Abstract
Despite substantial progress in measuring the anatomical and functional variability of the human brain, little is known about the genetic and environmental causes of these variations. Here we developed an automated system to visualize genetic and environmental effects on brain structure in large brain MRI databases. We applied our multi-template segmentation approach termed “Multi-Atlas Fluid Image Alignment” to fluidly propagate hand-labeled parameterized surface meshes, labeling the lateral ventricles, in 3D volumetric MRI scans of 76 identical (monozygotic, MZ) twins (38 pairs; mean age=24.6 (SD=1.7)); and 56 same-sex fraternal (dizygotic, DZ) twins (28 pairs; mean age=23.0 (SD=1.8)), scanned as part of a 5-year research study that will eventually study over 1000 subjects. Mesh surfaces were averaged within subjects to minimize segmentation error. We fitted quantitative genetic models at each of 30,000 surface points to measure the proportion of shape variance attributable to (1) genetic differences among subjects, (2) environmental influences unique to each individual, and (3) shared environmental effects. Surface-based statistical maps, derived from path analysis, revealed patterns of heritability, and their significance, in 3D. Path coefficients for the ‘ACE’ model that best fitted the data indicated significant contributions from genetic factors (A=7.3%), common environment (C=38.9%) and unique environment (E=53.8%) to lateral ventricular volume. Earlier-maturing occipital horn regions may also be more genetically influenced than later-maturing frontal regions. Maps visualized spatially-varying profiles of environmental versus genetic influences. The approach shows promise for automatically measuring gene-environment effects in large image databases.
Introduction
Imaging genetics is a rapidly growing research field, examining how genetic factors contribute to brain structure and function. Several recent special issues of neuroscience and psychiatry journals have been devoted to studies analyzing brain images with quantitative genetic models (e.g., Glahn et al., 2007a,b; Giedd et al., 2007; Blokland et al., 2008). Several studies have associated brain structural differences with variations in individual genes, in either diseased populations (Cannon et al., 2002) or healthy subjects (Chou et al., 2008). Maps of heritability coefficients (Thompson et al., 2001), or genetic and environmental components of variance (Chiang et al., 2008) have been applied not just to structural MRI, but also to diffusion tensor images (DTI), revealing genetic influences on aspects of fiber architecture that are correlated with intellectual performance (Chiang et al., 2008).
Morphometric analyses are highly automated — often combining images from hundreds of subjects — analyses using genetic statistical designs which can be performed with voxel-based morphometry (Hulshoff Pol et al., 2006), tensor-based morphometry (Brun et al., 2008), cortical surface modeling (Lenroot et al., 2007; Schmitt et al., in press, 2008), subcortical surface modeling (Chou et al., 2008) and DTI (Lee et al., 2008; Chiang et al., 2008). A key direction in this work is to understand how specific genes contribute to variations in cognition (Gray and Thompson, 2004), and to the risk for degenerative brain disorders such as Alzheimer’s disease (Hua et al., 2008) or psychiatric illnesses such as schizophrenia (Bearden et al., in press). Even so, with a few exceptions (e.g., the apolipoprotein E4 gene; Roses, 1998; Shaw et al., 2007; Burggren et al., 2008), specific genes that influence normal brain anatomy and function have been hard to identify. Single gene variations (also known as ‘polymorphisms’) are likely to influence the overall architecture of the brain only to a very minor extent. To expedite the search for specific genes, many researchers have first attempted to identify heritable features of brain structure, i.e., quantitative features in images that can be shown to be under strong genetic control. By exploiting the known genetic resemblance of family members, such as identical and fraternal twins, the relative contribution of genetic and environmental factors to a trait, such as the volume of a specific brain structure, can be identified. Twin studies have been invaluable for studying these questions because they provide naturally matched pairs where the confounding effects of a large number of potentially causal factors are removed by comparing twins who share them (see Thompson et al., 2002, and Schmitt et al., 2007, for reviews of twin studies using MRI).
The most common twin study design examines resemblances among twins raised in the same family environments (rather than twins raised apart). Monozygotic (identical) twins share all of their genes, while dizygotic (fraternal) twins share only half of them on average. Because they were born at the same time, and raised in the same family, DZ twins may be assumed to have roughly similar upbringings (although some research suggests that parents, teachers, peers and others may treat identical twins more similarly than fraternal twins; Richardson and Norgate, 2005). Modern twin studies try to quantify the effect of this shared environment, and that of the unique environment (individual experiences that shape a person’s life) on a trait of interest. In comparing the similarity between identical twins to that of fraternal twins, any additional likeness in the first group compared to the second group may be assigned to genes rather than shared environment. Twin studies have identified observable characteristics that are under strong genetic influence, including body height, eye color (Bito et al., 1997) and IQ (Plomin et al., 1994; Posthuma et al., 2002; Gray and Thompson, 2004).
In this study, we investigated the regional heritability of lateral ventricular shape using an automated approach that creates 3D maps of genetic parameters, on surface models of anatomy. This approach reveals whether a structure is under genetic control, and to what degree, and also plots the spatially-varying profile of genetic (and environmental) influences. We mapped genetic influences in 3D rather than analyzing specific numeric summaries, such as subvolumes of parcellated brain subregions, to allow for spatially varying profiles of genetic influences within a structure. This mapping approach is comparable to that of two prior studies (Thompson et al., 2001; Hulshoff Pol et al., 2006) that mapped heritability on a voxel-by-voxel basis.
To illustrate our approach, here we applied a surface extraction algorithm to brain MRI scans of 76 MZ twins (38 pairs) and 56 same-sex DZ twins (28 pairs), automatically extracting 3D anatomical surface models of the ventricles. No interactive human input is required for this step, other than the initial expert labeling of a small set of images. After a fluid segmentation using multiple propagated templates (Chou et al., 2008), a mesh containing 30,000 vertices is generated for each surface. We then performed a quantitative genetic analysis at each of the surface vertices, computing maps of heritability using the classical Falconer method (Falconer, 1989), based on twice the difference between the intraclass correlations (ICC) of MZ and DZ twin pairs. At each surface location on the ventricles, we also fitted a structural equation model (Neale and Cardon. 1992), to estimate the proportion of local anatomical variation attributable to genetic versus common and shared environmental effects. This starts from a set of covariances empirically estimated between twin pairs on the ventricular maps. The overall goal of this work is to zero in on promising anatomical measures that may be used in the future to investigate the effects of genes on brain morphology. We hypothesized that the occipital horns of the ventricles would be more strongly genetically influenced, as the white matter that surrounds them matures rapidly in early infancy. We hypothesized that the frontal horns might be more environmentally influenced, in line with prior findings that frontal brain regions have a more protracted developmental course, and might be more susceptible to environmental variations.
Materials and methods
Subjects
Subjects included a total of 76 identical (monozygotic, MZ) twins (38 pairs; mean age=24.6, SD=1.7; age range=21-27; 21 males/17 females) and 56 same-sex fraternal (dizygotic, DZ) twins (28 pairs; mean age=23.0, SD=1.8; age range=20-26; 10 males/18 females; p=0.0032, t-test) who received high-resolution MRI scans as part of a 5-year research study of over 1000 individuals (NIH grant: 100 MZ pairs, 150 DZ pairs, 200 siblings; NHMRC: 75 MZ pairs, 75 DZ pairs, 150 siblings; the pilot sample consists of 132 twins as data collection is ongoing), and had undergone comprehensive neurocognitive evaluation at age 16. There were no significant differences in means or variances for MZ and DZ twins, for any of the cognitive measures. Cognitive ability was found to be moderate-highly heritable. High heritability was not just found for the broadest index of cognition (FIQ) but also for measures of specific cognitive processes (Wright and Martin, 2004). Zygosity was established objectively by typing nine independent DNA microsatellite polymorphisms (PIC>0.7) by using standard polymerase chain reaction (PCR) methods and genotyping. These results were cross-checked with blood group (ABO, MNS and Rh), and phenotypic data (hair, skin and eye color), giving an overall probability of correct zygosity assignment of greater than 99.99%. All subjects underwent physical and psychological screening to exclude cases of pathology known to affect brain structure. Twins were excluded if either twin reported a history of significant head injury, a neurological or psychiatric illness, substance abuse or dependence, or if they had a first-degree relative with a psychiatric disorder. All twins had previously participated in a study assessing cognition when they were 16 years old, for which the exclusion criteria were the same, but with assessment by parental report.
Image acquisition and preprocessing
All MR images were collected using a 4 T Bruker Medspec whole body scanner (Bruker Medical, Ettingen, Germany) at the Center for Magnetic Resonance (University of Queensland, Australia). Three-dimensional T1-weighted images were acquired with an inversion recovery rapid gradient echo (MP-RAGE) sequence to resolve anatomy at high resolution. Acquisition parameters were: inversion time (TI)/repetition time (TR)/echo time (TE)=1500/2500/3.83 ms; flip angle=15°; slice thickness=0.9 mm with a 256×256×256 acquisition matrix. All images were spatially normalized to the ICBM-53 standard template (Mazziotta et al., 2001) with a 6-parameter (3 translations, 3 rotations) rigid-body transformation using the Minctracc algorithm (Collins et al., 1994) for the correction of head tilt and alignment, and resampled to 1-mm isotropic voxels. In this way, each individual’s brain was approximately matched in space, but global differences in brain size and shape remained intact. To equalize image intensities across subjects, registered scans were histogram-matched.
Automated lateral ventricular segmentation
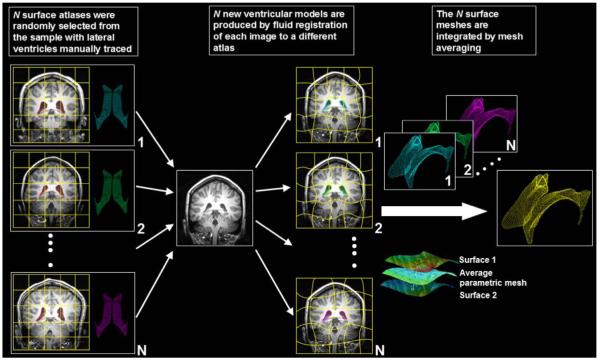
Lateral ventricles were automatically segmented in all scans using a technique that we recently validated and described in a previous study (Chou et al., 2008). Fig. 1 shows the steps used to map multiple surface-based atlases into single average surface mesh via fluid registration. Briefly, a small subgroup of 4 images (2 males — 1 MZ, 1 DZ, and 2 females — 1MZ, 1 DZ) was randomly chosen and the lateral ventricles were manually traced in contiguous coronal brain sections using the software MultiTracer (Woods, 2003). To make it easier to create well-defined surface models, the lateral ventricles were divided into anterior, posterior and inferior horns as in our prior studies (Narr et al., 2001). Lateral ventricular surface models were created in these images and converted into parametric meshes (Thompson et al., 1996; we will call these 4 labeled image ‘atlases’). We fluidly registered each atlas and the embedded mesh models to all other subjects using a nonlinear image registration algorithm (Lepore et al., 2008; based on Gramkow, 1996), treating the deforming image as a viscous fluid governed by the Navier-Stokes equation, as pioneered by (Christensen et al. 1996). The summed squared intensity difference was chosen as the cost function, which is reasonable given the high contrast of ventricular CSF and comparable image intensities across subjects scanned with the same imaging protocol. Transformations resulting from the fluid registration were also applied to the manually traced ventricular boundary using a tri-linear interpolation, generating a propagated contour on the unlabeled images. A mesh averaging technique combined the resulting fluidly propagated surface meshes for each image. Volumes obtained from the ventricular surface tracings were retained for statistical analyses.
Fig. 1.
Mapping multiple surface-based atlases into new subjects’ scans via fluid registration. N images (subsequently called atlases) were randomly selected from the sample and the lateral ventricles are manually traced and converted into surface mesh models. N new ventricular models were then produced by fluid registration of each image to a different atlas. The N surface meshes per subject were integrated by simple mesh averaging for each individual subject (see Chou et al., 2008, for details). In this study, we set N=4, which gave good results in our prior work.
Ventricular shape modeling and statistical maps
Ventricular surface meshes were constructed using a surface-based anatomical modeling approach as previously detailed in Thompson et al. (1996). Sets of points representing the tissue boundaries from each region were resampled and made spatially uniform by stretching a regular parametric grid (100×150 surface points) over each surface. Grid-points from corresponding surfaces were then matched across subjects to obtain group average parametric meshes. For each surface model, a medial curve was derived from the line traced out by the centroid of the boundary for each ventricular surface model (illustrated in Fig. 2a; red curves). The local radial size was defined as the radial distance between a boundary point and its associated medial curve (Fig. 2b). This allows statistical comparisons of local surface geometry at equivalent 3D surface locations across subjects for subsequent analysis of genetic variance. Statistical maps were generated indicating the intraclass correlation coefficient (ICC) values for MZ and DZ pairs at each ventricular surface point producing a color-coded map. At each point, a p value was also computed describing the significance of the ICC values, with the null hypothesis that the ICC value was zero. An advantage of the ventricular mapping analysis, relative to a simple volumetric analysis, is the ability to localize effects on brain structure in the form of a map, allowing for heritability estimates to vary spatially across a structure.
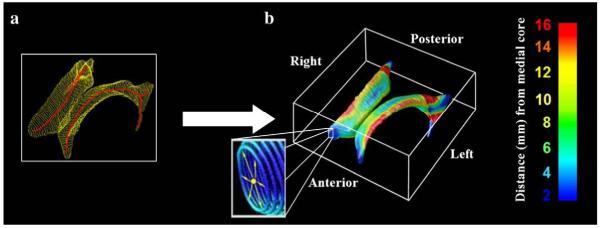
Fig. 2.
Ventricular Surface Modeling: (a) Medial curves (red), equidistant from each surface, were derived from mesh representations of the anterior, posterior and inferior horns of the lateral ventricles. (b) The radial distance of each ventricular boundary point to a medial curve can be interpreted as a local thickness and plotted in color with blue (red) points being closer to (farther from) the medial core. The parts of the lateral ventricles discussed in the paper follow the terminology indicated in (b): left, right, anterior, posterior are considered from the patient’s point of view (see Thompson et al., 2004a,b, and Chou et al., 2008, for additional details).
Heritability analyses
Heritability can be defined as the proportion of phenotypic variation that is attributable to genetic variation in a population (Falconer, 1989). Variation among individuals may be due to genetic and/or environmental factors. Heritability analyses estimate the relative contributions of differences in genetic and non-genetic factors to the total phenotypic variance in a population. To determine the proportion of variance attributable to genetic factors, heritability analyses were performed for lateral ventricular shape and volume, using two different statistical approaches: (i) classical heritability analysis (using Falconer’s method; Falconer, 1989) and (ii) maximum likelihood estimation (MLE) using structural equation modeling, also referred to as path analysis (Neale and Cardon, 1992). Both methods are described below; both are widely used in twin studies. Assuming a polygenic model, a heritability estimate of 0% implies no genetic effects; values close to 100% imply strong genetic influences.
Intraclass correlation and Falconer’s estimate
Intraclass correlation (ICC; Scout and Fleiss, 1979) is a measure of the correlations between pairs of observations, and is defined as:
| (1) |
Here is the pooled variance between pairs and is the variance of the traits within pairs, which is the mean-square estimate of within-pair variance (MSwithin) if reinterpreted in terms of the mean square in ANOVA. is the total variance of the measures. If a group is composed of k ratings, then the mean-square estimate of between-pair variance (MSbetween) equals . From this we get:
| (2) |
and the expression for ICC is:
| (3) |
The case of twin pairs, k=2, leads to the following formula for the intraclass correlation:
| (4) |
In this study, ICCs were calculated in order to determine the degree of concordance in lateral ventricular shapes in both the MZ and DZ twin pairs, and we applied the restricted maximum-likelihood (ReML) method to estimate variance components. The non-negative ReML estimator of the intraclass correlation is:
| (5) |
where n is the number of twin pairs.
The advantage of non-negative ReML compared to traditional regression analyses is that it forces r(MZ) and r(DZ) to lie in the range 0 to 1. Falconer’s method (Falconer, 1989) estimates the heritability as twice the difference in correlation between MZ and DZ twins, h2=2(r(MZ)-r(DZ)).
Structural equation modeling
Heritability of the lateral ventricular volumes was also estimated using structural equation modeling, implemented in the genetic analysis software, Mx GUI (version 1.3.65; Neale and Cardon, 1992). In contrast to Falconer’s method, in Mx, the phenotypic score is expressed as a linear function of three factors in a structural equation model: genetic (A), common environmental (C), and unique environmental variance (E), i.e., the environmental variations that are specific to each individual — the so-called ACE model. The logic behind the design is as follows: MZ twins are genetically identical (except for rare somatic mutations) while DZ twins, like ordinary full siblings, share on average only half of their genetic polymorphisms. As such, when the similarity of MZ twins is greater than that of DZ twins, this is interpreted as evidence for a genetic influence on a trait. In model fitting, this yields a significant variance component called A (additive genetic variance). Heritability is approximately twice the difference in correlation between MZ and DZ twins. Therefore, if only genes were influencing their traits, and trait similarity was proportional to genetic similarity, MZ twins’ measures should be twice as similar as DZ twins’. If not, and if DZ twins’ measures are less than half those of MZ twins’, this indicates that their resemblance is enhanced by shared environmental effects, which yields a significant variance component called C (common or shared environmental variance) in the model fitting. If MZ twins, despite sharing all their genes, are not perfectly identical in their measured traits, this indicates that experiences unique to each twin, and not shared with their co-twin, have reduced the twins’ resemblance. In model fitting, this yields a significant variance component called E (unique environmental variance), which also includes all sources of variance due to measurement errors. The observed correlation between MZ twins (rMZ) provides an estimate of A+C, and the correlation between DZ twins rDZ is an estimate of (1/2)A+C. These two equations allow us to derive A and C. E is estimated directly by how much the MZ twin correlation deviates from 1 (E=1-rMZ).
We modeled the covariance structure using the possible combinations of these model parameters (ACE, CE, AE and E). The maximum likelihood method was used to estimate the model parameters and expected covariance matrices for both MZ and DZ twins. Then χ2 values were computed representing the agreement between the observed and expected covariance matrices. These were first used as goodness of fit indices. A p value of 0.05 or smaller indicated a lack of fit to the data, and led to rejection of the model. The Akaike Information Criterion (AIC), computed as χ2-2 df, was used to compare the relative fit of different models. Genetic factors tend to increase correlations between MZ twin pairs, common environmental factors increase intrapair correlations for both MZ and DZ twin pairs, and unique environmental factors tend to decrease intrapair correlations for both MZ and DZ twin pairs. The full twin model can be depicted in a path diagram (Fig. 3).
Fig. 3.
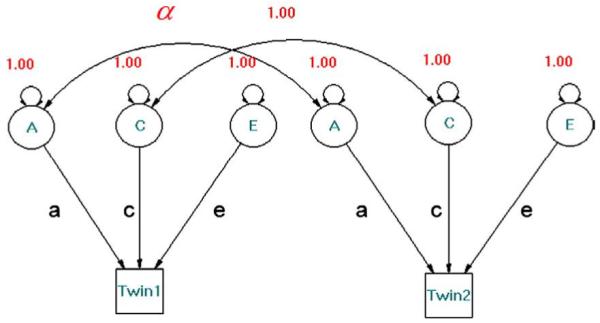
Structural equation model for the classical ACE design. Three latent variables (circles) are denoted by A (additive genetic), C (shared environment), and E (unique environment); phenotypes are shown as squares. Single-headed arrows represent causal paths; double-headed arrows represent correlations (a double-headed arrow to the same variable denotes its variance). α represents the genetic correlation between twin pairs (1 for MZ pairs; 0.5 for DZ pairs). Genetic factors tend to increase correlations within MZ twin pairs; common environmental factors tend to increase intrapair correlations for both MZ and DZ twin pairs; unique environmental factors tend to decrease intrapair correlations for both MZ and DZ twin pairs.
Results
Shape analysis of the lateral ventricles
Fig. 4 shows a boxplot of the intraclass correlations for the radial thicknesses, at all 30,000 points on each of the left and right lateral ventricles, and for both left and right pooled, without any adjustment (by registration) for the effects of brain scale. Mean MZ ICCs were generally higher (0.38 [L], 0.33 [R], 0.36 [Total]) than the DZ ones (0.13 [L], 0.21 [R], 0.18 [Total]). DZ twins resemble each other less than MZ twins do for both the left and right lateral ventricle measures, which gives some initial evidence that genetic factors may influence ventricular shape differences. Color-coded maps (Fig. 5) show local intraclass correlations between MZ and DZ co-twins and associated statistical significance maps for radial thickness at each vertex. To assess the significance of the intraclass correlation coefficients, the expected value of coefficient (r) for each twin pair was transformed into a z-score using Fisher’s r-to-z transformation (yielding z(r)). These p values were computed pointwise by pairing the twins with other twins who were not their actual pair to create some scrambled pairings, i.e. by generating a nonparametric null distribution for z(r) at each ventricular surface point to confirm that the within-pair correlations differ statistically from zero. To correct for multiple comparisons implicit in performing statistical tests at a large number of points across the ventricular surface, we computed the false discovery rate (FDR) when thresholding the resulting maps at p=0.05. The false discovery rate was q=0.0366 for the left ventricles and q=0.0274 for the right ventricles in the monozygotic twins, and the false discovery rate was q=0.0019 for the left ventricles and q=0.0007 for the right ventricles in the dizygotic twins. This confirms that the pattern of observed intra-twin correlations would not be expected to occur by chance. We found that for many regions the MZ and DZ twin correlations were of similar value, suggesting the common environmental factors may be important. Falconer’s estimate, which computes heritability (h2) as twice the difference between MZ and DZ correlations, is plotted at each surface point (Fig. 6a) to reveal the spatial patterns of genetic influences. Heritability estimates are highest in the mid-portion of anterior horn and in the posterior horn. Figs. 6b and c show the significance of additive genetic variance (A), and common environmental variance (C) by fitting the ACE model at each vertex.
Fig. 4.
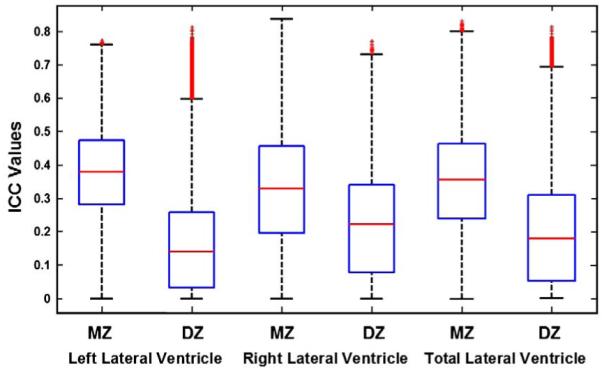
ICC values of radial ventricular thickness for the left, right and total lateral ventricles in MZ and DZ twin pairs. Box plots are formed by drawing a line at the median and a box between the lower and upper quartiles. Any data observation that lies more than 1.5 times the interquartile range (IQR) lower than the first quartile or 1.5 times the IQR higher than the third quartile is considered an outlier; whiskers indicate the most extreme values, on either side of the median, that are not outliers. Thus the box plot identifies the middle 50% of the data, the median, and the extreme points.
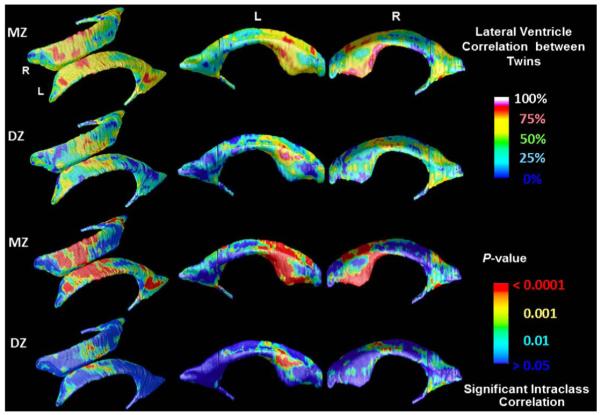
Fig. 5.
Color-coded maps show regional radial thickness correlations in MZ and DZ twins for the left and right lateral ventricles, and their significance at each surface point. Note the high resemblance among identical twins, and weaker resemblance among fraternal twins.
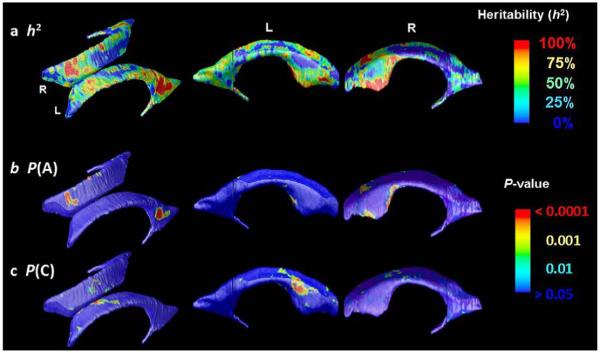
Fig. 6.
Color-coded maps for (a) heritability, h2, of radial thickness at each vertex between MZ and DZ twin pairs reveal the spatial patterns of genetic influences measured by Falconer’s formula. (b) and (c) show the significance of the additive genetic variance term (A), and the common environmental variance term (C) respectively.
Volumetric analysis of lateral ventricles
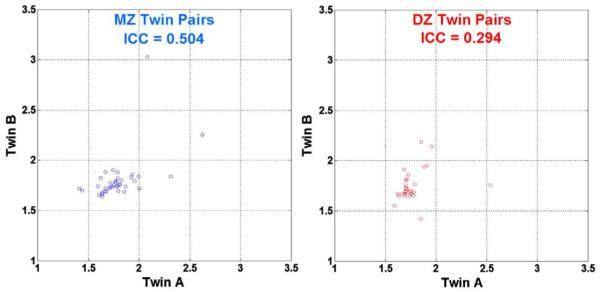
As a measure of the overall similarity in ventricular volume between the twins, we found that the percentage volumetric differences between twin pairs was, on average, 5.9% for the MZ twins and 6.1% for the DZ twins. These percent differences were computed from the formula |(A-B)|/{(A+B)/2}, where A and B are the ventricular volumes in each twin, respectively, |A-B| is the absolute value, and the divisor, (A+B)/2, is the average volume for the twins in the pair. The fact that the DZ similarity is very similar to the MZ twin similarity suggests the lack of a strong genetic effect, which, if present, would tend to make the MZ percentage volumetric differences smaller than the DZ percentage volumetric differences. Also, the average percent difference between unrelated people was found to be around 10%, based on random permutations in MZ twins (9.9%) and DZ twins (10.5%). MZ and DZ groups did not differ in average volume (P=0.29, two-tailed test). Fig. 7 shows a scatter plot of the lateral ventricular volumes for the MZ and DZ twin pairs. Each circle in the diagram represents a single twin pair and the order of the first twin and the second twin is random. Volumes are somewhat more highly correlated in MZ twins (ICC=0.50, P=0.01) than in DZ twin pairs (ICC=0.29, P=0.05). The greater resemblance of MZ twins provides initial support for the contribution of genetic factors to differences in lateral ventricular volumes.
Fig. 7.
Scatter plot of ventricular volumes (cm3) in MZ and DZ twins. Each circle represents a twin pair (the order is random). The correlation within MZ twin pairs is higher than that in DZ twin pairs as measured by ICC coefficients.
Statistical power to detect additive genetic variation under the ACE model was derived analytically for least-squares goodness-of-fit and maximum likelihood-based test statistics. The ACE model includes three parameters, and the significance of each is tested by comparing the reduced models (AE and CE) to the full ACE model to determine whether the removal of one or more parameters significantly increases the chi-square goodness-of-fit statistic and thus decreases model fit. The full ACE model was the best-fitting model (χ2=4.677, P=0.197, AIC=-1.323, df=3) compared to the AE model (χ2=5.451, P=0.142, AIC=-2.529, df=4), CE model (χ2=4.716, P=0.194, AIC=-3.284, df=4) and E model (χ2=19.481, P=0.002, AIC=9.481, df=5; see Table 1). Path coefficients for the ACE model that best fitted the lateral ventricular volume data indicated contributions from genetic factors (A=7.3%), common environment (C=38.9%) and unique environment (E=53.8%).
Table 1.
Parameters estimates for the ACE, CE, AE and E model on the observed ventricular volume values
| Heritability estimates (%) |
Model fit |
||||||
|---|---|---|---|---|---|---|---|
| A | C | E | χ2 | p | AIC | df | |
| 1. ACE | 7.3 | 38.9 | 53.8 | 4.677 | 0.197 | - 1.323 | 3 |
| 2. CE | 0 | 45.4 | 54.6 | 4.716 | 0.218 | - 3.284 | 4 |
| 3. AE | 47.5 | 0 | 52.5 | 5.451 | 0.24 | - 2.549 | 4 |
| 4. E | 0 | 0 | 100 | 19.481 | 0.002 | 9.481 | 5 |
Sex differences
To determine whether there were sex differences in ventricular shape and volume, a two-tailed alpha level of P<0.05 was used as the threshold for statistical significance and plotted onto the surface at each point on the ventricles with a color code to produce a statistical map, with sex modeled as a binary covariate (male, 0; female, 1). Overall P values were assigned to the left and right lateral ventricles using permutation testing approach for the observed effects that is corrected for multiple comparisons. As shown in Fig. 8, significant gender differences were found in ventricular shape and volume (L: P=0.0249; R: P=0.0136). A parallel analysis of sex effects on total ventricular volumes failed to detect an effect, when run in MZ groups and DZ group separately, and when MZ and DZ groups were pooled (P>0.05). Due to the small sample available to fit many simultaneous coefficients, we did not attempt to fit a sex interaction in the ACE model. It may require a very large sample to distinguish any sex differences, if present, in the degree of genetic or environmental influences.
Fig. 8.
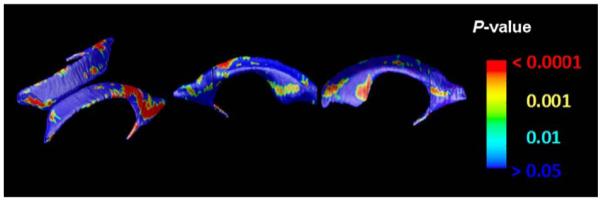
Maps of Sex Differences. These maps show the significance of differences in ventricular shape associated with sex at each surface point. In the red colored regions, ventricles are thinner in women, when data are not adjusted for overall differences in brain scale. The sex difference is significant on both left and right by permutation testing, but not detected using volume measures.
Discussion
In this study, we found significant correlation in ventricular shape and volume in both DZ and MZ twins (all p<0.05, after FDR-based multiple comparisons correction). Although, volumes were somewhat more highly correlated in MZ twins (ICC=0.50, P=0.01) than in DZ twin pairs (ICC=0.29, P=0.05), the percentage intrapair differences in volume were around 5.9% for the MZ twins and 6.1% for the DZ twins, suggesting evidence for an environmental effect. The maps of shared environmental components of variance indicated a region of environmental contribution mainly cluster in ventricular areas adjacent to the caudate head, an area in which rapid reductions in gray matter volume have been observed during development (Thompson et al., 2000). Path coefficients for the ACE model that best fitted the lateral ventricular volume data indicated contributions from genetic factors (A=7.3%), common environment (C=38.9%) and unique environment (E=53.8%). This is largely in line with prior studies that have included assessments of ventricular volume heritability. To our knowledge, the largest study of ventricular volume in twins to date was conducted by Schmitt et al. (2007) in 127 MZ twins, 36 DZ twins, and 158 additional unrelated individuals: after adjusting for total brain volume they reported contributions to overall ventricular volume from genetic factors (A=17%), common environment (C=43%) and unique environment (E=40%). Although their study did not provide spatially detailed maps of these influences, their variance components for overall ventricular volume are very similar to ours (though it is worth noting that their study was in pediatric sample of twins who were on average around 10 years younger than our sample).
The mapping technique implemented here allows for spatially precise mapping of heritability estimates on the ventricular surface. The mapping approach differentiates, in principle, the effects of multiple overlying structures with different functions. For example, the evidenced environmental effects mainly cluster in ventricular areas adjacent to the caudate head, and this effect may contribute to the environmental component of variance in ventricular volumes, observed in past studies (Baare et al., 2001; Wright et al., 2002; Schmitt et al., 2007).
As noted in Table 2, most prior twin studies that included examinations of ventricular morphology have observed environmental effects (Baare et al., 2001; Wright et al., 2002; Schmitt et al., 2007) but genetic effects have tended to be found only in the larger studies (Pfefferbaum et al., 2000), and not in all larger studies. Baare et al. (2001), for example, performed one of the larger studies (of 54 MZ twins, 58 DZ twins, and 34 siblings, and found values of A (0%), C (59%), and E (41%) that support environmental but not genetic influences. In addition to small sample size, our maps further suggest that the high intraclass correlation for DZ twins, as well as a possibly spatially varying profile for these environmental effects, may have diminished the power to detect distinct genetic and environmental effects in most samples.
Table 2.
Twin studies of lateral ventricular volumes
| Authors | Subjects | Heritability |
|---|---|---|
| Pfefferbaum et al. (2000) | MZ: 45, DZ: 40 | A: 79% |
| Baare et al. (2001) | MZ: 54, DZ: 58, siblings: 34 | A: 0%, C: 59%, E: 41% |
| Wright et al. (2002) | MZ: 10, DZ: 10 | A: 0%, C: 48%, E: 50% |
| Pfefferbaum et al. (2004) | MZ: 37, DZ: 37; 4 year longitudinal study |
(T1) A: 79% |
| (T2) A: 88% | ||
| Wallace et al. (2006) | MZ: 90, DZ: 37 | A: 31%, C: 24%, E: 45% A: 32%, C: 28%, E: 39% |
| Schmitt et al. (2007) | MZ: 127, DZ: 36, singletons: 158 | After adjusting for total brain volume: A: 17%, C: 43%, E: 40% |
Our findings supported some of our a priori hypotheses, but not others. One hypothesis, which takes into account to the ontogenetic sequence in which structures develop in the embryonic brain, suggests that the occipital horns of the ventricles may be more strongly genetically influenced, as the white matter that surrounds them matures rapidly in early infancy. Our experiments did not find such a genetic effect in the occipital regions, although the maps in Styner et al. (2005) tend to suggest that there is very high similarity in the occipital horns between monozygotic twins, and a lesser degree of similarity between normal pairs of subjects. Even so, neither our study nor that of Styner et al. (2005) could confirm a genetic influence in the occipital horn region. This may be due to the extremely wide variations in the shape of the occipital horns across subjects — during development, there is a progressive indentation of the occipital horns by the calcar avis and primary visual (calcarine) cortex, which causes impressions on the ventricular surfaces and makes them more curved on their medial surfaces. A genetic effect, if present, may therefore be more readily detected by modeling the visual cortex directly or by greatly expanding the sample size (which will be possible in the future). The same “ontogeny hypothesis” suggests that the frontal horns might be more environmentally influenced, in line with prior findings that frontal brain regions have a more protracted developmental course, and might be more susceptible to environmental variations. This result is partially supported by our findings, as we did find environmental influences in regions of the frontal horns. Even so, a far larger sample would be required to have sufficient power to detect whether environmental influences were proportionally greater in posterior-to-anterior gradient. We suggest that this hypothesis, of greater environmental influence on structures with a more protracted maturational time-course, has been invoked in the schizophrenia research literature to suggest why frontal brain regions might be more vulnerable to psychosis (Cannon et al., 2002; Vidal et al., 2006).
A minor limitation of this study is that the mean age of the DZ twins (23.0 years) is roughly one standard deviation below the mean age of the MZ twins (24.6 years; p=0.0032, t-test), a difference of 1.6 years. This is a significant but very small difference and is unlikely to impact on the estimation of the different genetic and environmental variance components derived from structural equation modeling. We recently estimated the rate of changes in brain morphology during mid-adolescence in healthy normal subjects (age 13-18), and found that the rate of ventricular expansion is around 0-1% per year (Gogtay et al., in press), although it may be accelerated in subjects with psychiatric illness. In line with intuition, the rates of change in brain morphometry also slow down and plateau between the teenage years and the early twenties (Hua et al., in press), the age range examined here (20-26 years). As such the difference in mean ventricular volume associated with age effects would be expected to be less than 1%, and not likely to substantially affect the variance components estimated here. We also regressed age versus ventricular volume and found that there was no detectable effect of age across our sample (p=0.77).
In our surface-based maps, we also found that there were sex differences in ventricular shape, with several regions (indicated in red in Fig. 8), where the ventricles are thinner in women, when data are not adjusted for overall differences in brain scale. This sex difference is significant on both left and right by permutation testing, but not detected using volume measures (Giedd et al., 1996 and Lenroot et al., 2007). Because our analyses considered same sex twin pairs only, these sex differences did not confound the analysis of intra-pair differences that underlies the covariance estimation and genetic analyses. Even so, future analyses in samples large enough to assess sex×heritability interactions may be of interest. Given that structural brain development has a marginally different trajectory and time-course in men and women, and may be influenced by levels of gonadal steroids that differ between the sexes, it is possible that the pattern of environmental or genetic influences may show sex differences as well.
Unfortunately, our deformation-based segmentation method may not be optimal for mapping the inferior horns of the ventricles, since healthy subjects typically have very low volumes of ventricular CSF in the inferior horns, and their geometry appears extremely narrow on coronally resliced MRI (typically a millimeter or less in width). When a structure such as the inferior horn is less than a millimeter thick, at the standard resolution of MRI, each component voxel tends to contain gray and/or white matter as well as CSF. Both voxel-based tissue classifiers and deformation-based models may have difficulty in recovering the geometry accurately. In other studies, the superior and posterior ventricular horns tend to show higher effect sizes for disease (Thompson et al., 2004a,b; Thompson and Apostolova, 2008) and genetic (e.g., ApoE) effects (Chou et al., 2008), perhaps partly because the error in segmenting them is much less as a proportion of their overall volume and the percentage difference in volume in disease is extremely large. In theory, an inferior horn template could be propagated into new scans using a fluid registration approach, but it would be hard to propagate accurately onto controls in regions with very limited CSF, as large localized contractions (i.e., compressions of the template) would be needed. The fluid prior in the registration enforces a spatial smoothness in the deformation that limits very large localized compressions. In our tensor-based morphometry (TBM) studies, we have found that the smoothness of the deformation fields may limit the accuracy of the fluid registration for relatively small-scale structures (Hua et al., in press).
Ironically, the fact that ventricular structure is not heavily genetically influenced in young adulthood does not mean that it is not heavily genetically influenced in old age. The apolipoprotein E (ApoE) gene is located on chromosome 19 with three alleles (ApoE2, ApoE3, ApoE4) (Zannis et al., 1982), and ApoE4 increases and ApoE2 decreases susceptibility to AD (Corder et al., 1993; Saunders et al., 1993; Blacker et al., 2007). We recently found that in elderly normal subjects, ApoE4 carriers had greater ventricular expansion that non-carriers (Chou et al., 2008), and normal ApoE2 carriers — which make up around 1/6 of the healthy elderly population — showed lesser ventricular expansion than carriers of the commonest genotype, ApoE3/3 (Hua et al., 2008).
As progressive ventricular enlargement is typical during both normal aging and dementia, degenerative brain changes in later life may be slowed or accelerated by genes in regions where those same genes may not have a profound influence on early brain morphology. Even so, Shaw et al. (2007) found an effect of ApoE genetic variants on entorhinal cortex morphology in young normal subjects, suggesting early gene-dependent differences in regions susceptible to neurode-generation up to 50 years later.
Relevance of ventricular measures
Ventricular changes reflect atrophy in surrounding structures, and ventricular measures and surface-based maps provide sensitive assessments of tissue reduction. These measures correlate with cognitive deterioration in several illnesses such as Alzheimer’s disease (AD), (Chou et al., 2008 and Thompson et al., 2004a), HIV/AIDS (Thompson et al., 2006) and schizophrenia (Narr et al., 2001), and offer a potential approach to evaluate disease progression in large-scale drug trials. However, this morphometric feature is non-specific and occurs in many other brain diseases, and its variability in healthy controls is not sufficiently understood. If ventricular structure was highly heritable, for example, it would motivate the use of family or discordance designs when studying ventricular measures of disease, because siblings who differ in disease status but share around half of their genes would tend to have similar morphology, allowing disease-specific differences to be detected more powerfully given the smaller background noise. This ‘sibling subtraction’ approach has been successful in identifying non-genetic deficits and characterizing the effects of risk genes and their associated morphometric correlates in schizophrenia (Cannon et al., 2002, 2005). In studies where ventricular variation is linked with disease, it is important to know if the sources of variation are at least partly genetic, because family or twin designs would then be useful in adjusting out this type of non-disease-related variation to obtain a more powerful effect size when aiming to detect a disease-specific effect, or a factor that might modulate disease. Some diseases that are highly heritable, such as autism, also show ventricular differences (Vidal et al., 2008), so it is of interest to know whether normal ventricular morphology is genetically influenced. Styner et al. (2005), performed a landmark study of lateral ventricular shape in twins discordant for schizophrenia, and in healthy twins, examined the effects of disease and whether they were genetically influenced. Because they detected an effect of genetics but not of disease on lateral ventricular shape, they suggested that genetics may have a stronger influence on the shape of lateral ventricles than do the disease-related changes in twins with schizophrenia. They also created average distance maps that suggested that MZ pairs and DZ pairs both showed greater similarity than normal pairings created by randomly sampling unrelated individuals from the MZ and DZ groups. It must be conceded that ventricular measures were more popular in the early days of computed tomography and MRI, while morphometry now focuses more on specific structures such as the hippocampus and cortex, often using voxel-based approaches to create maps of structural differences. Even so, a recent editorial (Weiner, 2008) and several large studies of epidemiological cohorts (Carmichael et al., 2007a,b) have underscored the value of ventricular change as a very powerful predictor of future clinical decline in the prodrome of Alzheimer’s disease, and as a potential surrogate marker for drug trials. As a result it has practical value for neuroscience to disentangle the sources of morphological variation in the ventricles, as any phenotypes showing genetic variance could be analyzed for allelic effects (e.g. ApoE4; Chou et al., 2008; Hua et al., 2008), or genetic variance could be adjusted out using genetically informative designs (e.g., twin or family studies).
A multitude of potential global and local, genetic and non-genetic factors are likely to impact on the adjacent gray and white matter structures of the lateral ventricles, over the human lifespan. As such, a more direct modeling of the caudate nucleus, basal ganglia, and thalamus would be needed to detect whether they share the same genetic influences as the adjacent regions on the ventricles. Any common genetic influences on ventricles and surrounding white matter, for example, could be identified using a cross-twin cross-trait design, which can identify common genetic influences of different structures (or different traits in general). In normal aging, gray and white matter reductions occur with cellular atrophy and myelin reduction over the human lifespan, and follow a stereotypical temporal trajectory (Sowell et al., 2003). As the twins here are all young adults, so we note that the effect of aging on the ventricles, which is the most prominent known predictor of ventricular morphology, may skew or modify the pattern of genetic versus environmental influences. Some studies of twins cover a broad enough age range to begin to seek out interactions between age and heritability, but very large samples are required to detect age×heritability interactions (e.g., 600 subjects including 308 twins in Lenroot et al., 2007, Schmitt et al., in press, 2008).
Several specific factors might alter ventricular morphology over the lifespan. In utero, the pattern of occipital horn asymmetry is already observable on ultrasound and is thought to be caused by an asymmetric structural development of the overlying left hemisphere language areas, causing the occipital horn to protrude further posteriorly in the left hemisphere. Such a process in utero is likely to be more heavily influenced by neurodevelopmental genetic programs than by environmental factors, although the distinction is blurred as such processes are regulated by growth factors whose production may also respond to environmental factors (e.g., nutrition). It is also plausible, but not confirmed empirically, that environmental factors that have a bearing on cognition (education, nutrition, childhood illness or infection, or exercise) may also influence brain morphology. We previously examined the profile of caudate growth in childhood (Thompson et al., 2000) as well as cortical development from ages 4 to 21 (Gogtay et al., 2004), and found that the pattern of gray matter reduction in some regions is accompanied by white matter growth. Since synaptic pruning, dendritic simplification, myelination, and glial effects contribute to these changes, it is likely that the ventricular morphology is affected by all of these mechanisms.
Most earlier genetic studies using MRI in twins found that many aspects of neuroanatomy are highly heritable. Area measurements of the corpus callosum showed heritability estimates between 79 and 94% (Oppenheim and Gazzaniga, 1989; Pfefferbaum et al., 2000; Scamvougeras et al., 2003; Hulshoff Pol et al., 2006). Pfefferbaum et al. (2001) also used diffusion imaging on a data set of 15 MZ and 18 DZ twin pairs of elderly men, and reported that the microstructural coherence of callosal white matter fibers showed significant heritability of FA (65%). Cerebral lobe volumes were found to be 65% heritable using an MRI dataset comprising 72 MZ and 67 DZ twin pairs (Geschwind et al., 2002). Measures of surface morphology, including gyral and surface curvature, surface area and cortical depth were also found to be highly correlated in MZ twin pairs (White et al., 2002). Furthermore, the volume of the cerebellum (Posthuma et al., 2000; Wright et al., 2002), the hippocampus (Sullivan et al., 2001) and Wernicke’s region (Thompson et al., 2001) were all shown to be particularly influenced by genetic factors. These findings suggest that genetic factors account for a large part of the individual variance in brain structures. Highly heritable brain morphometric measures provide biological markers for inherited neuroanatomical phenotypes, that may serve as targets for genetic studies and offer a compelling alternative to categorically defined disease syndromes.
Some genetic studies have gone beyond heritability analyses to association or linkage designs that implicate specific genes in structural variations. To date, these have all been studies of subjects with brain disorders rather than healthy individuals (Pietiläinen et al., in press). It is easier to identify candidate genes that may associate with a deficit by first determining which genetic polymorphisms are commonly overexpressed in specific patient populations.
A third and complementary approach to elucidate gene effects on anatomy and function is to study monogenic disorders, in which a known genetic deletion can profoundly alter brain development. Fragile X syndrome is an example of a disease associated with a single genetic mutation, which is responsible for the overproduction of a protein that regulates dendritic pruning (FMRP), and is associated with abnormally enlarged basal ganglia, immediately adjacent to the lateral ventricles (Lee et al., 2006; Gothelf et al., 2007). The specific genetic difference in the case of Fragile X is not necessarily germane to understanding genetic variations in healthy subjects, but it is notable as a known genetic cause for a very prominent difference in caudate anatomy. Characteristic morphometric signatures associated with other genetic disorders of development, such as 22q11 deletion syndrome, are reviewed in (Bearden et al., 2007b; see also Bearden et al., 2007a, in press).
Finally, we note that the surface-based genetic mapping approach proposed here is general and could be adapted to other methods for automated structure extraction (e.g., Morra et al., in press), to reveal environmental effects on the hippocampus, basal ganglia, and even fiber pathways (Chiang et al., 2008).
Acknowledgments
This work was generously supported by NIH grant R01 HD050735 and National Health and Medical Research Council, Australia grant 496682. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of The National Institute of Child Health and Human Development or The National Institutes of Health.
References
- Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb. Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Bearden CE, van Erp TGM, Dutton RA, Tran H, Zimmerman L, Sun D, Geaga JA, Simon TJ, Glahn DG, Cannon TD, Emanuel BS, Toga AW, Thompson PM. Mapping cortical thickness in children with 22q11.2 deletions. Cerebral Cortex. 2007;17(8):1889–1898. doi: 10.1093/cercor/bhl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, van Erp TGM, Thompson PM, Toga AW, Cannon TD. Cortical mapping of genotype-phenotype relationships in schizophrenia. Hum. Brain Mapp. 2007;28:519–532. doi: 10.1002/hbm.20404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, van Erp TGM, Dutton RA, Zimmerman L, Simon TJ, Glahn DG, Cannon TD, Emanuel BS, Toga AW, Thompson PM. Alterations in midline cortical thickness and complexity mapped in children with 22q11.2 deletions. Cereb. Cortex. in press. published online on May 14, 2008 (Electronic publication ahead of print) [Google Scholar]
- Bito LZ, Matheny A, Cruickshanks KJ, Nondahl DM, Carino OB. Eye color changes past early childhood. The Louisville Twin Study. Arch. Ophthalmol. 1997;115(5):659–663. doi: 10.1001/archopht.1997.01100150661017. [DOI] [PubMed] [Google Scholar]
- Blacker D, Lee H, Muzikansky A, Martin EC, Tanzi R, McArdle JJ, Moss M, Albert M. Neuropsychological measures in normal individuals that predict subsequent cognitive decline. Arch. Neurol. 2007 June 1;64(6):862–871. doi: 10.1001/archneur.64.6.862. [DOI] [PubMed] [Google Scholar]
- Blokland GAM, McMahon KL, Hoffman J, Zhu G, Meredith M, Martin NG, Thompson PM, de Zubicaray GI, Wright MJ. Quantifying the heritability of task-related brain activation and performance during the N-back working memory task: a twin fMRI Study. Biol. Psychol., Special Issue on Imaging Genomics. 2008;79(1):70–79. doi: 10.1016/j.biopsycho.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun CA, Lepore N, Pennec X, Chou Y-Y, Lee AD, McMahon K, de Zubicaray GI, Meredith M, Wright MJ, Barysheva M, Toga AW, Thompson PM. A new registration method based on log-Euclidean metrics and its application to genetic studies; International Workshop on Biomedical Imaging (ISBI); 2008; pp. 1115–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren AC, Zeineh MM, Ekstrom AE, Braskie MN, Thompson PM, Small GW, Bookheimer SY. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. NeuroImage. 2008;41(4):1177–1183. doi: 10.1016/j.neuroimage.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Thompson PM, van Erp TG, Toga AW, Poutanen VP, Huttunen M, Lonnqvist J, Standerskjold-Nordenstam CG, Narr KL, Khaledy M, Zoumalan CI, Dail R, Kaprio J. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3228–3233. doi: 10.1073/pnas.052023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TGM, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta JC, Peltonen L. DISC1/TRAX haplotypes associate with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry. 2005 Nov;62(11):1205–1213. doi: 10.1001/archpsyc.62.11.1205. 2005. [DOI] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SH, Lee JY, Aizenstein HA, Meltzer CC, Liu Y, Toga AW, Becker JT. Acceleration of cerebral ventricular expansion in the Cardiovascular Health Study. Neurobiol. Aging. 2007;28(9):1316–1321. doi: 10.1016/j.neurobiolaging.2006.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SH, Lee JY, Aizenstein HA, Meltzer CC, Liu Y, Toga AW, Becker JT. Cerebral ventricular changes associated with transitions between normal cognitive function, mild cognitive impairment, and dementia. Alzheimer Dis. Assoc. Disord. 2007 January/March;21(1):14–24. doi: 10.1097/WAD.0b013e318032d2b1. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Lee AD, Madsen S, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Srivastava A, Balov N, Thompson PM. Mapping genetic influences on brain fiber architecture with high angular resolution diffusion imaging (HARDI); International Workshop on Biomedical Imaging (ISBI); 2008; pp. 871–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y, Leporé N, de Zubicaray G, Carmichael O, Becker J, Toga A, Thompson P. Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. NeuroImage. 2008;40:615–630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans. Image Process. 1996;5:1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmacher WJ, Schmechel DE, Gaskell PC, Small GW. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Falconer DS. Introduction to Quantitative Genetics. third ed. Longman; Essex, UK: 1989. [Google Scholar]
- Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Snell JW, Lange N, Rajapakse JC, Casey BJ, Kozuch PL, Vaituzis AC, Vauss YC, Hamburger SD, Kaysen D, Rapoport JL. Quantitative magnetic resonance imaging of human brain development: ages 4-18. Cereb. Cortex. 1996;6:551–560. doi: 10.1093/cercor/6.4.551. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Schmitt JE, Neale MC. Structural brain magnetic resonance imaging of pediatric twins. Hum. Brain Mapp. 2007;28:474–481. doi: 10.1002/hbm.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: strategies for finding genes influencing brain structure and function. Hum. Brain Mapp. 2007;28(6):488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Paus T, Thompson PM. Imaging genomics: mapping the influence of genetics on brain structure and function. Hum. Brain Mapp. 2007;28(6):461–463. doi: 10.1002/hbm.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaitiuzis C, Nugent TF, Herman DH, Classen L, Toga AW, Rapoport JL, Thompson PM. Dynamic mapping of human cortical development during childhood and adolescence. Proc. Natl. Acad. Sci. 2004 May 25;101(21):8174–8179. doi: 10.1073/pnas.0402680101. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Lu A, Leow AD, Klunder AD, Lee AD, Chavez A, Greenstein D, Giedd JN, Toga AW, Rapoport JL, Thompson 3D Brain Growth Abnormalities in Childhood-Onset Schizophrenia visualized using Tensor Based Morphometry. Proceedings of the National Academy of Sciences. doi: 10.1073/pnas.0806485105. in press. Electronic publication ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothelf D, Furfaro JA, Eckert MA, Hall SS, Hara R, Erba HW, Ringel J, Hayashi KM, Patnaik S, Golianu B, Kraemer HC, Thompson PM, Piven J, Reiss AL. Neuroanatomy of Fragile X Syndrome is associated with aberrant behavior and FMRP. Ann. Neurol. 2008;63(1):40–51. doi: 10.1002/ana.21243. O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramkow C. Registration of 2D and 3D medical images, Master’s thesis. Danish Technical University; Copenhagen, Denmark: 1996. [Google Scholar]
- Gray JR, Thompson PM. Neurobiology of intelligence: science and ethics. Nat. Rev., Neurosci. 2004;5:1–13. doi: 10.1038/nrn1405. [DOI] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Weiner MW, Thompson PM. Tensor-Based Morphometry as a Neuroimaging Biomarker for Alzheimer’s Disease: An MRI Study of 676 AD, MCI, and Normal Subjects. NeuroImage. 2008;43(3):458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Levitt JG, Caplan R, Thompson PM, Toga AW. Detecting Brain growth patterns in normal children using tensor-based morphometry. Hum. Brain Mapp. doi: 10.1002/hbm.20498. in press. Electronic publication ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RC, Baare WF, van Oel C, van Haren NE, Collins L, Evans AC, Amunts K, Burgel U, Zilles K, de Geus EJ, Boomsma DI, Kahn RS. Genetic contributions to human brain morphology and intelligence. J. Neurosci. 2006;26:10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AD, Leow AD, Lu A, Reiss AL, Hall S, Toga AW, Thompson PM. 3D pattern of brain abnormalities in Fragile X Syndrome visualized using tensor-based morphometry. NeuroImage. 2006;34(3):924–938. doi: 10.1016/j.neuroimage.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AD, Lepore N, Barysheva M, Chou YY, Brun CA, Madsen SK, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Toga AW, Thompson PM. Comparison of Fractional and Geodesic Anisotropy in Diffusion Tensor Images of 90 Monozygotic and Dizygotic Twins; International Workshop on Biomedical Imaging (ISBI); 2008; pp. 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum. Brain Mapp. 2007 Nov 27; doi: 10.1002/hbm.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore N, Chou YY, Lopez OL, Aizenstein HJ, Becker JT, Toga AW, Thompson PM. Fast 3D Fluid Registration of Brain Magnetic Resonance Images. Proc. SPIE. 2008 doi: 10.1117/12.774338. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM. Automated 3D Mapping of Hippocampal Atrophy and its Clinical Correlates in 400 Subjects with Alzheimer’s Disease, Mild Cognitive Impairment, and Elderly Controls. Human Brain Mapping. doi: 10.1002/hbm.20708. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Thompson PM, Sharma T, Moussai J, Blanton RE, Anvar B, Edris A, Krupp R, Rayman J, Khaledy M, Toga AW. 3D shape characterization and mapping of temporo-limbic regions and the lateral ventricles in schizophrenia. Biol. Psychiatry. 2001;50:84–97. doi: 10.1016/s0006-3223(00)01120-3. [DOI] [PubMed] [Google Scholar]
- Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Kluwer Academic; Boston: 1992. [Google Scholar]
- Oppenheim JS, Gazzaniga MS. Magnetic resonance imaging morphology of the corpus callosum in monozygotic twins. Ann. Neurol. 1989;26(1):100–104. doi: 10.1002/ana.410260117. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Swan GE. Brain structure in men remains highly heritable in the seventh and eighth decades of life. Neurobiol. Aging. 2000;21:63–74. doi: 10.1016/s0197-4580(00)00086-5. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Carmelli D. Genetic regulation of regional microstructure of the corpus callosum in late life. Neuroreport. 2001;12:1677–1681. doi: 10.1097/00001756-200106130-00032. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Carmelli D. Morphological changes in aging brain structures are differentially affected by time-linked environmental influences despite strong genetic stability. Neurobiol. Aging. 2004;25:175–183. doi: 10.1016/s0197-4580(03)00045-9. [DOI] [PubMed] [Google Scholar]
- Pietiläinen OPH, Paunio T, Loukola A, Tuulio-Henriksson A, Kieseppä T, Thompson PM, Toga AW, van Erp TGM, Soronen P, Hennah W, Turunen JA, Peltonen JO, Palo OM, Silander K, Lönnqvist J, Kaprio J, Cannon TD, Peltonen L. Association of AKT1 with verbal learning, verbal memory and regional cortical grey matter density in twins. Neuropsychiatric Genetics. doi: 10.1002/ajmg.b.30890. in press. Electronic publication ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Pedersen NL, Lichtenstein P, McClearn GE. Variability and stability in cognitive abilities are largely genetic later in life. Behav. Genet. 1994;24(3):207–215. doi: 10.1007/BF01067188. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJC, Neale MC, Pol HEH, Baare WEC, Kahn RS, Boomsma D. Multivariate genetic analysis of brain structure in an extended twin design. Behav. Genet. 2000;30:311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJ, Baare WF, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat. Neurosci. 2002;5(2):83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- Richardson K, Norgate S. The equal environments assumption of classical twin studies may not hold. Br. J. Educ. Psychol. 2005;75(3):339–350. doi: 10.1348/000709904X24690. [DOI] [PubMed] [Google Scholar]
- Roses AD. Apolipoprotein E and Alzheimer’s disease: the tip of the susceptibility iceberg. Ann. N.Y. Acad. Sci. 1998;855:728–743. doi: 10.1111/j.1749-6632.1998.tb10653.x. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Stritimatter WJ, Schmechel D, ST. George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-Maclachlan DR, Alberts MJ, Hulette C, Crain B, Goldgaber D, Roses AD. Association of apolipoprotein E allele e4 with late-onset familial and sporadic Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8098–8102. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Scamvougeras A, Kigar DL, Jones D, Weinberger DR, Witelson SF. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci. Lett. 2003;338:91–94. doi: 10.1016/s0304-3940(02)01333-2. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Eyler LT, Giedd JN, Kremen WS, Kendler KS, Neale MC. Review of twin and family studies on neuroanatomic phenotypes and typical neurodevelopment. Twin Res. Hum. Genet. 2007 Oct;10(5):683–694. doi: 10.1375/twin.10.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Lenroot R, Ordaz SE, Wallace GL, Lerch JP, Evans AC, Prom EC, Kendler KS, Neale MC, Giedd JN. Variance decomposition of MRI-based covariance maps using genetically informative samples and structural equation modeling. Neuroimage. doi: 10.1016/j.neuroimage.2008.06.039. in press. Electronic publication ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Lenroot RK, Wallace GL, Ordaz S, Taylor KN, Kabani N, Greenstein D, Lerch JP, Kendler KS, Neale MC, Giedd JN. Identification of genetically mediated cortical networks: a multivariate study of pediatric twins and siblings. Cereb. Cortex. 2008;18(8):1737–1747. doi: 10.1093/cercor/bhm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;2:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D, Clasen L, Evans A, Rapoport JL, Giedd JN. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 2007 Jun;6(6):494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human lifespan. Nat. Neurosci. 2003 March;6(3):309–315. doi: 10.1038/nn1008. 2003. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4872–4877. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D. Heritability of hippocampal size in elderly twin men: equivalent influence from genes and environment. Hippocampus. 2001;11:754–762. doi: 10.1002/hipo.1091. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Apostolova LG. Computational Anatomical Methods as Applied to Aging and Dementia. British Journal of Radiology. 2008 Feb 14; doi: 10.1259/BJR/20005470. in press. 2008. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Schwartz C, Toga AW. High-resolution random mesh algorithms for creating a probabilistic 3D surface atlas of the human brain. NeuroImage. 1996;3:19–34. doi: 10.1006/nimg.1996.0003. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum-mechanical tensor maps. Nature. 2000 March 9;404(6774):190–193. doi: 10.1038/35004593. 2000. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M, Dail R, Zoumalan CI, Toga AW. Genetic influences on brain structure. Nat. Neurosci. 2001;4(12):1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Toga AW. Mapping genetic influences on human brain structure. Ann. Med. 2002;34:523–536. doi: 10.1080/078538902321117733. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Hong MS, Herman D, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer’s disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Simon S, Geaga J, Hong MS, Sui Y, Lee JY, Toga AW, Ling WL, London ED. Structural abnormalities in the brains of human subjects who use methamphetamine. J. Neurosci. 2004;24:6028–6036. doi: 10.1523/JNEUROSCI.0713-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Dutton RA, Hayashi KM, Lu A, Lee SE, Lee JY, Toga AW, Lopez OL, Aizenstein HJ, Becker JT. 3D Mapping of ventricular and corpus callosum abnormalities in HIV/AIDS. Neuroimage. 2006 Jan;31(1):12–23. doi: 10.1016/j.neuroimage.2005.11.043. 2006. [DOI] [PubMed] [Google Scholar]
- Vidal CN, Hayashi KM, Geaga JA, Sui Y, McLemore LE, Alaghband Y, Giedd JN, Gochman P, Blumenthal J, Gogtay N, Nicolson R, Toga AW, Rapoport JL, Thompson PM. Dynamically spreading frontal and cingulate deficits mapped in adolescents with schizophrenia. Arch. Gen. Psychiatry. 2006 Jan;63(1):25–34. doi: 10.1001/archpsyc.63.1.25. 2006. [DOI] [PubMed] [Google Scholar]
- Vidal CN, Nicolson R, Boire JY, Barra V, DeVito TJ, Hayashi KM, Geaga JA, Drost DJ, Williamson PC, Rajakumar N, Toga AW, Thompson PM. Three-dimensional mapping of the lateral ventricles in autism. Psychiatry Research. Neuroimaging. 2008;163(2):106–115. doi: 10.1016/j.pscychresns.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GL, Eric SJ, Lenroot R, Viding E, Ordaz S, Rosenthal MA, Molloy EA, Clasen LS, Kendler KS, Neale MC, Giedd JN. A pediatric twin study of brain morphometry. J. Child Psychol. Psychiatry. 2006;47:987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- Weiner MW. Expanding ventricles may detect preclinical Alzheimer disease. Neurology. 2008;70:824–825. doi: 10.1212/01.wnl.0000304743.72127.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Andreasen MC, Nopoulos P. Brain Volume and surface morphology in monozygotic twins. Cerebral Cortex. 2002;12(5):486–493. doi: 10.1093/cercor/12.5.486. [DOI] [PubMed] [Google Scholar]
- Woods RP. Multitracer: a Java-based tool for anatomic delineation of grayscale volumetric images. NeuroImage. 2003;19:1829–1834. doi: 10.1016/s1053-8119(03)00243-x. [DOI] [PubMed] [Google Scholar]
- Wright MJ, Martin NG. Brisbane Adolescent Twin Study: outline of study methods and research projects. Aust. J. Psychol. 2004;56(2):65–78. [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- Zannis VI, Breslow JL, Utermann G, Mahley RW, Weisgraber KH, Havel RJ, Goldstein JL, Brown MS, Schonfeld G, Hazzard WR, Blum C. Proposed nomenclature of apoE isoproteins, apoE genotypes and phenotypes. J. Lipid Res. 1982;23:911–914. [PubMed] [Google Scholar]