Abstract
Common fragile sites (CFSs) represent large, highly unstable regions of the human genome. CFS sequences are sensitive to perturbation of replication; however, the molecular basis for the instability at CFSs is poorly understood. We hypothesized that a unique epigenetic pattern may underlie the unusual sensitivity of CFSs to replication interference. To examine this hypothesis, we analyzed chromatin modification patterns within the six human CFSs with the highest levels of breakage, and their surrounding non-fragile regions (NCFSs). Chromatin at most of the CFSs analyzed has significantly less histone acetylation than that of their surrounding NCFSs. Trichostatin A and/or 5-azadeoxycytidine treatment reduced chromosome breakage at CFSs. Furthermore, chromatin at the most commonly expressed CFS, the FRA3B, is more resistant to micrococcal nuclease than that of the flanking non-fragile sequences. These results demonstrate that histone hypoacetylation is a characteristic epigenetic pattern of CFSs, and chromatin within CFSs might be relatively more compact than that of the NCFSs, indicating a role for chromatin conformation in genomic instability at CFSs. Moreover, lack of histone acetylation at CFSs may contribute to the defective response to replication stress characteristic of CFSs, leading to the genetic instability characteristic of this regions.
INTRODUCTION
Chromosomal common fragile sites (CFSs) are specific loci that show non-random gaps, breaks, or rearrangements in metaphase chromosomes when cells are cultured under conditions that inhibit or impair DNA replication, such as in the presence of aphidicolin (APH) (1). CFSs are highly unstable regions of the genome, and molecular deletions and rearrangements within CFSs have been identified in a variety of human tumors. Moreover, a high frequency of loss of heterozygosity at known CFSs, likely mediated by replication stress, occurs during the pre-malignant and pre-invasive stages of many types of human tumors (2). These features have led us and others to hypothesize that CFSs play a mechanistic role in the recurring chromosomal rearrangements, deletions and somatic recombination observed in tumor cells.
Although extensive effort has been invested in cloning CFSs and characterizing genetic rearrangements of CFSs in cancer cells, little progress has been made in elucidating the mechanism(s) of fragile site induction. To date, 89 CFSs have been identified in humans, among which 13 have been cloned and characterized at the molecular level (1). On the basis of sequence analysis of the cloned CFSs, a number of molecular features have been identified, including high A/T content, low gene content, high-flexibility, and high content of Long Interspersed Nuclear Elements (LINE) and Medium Reiterated (MER) repeats. Several lines of evidence suggest that DNA replication is involved in the induction of fragile sites (3). We and other investigators have shown that CFSs replicate in mid-late S phase, and that exposure to APH further delays the timing of replication (3–5). Furthermore, expression of CFSs is induced by conditions that impair replication, such as culturing cells in the presence of the DNA polymerase inhibitor, APH, and expression is enhanced by G2/M checkpoint inhibitors, such as caffeine. By examining the location of APH-induced breaks, CFSs have been demonstrated to lie at the interface of R- and G-bands, suggesting that CFSs are regions of unusual chromatin conformation, that replicate late in S phase (6).
Chromatin conformation influences DNA replication in at least two ways. First, as demonstrated in the Xenopus egg extract system, binding of the origin recognition complex to DNA is negatively regulated by DNA methylation (7). Second, replication origin activity, including origin assembly and origin activation timing, can be positively regulated by histone acetylation in a variety of systems (8,9). However, whether histone acetylation is required for origin selection or replication-timing specification remains an open question (10).
On the basis of these observations, we hypothesized that CFSs represent sequences that are inherently difficult to replicate. Moreover, perturbation of DNA replication within CFSs by APH treatment may be mediated, in part, by specific epigenetic patterns at CFSs, resulting in incomplete DNA replication and, ultimately, leading to the formation of gaps, breaks, or rearrangements in metaphase chromosomes (referred to as ‘CFS expression’ in this report). To test these hypotheses, we examined the chromatin modification pattern within six of the most highly expressed human CFSs in a human lymphoblastoid cell line, using a Chromatin immunoprecipitation (ChIP)-on-chip assay described previously (11). Here, we show that the majority of these CFSs are characterized by less acetylation, a feature typical of a more condensed chromatin structure, than their flanking non-fragile sequences (NCFSs). By using FRA3B as an example, we demonstrate that chromatin within the fragile site sequences is relatively more compact than that of the flanking NCFSs. Furthermore, we demonstrate that modifying chromatin structure at CFSs by treatment with trichostatin A (TSA) and/or 5-azadeoxycytidine (5-Aza) increases histone acetylation, decreases CpG methylation at CFSs and, notably, reduces fragile site expression. Taken together, our results demonstrate that histone hypoacetylation is a feature of the chromatin at CFSs, and that there is a link between histone acetylation, chromatin compaction, and the level of chromosome breakage observed at CFSs.
RESULTS
Most CFSs have features of condensed/repressive chromatin
To examine the potential role of chromatin modification in fragile site expression, we analyzed several epigenetic modifications associated with open versus condensed chromatin at six CFS and their flanking NCFS sequences.
Histone acetylation pattern at CFSs
We first examined the acetylation level of the chromatin, a mark that correlates with the level of compaction of the chromatin. We analyzed a human lymphoblastoid cell line (11 365) with a normal female karyotype using ‘ChIP-on-chip’ technology to determine the distribution of acetylated histone H3K9/14 (Ac-H3K9/14) within six CFSs (FRA3B, FRA16D, FRA7G, FRAXB, FRA2G, FRA7H), and their flanking NCFS sequences, covering almost 20 Mb of the human genome (Table 1 and Supplementary Material, Tables S1 and S2) (11). The iso-Tm oligonucleotide (45–65 nt) array platform we designed was highly tiled with a 35 nt start-to-start distance of the immediately adjacent probes. Repetitive sequences that were represented more than 35 times in the human genome were masked during the probe design. These six CFSs have been previously mapped by fluorescence in situ hybridization (FISH) with BAC and PAC clones to define the regions of highest breakage, designated as CFS sequences and the flanking NCFS sequences (12–18). Breakage studies have shown that these six CFSs are among the most commonly expressed CFSs induced by APH, among which, the FRA3B has the highest breakage frequency of all known CFSs, followed closely by the FRA16D. The level of fragile site expression in this cell line, and in other cells is FRA3B > FRA16D > FRA7G = FRAXB > FRA2G = FRA7H (19,20).
Table 1.
Summary of the Ac-H3K9/14 mapping results
| FRA3B |
FRA16D |
FRA7G |
FRAXB |
FRA2G |
FRA7H |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | |||
| Region size (Mb) | 0.46 | 4.57 | 0.27 | 1.94 | 0.51 | 4.91 | 0.83 | 0.53 | 0.97 | 0.48 | 0.16 | 4.34 | ||
| Gene density (/Mb) | 1 | 1.7 | 1 | 2.1 | 7.8 | 2.9 | 2.4 | 3.8 | 10.3 | 6.3 | 0 | 3.5 | ||
| Gene coverage (%) | 100 | 52 | 100 | 52 | 28 | 29 | 28 | 5 | 69 | 46 | 0 | 41 | ||
| Acetylation coverage | U | Average (%) | 0.3 ± 0.1 | 4.5 ± 1.2 | 1.9 ± 0.3 | 1.9 ± 0.2 | 1.6 ± 0.9 | 3.9 ± 1.3 | 8.5 ± 1.6 | 2.4 ± 0.4 | 2.4 ± 1.1 | 11.6 ± 2.8 | 40.5 ± 3.2 | 7.0 ± 1.7 |
| Shared (%) | 0.0 | 6.1 | 1.7 | 2.1 | 1.8 | 5.3 | 10.5 | 2.2 | 3.8 | 18.5 | 51.5 | 9.2 | ||
| A | Average (%) | 0.5 ± 0.3 | 6.2 ± 1.2 | 3.0 ± 0.6 | 2.9 ± 0.7 | 2.7 ± 0.7 | 5.4 ± 0.3 | 8.9 ± 0.7 | 3.1 ± 0.4 | 2.7 ± 1.1 | 17.6 ± 2.9 | 47.5 ± 1.6 | 7.7 ± 0.9 | |
| Shared (%) | 0.0 | 7.8 | 4.6 | 3.6 | 4.3 | 7.2 | 11.0 | 3.9 | 2.6 | 20.6 | 52.4 | 10.6 | ||
| T | Average (%) | 3.5 ± 0.7 | 7.2 ± 0.2 | 9.8 ± 1.3 | 4.1 ± 0.8 | 8.5 ± 0.4 | 6.0 ± 1.0 | 11.6 ± 1.3 | 3.0 ± 0.5 | 4.9 ± 0.7 | 23.5 ± 3.6 | 50.2 ± 8.4 | 10.5 ± 0.3 | |
| Shared (%) | 4.8 | 9.5 | 14.1 | 5.0 | 6.4 | 7.8 | 15.1 | 3.4 | 5.5 | 29.5 | 59.0 | 13.4 | ||
| T+A | Average (%) | 3.4 ± 1.2 | 7.2 ± 1.8 | 10.4 ± 4.4 | 4.5 ± 1.2 | 6.3 ± 0.7 | 4.1 ± 1.4 | 9.8 ± 1.4 | 2.7 ± 0.1 | 4.6 ± 1.0 | 21.7 ± 7.0 | 42.6 ± 13.2 | 8.4 ± 1.0 | |
| Shared (%) | 5.4 | 10.3 | 13.9 | 6.3 | 9.9 | 6.2 | 14.9 | 1.1 | 5.7 | 30.2 | 58.0 | 11.9 | ||
The acetylation coverage is displayed as the average of the acetylation coverage for all four data sets of both experiments combined (‘Average’ row), and the acetylation domains shared between the two independent experiments (‘Shared’ row). The shared acetylated chromatin domains correspond to the loci observed in a minimal of three of the four data sets. U: untreated; A: APH; T: TSA. Gene Coverage is defined as the percent of a region is covered by genes.
To identify the enriched regions of H3K9/14 acetylation, we used a permutation algorithm that was described previously (11). In a recent report, this algorithm was found to be one of the three most robust approaches in analyzing ChIP-on-chip data from NimbleGen tiling arrays hybridized with unamplified DNA samples mixed with spiked-in genomic DNA sequences (average length of 497 bp corresponding to predicted promoters in the human genome) (21). We applied this peak finding process to four data sets from two independent experiments, each containing two replicates, and obtained the map of the acetylated domains in the regions examined (Fig. 1A and C–E, and Supplementary Material, Fig. S1, panels iii–vi).
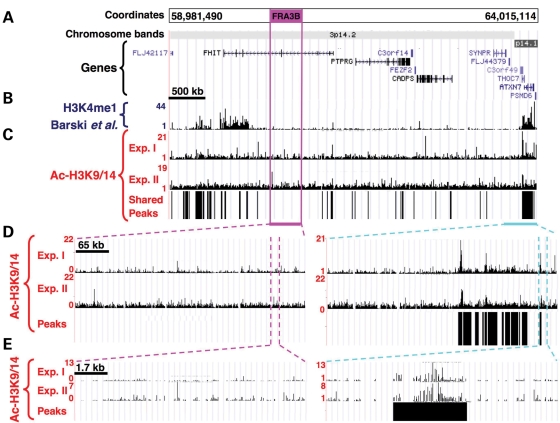
Figure 1.
Genomic map, and results of chromatin structure analysis within FRA3B. (A) Chromosome coordinates corresponding to the NCBI build 35, chromosome band and location of the annotated genes. The position of the CFS within the arrayed region is represented by the burgundy box. (B) Screenshot from the UCSC genome browser showing ChIP-Seq data with antibody for H3K4me1. (C) Screenshot from the UCSC genome browser showing ChIP-on-chip data for the lymphoblastoid cell line, 11 365, using anti-Ac-H3K9/14 antibody, displayed as the linear ratio of ChIP-on-chip sample fluorescence to input DNA fluorescence. The location of the mapped acetylated chromatin domains shared between two independent ChIP experiments is represented by vertical bars under the ChIP-on-chip data. (D, E) Screenshot of ChIP-on-chip data of zoomed-in sections within the CFS (left panels) or the NCFS sequences (right panels), with shared peaks (black bars) shown underneath.
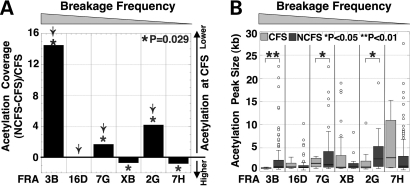
To compare the acetylation level of the different regions arrayed, we calculated the ‘acetylation coverage’, which corresponds to the sum of the width of all of the acetylated chromatin domains within a given region divided by the total length of the region, for each data set of each experiment (Table 1, Fig. 2A, the fragile sites are arranged in order of descending frequency of breakage in this and subsequent figures). Because of the large variation in acetylation coverage across the regions analyzed (0.3–40.5%), we asked whether there was a relationship between gene density and gene coverage (a measure of how much of a region is covered by genes) of the six fragile site regions studied here and their acetylation coverage (Table 1). No statistically significant correlation was found either between gene density and acetylation coverage (CFS: r = 0.20, P = 0.380; NCFS: r = 0.65, P = 0.053), or between gene coverage and acetylation coverage (CFS: r = 0.51, P = 0.11; NCFS: r = 0.10, P = 0.54). Histone acetylation at H3K9/14 has been shown to be a hallmark of the promoters of actively transcribed genes (22). To examine whether the acetylation peaks we identified are biased toward promoter regions, we calculated the extent of the acetylation coverage represented by promoters (2 kb upstream and downstream of the transcription start site). We found that although 65.5 ± 12.3% of the promoters were acetylated, they accounted for only 8.3 ± 3.7% of the total observed acetylated regions in untreated cells, suggesting that the majority of the H3K9/14 acetylation-enriched sequences are not promoter sequences (Supplementary Material, Fig. S2). Interestingly, five of the CFSs, FRA3B, FRA16D, FRA7G, FRAXB and FRA2G, were characterized by a relatively low acetylation coverage (0.30–8.47%) as compared with the level of acetylation estimated for the human genome based on the results of the ENCODE project ChIP-on-chip experiments (11.88%) (22) (Table 1, Fig. 2A). Of note is that the location of the acetylated sequences we identified is consistent with that of the acetylated domains mapped by the ENCODE project within the same chromosome region (Supplementary Material, Fig. S3A).
Figure 2.
Acetylation coverage at CFSs and flanking NCFS regions. (A) The graph illustrates the fold change in acetylation coverage of flanking NCFS regions versus each respective CFS. The arrow indicates those CFSs with less than 4% acetylation coverage. The order of the CFSs on the X-axis is shown in decreasing order of the frequency of fragile site expression, which is used in the figures throughout. (B) The distribution of the size of the acetylated chromatin domains (in kb) is represented as a box plot for each CFS (light gray plots) and NCFS (dark gray plots) region. The circles correspond to outlying data points, i.e. more than 1.5 times the inter-quartile range higher than the third quartile value. Of note, for presentation purposes, one outlier data point corresponding to a 55 kb domain within FRA7H CFS is not represented on the graph. The asterisks indicate a significant difference in the distribution of the size of the acetylated chromatin domains between CFSs and NCFSs (*, P < 0.05; **, P < 0.01).
In addition, we analyzed our data with TAMAL (23), a method that has also been shown to be very robust in analyzing data from NimbleGen tiling arrays hybridized with unamplified DNA samples (21). In comparison, TAMAL identified fewer peak regions and the size of individual peaks was smaller. However, the peaks mapped by TAMAL represent a subset of peaks mapped by the permutation algorithm, and were completely contained within the latter set, indicating that the discrepancy in peak number and size is a result of different stringencies set by these algorithms (Supplementary Material, Fig. S3B). TAMAL is designed to locate discrete transcription factor binding sites; thus, its ‘peak calling’ criteria is highly stringent requiring continuous runs of consecutive probes above a cutoff, such as 98th percentile threshold at P < 0.0001, to call a peak. In contrast, we sought to detect continuous regions that are enriched in Ac-H3K9/14. Hence, we applied several criteria with a lower stringency in our permutation method, such as a percentage of probes (not necessarily continuous) above the cutoff within a given window, and we combined peaks that were ≤2 kb apart. Nonetheless, both methods gave comparable results when we compared the acetylation coverage between the CFS and NCFS sequences as described below (Supplementary Material, Fig. S3C).
Exact permutation tests were used to compare acetylation coverage between CFSs and their flanking NCFSs. We found that the acetylation coverage at FRA3B, FRA7G and FRA2G were significantly lower within the CFS in comparison to their flanking regions (Fig. 2A). The converse was observed at FRAXB and FRA7H. However, it is notable that the acetylation level within FRAXB was very low within both the NCFS and CFS sequences, with the exception of five large acetylated domains (8–19 kb) within the CFS region overlapping with the HDHD1A gene, which is transcriptionally active in lymphoblastoid cells (Supplementary Material, Fig. S1J). Indeed, the acetylation coverage within the FRAXB CFS is only 2%, when the acetylated domains within HDHD1A are excluded, which is then comparable to that observed at FRA2G and FRA7G. Although the acetylation coverage at FRA16D (the second most highly expressed CFS) was not significantly lower than the flanking NCFS sequences, we observed a very low acetylation coverage (∼2%) within the entire arrayed region, indicating that the whole region is hypoacetylated. In addition, the majority of the acetylated domains were significantly smaller within the FRA3B, FRA7G and FRA2G, in comparison to those within their flanking regions (Fig. 2B). Of note, the size of the acetylated domains within all of the CFSs was also smaller (median size of 0.7–2.9 kb) than those of the ENCODE project (median size of 4.9 kb) (22). A caveat is that although both our study and the ENCODE project employed the same antibody for the ChIP DNA preparation, the array used by the ENCODE project was composed of probes that were 1024 bp (average) in length, significantly longer than the probes in our array (45–65 bp). Whether this variation introduces a difference in the results of the array experiments is unknown.
Overall, these results indicate that, with the exception of the FRA7H, which is expressed at low levels, hypoacetylated chromatin is a common characteristic of CFSs. Similar results were observed in APH-treated cells. Although APH treatment resulted in a statistically significant increase in acetylation coverage at several of the CFSs examined (FRA16D and FRA7H), and at the NCFS sequences of the FRA16D and FRA2G, the increase was one-half fold, indicating that APH treatment does not have a strong effect on the acetylation level of the chromatin in CFS and NCFS sequences (Supplementary Material, Fig. S4).
Histone methylation pattern at CFSs
To assess the chromatin structure at CFSs and NCFSs further, we examined the distribution of several histone methylation patterns characteristic of active chromatin by using the extensive epigenetic data analysis performed by Barski et al. (24). This large genome-wide study was performed using CD4+ human T cells, which also express CFSs (20). We aligned our acetylation maps with their methylation maps of lysine modifications that are markers of active chromatin, e.g. high levels of H3K4me1 (Fig. 1B and Supplementary Material, Fig. S1A–L, panels ii). Overall, the distribution of the active lysine methylation patterns observed within the six CFSs was very consistent with the Ac-H3K9/14 results obtained from our ‘ChIP-on-chip’ experiments.
In summary, analysis of histone acetylation and methylation patterns indicated that four of the most highly-expressed CFSs, FRA3B, FRA16D, FRA7G and FRA2G, are characterized by lower acetylation than their flanking NCFSs. The chromatin within the FRAXB CFS was also hypoacetylated for most of the region, with the exception of the HDHD1A gene located in the center of the CFS. Moreover, chromatin at these five CFSs has substantially lower acetylation than the average of the genome as predicted by the ENCODE project. Unexpectedly, we observed features of open chromatin structure within FRA7H, which corresponds to a gene-free region, and has the lowest level of expression of the six CFSs analyzed in this study. Taken together, these observations suggest that histone hypoacetylation may play a role in CFS expression by slowing or perturbing the replication process, making these regions more sensitive to APH-induced replication stress. Whether there are multiple subtypes of CFSs based on their chromatin conformation remains to be determined.
TSA decreases chromosomal breaks induced by APH/Caffeine
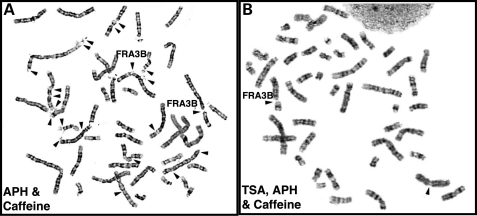
To examine whether increasing histone acetylation at fragile site sequences alters the frequency of chromosomal breaks and gaps, we treated phytohemagglutinin-stimulated peripheral blood lymphocytes (PHA-PBLs) isolated from three healthy individuals with a histone deacetylase (HDAC) inhibitor, TSA, in addition to the fragile site-inducing chemicals, APH and Caffeine, and analyzed metaphase cells for chromosomal breaks and gaps. We observed a dramatic decrease of fragile site expression in cells treated with TSA (Fig. 3). PHA-PBLs treated with TSA in addition to APH/Caffeine showed an average of 82 ± 14% decrease in total breaks in three human subjects as compared with cells treated with APH/Caffeine (P = 0.09) (Table 2 and Supplementary Material, Table S3). When we focused on individual fragile sites, we noted the same trend, i.e. an average of 82 ± 8% decrease for FRA3B (P = 0.02). We also performed similar studies on the 11 365, Blin (B cell ALL) and Molt-4 (T cell ALL) cell lines. Comparable with the observations in lymphocytes, TSA treatment resulted in a significant increase in acetylation on H3K9/14 (data not shown), and decreased fragile site expression (Table 2 and Supplementary Material, Table S3).
Figure 3.
TSA treatment decreases fragile site expression. PHA-stimulated lymphocytes were treated either with 0.4 µm APH for the last 24 h (left panel), or with 1.0 µm TSA and 0.4 µm APH for the last 24 h (right panel). Caffeine (1.5 µm) was added 4 h prior to metaphase cell preparation. Arrows identify breaks at CFSs.
Table 2.
TSA and/or 5-Aza treatment decreases fragile site expression
| Breakes per 25 cells | PHA stimulated human lymphocytes |
11365 |
Blin |
Molt-4 |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject 1 |
Subject 2 |
Subject 3 |
||||||||||||||||||||||
| T | A/C | T/A/C | T | A/C | T/A/C | T | A/C | T/A/C | T | A/C | T/A/C | T | A/C | T/A/C | Z | Z/A/C | T/Z/A/C | T | A/C | T/A/C | Z | Z/A/C | T/Z/A/C | |
| Total | 1 | 219 | 44 | 2 | 767 | 25 | 6 | 298 | 94 | 11 | 111 | 21 | 2 | 109 | 65 | 19 | 52 | 14 | 12 | 68 | 13 | 15 | 24 | 22 |
| FRA3B (3p14.2) | 0 | 18 | 4 | 0 | 33 | 3 | 0 | 26 | 6 | 0 | 10 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| FRA16D (16q23.2) | 1 | 17 | 3 | 0 | 17 | 0 | 0 | 9 | 3 | 0 | 23 | 5 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| FRA7G (7q31.2) | 0 | 11 | 0 | 0 | 14 | 0 | 0 | 6 | 1 | 0 | 6 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| FRAXB (Xp22.3) | 0 | 5 | 3 | 0 | 32 | 2 | 0 | 14 | 4 | 0 | 12 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| FRA2G (2q31) | 0 | 1 | 0 | 0 | 4 | 0 | 0 | 3 | 2 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| FRA7H (7q32.3) | 0 | 5 | 1 | 0 | 8 | 0 | 0 | 5 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
Fragile site breaks were scored in 25 Trypsin–Giemsa banded metaphase cells from PHA-stimulated lymphocytes, 11365, Blin, and Molt-4 cells cultured under various conditions (A: APH; C: Caffeine; T: TSA; Z: 5-Aza). The Molt-4 cell line is tetraploid, and both Molt-4 and Blin contain structural chromosomal abnormalities. Treatment with 5-Aza alone induced ∼0.6 breaks/metaphase cell for both Blin and Molt-4 cells, which is less than that reported previously (3 breaks/metaphase cell using a 10-fold higher concentration of 5-Aza) (18). Note that the number of breaks observed in untreated cells is typically zero.
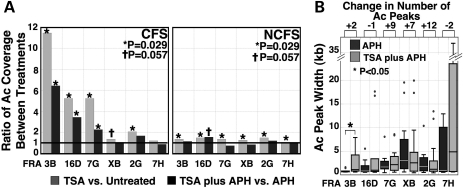
ChIP-on-chip analysis of the 11 365 lymphoblastoid cell line revealed that TSA treatment significantly increased acetylation coverage within the CFS portions of four of the six CFSs examined (FRA3B, FRA16D, FRA7G and FRA2G), and marginally increased acetylation coverage in one additional CFS (FRAXB) (Fig. 4A). Similarly, in cells treated with both TSA and APH, three of these CFSs (FRA3B, FRA16D and FRA7G) had an increase of Ac-H3K9/14 within the CFS portions, as compared with APH treatment alone. Although there was an increase of histone acetylation at the NCFS regions following treatment, the magnitude of the increase was lower than that of the CFS regions (Fig. 4A, right panel). The overall increase of acetylation coverage seen after TSA treatment was not due to the increase of Ac-H3K9/14 at the promoters, since there was no significant increase of promoter acetylation in cells treated with TSA. Moreover, promoter Ac-H3K9/14 accounted for significantly less of the total Ac-H3K9/14 in TSA-treated cells as compared with untreated cells (5.4 ± 2.9% in TSA versus 8.3 ± 3.7% in untreated, P = 0.01, Supplementary Material, Fig. S2B). Further comparison of the acetylated peaks at these CFSs revealed that, although the median length of the acetylation peaks was comparable in the TSA plus APH-treated cells as compared with cells treated with APH alone, the distribution of acetylated peak size in TSA plus APH-treated cells was shifted towards larger peak size indicated by the increased size of the box plot (i.e. greater range) for the FRA3B (P < 0.05), and by the large peak size of the outliers in the FRA16D (Fig. 4B). We also observed a substantial increase in the number of acetylated peaks in the FRA2G and FRA7G. Thus, the increase of acetylation coverage following TSA treatment is the result of the combined effects of an increase in the number of acetylation peaks and an increase in the size of large peaks within CFS sequences. Taken together, our results indicate that histone modification and, by extension, chromatin conformation likely play a role in fragile site induction.
Figure 4.
TSA increases histone H3K9/14 acetylation within CFSs. (A) H3K9/14 acetylation coverage increases within CFSs following TSA treatment. The ratios of the acetylation coverage between various treatments were calculated for the CFS sequences (left panel) and the flanking NCFS sequences (right panel). Statistical significance is calculated based on the exact permutation test (*, P = 0.029; †, P = 0.057). (B) Comparison of the distribution of acetylation peak width in APH-treated and TSA plus APH-treated cells. Box plots show the distribution of the acetylation peak width for each CFS following the indicated treatment (*, P < 0.05). The absolute gain (or loss) in the number of the acetylated peaks for each CFS is listed above the figure. For presentation purposes, an outlier in the FRA7H in the APH-treated sample is omitted due to its large size (∼55 kb).
5-Aza decreases chromosomal breaks induced by APH/Caffeine
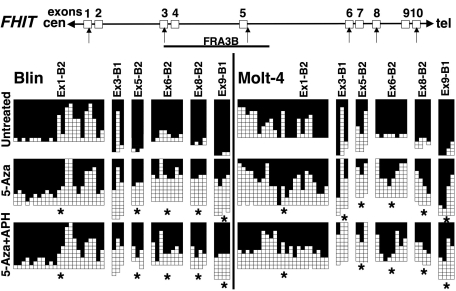
Chromatin conformation is thought to be regulated not only by histone modifications, but also by DNA methylation. To assess whether DNA methylation affects fragile site expression in a manner similar to histone acetylation, we treated Blin and Molt-4 cells with 5-Aza, an inhibitor of DNA methylation, and examined the frequency of chromosomal breaks at CFSs. These cell lines were selected because expression of fragile histidine triad gene (FHIT), which contains the FRA3B, is undetectable due to CpG methylation of its promoter region (25). Treatment with 5-Aza alone induced about 0.6 breaks per 25 metaphase cells for both Blin or Molt-4 cells, which is less than that reported previously in T lymphoblasts (three breaks per metaphase cells using a 10-fold higher concentration of 5-Aza) (19). Treatment with 5-Aza reduced chromosomal breakage at CFSs in APH-treated Blin and Molt-4 cells by 52 and 65%, respectively (Table 2). Moreover, Blin and Molt-4 cells treated with both 5-Aza and TSA were further protected from APH-induced chromosomal breaks with an 87 and 68% decrease in breaks, respectively.
To investigate whether this reduction of chromosomal instability was associated with an effect of 5-Aza on DNA CpG methylation at CFSs, we examined the genomic CpG methylation pattern within the FRA3B using bisulfite-sequencing with six sets of primers amplifying 43 CpG dinucleotides (Fig. 5). CpG methylation was significantly reduced by 5-Aza treatment within most of the regions examined (P < 0.05, Fig. 5). FRA3B is embedded within the FHIT gene, raising the possibility that FHIT transcription interferes with DNA replication within the FRA3B, leading to impaired replication at CFSs. We found that the level of fragile site expression after 5-Aza treatment was unrelated to the level of FHIT expression, since we observed a reduction of FRA3B breaks rather than an increase, whereas 5-Aza alone or with TSA only minimally reactivated FHIT expression in Blin cells, but not in Molt-4 cells (Supplementary Material, Fig. S5).
Figure 5.
5-Aza treatment decreases CpG methylation of FRA3B sequences. Bisulfite sequencing of the FHIT/FRA3B locus in untreated Blin and Molt-4 cells, and cells treated with 0.2 µm 5-Aza for 72 h, or 5-Aza and APH (added 24 h prior to harvest). Each row presents a clone. Filled squares indicate methylated CpGs, whereas open squares refer to unmethylated CpGs. Primer sequences are listed in Supplementary Material, Table S4. The genomic structure of the FHIT/FRA3B locus is illustrated at the top of the panel, and the regions examined are indicated by arrows. Stars indicate regions that had a significant decrease (P < 0.05) of CpG methylation in treated cells.
Taken together, our results suggest that altering the epigenetic pattern by increasing histone acetylation and/or decreasing DNA methylation results in a reduction in the incidence of breakage at CFSs.
FRA3B chromatin is less sensitive to micrococcal nuclease
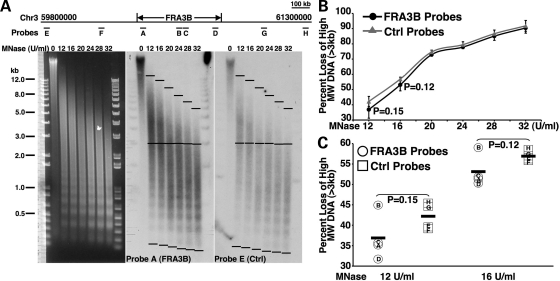
Given our observation that histones within the CFS sequences are less acetylated than those of the NCFS sequences, and since hypoacetylation is a mark for condensed chromatin structure, we sought to determine whether chromatin at CFSs is more compact than that of the NCSFs. To address this question, we examined the micrococcal nuclease (MNase) sensitivity of chromatin from the FRA3B and its flanking NCFSs. Loss of the high molecular weight (MW, >3 kb) DNA of the selected regions within either the FRA3B or its flanking sequences after treatment with various doses of MNase was assessed by Southern blotting followed by quantification using ImageJ (Fig. 6A). The average loss of the high MW DNA of the four FRA3B regions (regions A–D) and the four control regions (regions E–H) were similar when 11 365 nuclei were treated with high doses of MNase (≥20 U/ml) (Fig. 6B). However, the FRA3B regions exhibited a smaller loss of the high MW DNA as compared with that of the control regions when 11 365 nuclei were treated with low doses of MNase (Fig. 6B). Although the differences were not statistically significant (two-tailed student t-test), we noted that compared with the control regions, three regions within the FRA3B (regions A, C and D) showed substantially less loss of the high MW DNA at 12 and 16 U/ml MNase treatments (Fig. 6C), indicating that the majority of the chromatin within the FRA3B sequences were less sensitive to MNase treatment and, hence, had a relatively more compact chromatin structure than the chromatin of the flanking NCFSs. Interestingly, chromatin at region B within the FRA3B displayed a similar MNase sensitivity to that of the control regions (Fig. 6C), raising the possibility that there are pockets of comparatively more open chromatin within the more compact FRA3B locus.
Figure 6.
Chromatin within the FRA3B is less sensitive to MNase. (A) Ethidium bromide-stained gel and Southern blots probed with probe A (FRA3B probe) or probe E (control probe) on DNA prepared from nuclei of 11 365 cells following treatment with increasing doses of MNase. The locations of the probes are illustrated at the top of the panel. (B) The percentage loss of high MW (>3 kb) signal for each lane was calculated by measuring the signal between the top and center black bars, and the total signal between the top and bottom black bars using ImageJ. The measured signals were normalized to the background signals of the blots. The average percentage loss of high MW DNA for either the FRA3B regions (black line) or the control flanking non-fragile sequences (gray line) was graphed against MNase concentrations. (C) The percentage loss of high MW DNA of each individual region when the nuclei were treated with either 12 or 16 U/ml MNase. The black bars indicate the averages.
DISCUSSION
CFSs are characterized by histone hypoacetylation and compact chromatin structure
The cloned CFSs have characteristics of G-bands, i.e. they are gene-poor, AT-rich, high in LINE and MER repeats. Furthermore, the analysis of the flexibility of DNA based on the twist angle of the double helix has revealed that CFSs contain regions of high flexibility and low DNA stability (26,27). A number of observations have linked DNA replication to CFS expression; however, the precise mechanism of CFS expression remains unclear. We hypothesized that a unique chromatin modification pattern at CFSs may slow or perturb the normal progression of replication forks, contributing to the high sensitivity of CFSs to DNA replication stress. By using a ChIP-on-chip assay to examine the epigenetic characteristics of six well-characterized, cloned CFSs, we demonstrate that the chromatin of five of six CFSs assayed is hypoacetylated, and potentially more compact than the flanking NCFSs (as in the example of the FRA3B), further supporting the observation made by El Achkar et al. (6) that CFSs lie at the interface of R- and G-bands with unusual chromatin structure.
Two of the CFSs examined in this study, the FRA3B and FRA16D, are embedded within large genes, FHIT and WWOX, respectively. Each of these genes spans more than 1 Mb; however, their transcripts are only ∼1 kb in length. Similar features are found in several other CFSs, including the FRA2F (LRP1B), FRA4G (GRID2), FRA6E (PARKIN), FRA13A (NBEA) and FRA15A (RORA), raising the possibility that CFSs may be associated with large genes (28). We examined the Ac-H3K9 acetylation pattern of large genes (>1 Mb) in the genome by using the publicly-available ENCODE ChIP-Seq data (Bernstein Lab, ENCODE Chromatin Group, Massachusetts General Hospital/Harvard Medical School and Broad Institute), and found that the acetylation coverage of these genes is substantially lower than that of the human genome on average (Supplementary Material, Fig. S6). This result suggests that the location of CFSs may affect their histone acetylation pattern, and that a proportion of the CFSs that have not yet been cloned may be embedded within large genes. However, more than half of the CFSs cloned to date are not located within large genes, yet the chromatin of these CFSs (e.g. the FRA7G, FRAXB and FRA2G) is predominantly hypoacetylated as compared with the genome on average, and to their flanking NCFSs. These observations suggest that hypoacetylation is an inherited characteristic of the majority of CFSs, and that physical location within large genes further exacerbates the effect of hypoacetylation on CFS expression, exemplified by the FRA3B and FRA16D.
We examined the compaction of the chromatin at the FRA3B more directly by measuring its sensitivity to MNase. Three of four randomly-selected regions within the FRA3B demonstrated greater resistance to MNase treatment than control regions within the flanking NCFSs. Together with the observation of histone hypoacetylation within the FRA3B, these results clearly indicate that chromatin structure of the FRA3B is relatively more compact than that of the flanking NCFSs. Although the difference is subtle, tighter chromatin structure may affect the access of the DNA replication and/or DNA repair machinery to fragile site sequences, especially under replication stress, resulting in incomplete DNA replication at CFSs and, ultimately, contributing to the expression of CFSs.
CFS expression is influenced by epigenetics
Our results suggest that CFS expression is influenced by epigenetic marks. In the FRA16D, AT-repeats have been implicated in CFS expression by stalling replication forks (29). We observed that all 12 AT-repeat-rich sequences in the FRA16D, identified by the TwistFlex program were located in regions with low acetylation in untreated cells; however, there was a 78% decrease of FRA16D expression after TSA treatment, even though only one AT-repeat-rich sequence became acetylated (Table 2, data not shown). These results raise the possibility that in addition to AT-repeats, other factors such as histone hypoacetylation may impact on genetic instability at CFSs, with the common theme that all of these factors affect DNA replication within CFSs.
High levels of acetylation have been associated with increased permissiveness for DNA replication (30). For example, the Hbo1 histone acetyltransferase associates with both origin recognition complexes and mini-chromosome maintenance complexes to promote access of pre-replication complexes to origins (31). Studies in Drosophila have also revealed histone hyperacetylation at active origins, a modification that is conserved among different Drosophila species (32). Histone acetylation may also facilitate DNA double strand break (DSB) repair by creating a binding platform to promote recruitment of remodeling complexes at the sites of DSBs [reviewed in (33)]. Similarly, decreasing CpG methylation can reduce/eliminate the recruitment of the methyl-CpG-binding proteins and other chromatin remodeling complexes, such as HDACs and H3K9 methylase which mediate chromatin compaction (34). Hence, the increase of histone H3 acetylation at CFSs after TSA treatment, and the decrease of CpG methylation after 5-Aza treatment, would be expected to relax chromatin at CFSs, potentially alleviating the structural effects that result in CFS expression during DNA replication stress, and/or facilitating the repair of the DSBs occurring within CFSs in response to APH treatment. However, we cannot exclude the possibility that TSA may increase expression and/or acetylation of certain non-histone proteins, which may also facilitate replication or repair processes, leading to a protective effect on CFS expression induced by replication stress.
It has also been hypothesized that the collision of transcription and replication complexes may result in replication fork stalling, DSBs and, ultimately, chromosome breaks at CFSs. Although we did not address this question directly, our data suggested that transcription does not play a role in fragile site expression. In general, treatment with TSA and 5-Aza augments transcription. Instead of an increase of CFS breakage, we observed a significant decrease of CFS breakage in all cell lines examined. Furthermore, we found that the level of FRA3B expression after 5-Aza treatment was unrelated to the level of FHIT expression (Table 2, Supplementary Material, Fig. S5). The issue of whether transcription complexes colliding with DNA replication complexes plays a role in inducing CFS expression is only relevant if genes containing CFSs are transcribed in S phase. Therefore, further studies are needed to address whether the genes in CFSs are transcribed during S phase, and whether there is a correlation between the expression levels of these genes and CFS expression.
Subtypes of CFSs
We observed a decrease in the frequency of chromosomal breaks and gaps at each of the six CFSs analyzed following TSA plus APH treatment as compared with APH treatment alone, which was coincident with a significant increase in Ac-H3K9/14 coverage within four of these CFSs. One exception, the FRA7H, has higher acetylation coverage than do the other five CFSs, as well as higher acetylation coverage than its flanking NCFSs. Thus, these observations raise the possibility that the FRA7H differs mechanistically from other CFSs, such as the FRA3B and FRA16D. Extensive analysis using FISH revealed multiple breakage ‘hot spots’ within the FRA7H, and some FISH signals showed an orientation that was the opposite of that predicted by physical maps, prompting the investigators to propose that the FRA7H may contain an unusual chromatin organization (16). Whether hyperacetylation contributes to this unusual chromatin organization remains to be determined.
Nonetheless, it is also possible that other types of histone modifications, such as acetylation on histone H4, may play a more determinant role in chromatin conformation of the FRA7H. In addition to the six CFSs examined extensively in this study, there are seven other CFSs that have been cloned (1). On the basis of the histone methylation map reported by Barski et al. (24), the majority of these CFSs also lie within predicted compact chromatin regions (data not shown). However, further ChIP-on-chip studies are needed to verify these observations.
Mechanism of CFS expression
In summary, we have demonstrated that histone hypoacetylation is a common feature of the subset of CFSs with the highest expression frequency. Moreover, we have shown that relaxing chromatin conformation at CFSs by TSA and/or 5-Aza treatments reduces fragile site expression. In addition, our data suggest that the relatively compact chromatin structure of the FRA3B, and by extension other CFSs, may affect the process of replication of CFS sequences, such as replication origin firing and replication fork progression, under DNA replication stress. Thus, CFSs may represent sequences with low histone acetylation that replicate very slowly, and are unable to recover from a further delay in DNA synthesis following replication stress.
MATERIALS AND METHODS
Cell culture and drug treatments
Cell lines (the human Epstein Barr Virus-transformed lymphoblastoid cell line 11 365 with a normal 46,XX karyotype, and leukemia cell lines, Blin and Molt-4) were maintained in RPMI 1640 medium supplemented with 10 mm HEPES, 100 units/ml Penicillin/100 µg/ml Streptomycin, and 10% fetal bovine serum (all reagents from Invitrogen, CA, USA) at 37°C in a humidified 5% CO2/95% air atmosphere. Peripheral blood lymphocytes from healthy individuals were cultured in the same medium supplemented with phytohemagglutinin (PHA, 0.09 mg/ml, Remel Inc., KS, USA) for 72 h. To induce fragile site expression, cells were treated with 0.4 µm APH (Sigma, MO, USA) for 24 h with the addition of 1.5 mm of caffeine (Sigma) during the last 4 h of culture. Cells in log-phase growth were cultured with TSA (Sigma) at a final concentration of 1.0 µm for 24 h, or 5-Aza (Sigma) at a final concentration of 0.2 µm for 72 h, with replenishment of the media and the drug every 24 h.
Chromatin immunoprecipitation
ChIP assays were performed according to the protocol described by the manufacturer (Millipore, http://www.millipore.com, MA, USA) with antibody against acetylated histone H3 K9/14 (Millipore). Minor modifications of the protocol were described in Lucas et al. (11).
Cytogenetic analysis
Metaphase cells were prepared using standard cytogenetic techniques, and air-dried slides were prepared and stained using Trypsin–Giemsa banding techniques as described previously (35). For the analysis of fragile site expression, chromosome breaks, gaps, rearrangements and aberrations were scored in 25 metaphase cells for each culture condition.
Bisulfite treatment and DNA sequencing
DNA (1 µg) was diluted in 50 µl water, and incubated with 5.5 µl 2 N NaOH at 37°C for 10 min, followed by treatment with 30 µl 10 mm hydroquinone and incubation with 520 µl 3.6 m Sodium Bisulfite at 50–55°C for 16–22 h (all chemicals from Sigma). During incubation, 5–6 drops of mineral oil was added to avoid evaporation. Bisulfite-treated DNA was collected by using the Wizard™ DNA Clean-up system. The eluted DNA was treated with 5.5 µl 3 N NaOH, incubated at RT for 5 min, and precipitated by EtOH precipitation.
Touchdown PCR (95°C, 10 min; 10 cycles of 95°C for 30 s, 65°C for 30 s and 72°C for 45 s, with a decrease in the annealing temperature of one-half degree per cycle; 35 cycles of 95°C for 30 s, 55°C or 50°C for 30 s and 72°C for 45 s, 3.75 or 2.5 mm final Mg2+ concentrations) was performed on bisulfite-treated DNA with primers designed using the regions that flank the CpG-containing sequences. Primer sequences, annealing temperatures and Mg2+ concentrations for each pair of primers are listed in the Supplementary Material, Table S4. PCR products were TOPO TA cloned and transformed into One-Shot cells (Invitrogen). Clones were randomly picked and sequenced. Sequences were analyzed by the DNAStar MegAlign program to assess the CpG methylation level.
MNase treatment and Southern blotting
Nuclei preparation and MNase (Worthington, NJ, USA) treatment were performed as previously described (36). DNAs were purified and fractionated on a 0.8% agarose gel in TPE buffer (Sigma). Gels were transferred onto Hybond N (GE Healthcare, NJ, USA) in 20× SSC, and the subsequent membranes were probed with 32P-labeled probes for either the FRA3B regions or control regions. The coordinates of the probes are listed in Supplementary Material Table S5. Probe sequences were free of repetitive sequences as analyzed by RepeatMasker.
Microarray construction and hybridization
To identify the CFS sequences assayed in this study, we used previously published reports of FISH analysis of BAC or PAC clones to determine the genomic sequences that exhibited the highest level (peak) of split signal patterns relative to breaks at the corresponding CFSs. The sequences surrounding these CFS sequences were identified as the NCFS sequences. It has been proposed that some CFSs, e.g. the FRA3B, may be larger than predicted previously based on the observation of split signals in a low percentage of cells using probes extending several megabases from the peak region of breakage (37). We classified these sequences as NCFSs in this study (Supplementary Material, Table S1). In addition, some CFSs may have an abnormal genomic structure, as suggested by the unusual hybridization pattern observed by FISH of genomic clones from the FRA7G (38), and the FRA7H (16), making it difficult to define the boundaries of these CFSs. We also included these sequences on our array, and categorized them as NCFSs in this study (Supplementary Material, Table S1). All regions were tiled with isothermal (Tm = 76°C) oligonucleotides of variable length (45–65 bp). To avoid repetitive regions, the set of probes were compared against a pre-computed frequency histogram of all possible 15mer probes in the human genome. For each probe, the maximum average frequency of all of the 15-mers comprising the probe was set at 35. The minimum and median tiling intervals were 35 nt (start-to-start). The construction of the arrays (385 000 oligonucleotides), the labeling of DNAs, the microarray hybridization, and scanning were performed by the Roche NimbleGen, Inc. Core Facility (WI, USA) as described previously (11,39).
Microarray data analysis
The microarray data analysis process was performed following the protocol described previously (11). Briefly, after the Z-score filtration (cutoff = 3) of the original data to eliminate the non-reproducible replicates on the array, we applied a sliding-window approach to identify potential enrichment regions. Within the window, a probe was considered to ‘qualify’ if its log2 ratio was above a set cutoff ri. A peak was called when the number of qualifying probes was above a set percentage of the probes within the window. The start position of the first qualifying probe and the end position of the last qualifying probe were set as the start and end positions of the peak. This procedure was repeated using a series of log2 ratio cutoff values, r1, r2, … ri, where r1> r2> … > ri, and r1 equals the ‘hypothetical maximum (mean+6 S.D.)’ for each data set of each region to minimize the effects of outliers. To assess the likelihood of any peak as representing a true enrichment region, we estimated the false-positive rate (FPR) for each identified region. The FPR was defined as the ratio of the average number of peaks found using the log2 ratio cutoff value ri for the randomized (20 times) and the non-randomized data. This FPR value was assigned to the peaks that were present at the log2 ratio cutoff value ri, but absent at the value ri−1. We chose peaks that had an FPR less than 15% for further analysis. After a set of enrichment regions were identified for each data set of each arrayed region at each individual cutoff, we merged any two peaks 2 kb or less apart on the same set and then combined merged peaks from different cutoff sets to obtain a final set of enrichment regions for each data set. Because the added component of variability between experiments was very low relative to the variation within experiments, we used the four data sets of both independent experiments to obtain the definitive map of the acetylated domains in the regions examined.
Statistical analysis of the microarray data
For statistical purposes, the two data sets (FWD1 and FWD2) were combined separately for each percentage of qualifying probes (20–100%), creating two new data sets, FWD1(20–100%) and FWD2(20–100%), which were used for the statistical analysis. The data were also replicated across two separate experiments. Due to the small sample sizes, acetylation coverage was analyzed using exact permutation tests. Acetylation peak size and peak width were analyzed using two-sample t-test for unequal variances (Satterthwaite approximation to degrees of freedom).
TwistFlex
AT-repeat-rich regions were identified by analyzing the FRA16D (Chr16: 77,209,000-77,479,000, NCBI, Build 35) sequences using the TwistFlex program online at http://margalit.huji.ac.il/TwistFlex/Home.html using parameters according to Mishmar et al. (16).
FHIT expression
RNA was isolated and cDNA was made following standard protocols. Real-time PCR was performed using a StepOnePlus (Applied Biosystems, CA, USA) thermocycler with primers for either FHIT spanning exons 1–4 (Forward: 5′-CAGCTGTCAACATCCTGGAA-3′; Reverse: 5′-CTCTTCGGAGTCCTCAGTGG-3′) or GAPDH (Forward: 5′-CGGAGTCAACGGATTTGGTCGTAT-3′; Reverse: 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′). The expression level of FHIT is normalized relative to the expression level of GAPDH.
Data deposition
The data discussed in this publication have been deposited in EMBL-EBI's ArrayExpress (http://www.ebi.ac.uk/microarray-as/aer/entry, accession number E-TABM-347).
SUPPLEMENTARY MATERIAL
Supplementary material is available at HMG online.
FUNDING
This work was supported by National Institutes of Health (CA41644 to M.M.L.), and was facilitated by the DNA Sequencing Facility of the University of Chicago Cancer Research Center.
ACKNOWLEDGEMENTS
We thank Drs Lucy Godley, Ursula Storb, Eileen Dolan and Douglas Bishop, as well as members of the Le Beau laboratory for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Glover T.W. Common fragile sites. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Jr, Kastrinakis N.G., Levy B., et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. (see comment) [DOI] [PubMed] [Google Scholar]
- 3.Lucas I., Palakodeti A., Le Beau M.M. The implications of replication in common fragile site induction. In: Arrieta I., Penagarikano O., Télez M., editors. Fragile Site. Hauppauge, USA: Nova Science Publishers Inc; 2007. pp. 67–98. [Google Scholar]
- 4.Hellman A., Rahat A., Scherer S.W., Darvasi A., Tsui L.C., Kerem B. Replication delay along FRA7H, a common fragile site on human chromosome 7, leads to chromosomal instability. Mol. Cell. Biol. 2000;20:4420–4427. doi: 10.1128/mcb.20.12.4420-4427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Beau M.M., Rassool F.V., Neilly M.E., Espinosa R., 3rd, Glover T.W., Smith D.I., McKeithan T.W. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 6.El Achkar E., Gerbault-Seureau M., Muleris M., Dutrillaux B., Debatisse M. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc. Natl. Acad. Sci. USA. 2005;102:18069–18074. doi: 10.1073/pnas.0506497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey K.J., Newport J. CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol. Cell. Biol. 2003;23:6769–6779. doi: 10.1128/MCB.23.19.6769-6779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelauer M., Rubbi L., Lucas I., Brewer B.J., Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 9.Kemp M.G., Ghosh M., Liu G., Leffak M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 2005;33:325–336. doi: 10.1093/nar/gki177. [erratum appears in Nucleic Acids Res. 2005;33(2):805] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dazy S., Gandrillon O., Hyrien O., Prioleau M.N. Broadening of DNA replication origin usage during metazoan cell differentiation. EMBO Rep. 2006;7:806–811. doi: 10.1038/sj.embor.7400736. (see comment) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucas I., Palakodeti A., Jiang Y., Young D.J., Jiang N., Fernald A.A., Le Beau M.M. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilke C.M., Hall B.K., Hoge A., Paradee W., Smith D.I., Glover T.W. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum. Mol. Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- 13.Rassool F.V., Le Beau M.M., Shen M.L., Neilly M.E., Espinosa R., 3rd, Ong S.T., Boldog F., Drabkin H., McCarroll R., McKeithan T.W. Direct cloning of DNA sequences from the common fragile site region at chromosome band 3p14.2. Genomics. 1996;35:109–117. doi: 10.1006/geno.1996.0329. [DOI] [PubMed] [Google Scholar]
- 14.Arlt M.F., Miller D.E., Beer D.G., Glover T.W. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer. 2002;33:82–92. doi: 10.1002/gcc.10000. [DOI] [PubMed] [Google Scholar]
- 15.Krummel K.A., Roberts L.R., Kawakami M., Glover T.W., Smith D.I. The characterization of the common fragile site FRA16D and its involvement in multiple myeloma translocations. Genomics. 2000;69:37–46. doi: 10.1006/geno.2000.6321. [DOI] [PubMed] [Google Scholar]
- 16.Mishmar D., Rahat A., Scherer S.W., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H., Qian C., Jenkins R.B., Smith D.I. Fish mapping of YAC clones at human chromosomal band 7q31.2: identification of YACS spanning FRA7G within the common region of LOH in breast and prostate cancer. Genes Chromosomes Cancer. 1998;21:152–159. doi: 10.1002/(sici)1098-2264(199802)21:2<152::aid-gcc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 18.Limongi M.Z., Pelliccia F., Rocchi A. Characterization of the human common fragile site FRA2G. Genomics. 2003;81:93–97. doi: 10.1016/s0888-7543(03)00007-7. [DOI] [PubMed] [Google Scholar]
- 19.Yunis J.J., Soreng A.L., Bowe A.E. Fragile sites are targets of diverse mutagens and carcinogens. Oncogene. 1987;1:59–69. [PubMed] [Google Scholar]
- 20.Glover T.W., Berger C., Coyle J., Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 21.Johnson D.S., Li W., Gordon D.B., Bhattacharjee A., Curry B., Ghosh J., Brizuela L., Carroll J.S., Brown M., Flicek P., et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch C.M., Andrews R.M., Flicek P., Dillon S.C., Karaoz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bieda M., Xu X., Singer M.A., Green R., Farnham P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Zheng S., Ma X., Zhang L., Gunn L., Smith M.T., Wiemels J.L., Leung K., Buffler P.A., Wiencke J.K. Hypermethylation of the 5′ CpG island of the FHIT gene is associated with hyperdiploid and translocation-negative subtypes of pediatric leukemia. Cancer Res. 2004;64:2000–2006. doi: 10.1158/0008-5472.can-03-2387. [DOI] [PubMed] [Google Scholar]
- 26.Toledo F., Coquelle A., Svetlova E., Debatisse M. Enhanced flexibility and aphidicolin-induced DNA breaks near mammalian replication origins: implications for replicon mapping and chromosome fragility. Nucleic Acids Res. 2000;28:4805–4813. doi: 10.1093/nar/28.23.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith D.I., McAvoy S., Zhu Y., Perez D.S. Large common fragile site genes and cancer. Seminars Cancer Biol. 2007;17:31–41. doi: 10.1016/j.semcancer.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., Freudenreich C.H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabancay A.P., Jr, Forsburg S.L. Eukaryotic DNA replication in a chromatin context. Curr. Top. Dev. Biol. 2006;76:129–184. doi: 10.1016/S0070-2153(06)76005-7. [DOI] [PubMed] [Google Scholar]
- 31.Iizuka M., Matsui T., Takisawa H., Smith M.M. Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvi B.R., Byrnes B.A., Kolpakas A.J. Conservation of epigenetic regulation, ORC binding and developmental timing of DNA replication origins in the genus Drosophila. Genetics. 2007;177:1291–1301. doi: 10.1534/genetics.107.070862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandita T.K., Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Roulston D., Le Beau M. Cytogenetic analysis of hematologic malignant diseases. In: Barch M.J., Knutsen T., Spurbeck J.L., editors. The AGT Cytogenetics Laboratory Manual. Philadelphia, PA: Lippincott-Raven; 1997. [Google Scholar]
- 36.Gilbert N., Boyle S., Sutherland H., de Las Heras J., Allan J., Jenuwein T., Bickmore W.A. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker N.A., Thorland E.C., Denison S.R., Phillips L.A., Smith D.I. Evidence that instability within the FRA3B region extends four megabases. Oncogene. 2002;21:8713–8722. doi: 10.1038/sj.onc.1205950. [DOI] [PubMed] [Google Scholar]
- 38.Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 39.Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.