes present a wide variation in their messenger RNA expression patterns. Their transcripts were mainly detected in developing and germinating seeds, and some of them were also expressed in leaves and roots. Subcellular localization of cystatins and cathepsin L-like proteases fused to green fluorescent protein demonstrated the presence of both protein families throughout the endoplasmic reticulum and the Golgi complex. Proteases and cystatins not only colocalized but also interacted in vivo in the plant cell, as revealed by bimolecular fluorescence complementation. The functional relationship between cystatins and cathepsin L-like proteases was inferred from their common implication as counterparts of mobilization of storage proteins upon barley seed germination. The opposite pattern of transcription expression in gibberellin-treated aleurones presented by inhibitors and enzymes allowed proteases to specifically degrade B, C, and D hordeins stored in the endosperm of barley seeds.
Phytocystatins (PhyCys) are plant proteinaceous inhibitors of Cys-proteases of the papain C1A family integrated in an independent subfamily on the cystatin phylogenetic tree (Margis et al., 1998; Martinez and Diaz, 2008; Rawlings et al., 2008). The cystatin inhibitory mechanism is produced by a tight and reversible interaction with their target enzymes. This mechanism involves a tripartite wedge formed by the partially flexible N terminus containing a Gly residue and two hairpin loops carrying a conserved QxVxG motif and a Trp residue, respectively (Stubbs et al., 1990). In addition to these three motifs, which are common to all cystatins, PhyCys contain a particular consensus sequence ([LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-x-[EDQV]-[HYFQ]-N) that conforms to a predicted secondary α-helix structure (Margis et al., 1998). Most PhyCys are small proteins with a molecular mass in the 12- to 16-kD range that are devoid of both disulfide bonds and putative glycosylation sites. Some PhyCys that have a molecular mass of 23 kD contain a C-terminal extension that is involved in the inhibition of a second family of Cys-proteases, the C13 legumain peptidases (Martinez et al., 2007; Martinez and Diaz, 2008). Several 85- to 87-kD multicystatins, with eight cystatin domains, have also been described (Girard et al., 2007; Nissen et al., 2009).
PhyCys have mainly been purified from seeds but have also been detected in other plant tissues (Pernas et al., 1998; Gaddour et al., 2001; Abraham et al., 2006). The subcellular location of these proteins is still unknown, with the exception of the multicystatins from potato (Solanum tuberosum) and tomato (Solanum lycopersicum) found in vacuoles and in cytoplasm and oryzacystatin OC-I from rice (Oryza sativa) detected in cytoplasm, vacuoles, and chloroplasts (Madureira et al., 2006; Prins et al., 2008; Nissen et al., 2009). Nevertheless, the signal peptide present in most of these inhibitors suggests a noncytosol location of these proteins (Martinez et al., 2005a; Abraham et al., 2006).
From a functional viewpoint, PhyCys have a dual role, as defense proteins and as endogenous regulators of protein turnover. The defense function has been inferred from the ability of PhyCys to inhibit the activity of digestive proteases from arthropods in vitro, in artificial diets as well as through bioassays on transgenic plants overexpressing PhyCys genes (Pernas et al., 1998; Atkinson et al., 2004; Ribeiro et al., 2006; Alvarez-Alfageme et al., 2007). Anti-mite activities and deleterious effects against phytopathogenic fungi and viruses have also been described for these inhibitors (Gutierrez-Campos et al., 1999; Pernas et al., 2000; Martinez et al., 2003; Abraham et al., 2006; Wang et al., 2008).
However, the principal function of PhyCys is perhaps related to the regulation of endogenous Cys-proteases, which are involved in multiple physiological processes and which prevent the breakdown of essential proteins during metabolism. Until now, PhyCys have been shown to have a role in plant growth and development, senescence, and programmed cell death (Solomon et al., 1999; Corre-Menguy et al., 2002; Belenghi et al., 2003; Kiyosaki et al., 2007; Weeda et al., 2009). However, these studies involved individual cystatin members from multiple plant species. Determining the functional diversity of all members of the cystatin gene family and identifying their target enzymes within a unique species would be very informative.
Plant proteolysis is a complex process involving many different pathways and cellular compartments, where protease activities are regulated at the transcriptional level by differential expression and at the protein level by the activation of inactive zymogens and by the binding of specific inhibitors and cofactors. Of the more than 800 proteases encoded by plant genomes, approximately 140 correspond to Cys-proteases that belong to 15 families in five clans (Rawlings et al., 2008). The main target of PhyCys are the papain-like proteases, C1A (family C1, clan CA), which comprise 38 and 44 genes in the Arabidopsis (Arabidopsis thaliana) and Populus genomes, respectively (Garcia-Lorenzo et al., 2006). Based on their similarity to mammalian proteases, the C1A proteases from plants have been grouped as cathepsin L-, B-, H-, or F-like (Martinez and Diaz, 2008). All of the C1A proteases contain several disulfide bonds and share three conserved catalytic residues (Cys, His, and Asn) in the catalytic triad, a trait that is common to all Cys-proteases, and they also contain a Gln involved in maintaining an active enzyme conformation. In addition, the GCNGG motif has been identified in all C1A protease members, and the noncontiguous ERFNIN motif has been found in cathepsin L- and H-like proteins (Grudkowska and Zagdanska, 2004; Zhang et al., 2009). C1A Cys-proteases from plants are synthesized as preproteins, processed either automatically or with the aid of processing enzymes, transported via the trans-Golgi network, and finally either are stored in the vacuoles and lysosomes or externally secreted (Grudkowska and Zagdanska, 2004). These plant proteases participate in protein degradation during senescence and abscission processes, programmed cell death, and the accumulation and mobilization of storage proteins in seeds and tubers (Grudkowska and Zagdanska, 2004; van der Hoorn, 2008; Shi and Xu, 2009; Weeda et al., 2009). Moreover, they also play an essential role in local and systemic defense responses against pathogens and pests (van der Hoorn, 2008; McLellan et al., 2009).
Cys-proteases are described as being the most abundant group of proteases responsible for the degradation and mobilization of storage proteins in seeds (Grudkowska and Zagdanska, 2004; Sreenivasulu et al., 2008). Seeds act as stores of starch, protein, and lipids that are used during the germination process. Seed germination begins with water uptake and ends with the elongation and protrusion of the radicle tip through the seed coat. Early postgermination events are also crucial for survival of the seedling until photosynthesis is fully established. Amylases and proteases are either stored during seed maturation or newly synthesized during early germination. Hydrolyzed reserves are the products of enzymes and are absorbed by the scutellum and utilized for seedling development. Zhang and Jones (1995) reported that 27 Cys-proteases are among the 42 proteases that are involved in the germination of barley (Hordeum vulgare) seed. Until now, few of these proteases have been characterized (Koehler and Ho, 1990; Davy et al., 1998), and there are currently only limited data on the effects of the inhibitors required for their regulation (Martinez et al., 2005b). In contrast, sets of genes encoding transcriptional factors synthesized in response to phytohormones have been implicated in controlling hydrolase activity during barley seed germination (Gubler et al., 1999; Sreenivasulu et al., 2008).
In barley, we have identified 32 Cys-proteases from the papain-like family (Martinez and Diaz, 2008) and have characterized seven genes, encoding cystatins HvCPI-1 to HvCPI-7, as being specific inhibitors of Cys-proteases (Abraham et al., 2006). These cystatins have shown a different gene structure, variations in the mRNA expression patterns, and important changes in the deduced amino acid sequences that affect their inhibitory properties (Martinez et al., 2005a; Abraham et al., 2006). This paper presents the molecular characterization of the entire barley cystatin gene family, which includes a total of 13 members, and the barley cathepsin L-like Cys-proteases from the C1A peptidase class, putative targets of cystatins. Enzyme-inhibitor interactions that are thought to be important to the mobilization of storage proteins in the seed during the germination process are also shown.
RESULTS
Phylogenetic and Sequence Analysis of the PhyCys within the Poaceae
In barley, 13 cystatins have been identified previously (Martinez and Diaz, 2008). In order to establish their phylogenetic relationships, the entire amino acid sequences of the 13 barley cystatins were compared with the amino acid sequences of the PhyCys proteins from the Poaceae family found in the data banks. From the alignment, an unrooted phylogenetic tree was constructed using the PhyML method (Fig. 1). The proteins were grouped into three major clusters (A, B, and C), as in previous studies by Martinez et al. (2005a) and Abraham et al. (2006), which are supported by high approximate likelihood-ratio test values (data not shown). Now, the availability of all members of the barley cystatin family has allowed the C group to be subdivided in two subgroups: the C1 subgroup formed by closely related barley, corn (Zea mays), sorghum (Sorghum bicolor), and rice proteins; and the C2 subgroup, which includes divergent cystatins from barley, corn, rice, sorghum, and wheat (Triticum aestivum). Interestingly, putative orthologous relationships were detected for the barley and rice cystatins belonging to groups A and B and from subgroup C1 of the proteins. No orthology could be detected among the barley and rice cystatins belonging to subgroup C2, where the barley cystatins with major sequence variations in sequence are located (Fig. 2 shows a schematic alignment of amino acid sequences). In relation to the protein motifs responsible for the papain-like Cys-protease inhibitory properties, one or two Glys in the N-terminal part of the molecule were conserved in the 13 barley cystatins. The QxVxG reactive site located in the loop between β2 and β3 sheets was present in all of the sequences with a few exceptions. In the group B HvCPI-9, the Val in the third position is changed to Ile, while in HvCPI-7 and HvCPI-12 in group C2, the Gly in the fifth position is changed to Glu and Ser, respectively. The conserved Trp present in the loop between β4 and β5 sheets was absent in the group C2 cystatins HvCPI-7 and HvCPI-11. In addition, a long C-terminal extension was observed in HvCPI-4, which has been involved in the properties inhibiting legumain-like Cys-proteases observed for this cystatin (Martinez et al., 2007). To show how differences in sequence determined changes in the three-dimensional structure, the known crystal structure of the rice OC-I cystatin (Nagata et al., 2000) was used as a molecular model to establish the predicted structures of the barley cystatins (Supplemental Fig. S1). The overall disposition of secondary structures (β-sheets and α-helixes) of the models for barley cystatins HvCPI-8 to HvCPI-13 was very similar to that observed in rice OC-I cystatin and to the models reported previously for cystatins HvCPI-1 to HvCPI-7 (Abraham et al., 2006). However, specific features were observed for some of the barley cystatins. The three-dimensional structure of group C2 HvCPI-11 could only be accurately modeled up to the β3 sheet, which suggests important structural changes in the loop between β4 and β5 sheets, where the conserved Trp is located. Changes in this region were also observed in group C1 HvCPI-6, which presented a shorter loop between β4 and β5 sheets, and group C2 HvCPI-7, which showed some distortions in the region of the β4 and β5 sheets, including a longer loop between these sheets.
Figure 1.
Unrooted phylogram of the Poaceae cystatins constructed by the PhyML method, corresponding to the 13 proteins from barley (gray boxes), 12 from rice, 12 from sorghum, 10 from maize, seven from wheat, and one each from sugarcane (Saccharum officinarum) and Job's tears (Coix lacryma-jobi). GenBank accession/model numbers are as follows: for barley, HvCPI-1 (Y12068), HvCPI-2 (AJ748337), HvCPI-3 (AJ748338), HvCPI-4 (AJ748344), HvCPI-5 (AJ748340), HvCPI-6 (AJ748341), HvCPI-7 (AJ748345), HvCPI-8 (AJ748343), HvCPI-9 (AJ748339), HvCPI-10 (AJ748342), HvCPI-11 (AJ748346), HvCPI-12 (AJ748347), and HvCPI-13 (AJ748348); for rice, OC-I (Os01g58890), OC-II (Os05g41460), OC-III (Os05g3880), OC-IV (Os01g68660), OC-V (Os01g68670), OC-VI (Os03g11180), OC-VII (Os03g11170), OC-VIII (Os03g31510), OC-IX (Os01g11160), OC-X (Os04g2250), OC-XI (Os09g08110), and OC-XII (Os01g16430); for corn, CCI (D63342), CCII (D38130), and CC3-CC10 (BN000508-BN000514); for sorghum, SbCPI-1 (estExt_fgenesh1_kg.C_chr_30300), SbCPI-2 (fgenesh1_pg.C_chr_9001840), SbCPI-3 (Sb09g024230), SbCPI-4 (Sb03g010730), SbCPI-5 (estExt_fgenesh1_pg.C_chr_91844), SbCPI-6 (estExt_fgenesh1_kg.C_chr_90155), SbCPI-7 (e_gw1.1.18252.1), SbCPI-8 (e_gw1.1.14914.1), SbCPI-9 (fgenesh1_pg.C_chr_1003205), SbCPI-10 (e_gw1.3.21020.1), SbCPI-11 (gw1.1.4616.1), and SbCPI-12 (fgenesh1_pg.C_chr_1003206); for wheat, WC1 (AB038392), WC2 (AB038395), WC3 (AB038394), WC4 (AB038393), gWC2 (AB038391), WC5 (AF364099), and MDC1 (AB223039); for sugarcane, SOF (AY119689); and for Job's tears, CLA (AB037156).
Figure 2.
A schematic of alignment of the amino acid sequences of the barley cystatins. Amino acid sequences responsible for the inhibitor-enzyme interaction and the LARFAV motif are indicated. The predicted signal peptide (SP) and the C-terminal tails (C-term) are shaded. The approximate locations of the five β-sheets (yellow arrows) and the single α-helix (purple segment) are indicated.
Cystatins Primarily Inhibit Cathepsin L-Like Cys-Protease Activity in Protein Extracts from Barley Tissues
The differences in amino acid sequence and the corresponding diversity in their predicted three-dimensional structures suggested that each barley cystatin has distinct biochemical properties. To study their inhibitory capability, the 13 cystatins were purified as recombinant proteins from Escherichia coli cultures. Purified cystatins were then used to inhibit enzyme activities in barley extracts, using substrates susceptible to degradation by cathepsin L-like, B-like, and H-like proteases (Table I). The assays were carried out by adding the same concentration of each cystatin (1 μm) to 1 μg of soluble protein extracts from the barley embryo and leaf. The results indicated that cystatins primarily inhibited the cathepsin L-like activity in both embryos and leaf tissues. In fact, 10 of the 13 recombinant cystatins showed a significant reduction (ranging from approximately 25% to 75%) in the cathepsin L-like activity present in barley protein extracts. Endogenous cathepsin B- and H-like activities were also inhibited by some cystatins, and the recombinant HvCPI-4, HvCPI-5, and HvCPI-6 proteins were particularly efficient. In contrast, cystatins HvCPI-7, HvCPI-12, and HvCPI-13 failed to inhibit all endogenous Cys-protease activities assayed.
Table I.
In vitro inhibitory activity of the recombinant barley cystatins (HvCPI-1–HvCPI-13) against extracts from different barley tissues
Samples were 16-h germinating embryos (1 μg) and 7-d-old leaves (1 μg) using the specific substrates Z-FR-AMC, Z-RR-AMC, and Bz-FVR-AMC (for cathepsin L-like, B-like, and H-like, respectively) and 1 mm cystatin. Values are expressed as percentages and are means ± se of triplicate measurements from a unique pool of extracts treated with the inhibitor versus its corresponding control without it. No effect (ne) was considered for inhibition less than 20%.
| Cystatin | Leaf |
Embryo |
Leaf |
Embryo |
Leaf |
Embryo |
|---|---|---|---|---|---|---|
| Z-FR-AMC Substrate (Cathespin L-Like) | Z-RR-AMC Substrate (Cathespin B-Like) | Bz-FVR-AMC Substrate (Cathespin H-Like) | ||||
| HvCPI-1 | 64.6 ± 4.7 | 56.8 ± 0.3 | ne | ne | 28.5 ± 1.2 | ne |
| HvCPI-2 | 47.3 ± 2.6 | 52.1 ± 1.7 | ne | ne | ne | ne |
| HvCPI-3 | 51.0 ± 4.7 | 32.2 ± 2.1 | ne | ne | ne | ne |
| HvCPI-4 | 71.8 ± 4.1 | 60.8 ± 2.2 | 28.1 ± 1.9 | ne | 46.6 ± 0.1 | 56.3 ± 1.9 |
| HvCPI-5 | 70.7 ± 3.2 | 64.8 ± 0.1 | 36.5 ± 3.0 | 33.2 ± 5.0 | 46.8 ± 3.2 | 44.2 ± 4.9 |
| HvCPI-6 | 74.6 ± 0.5 | 70.5 ± 1.2 | 36.3 ± 3.0 | 27.3 ± 2.5 | 59.7 ± 0.8 | 56.7 ± 1.4 |
| HvCPI-7 | ne | ne | ne | ne | ne | ne |
| HvCPI-8 | 60.2 ± 8.0 | 47.4 ± 1.3 | 25.6 ± 3.0 | ne | ne | ne |
| HvCPI-9 | 44.3 ± 1.7 | 27.1 ± 1.5 | ne | ne | ne | ne |
| HVCPI-10 | 26.5 ± 2.7 | ne | ne | ne | ne | ne |
| HvCPI-11 | 50.1 ± 3.3 | ne | 24.6 ± 3.3 | ne | ne | ne |
| HvCPI-12 | ne | ne | ne | ne | ne | ne |
| HvCPI-13 |
ne |
ne |
ne |
ne |
ne |
ne |
Barley Cathepsin L-Like Proteases and Cystatins Show a Wide Variation in Their Expression Pattern
The inhibitory effects of barley protein extracts discussed above have focused our attention on the endogenous cathepsin L-like Cys-proteases. Among the 32 Cys-protease sequences of the C1A class found in barley, we have recently identified 22 cathepsin L-like sequences that were classified into five different groups (Martinez and Diaz, 2008). To explore the relationship between barley cystatins and cathepsin L-like peptidases in detail, we selected one member (HvPap4, HvPap6, HvPap10, HvPap16, and HvPap17 genes) of each group (see Fig. 4A below). Only the HvPap10 gene encoding the EP-B2 Cys-protease had been characterized previously (Koehler and Ho, 1990).
Figure 4.
Analysis of the expression and localization of barley cathepsin L-like peptidases. A, Groups of genes encoding cathepsin L-like peptidases from barley, derived from the complete cladogram of Cys-proteases published by Martinez and Diaz (2008). Underlined genes were selected for further assays. B, Barley genes (HvPap14, HvPap6, HvPap10, HvPap16, and HvPap17) encoding cathepsin L-like peptidases show differential expression patterns in several barley tissues by real-time quantitative PCR. Values expressed as the relative mRNA contents of barley cathepsin L-like genes were standardized to the barley Actin2 mRNA level. RNA was extracted from developing endosperms at 10 (E1), 14 (E2), 18 (E3), and 22 (E4) daf, from immature embryos (Ie), mature embryos (Me), and germinating embryos collected at 8 (G1), 16 (G2), 24 (G3), and 48 (G4) hai, and from 7-d-old leaves (L) and roots (R).
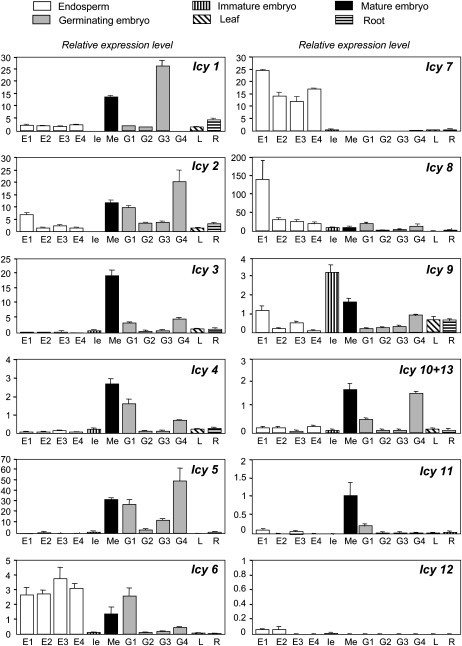
The transcript accumulation patterns of the 13 barley cystatins and the five selected cathepsin L-like proteases were first investigated by real-time quantitative reverse transcription (qRT)-PCR analysis in the major barley tissues. RNA samples were prepared from developing endosperms at 10, 14, 18, and 22 d after flowering (daf), from immature (18 daf), mature, and germinating embryos at 8, 16, 24, and 48 h after incubation (hai), and from 7-d-old leaves and roots. For cystatins, specific primers were designed to amplify each gene with the exception of Icy10 and Icy13 genes, whose nucleotide sequences shared 97% identity. The results, expressed as cystatin mRNA content normalized to barley Actin2 mRNA levels, indicated that the expression patterns varied widely across the 13 cystatin genes tested (Fig. 3). Most cystatin genes were expressed in seeds, although at different relative concentrations. Transcripts of genes Icy6, Icy7, and Icy8 were mainly accumulated in developing endosperms, while Icy1, Icy2, Icy3, Icy4, Icy5, and Icy10/13 mRNAs were preferentially detected in mature and germinating embryos. In contrast, the highest accumulation of Icy9 messengers was observed in the immature embryo. Moreover, most cystatin genes showed low expression levels in vegetative tissues such as leaves and roots under the growth conditions used. The presence of mRNAs of Icy11 and Icy12 genes was low, and sometimes undetectable, in most of the tissues tested.
Figure 3.
Expression of barley cystatin genes (Icy1–Icy13) in different barley tissues analyzed using real-time quantitative PCR. Values were expressed as relative mRNA levels of barley cystatin genes and were standardized using the barley Actin2 mRNA content. RNA was extracted from developing endosperms at 10 (E1), 14 (E2), 18 (E3), and 22 (E4) daf, from immature embryos (Ie), mature embryos (Me), and germinating embryos collected at 8 (G1), 16 (G2), 24 (G3), and 48 (G4) hai, and from 7-d-old leaves (L) and roots (R).
The expression pattern of barley cathepsin L-like genes, referred to as the relative content of the barley Actin2 mRNA levels, showed that the transcripts of four of the five cathepsin L-like genes (HvPap4, HvPap6, HvPap10, and HvPap17) were primarily accumulated in germinating embryos (Fig. 4). The most abundant mRNAs were those of HvPap10, which had an embryo-specific pattern of expression, followed by HvPap6, HvPap4, and HvPap17 genes, although qRT-PCR assays are limited by the primer sequences to compare transcript levels of different genes. None of the cathepsin L-like genes studied were expressed in developing barley endosperms. Messengers of HvPap4, HvPap6, and HvPap17 genes were also detected in leaves and roots. The fifth gene analyzed, HvPap16, was expressed exclusively in leaves (Fig. 4).
Cystatin and Cathepsin L-Like Proteases Colocalize in Endoplasmic Reticulum and Golgi Bodies
To establish the relationship between cystatins and cathepsin L-like proteases, we determined whether these proteins localized in the same subcellular compartment. The open reading frames (ORFs) of cystatin and cathepsin L-like genes were cloned into an expression vector fused to the GFP. Onion (Allium cepa) epidermal layers were transiently transformed with these constructs and cobombarded with the plasmid pRTL2ΔNS/ss-RFP-HDEL containing the C-terminal HDEL signal (Shockey et al., 2006), which specifically localizes to the endoplasmic reticulum (ER). As shown in Figure 5 and Supplemental Figure S2, both cystatin and proteases were detected throughout the entire ER network, which is continuous with the nuclear envelope. This was revealed by their colocalization with the red fluorescence emitted by the ER-plasmid control in the same cells. As an additional control, coexpression assays of either cystatin-GFP or cathepsin-GFP constructs with the Golgi marker CONST1-YFP (for yellow fluorescent protein) plasmid (Tse et al., 2006) were also performed to assess whether cystatins also localized in the Golgi stacks. Typical punctate fluorescent signals were detected in cells expressing the CONST1-YFP, which overlapped with the cystatin-GFP expression and confirmed the association of cystatins with the Golgi complex (Supplemental Fig. S3).
Figure 5.
Subcellular localization of barley cystatins (HvCPI-1–HvCPI-13) and cathepsin L-like proteases in onion epidermal cells. Cystatins (HvCPI-1, HvCPI-4, and HvCPI-5) and cathepsin L-like proteases (HvPap-4, HvPap-6, HvPap-10, and HvPap-16) fused to GFP at the N terminus were transiently expressed in onion epidermal layers. Green fluorescence was observed after incubation for 24 h with a confocal microscope (LEICA-Sp2-AOBS-UV) using appropriate filters. The plasmid pRTL2ΔNS/ss-RFP-HDEL containing the C-terminal HDEL ER retrieval signal gene (which emits red fluorescence specifically in the ER) was used as a control. The overlap of both fluorescence images is shown. Bars are as indicated.
One exception to the protein localization pattern among the 13 cystatins tested occurred with the HvCPI-1 protein, the only barley cystatin lacking an N-terminal signal peptide. This protein, aside from being localized in ER/Golgi structures, presented a diffuse fluorescence in the cytoplasm. This is the common appearance of proteins distributed in this compartment. In addition, cystatins HvCPI-1 and HvCPI-4 also were targeted to the nucleus. No fluorescence signal was observed when the HvCPI-12 cystatin and the HvPap-17 cathepsin fused to the GFP marker were analyzed. Interestingly, the localization pattern showed by the barley cystatins was coincident with that presented by the cathepsin L-like proteases (Fig. 5; Supplemental Figs. S2 and S3).
Barley Cystatins Interact with and Inhibit Barley Cathepsin L-Like Cys-Proteases
To study the cystatin specificity to cathepsin L-like peptidases, both inhibitors (HvCPI-1–HvCPI-13) and proteases (HvPap-4, HvPap-6, HvPap-10, HvPap-16, and HvPap-17) were expressed as fusion proteins with a His tail in E. coli cultures. The recombinant proteins were purified to homogeneity by affinity chromatography to a Ni2+ column and were finally eluted from the column. Inhibitory in vitro assays were conducted using a substrate that can be degraded by cathepsin L-like enzymes. Papain, a plant cathepsin L-like protease of commercial origin, was also included in the assay. Kinetic analyses revealed that barley cystatins exhibited a noncompetitive inhibition against all of the proteases tested (Supplemental Fig. S4B). The inhibition constant (Ki) values against the cathepsin L-like proteases showed different target specificities (Table II). The cystatin HvCPI-2 was the strongest inhibitor of HvPap-6 and HvPap-10 peptidases, with Ki values of 2.3 × 10−9 m and 3.4 × 10−10 m, respectively. The HvPap-4 protease was mainly inhibited by the HvCPI-5 cystatin (Ki = 8.1 × 10−9 m), and the HvPap-16 protease was inhibited by the protein HvCPI-6 (Ki = 2.5 × 10−9 m). There are no inhibitory results for the HvPap-17 protease, because the purified protein was inactive. Most barley cystatins also were good inhibitors of papain, as shown by the estimated 10−6 to 10−9 m values of Ki. When the inhibitory profiles among all barley cystatins were compared, recombinant proteins HvCPI-1, -2, -4, -5, -6, -8, -9, and -11 were able to inhibit all cathepsin L-like proteases tested, whereas HvCPI-3, -10, and -13 were not effective at inhibiting cathepsin L-like from barley. Cystatins HvCPI-7 and HvCPI-12 were completely inactive against all proteases analyzed under the tested conditions.
Table II.
Ki values of the barley cystatins against cathepsin L-like proteases from commercial origin (papain) and from barley expressed as recombinant proteins in E. coli cultures (HvPap-4, HvPap-6, HvPap-10, and HvPap-16)
No inhibitory activity (ni) was observed at 5 mm concentration of the cystatin.
| Cystatin |
Ki |
||||
|---|---|---|---|---|---|
| Papain | HvPap-4 | HvPap-6 | HvPap-10 | HvPap-16 | |
| m | |||||
| HvCPI-1 | 2.0 × 10−8 | 1.2 × 10−7 | 1.0 × 10−7 | 4.1 × 10−9 | 1.2 × 10−7 |
| HvCPI-2 | 5.1 × 10−8 | 1.0 × 10−8 | 2.3 × 10−9 | 3.4 × 10−10 | 6.2 × 10−9 |
| HvCPI-3 | 4.0 × 10−8 | ni | 1.6 × 10−8 | 1.9 × 10−6 | 6.4 × 10−7 |
| HvCPI-4 | 2.2 × 10−7 | 1.4 × 10−7 | 2.2 × 10−8 | 7.2 × 10−8 | 4.0 × 10−7 |
| HvCPI-5 | 6.3 × 10−8 | 8.1 × 10−9 | 6.6 × 10−8 | 1.3 × 10−8 | 8.2 × 10−8 |
| HvCPI-6 | 1.7 × 10−9 | 1.3 × 10−7 | 9.4 × 10−8 | 2.8 × 10−9 | 2.5 × 10−9 |
| HvCPI-7 | ni | ni | ni | ni | ni |
| HvCPI-8 | 1.0 × 10−8 | 4.9 × 10−8 | 9.3 × 10−8 | 4.3 × 10−9 | 2.9 × 10−8 |
| HvCPI-9 | 1.4 × 10−9 | 4.7 × 10−8 | 1.3 × 10−8 | 2.1 × 10−7 | 1.3 × 10−7 |
| HvCPI-10 | 3.3 × 10−7 | 6.9 × 10−7 | 3.3 × 10−6 | ni | 1.0 × 10−5 |
| HvCPI-11 | 8.2 × 10−8 | 7.0 × 10−7 | 2.9 × 10−7 | 7.0 × 10−7 | 4.3 × 10−6 |
| HvCPI-12 | ni | ni | ni | ni | ni |
| HvCPI-13 |
1.4 × 10−6 |
1.9 × 10−6 |
1.2 × 10−6 |
ni |
ni |
To assess whether the interaction between barley cystatins and barley cathepsin L-like proteases observed in vitro was also produced in vivo, we used the bimolecular fluorescence complementation (BiFC) assay. For the BiFC experiments, we selected four barley proteases and two cystatins. HvPap-4, HvPap-6, HvPap-10, and HvPap-16 were fused to the C-terminal fragment of the GFP. The HvPap17 cathepsin was excluded because it could not be localized at the subcellular level, as mentioned above. Cystatins HvCPI-6 and HvCPI-7 were fused to the N-terminal fragment of the GFP. These two cystatins were selected based on their opposing ability to inhibit the four barley proteases. The HvCPI-6 protein was one of the strongest protease inhibitors, while HvCPI-7 did not inhibit any of the proteases tested in vitro (Tables I and II). Onion epidermal layers were cobombarded with combinations of constructs containing the entire ORFs of the proteases and cystatins that fused translationally to the corresponding GFP fragments. When interactions between cystatin and cathepsin L-like took place, the fluorophore was reconstituted and fluorescence was emitted. Microscope observations from the different combinations in the appropriate fusions to the GFP portions showed interactions between the HvCPI-6 cystatin and the four cathepsin L-like proteases in plant cells. As expected, the GFP fluorescence was targeted to the ER network and to the Golgi complex (Fig. 6). In contrast, no fluorescence was observed when the HvCPI-7 protein was used, both when the two GFP fragments were alone and when they used simultaneously. An example of the absence of fluorescence is shown in Figure 6. These results revealed the specificity in the enzyme-inhibitor interaction and corroborate the in vitro inhibitory data presented in Table II.
Figure 6.
Subcellular localization of reconstituted GFP complexes in transiently transformed onion epidermal cells. Fluorescence was analyzed after cobombardment with HvCPI-6 and HvCPI-7 cystatins fused to the N-terminal GFP fragment (N-term) and cathepsin L-like proteases HvPap-4, HvPap-6, HvPap-10, and HvPap-16 fused to the C-terminal GFP fragment (C-term). Samples were observed with a confocal ultraspectral microscope (left panels). Observations with a Nomarski bright field are also shown (right panels). Bars are as indicated.
Cystatins and Their Target Cathepsin L-Like Proteases Are Involved in the Mobilization of Hordeins in the Barley Seed
The functional relevance of the interaction observed among barley cystatins and cathepsin L-like proteases was further investigated in barley seed germination, an endogenous physiological process. For this study, we selected those cathepsin L-like proteases (HvPap-4, HvPap-6, and HvPap-10) that showed detectable levels of gene expression in germinating embryos (Fig. 4). Similarly, a set of cystatins with different inhibitory capacities against the selected proteases was used (HvCPI-1, HvCPI-2, HvCPI-5, HvCPI-6, and HvCPI-11). We attempted to include at least one member of each phylogenetic cluster shown in Figure 1 in this set.
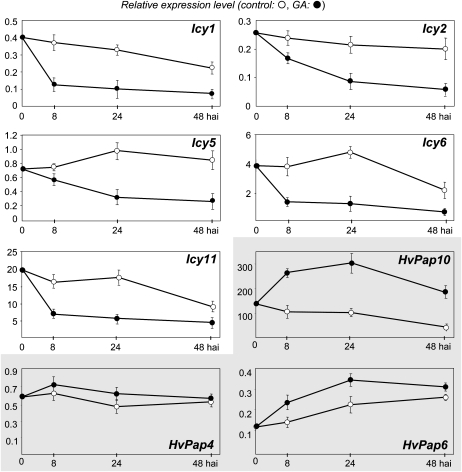
First, we analyzed the expression pattern of all selected genes in germinating barley aleurones and their responses to gibberellin (GA) treatment by qRT-PCR assays. These aleurone layers do not synthesize GA but are able to respond to external hormone treatment. Aleurone layers were isolated from deembryonated seeds of barley after 36 h of water imbibition and were then incubated both in the presence and in the absence of GA over different time periods (8, 24, and 48 h). As shown in Figure 7, both cystatin and protease genes were expressed in aleurone layers, although at different levels, with variable temporal expression patterns. Interestingly, when the barley aleurone layers were incubated in the buffer containing 1 μm GA, the accumulation of the five cystatin transcripts was drastically reduced. In contrast, the GA incubation increased the quantities of mRNA from L-like cathepsin HvPap6 and HvPap10 and had no effect on the expression level of the HvPap4 gene.
Figure 7.
Expression of barley cystatin genes (Icy1, Icy2, Icy5, Icy6, and Icy11) and barley cathepsin L-like genes (HvPap4, HvPap6, and HvPap10) in aleurone layers after 8, 24, and 48 h of incubation in the presence or absence of 1 μm GA3 as determined by real-time quantitative PCR. Values are expressed as relative mRNA levels of barley cystatin genes and are standardized using barley Actin2 mRNA content. Panels corresponding to the protease data have gray backgrounds.
We then explored whether cathepsin L-like proteases versus cystatins were involved in the breakdown of storage proteins accumulated in the dead starchy endosperm, causing nutrients to be released for seedling development. We chose the HvPap-6 and HvPap-10 cathepsin L-like proteases because they were activated by GA and their transcript levels increased during germination in the aleurone layer and embryo (Figs. 4 and 7). Total hordeins, the major prolamins accumulated in the mature barley seed, were purified and incubated with HvPap-6 and HvPap-10 cathepsin L-like proteases. Figure 8 shows the different abilities of these two proteases to degrade hordeins. All electrophoretic bands corresponding to B, C, and D hordeins were hydrolyzed by the HvPap10 protease, while HvPap6 only partially reduced the presence of those bands. In addition, protease activities were differentially inhibited by the addition of cystatins. HvCPI-1, HvCPI-2, HvCPI-5, and HvCPI-6 cystatins differentially inhibited the activities of both barley proteases, as shown by the presence of the whole hordein band profile. Cystatin HvCPI-11 only partially inhibited the activity of the HvPap-6 protease and did not inhibit the protease activity of HvPap-10. These results were consistent with the high Ki values that resulted from the in vitro inhibitory assays performed with recombinant proteins (Table II).
Figure 8.
Effects of barley cathepsin L-like and cystatin proteins on the degradation of seed hordeins shown by SDS-PAGE gels stained with Coomassie Brilliant Blue. Alcohol-extracted soluble proteins from barley (cv Bomi) dry seeds (H, lane 2) were incubated with the protease without inhibitor (−, lane 3) or with the protease and 1 μm barley cystatins HvCPI-1, -2, -5, -6, or -11 (lanes 4–9). Molecular markers (M, lane 1; molecular masses in kD) and the location of B, C, and D hordeins are indicated.
DISCUSSION
PhyCys have been studied in many plants. At the time of this work, nearly 90 PhyCys have been identified from multiple origins, but this work provides, to our knowledge, the first molecular analysis and description of the functional diversity of a whole gene family within a single plant species, barley. A total of 13 nonredundant genes from barley encoding cystatins were found. We assume that all barley members have been identified based on estimates from known plant genomes, ranging from seven cystatin genes for Arabidopsis and nine for poplar to 12 genes in rice and sorghum. In addition, although the barley genome has not been fully sequenced, an available collection contains almost 450,000 ESTs covering different cDNA libraries from various stages of plant development and tissues facing different biotic and abiotic stresses.
The alignment of the amino acid sequences among barley cystatins indicates a high conservation of motifs within this species, which is also reflected in the corresponding predicted three-dimensional structures of the cystains. Aside from the LARFAV sequence of unknown function, which is conserved with minor changes in all barley members, most of the cystatins contain the three motifs forming the tripartite wedge that enters the active site responsible for inhibiting papain-like Cys-proteases. Probably, the most important motif involved in this inhibition process corresponds to the QxVxG sequence, which physically interacts with the active site of target enzymes. This is supported by the inhibitory in vitro results obtained with the purified cystatins HvCPI-7 and HvCPI-12, whose Gly in the fifth position is substituted by Glu and Ser, respectively. Neither of these two proteins inhibited any of the Cys-proteases tested. Moreover, both cystatins also lacked the inhibitory effect against protein extracts from the barley tissues assayed. In contrast, the presence of Ile instead of Val in the third position of the reactive site in the HvCPI-9 protein does not appear to be crucial for the inhibition process. These results are in agreement with the changes in the inhibition activity toward papain reported for the oryzacystatin OC-I and the variants derived from barley cystatin HvCPI-1, generated by direct mutagenesis of the Gln-x-Val-x-Gly region (Arai et al., 1991; Martinez et al., 2003). Similarly, the Trp present in the loop between the β4 and β5 sheets is probably not essential to the inhibitory capability either, as shown by the Ki values obtained with the HvCPI-11 cystatin against cathepsin L-like proteases (Table II).
In order to analyze cystatin functions in the plant, it is crucial to identify their target proteases. Because most cystatins showed preferential affinity to cathepsin L-like proteases, five genes (one gene per group) encoding these proteases were selected from the 22 sequences identified in barley (Martinez and Diaz, 2008). The main goal was to study the relationship between cystatin and the cathepsin L-like proteases. Interestingly, the barley proteases colocalized with cystatins throughout the entire ER network and the Golgi complex. It is known that secretory proteins pass through a series of membrane-enclosed organelles, including the ER and the Golgi complex, on their way to being secreted (Bonifacino and Glick, 2004). Moreover, most of the cystatins are recognized as secretory proteins, as they contain signal peptides that direct them to the ER. From the ER, they are finally targeted to intracellular or extracellular locations (Tian et al., 2009). In addition, most C1A proteases have specific localization sequences that allow them to be delivered to different compartments or to be secreted (Grudkowska and Zagdanska, 2004). In this way, cystatins can interact and neutralize their cognate cathespin L-like Cys-proteases and thus participate in a common physiological process. In fact, the BiFC approach has allowed us to demonstrate that the cystatin-cathepsin L-like interaction takes place in the ER and in the Golgi complex in vivo and in real time, although these observations were made in heterologous plant cells. Additionally, the in vitro inhibitory assays performed with the recombinant barley cystatins have shown the different inhibitory capabilities of each cystatin against recombinant barley cathepsin L-like proteases.
The classification of the barley cystatins based on phylogenetic groups does not necessarily provide the most biologically accurate means of addressing their function. However, several cystatins allocated in each of the clusters/subclusters (A, B, C1, and C2) seem to share a functional homology. Barley genes included in cluster A (Icy1, Icy2, Icy3, and Icy4) show a clear correlation between their gene expression patterns and their functional activity as protease inhibitors. These cystatins are preferentially expressed in dry and germinating seeds and are efficient inhibitors, primarily of cathepsin L-like Cys-proteases. This behavior suggests that their main roles are as specialized endogenous regulators of enzymes involved in the mobilization of stored proteins upon germination, which is crucial for the seedling growth until the photosynthesis is fully established. A similar role has been described for cystatins from rice and wheat, also clustered in group A, which are able to inhibit Cys-proteases such as oryzains and gliadains involved in turnover functions in rice and wheat aleurone layers, respectively (Arai et al., 2002; Kiyosaki et al., 2007). The two barley cystatins (Icy5 and Icy9 genes) clustered in group B present different expression patterns. The Icy5 gene shares a similar expression pattern with group A members and has anti-cathepsin L-, B-, and H-like activities. However, Icy9 transcripts are also detected in other barley tissues, as in the immature embryo and the developing endosperm. The two subclusters of group C differ in their mRNA accumulation patterns as well as in their inhibitory capabilities. Icy6 and Icy8 genes (group C1) are preferentially expressed in seeds, particularly in developing endosperms, and as a result they are good inhibitors of C1A peptidases. These two cystatins and the Icy9 gene likely regulate protein accumulation in the endosperm during seed development. In contrast, genes allocated in group C2 show expression levels that are almost undetectable, and the gene products have a low inhibitory capacity or even fail to inhibit most of the endogenous Cys-protease activities assayed. The exception is the Icy7 gene, which is only expressed in endosperm and could be related to the processing of other proteins due to its anti-legumain activity (Martinez and Diaz, 2008; Carrillo, 2009). Nevertheless, cystatins of all clusters could also participate as defense proteins, as has been shown previously for HvCPI-1 cystatin (Martinez et al., 2003; Alvarez-Alfageme et al., 2007; Carrillo, 2009).
The functional relevance of the cystatin-protease interaction can be inferred from their preferential expression in seeds and from the roles of both enzyme inhibitors in the mobilization of storage proteins during barley seed germination. Upon seed germination, aleurone cells respond to GA3 synthesized by the embryo and trigger a signal transduction pathway that prompts the activation of α-amylases and proteases. These hydrolases are secreted and break down the stored reserves in the endosperm, releasing nutrients that support the embryo growth until the onset of photosynthesis (Muntz et al., 2001; Sreenivasulu et al., 2008). In this biological context, the reduction in the level of cystatin transcripts in the aleurone cells treated with GA, together with the GA induction of those genes encoding cathespin L-like proteases, indicate their respective functions as counterparts in the same biological process. The lack of cystatin expression allows proteases to degrade storage proteins, primarily B and C hordeins that are accumulated in the endosperm and subaleurone layers of mature barley seeds (Shewry and Halford, 2002). In parallel, other seed storage proteins such as D hordeins, globulins, and albumins, also putative targets of Cys proteases, are accumulated in the embryo and starchy endosperm (Shewry and Halford, 2002; Shi and Xu, 2009). The differential spatial distribution of storage proteins in the seed is congruent with the presence of both cathepsin L-like and cystatin mRNAs in germinating embryo and aleurone tissues. Moreover, the protein mobilization in storage tissues occurs on a gradient based on both stored proteases and proteases that are formed de novo whose activity must be regulated by inhibitors and/or activation of inactive zymogens (Sreenivasulu et al., 2008). The differential specificity of tested cystatins by HvPap-6 and/or HvPap-10 proteases shown here is likely to be crucial in this process. In addition, prolamins can be synthesized on rough ER membranes and accumulated into the ER lumen, leading to the formation of protein bodies as other storage proteins do (Shewry and Halford, 2002). In this case, the colocalization and interactions of cystatin and L-like cathepsin proteases in the ER/Golgi secretory system demonstrated in this study could preserve the storage protein integrity upon their deposition in the seed. Finally, the success of seed germination depends on the transcriptional regulatory network's ability to control the expression of genes encoding either hydrolytic enzymes and/or their inhibitors. This process is mediated by hormones, not only in seed germination but also during seed development.
CONCLUSION
Barley cathepsin L-like proteases and their proteinaceous inhibitors, cystatins, interact in vitro and in plant cells. The specificity and strength of this interaction is variable and depends on the cystatin and protease family member. The combinatorial expression of members from both families in a particular tissue and time could force the regulation of the activity exerted by the proteases. We have also described the role of several cystatins and Cys-proteases in the physiological process of mobilizing storage proteins upon germination of the barley seed. Likewise, both protein families must have a role in regulating some other important processes in the plant. Future work will be conducted on these subjects to discover new physiological pathways controlled by the interaction of cystatins and Cys-proteases.
MATERIALS AND METHODS
Cystatin Sequence Comparisons and Analyses
We have previously isolated cDNAs encoding HvCPI-1 to HvCPI-13 from a barley (Hordeum vulgare) cDNA library and EST collections (Gaddour et al., 2001; Abraham et al., 2006; Martinez and Diaz, 2008). Analysis of DNA and comparisons of deduced protein sequences were conducted with bioinformatics tools. Alignment of protein sequences within the Poaceae was performed with the default parameters of MUSCLE version 3.6 (Edgar, 2004). Alignment ambiguities and gaps were excluded from phylogenetic analysis using GBLOCKS version 0.91b (Castresana, 2000). Phylogenetic and molecular evolutionary analyses were conducted using the programs PhyML (Guindon and Gascuel, 2003) and MEGA version 4.0 (http://www.megasoftware.net; Guindon and Gascuel, 2003; Tamura et al., 2007). The displayed tree was derived using a maximum likelihood PhyML method and the WAG substitution model with a BIONJ starting tree. The approximate likelihood-ratio test based on a Shimodaira-Hasegawa-like procedure was used as the statistical test for nonparametric branch support (Anisimova and Gascuel, 2006). Signal peptide analysis was performed using SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP; Bendtsen et al., 2004).
The three-dimensional structures of the barley cystatins were modeled by the automated SWISS-MODEL program (Peitsch, 1995, 1996). The known crystal structure of the rice OC-I (Protein Data Bank identifier 1EQK) was used to construct the homology-based models. Structure analysis was performed using the RasMol 2.7 program (Sayle and Milner-White, 1995).
Expression and Purification of Recombinant Cystatins and Cathepsin L-Like Peptidases from Escherichia coli
The cDNA fragments spanning the whole cystatin ORFs, aside from their signal peptide sequences (HvCPI-8–HvCPI-13 proteins), were amplified by PCR and inserted in-frame into the expression vector pRSETB (Invitrogen). The recombinant plasmids and the constructs for barley HvCPI-1 to HvCPI-7 (Abraham et al., 2006) were introduced in E. coli BL21 CodonPlus (Stratagene). Bacterial cells were grown at 37°C to an optical density at 550 nm of approximately 0.5 and induced with 1 mm isopropyl β-d-thiogalactopyranoside for 2 h before being harvested and processed. The fusion proteins with a His tail were purified using His-Bind Resin (Novagen) following the manufacturer's instructions, and the purification was verified by SDS-PAGE. Expression and purification of barley cathepsin L-like peptidases were performed according to Bethune et al. (2006) with minor modifications. Briefly, the complete amino acid sequences lacking their signal peptides were cloned into the expression vector pRSETB (Invitrogen) to yield a construct encoding the proteases with both N- and C-terminal His6 tags. The resulting plasmid was introduced into E. coli BL21 CodonPlus (Stratagene). Bacterial cells were grown at 37°C to an optical density at 550 nm of approximately 0.5 and induced with 0.25 mm isopropyl β-d-thiogalactopyranoside for 20 h before being harvested and processed using a nickel-nitrilotriacetic acid agarose resin (Qiagen) as described by Bethune et al. (2006). After dialysis, the purification processes were checked by SDS-PAGE. An example is shown in Supplemental Figure S4A. The final protein concentrations were quantified using a Bio-Rad kit, with bovine serum albumin as a standard. The proenzyme preparations were stored at −80°C. Activation of HvPap-6, HvPap-10, and HvPap-16 was achieved by diluting the proteases by a ratio of 1:4 in a buffer containing 100 mm sodium acetate at pH 4.0. The HvPap-4 cathepsin was diluted in the same buffer and activated with the addition of pepsin at a concentration of 0.6 mg mL−1.
Inhibitory Activity of Cystatins against Barley Protein Extracts
For inhibitory endogenous protease assays, seeds of barley (cv Bomi) were germinated at 22°C in the dark for 7 d and used to collect leaf samples. Germinating embryos were obtained from imbibed seeds at 16 h of incubation at 22°C in the dark. All samples were frozen into liquid N2 and stored at −70°C until being used for protein extraction. Samples were ground with mortar and pestle and resuspended in 0.15 m NaCl, 50 mm sodium phosphate, pH 6.0, and 2 mm EDTA for 1 h at 4°C. After centrifugation for 15 min at 10,000 rpm, supernatants were recovered and their protein content was quantified using a Bio-Rad kit with bovine serum albumin as a standard. The inhibitory activity of recombinant cystatins was tested by the hydrolysis of substrates containing the 7-amino-4-methylcoumarin (AMC) fluorophore in microliter plate format. After testing different concentrations of cystatins, 1 μm of each inhibitor was incubated with 1 μg of a soluble leaf or embryo protein extract at 25°C for 10 min, prior to the addition of 25 μm Z-FR-AMC (N-carbobenzoxyloxy-Phe-Arg-7-amido-4-methylcoumarin), Z-RR-AMC (N-carbobenzoxyloxy-Arg-Arg-7-amido-4-methylcoumarin), and Bz-FVR-AMC (benzoyl-Phe-Val-Arg-7-amido-4-methylcoumarin) substrates susceptible to degradation by cathepsin L-, B-, and H-like proteases, respectively. Mixtures were then incubated for 1 h at 30°C, and emitted fluorescence was measured with a 365-nm excitation wavelength filter and a 465-nm emission wavelength filter. Results were expressed as a percentage of protease activity relative to that in the absence of the inhibitor. All assays were carried out in triplicate, and blanks were used to account for the spontaneous breakdown of substrates. As negative controls, proteins from E. coli transformed with the empty expression vector were used.
Real-Time qRT-PCR Analysis
For real-time qRT-PCR studies, seeds of barley (cv Bomi) were germinated at 22°C in the dark for 7 d and used to collect samples of leaves and roots. Developing endosperms of 10 to 22 daf and immature embryos (18 daf) were prepared from kernels of plants grown in a greenhouse at 18°C under an 18-h-light/6-h-dark photoperiod. Mature embryos were isolated from dry barley (cv Bomi) seeds. Germinating embryos were obtained from imbibed seeds after 8, 16, 24, and 48 hai at 22°C in the dark. All samples were frozen in liquid N2 and stored at −80°C until being used for RNA extraction. Isolated aleurone layers from embryoless barley seeds (cv Himalaya) were prepared as described by Moreno-Risueno et al. (2007), incubated in the presence or absence of 1 μm GA3 for 8, 24, or 48 h in a calcium succinate buffer (20 mm Na-succinate, 20 mm CaCl2, pH 5.2), and then frozen.
Total RNA was extracted from frozen samples by the phenol/chloroform method, followed by precipitation with 3 m LiCl (Lagrimini et al., 1987) and digestion with DNase. cDNAs were synthesized from 1 μg of RNA using the High Reverse Transcription kit (Applied Biosystems) following the manufacturer's instructions. qRT-PCR analyses were performed for duplicate samples in a 7300 Real-Time PCR System (Applied Biosystems) using the SYBR Green detection system. Quantification was standardized to barley Actin2 mRNA levels. The primers used for PCR amplification are shown in Supplemental Table S1.
Inhibitory Activity of Cystatins against Cathepsin L-Like Peptidases from Barley
The recombinant cystatin proteins were also assayed against the barley HvPap-4, HvPap-6, HvPap-10, and HvPap-16 cathepsin L-like proteases purified from E. coli cultures as well as against commercial papain (EC 3.4.22.2). Briefly, different concentrations of inhibitor plus the corresponding amount of the enzymes with a similar enzyme activity (800 ng of HvPap-4, 80 ng of HvPap-6, 20 ng of HvPap-10, 80 ng of HvPap-16, and 10 ng of papain), were incubated at room temperature in a buffer containing 100 mm sodium phosphate, pH 6.0, 10 mm l-Cys, 10 mm EDTA, and 0.01% (v/v) Brij35. Then, the Z-FR-AMC substrate was added at different concentrations (10–30 μm) and the reactions were incubated for 1 h at 30°C. Emitted fluorescence was measured as above. The system was calibrated with known amounts of AMC hydrolysis product in a standard reaction mixture. All assays were carried out in triplicate, and blanks were used to account for the spontaneous breakdown of substrates. As negative controls, proteins from E. coli transformed with the empty expression vector were used. The Ki values were determined from Lineweaver-Burk plots (1/V versus 1/[S]).
Subcellular Localization of Cystatins and Cathepsin L-Like Proteases
The ORFs of the barley cystatin genes (Icy1–Icy13) and the barley cathepsin L-like genes (HvPap4, HvPap6, HvPap10, HvPap16, and HvPap17) were translationally fused to the N terminus of the whole GFP reporter gene. The ORFs were cloned into BamHI restriction sites of the psmRS-GFP plasmid containing the cauliflower mosaic virus 35S promoter (Davies and Vierstra, 1998). As a control, two plasmids were used as localization markers. The plasmid pRTL2ΔNS/ss-RFP-HDEL containing the Arabidopsis (Arabidopsis thaliana) chitinase signal sequence and the C-terminal HDEL ER retrieval signal, whose protein specifically localized in the endoplasmic reticulum, and the Golgi reporter CONST1-YFP plasmid (Shockey et al., 2006; Tse et al., 2006). Transient transformations of onion (Allium cepa) epidermal cells were performed by particle cobombardment with a biolistic helium gun device (DuPont PDS-1000; Bio-Rad) as described by Diaz et al. (2005). Fluorescence images were acquired after 24 h of incubation at 22°C in the dark using a confocal microscope (LEICA-Sp2-AOBS-UV) with appropriate filters (Leica; http://www.leica.com).
BiFC
HvPap4, HvPap10, HvPap14, and HvPap16 genes were fused in-frame to the C-terminal nonfluorescent fragment of the Gfp gene (corresponding to the last 85 amino acid residues of the GFP protein), using plasmids and procedures described previously by Diaz et al. (2005). Similarly, the Icy6 and Icy7 cystatin genes were fused to the N-terminal nonfluorescent fragment of the Gfp gene (corresponding to the first 154 amino acid residues of the GFP protein). Cobombardment of inner epidermal layers isolated from onion was carried out according to Diaz et al. (2005). The fluorescence emission was observed after 48 h of incubation at 22°C in the dark using a confocal microscope (LEICA-Sp2-AOBS-UV) with appropriate filters (Leica; http://www.leica.com).
Assays of Hordein Digestion by Barley Cathepsin L-Like Peptidases
Dry seeds of barley (cv Bomi) were crushed and used to extract hordeins by 55% (v/v) propan-2-ol containing 1% (v/v) 2-mercaptoethanol according to Phillips and Wallace (1989). After incubation for 1 h at 60°C, samples were centrifuged at 12,000g for 10 min, and supernatants were recovered. Protein concentrations were determined using the Bio-Rad kit and bovine serum albumin as a standard.
To test the capacity of barley Cys-proteases to degrade hordeins, 6 μg of the final supernatant containing hordeins was incubated with 3 μg of proHvPap-10 or 1.5 μg of proHvPap-6 for 4 h. To determine the influence of cystatin inhibitors on hordein degradation, several cystatins (HvCPI-1, HvCPI-2, HvCPI-5, HvCPI-6, and HvCPI-11) at 1 μm concentration were incubated with the proteases for 30 min prior to the addition of the hordeins. Final mixtures were then electrophoretically separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Three-dimensional structures of barley cystatins.
Supplemental Figure S2. Subcellular localization of barley cystatins (HvCPI-2–HvCPI-13).
Supplemental Figure S3. Golgi localization of several barley cystatins and cathepsin L-like peptidases.
Supplemental Figure S4. Examples of protein purification and Lineweaver-Burk plot.
Supplemental Table S1. Primer sequences used for the amplification of barley genes in qRT-PCR assays.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. P. Dupree (University of Cambridge) for providing the plasmids pRTL2ΔNS/ss-RFP-HDEL and CONST1-YFP. We also thank Dr. P. González-Melendi (Centro de Biotecnología y Genómica de Plantas) for critically reading the manuscript.
This work was supported by the Ministerio de Educacion y Ciencia from Spain (project no. BFU2005–00603) and the Ministerio de Ciencia e Innovación (project no. BFU2008–01166).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Isabel Diaz (i.diaz@upm.es).
The online version of this article contains Web-only data.
References
- Abraham Z, Martinez M, Carbonero P, Diaz I (2006) Structural and functional diversity within the cystatin gene family of Hordeum vulgare. J Exp Bot 57 4245–4255 [DOI] [PubMed] [Google Scholar]
- Alvarez-Alfageme F, Martinez M, Pascual-Ruiz S, Castañera P, Diaz I, Ortego F (2007) Effects of potato plants expressing a barley cystatin on the predatory bug Podisus maculiventris via herbivorous prey feeding on the plant. Transgenic Res 16 1–13 [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55 539–552 [DOI] [PubMed] [Google Scholar]
- Arai S, Matsumoto I, Emori Y, Abe K (2002) Plant seed cystatins and their enzymes of endogenous and exogenous origin. J Agric Food Chem 50 6612–6617 [DOI] [PubMed] [Google Scholar]
- Arai S, Watanabe H, Kondo H, Emori Y, Abe K (1991) Papain inhibitory activity of oryzacystatin, a rice seed cysteine-protease inhibitor, depends on the central Gln-Val-Val-Ala-Gly region conserved among cystatin superfamily members. J Biochem 109 294–298 [PubMed] [Google Scholar]
- Atkinson HJ, Grimwood S, Johnston K, Green J (2004) Prototype demonstration of transgenic resistance to the nematode Rodopholus simies conferred on banana by a cystatin. Transgenic Res 13 135–142 [DOI] [PubMed] [Google Scholar]
- Belenghi B, Acconcia F, Trovato M, Perazzolli M, Bocedi A, Polticelli F, Ascenzi P, Delledonne M (2003) AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur J Biochem 270 2593–2604 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptide: SignalP 3.0. J Mol Biol 34 783–795 [DOI] [PubMed] [Google Scholar]
- Bethune MT, Strop P, Tang Y, Sollid LM, Khosia C (2006) Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem Biol 13 637–647 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 23 153–166 [DOI] [PubMed] [Google Scholar]
- Carrillo L (2009) Cistatinas de cebada: proteínas de defensa contra artrópodos. PhD thesis. Escuela Técnica Superior de Ingenieros Agrónomos-Universidad Politécnica de Madrid, Madrid
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17 540–552 [DOI] [PubMed] [Google Scholar]
- Corre-Menguy F, Cejudo FJ, Mazubert C, Vidal J, Lelandais-Briere C, Torres G, Rode A, Hartmann C (2002) Characterization of the expression of a wheat cystatin gene during caryopsis development. Plant Mol Biol 50 687–698 [DOI] [PubMed] [Google Scholar]
- Davies SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36 521–528 [DOI] [PubMed] [Google Scholar]
- Davy A, Svendsen I, Sorensen SO, Sorensen MB, Rouster J, Meldal M, Simpson D, Cameron-Mills V (1998) Substrate specificity of barley cysteine endoproteases EP-A and EP-B. Plant Physiol 117 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P (2005) The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 42 652–662 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddour K, Vicente-Carbajosa J, Lara P, Isabel-Lamoneda I, Díaz I, Carbonero P (2001) A constitutive cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol Biol 45 599–608 [DOI] [PubMed] [Google Scholar]
- Garcia-Lorenzo M, Sjodin A, Jansson S, Funk C (2006) Protease gene family in Populus and Arabidopsis. BMC Plant Biol 6 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Rivard D, Kiggundu A, Kunert K, Gleddie SC, Cloutier C, Michaud D (2007) A multicomponent, elicitor-inducible cystatin complex in tomato, Solanum lycopersicum. New Phytol 173 841–851 [DOI] [PubMed] [Google Scholar]
- Grudkowska M, Zagdanska B (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51 609–624 [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17 1–9 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52 696–704 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Campos R, Torres Acosta J, Saucedo-Arias LJ, Gomez-Lim MA (1999) The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat Biotechnol 17 1223–1226 [DOI] [PubMed] [Google Scholar]
- Kiyosaki T, Matsumoto I, Asakura T, Funaki J, Kuroda M, Misaka T, Arai S, Abe K (2007) Gliadain, a gibberellin-inducible cysteine proteinase occurring in germinating seeds of wheat, Triticum aestivum L., specifically digests gliadin and is regulated by intrinsic cystatins. FEBS J 164 470–477 [DOI] [PubMed] [Google Scholar]
- Koehler SM, Ho THD (1990) Hormonal regulation, processing, and secretion of cysteine proteinases in barley aleurone layers. Plant Cell 2 769–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rosthein S (1987) Molecular cloning of complementary DNA encoding the lignin forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA 84 7542–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madureira HC, Da Cunha M, Jacinto T (2006) Immunolocalization of a defense-related 87KDa cystatin in leaf blade of tomato plants. Environ Exp Bot 55 201–208 [Google Scholar]
- Margis R, Reis EM, Villeret V (1998) Structural and phylogenetic relationships among plant and animal cystatins. Arch Biochem Biophys 359 24–30 [DOI] [PubMed] [Google Scholar]
- Martinez M, Abraham Z, Carbonero P, Diaz I (2005. a) Comparative phylogenetic analysis of cystatin gene families from Arabidopsis, rice and barley. Mol Genet Genomics 273 423–432 [DOI] [PubMed] [Google Scholar]
- Martinez M, Diaz I (2008) The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol Biol 8 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Diaz-Mendoza M, Carrillo L, Diaz I (2007) Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and legumain cysteine proteinases. FEBS Lett 581 2914–2918 [DOI] [PubMed] [Google Scholar]
- Martinez M, Lopez-Solanilla E, Rodriguez-Palenzuela P, Carbonero P, Diaz I (2003) Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol Plant Microbe Interact 16 876–883 [DOI] [PubMed] [Google Scholar]
- Martinez M, Rubio-Somoza I, Carbonero P, Diaz I (2005. b) A cathepsin B-like cysteine protease gene from Hordeum vulgare (gene CatB) induced by GA in aleurone cells is under circadian control in leaves. J Exp Bot 54 951–959 [DOI] [PubMed] [Google Scholar]
- McLellan H, Gilroy EM, Yun B-W, Birch PRJ, Loake GJ (2009) Functional redundancy in the Arabidopsis cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol 183 408–418 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Diaz I, Carrillo L, Fuentes R, Carbonero P (2007) The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J 51 352–365 [DOI] [PubMed] [Google Scholar]
- Muntz K, Belozersky MA, Dunaesvsky YE, Schlereth A, Tiedemann J (2001) Stored proteinases and the initiation of storage protein mobilization in seeds during germination and seedling growth. J Exp Bot 52 1741–1752 [DOI] [PubMed] [Google Scholar]
- Nagata K, Kudo N, Abe K, Arai S, Tanakura M (2000) Three-dimensional solution of oryzacystatin-I, a cysteine proteinase inhibitor of rice, Oryza sativa L. japonica. Biochemistry 39 14753–14760 [DOI] [PubMed] [Google Scholar]
- Nissen MS, Kumar GNM, Youn B, Knowles DB, Lam KS, Ballinger WJ, Knowles NR, Kang CH (2009) Characterization of Solanum tuberosum multicystatin and its structural comparison with other cystatins. Plant Cell 21 861–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch M (1995) Protein modeling by E-mail. Biotechnology 13 658–660 [Google Scholar]
- Peitsch M (1996) ProMod and Swiss-Model: internet-based tools for automated comparative protein modeling. Biochem Soc Trans 224 274–279 [DOI] [PubMed] [Google Scholar]
- Pernas M, Sanchez-Monge R, Gomez L, Salcedo G (1998) A chestnut seed cystatin differentially effective against cysteine proteinase from closely related pests. Plant Mol Biol 38 1235–1242 [DOI] [PubMed] [Google Scholar]
- Pernas M, Sanchez-Monge R, Lombardero M, Arteaga C, Castañera P, Salcedo G (2000) Der p1 and Der f1, the highly related and major allergens from house mites, are differentially affected by a plant cystatin. Clin Exp Allergy 30 972–978 [DOI] [PubMed] [Google Scholar]
- Phillips HA, Wallace W (1989) A cysteine endopeptidase from barley malt which degrades hordein. Phytochemistry 28 3285–3290 [Google Scholar]
- Prins A, van Heerden PDR, Olmos E, Kunert KJ, Foyer CH (2008) Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves: a model for dynamic interactions with ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. J Exp Bot 59 1935–1950 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Barrett AJ (2008) MEROPS: the peptidase database. Nucleic Acids Res 36 D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro APO, Pereira EJC, Galvan TL, Picanzo MC, Picoli EAT, da Silva DJH, Fari MG, Otoni WC (2006) Effect of eggplant transformed with oryzacystatin gene on Myzus persicae and Macrosiphum euphorbiae. J Appl Entomol 130 84–90 [Google Scholar]
- Sayle R, Milner-White EJ (1995) RasMol: biomolecular graphics for all. Trends Biochem Sci 20 374. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot 53 947–958 [DOI] [PubMed] [Google Scholar]
- Shi C, Xu LL (2009) Characters of cysteine endopeptidases in wheat endosperm during seed germination and subsequent seedling growth. J Integr Plant Biol 51 52–57 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gigga SK, Chapital DC, Kuan J-C, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in programmed cell death in plants. Plant Cell 11 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MT, Laber B, Bode W, Huber R, Jerala R, Lenarcic B, Turk V (1990) The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J 9 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tian L, Zhang L, Zhang J, Song Y, Guo Y (2009) Differential proteomic analysis of soluble extracellular proteins reveals the cysteine protease and cystatin involved in suspension-cultured cell proliferation in rice. Biochim Biophys Acta 1794 459–467 [DOI] [PubMed] [Google Scholar]
- Tse YC, Lo SW, Hillmer S, Dupree P, Jiang L (2006) Dynamic response of pre-vacuolar compartments to brefeldin A in plant cells. Plant Physiol 142 1442–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RAL (2008) Plant proteases: from phenotypes to molecular mechanism. Annu Rev Plant Biol 59 191–223 [DOI] [PubMed] [Google Scholar]
- Wang KM, Kumar S, Cheng YS, Venkatagiri S, Yang AH, Yeh KW (2008) Characterization of inhibitory mechanism and antifungal activity between group-1 and group-2 phytocystatins from taro (Colocasia esculenta). FEBS J 275 4980–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda AM, Kumar GNM, Knowles NR (2009) Developmentally linked changes in proteases and protease inhibitors suggest a role for potato multicystatin in regulating protein content of potato tubers. Planta 230 73–84 [DOI] [PubMed] [Google Scholar]
- Zhang N, Jones BL (1995) Characterization of germinated barley endoproteolytic enzymes by two dimensional gel electrophoresis. J Cereal Sci 21 145–153 [Google Scholar]
- Zhang XM, Wang Y, Lv XM, Li H, Sun P, Lu H, Li FI (2009) NtCP56, a new cysteine protease in Nicotiana tabacum L., involved in pollen grain development. J Exp Bot 60 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.