Abstract
Recently, associations of several common genetic variants with height have been reported in different populations. We attempted to identify further variants associated with adult height in a self-contained population (the Sorbs in Eastern Germany) as discovery set. We performed a genome wide association study (GWAS) (∼390 000 genetic polymorphisms, Affymetrix gene arrays) on adult height in 929 Sorbian individuals. Subsequently, the best SNPs (P < 0.001) were taken forward to a meta-analysis together with two independent cohorts [Diabetes Genetics Initiative, British 1958 Birth Cohort, (58BC, publicly available)]. Furthermore, we genotyped our best signal for replication in two additional German cohorts (Leipzig, n = 1044 and Berlin, n = 1728). In the primary Sorbian GWAS, we identified 5 loci with a P-value < 10−5 and 455 SNPs with P-value < 0.001. In the meta-analysis on those 455 SNPs, only two variants in GPR133 (rs1569019 and rs1976930; in LD with each other) retained a P-value at or below 10−6 and were associated with height in the three cohorts individually. Upon replication, the SNP rs1569019 showed significant effects on height in the Leipzig cohort (P = 0.004, beta = 1.166) and in 577 men of the Berlin cohort (P = 0.049, beta = 1.127) though not in women. The combined analysis of all five cohorts (n = 6,687) resulted in a P-value of 4.7 × 10−8 (beta = 0.949). In conclusion, our GWAS suggests novel loci influencing height. In view of the robust replication in five different cohorts, we propose GPR133 to be a novel gene associated with adult height.
INTRODUCTION
The high genetic influence on body stature has been known for a long time and twin and full sibling studies estimated a heritability of 0.80 and higher (1,2). In the last decades, candidate gene approaches and linkage studies could disclose only little of the complex genetic background of height (3–7). However, several new genetic variants affecting human stature (e.g. in HMGA2, GDF5-UQCC) were detected in a first sweep of genome-wide association studies (GWAS) providing further insight into the genetic architecture of this polygenic trait (8–12). These GWAS revealed a total of 44 new loci associated with height in adults (13). Several variants clustered around biological candidate genes for height. Single nucleotide polymorphisms (SNPs) in four genes (HHIP, HMGA2, ZBTB38, GDF5) were significantly associated with height in three GWAS and seven variants (EFEMP1, CDK6, GPR126, TRIP11/ATXN3, LCORL, SH3GL3/ADAMTSL3, SOC2) in at least two out of three studies (8–10). With ∼0.4 cm per allele, the average effect-sizes were rather small (14). More recently, a second sweep of GWAS reported further associations (15–17) which lead to a total of 48 validated height loci (16,18). Interestingly, a GWAS in Korean cohorts confirmed several of the known height loci that were initially found in Caucasian samples (19). Despite considerably greater power of GWAS to detect alleles affecting height, only ∼5% of the variability of height can be explained by the variants discovered so far (18). Hence, the genetics of height remains poorly understood.
The majority of reported GWAS was carried out in outbred populations. To reduce genetic heterogeneity of the study cohorts as well as the phenotypic complexity of traits such as height, self-contained populations may be extremely helpful. Populations with a different mutational and/or demographic history may permit the detection of new allelic or haplotypic associations (20,21).
Here, we report the results of a GWAS for adult height primarily in the Sorbs as the discovery set. The Sorbs are a self-contained population of Slavonic origin resident in Germany.
RESULTS
Genome wide scan for height in the Sorbian sample
We examined the distribution of test statistics and the deviation from the expected distribution under the null hypothesis of no association in a quantile–quantile plot (Fig. 1). In a genome wide scan for association of SNPs with adult height in the Sorbian sample (N = 929), 5 SNPs reached a significance level of P < 10−5 (Table 1). Two of these signals map in intronic regions of MYBPC1 (rs11110932) and CTNNA2 (rs17018086). Additionally, two variants map 101 kb 5′ upstream of ATP2B2 (rs17033062) and 45 kb 5′ upstream of ATXN1 (rs7740575), respectively. The closest gene to rs9545880 is SPRY2, which maps 1498 kb 5′ upstream of the SNP. Furthermore, we compared the results of our GWAS with published GWA data on height. Eleven of the 48 published SNPs showed consistent effects on height with P-values < 0.05 in the Sorbs (Table 2). In regions of published linkage peaks (5,7,22–24), we did not find any evidence of association with height.
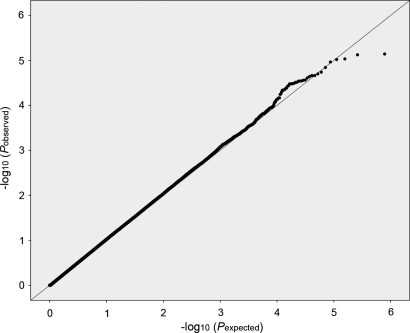
Figure 1.
Comparison of the distribution of observed test statistics with the distribution expected under the null in the quantile–quantile plot of 390 619 SNPs in the genome-wide association scan for height in the Sorbs.
Table 1.
SNPs associated with height (P < 10−5) in the genome-wide association scan in the Sorbs
| Chr | SNP | Position | A1 Sorbs | MAF Sorbs | Effect direction | P-value Sorbs | P-value DGI | P-value 58BC | Combined P-value | Nearby Genea |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | rs17033062 | 10564946 | T | 0.08 | −−− | 6.72 × 10−6 | 0.433 | 0.4215 | 0.0009 | ATP2B2 |
| 12 | rs11110932 | 100552643 | A | 0.26 | +−+ | 7.06 × 10−6 | 0.201 | 0.3686 | 0.0469 | MYBPC1 |
| 13 | rs9545880 | 81311370 | C | 0.23 | −++ | 7.11 × 10−6 | 0.785 | 0.7941 | 0.0408 | – |
| 2 | rs17018086 | 80026690 | A | 0.24 | +++ | 7.61 × 10−6 | 0.428 | 0.7293 | 0.0021 | CTNNA2 |
| 6 | rs7740575 | 16914552 | C | 0.24 | −−− | 8.17 × 10−6 | 0.040 | 0.0177 | 6.7 × 10−7 | ATXN1 |
SNPs with P < 10−5 in the genome-wide scan for association with adult height in the Sorbian sample (n = 929). P-values in the Sorbian cohort are corrected for age, gender, genomic control inflation factor (λ = 1.31) and the first four vectors of multidimensional scaling. A1 is the minor allele. +/− indicates the effect direction relative to the minor allele.
aNearby gene is defined as the closest gene to the SNP in a 200 kb window.
Table 2.
Previously replicated SNPs associated with height which show consistent effects in the Sorbian sample
| Closest genes | Chr | SNP | Position | A1 Sorbs | MAF Sorbs | Reference | Sorbs |
|
|---|---|---|---|---|---|---|---|---|
| P-value | Beta | |||||||
| DNM3 | 1 | rs4072117 (rs678962 r2 = 1) | 168940528 | C | 0.22 | (8) | 0.0823 (unadj. 0.038) | 0.6835 |
| ZNF462 | 9 | rs4743034 | 106711908 | A | 0.22 | (8) | 0.0353 | 0.7765 |
| ZBTB38 | 3 | rs6440003 | 142576907 | A | 0.42 | (10) | 0.0155 | 0.7796 |
| GDF5/UQCC | 20 | rs6060373 | 33377622 | G | 0.43 | (10) | 0.0070 | 0.8506 |
| rs6088792 | 33373198 | T | 0.27 | (8) | 0.0246 | 0.7868 | ||
| rs6060369 | 33370575 | C | 0.42 | (9) | 0.0134 | 0.7615 | ||
| LCORL | 4 | rs16896068 | 17621109 | A | 0.11 | (10) | 0.0473 | −0.9938 |
| rs6830062 | 17693999 | C | 0.11 | (8) | 0.0726 (unadj. 0.036) | −0.8894 | ||
| C6orf106 | 6 | rs2814993 | 34726871 | A | 0.16 | (10) | 0.0453 | 0.8427 |
| PTCH1 | 9 | rs10512248 | 95339258 | G | 0.35 | (10) | 0.0030 | 0.9685 |
| NOG/DGKE/ TRIM25/ COIL/RISK | 17 | rs4794665 | 52205328 | G | 0.43 | (8) | 0.0655 (unadj. 0.030) | 0.615 |
Published SNPs which show consistent effects in the Sorbian sample (n = 929). P-values in the Sorbian cohort are corrected for age, gender, genomic control inflation factor (λ = 1.31) and the first four vectors of multidimensional scaling.
Meta-analysis including Sorbian, DGI and 58BC cohorts
We filtered SNPs with a P-value < 0.001 (455 SNPs) and took them forward to a meta-analysis together with Diabetes Genetics Initiative (DGI) and 58BC. Twenty-nine SNPs showed a combined P-value of <0.001 (Tables 1 and 3 and Supplementary Material, Table S1). Our strongest associations were found for two variants in GPR133 (rs1569019 and rs1976930) (P-value at or below 10−6) and were significantly associated with height in the three cohorts individually. Since both variants were in nearly complete LD (r2 = 0.996), we selected rs1569019 for further replication in two additional independent German cohorts (Leipzig and Berlin).
Table 3.
Meta-analysis including Sorbian, DGI and 58BC cohorts for SNPs with a P-value < 0.001 in the Sorbs
| SNP | Chr | Position | A1 | Meta P-value | Z score | Direction | P-value Sorbs | P-value 58BC | P-value DGI | Nearby Genea |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1976930 | 12 | 130101325 | T | 3.37E−07 | 5.10 | +++ | 0.00049 | 0.035 | 0.001 | GPR133 |
| rs7740575 | 6 | 16914552 | C | 6.70E−07 | −4.97 | −−− | 8.17 × 10−6 | 0.018 | 0.040 | ATXN1 |
| rs1569019 | 12 | 130101071 | A | 1.02E−06 | 4.89 | +++ | 0.00075 | 0.059 | 0.002 | GPR133 |
| rs4557669 | 8 | 127375784 | C | 3.60E−06 | 4.63 | +++ | 0.00007 | 0.042 | 0.028 | – |
| rs2471943 | 8 | 127382447 | C | 8.46E−06 | 4.45 | +++ | 0.00004 | 0.031 | 0.085 | – |
| rs6984050 | 8 | 127333184 | C | 1.02E−04 | 3.89 | +++ | 0.00045 | 0.077 | 0.097 | – |
| rs13251380 | 8 | 127361613 | T | 1.04E−04 | 3.88 | +++ | 0.00047 | 0.057 | 0.121 | – |
| rs4560755 | 8 | 127361938 | A | 1.10E−04 | 3.87 | +++ | 0.00049 | 0.070 | 0.107 | – |
| rs9456801 | 6 | 162998701 | A | 1.53E−04 | −3.79 | −−− | 0.00003 | 0.093 | 0.292 | PARK2 |
| rs1542178 | 2 | 101053993 | A | 1.55E−04 | 3.78 | +++ | 0.00030 | 0.518 | 0.021 | NPAS2 |
| rs5771222 | 22 | 48748161 | C | 1.75E−04 | −3.75 | −−− | 0.00032 | 0.480 | 0.026 | FLJ41993 (IL17REL) |
| rs7450548 | 6 | 162964656 | T | 1.85E−04 | −3.74 | −−− | 0.00005 | 0.074 | 0.330 | PARK2 |
| rs4091546 | 6 | 162964604 | C | 1.89E−04 | −3.73 | −−− | 0.00004 | 0.077 | 0.350 | PARK2 |
| rs872683 | 2 | 64502097 | G | 2.39E−04 | −3.67 | −−− | 0.00017 | 0.872 | 0.016 | HSPC159 |
| rs10507349 | 13 | 25679528 | A | 3.64E−04 | 3.57 | +++ | 0.00093 | 0.115 | 0.138 | RNF6 |
| rs3908324 | 2 | 49529149 | C | 4.23E−04 | −3.53 | −+− | 0.00023 | 0.886 | 0.013 | – |
| rs817015 | 2 | 49512074 | G | 4.44E−04 | −3.51 | NA | 0.00050 | – | 0.084 | – |
| rs137866 | 22 | 48749841 | A | 4.50E−04 | −3.51 | −−− | 0.00058 | 0.624 | 0.031 | FLJ41993 (IL17REL) |
| rs152837 | 16 | 70675252 | C | 4.53E−04 | 3.51 | +++ | 0.00032 | 0.869 | 0.022 | TXNL4B |
| rs7138495 | 12 | 100517629 | G | 4.59E−04 | 3.50 | +++ | 0.00023 | 0.569 | 0.058 | MYBPC1 |
| rs371606 | 15 | 65043818 | T | 5.13E−04 | −3.47 | −+− | 0.00024 | 0.373 | 0.003 | SMAD3 |
| rs8056945 | 16 | 78319568 | T | 5.97E−04 | −3.43 | −−− | 0.00080 | 0.027 | 0.452 | MAF |
| rs7966378 | 12 | 93585744 | G | 7.03E−04 | 3.39 | +++ | 0.00038 | 0.989 | 0.024 | TMCC3 |
| rs11195417 | 10 | 112821984 | A | 8.16E−04 | −3.35 | −−− | 0.00071 | 0.424 | 0.087 | ADRA2A |
| rs6930532 | 6 | 162937507 | C | 8.36E−04 | −3.34 | −−− | 0.00062 | 0.062 | 0.407 | PARK2 |
| rs7966506 | 12 | 93585842 | T | 8.63E−04 | 3.33 | +++ | 0.00015 | 0.672 | 0.096 | TMCC3 |
| rs7966485 | 12 | 93585810 | T | 8.71E−04 | 3.33 | +++ | 0.00015 | 0.699 | 0.091 | TMCC3 |
| rs17690928 | 2 | 64520060 | A | 9.05E−04 | −3.32 | −+− | 0.00032 | 0.813 | 0.020 | HSPC159 |
| rs17033062 | 3 | 10564946 | T | 9.07E−04 | −3.32 | −−− | 6.72 × 10−6 | 0.422 | 0.433 | ATP2B2 |
| rs1795849 | 12 | 103413713 | C | 9.10E−04 | 3.32 | +++ | 0.00073 | 0.526 | 0.071 | CHST11 |
| rs11646174 | 16 | 78327565 | A | 9.93E−04 | −3.29 | −−− | 0.00088 | 0.040 | 0.490 | MAF |
SNPs with a combined P-value < 10−3 in the meta-analysis of three GWAS (Sorbs, N = 929, 58BC, N = 1490 and DGI, N = 1496). Only SNPs associated with P-values < 0.001 with adult height in the Sorbian sample were included in this analysis. P-values in the Sorbian cohort are corrected for age, gender, genomic control inflation factor (λ = 1.31) and the first four vectors of multidimensional scaling. A1 is the minor allele.
aNearby gene is defined as the closest gene to the SNP in a 200 kb window.
Association of rs1569019 with height in German cohorts (Leipzig and Berlin)
Rs1569019 was significantly associated with height in the Leipzig cohort (P = 0.004, beta = 1.166, n = 1044). In contrast, no significant effects were found in the Berlin cohort (P = 0.253, beta = 0.359, n = 1728). Upon sex-stratification, however, there was a significant association with height in men (P = 0.049, beta = 0.573, n = 577), but not in women (P = 0.965, beta = –0.017, n = 1151). The higher proportion of females in the Berlin cohort (1151 females versus 577 males) might therefore explain the lack of association in the entire cohort. The hypothesis of diverse effects in both genders was supported by the Leipzig cohort in which the effect of rs1569019 on height appeared to be stronger in males (P = 0.022, beta = 1.423, n = 524) than in females (P = 0.055, beta = 0.963, n = 520). In the Sorbs, rs1569019 showed evidence of association with height in both male (P = 0.034, beta = 1.524, n = 385) and female subjects (P = 0.005, beta = 1.67, n = 544).
Meta-analysis of rs1569019 in all five cohorts
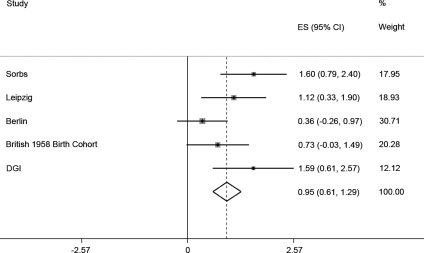
In all five cohorts (N = 6687), the estimated pooled effect size in the fixed effects model was 0.949 cm (95% CI: 0.608; 1.291), P = 4.7 × 10−8 (P = 5.87 × 10−5 in the random effects model) (Fig. 2).
Figure 2.
Forrest plot for the association of GPR133 variant rs1569019 on height in the three German cohorts (Leipzig, Sorbs, Berlin) and the DGI and British 58 Birth Cohort sample (total N = 6687). Error bars represent 95% CI.
DISCUSSION
To focus on variants with strong effects in one homogenous population might help identify loci of interest among SNPs not ranked in the top tier of a classical meta-analysis, but still of potential physiological significance. Therefore, we pre-selected SNPs only based on the Sorbian sample for subsequent meta-analysis. In our GWAS in the self-contained population of Sorbs, we could replicate several of the previously shown associations with adult height (e.g. GDF5, ZBTB38). This indicates that our cohort has sufficient power to pick up some of the signals found in larger meta-analyses and indirectly supports a possible physiological role of those variants. In the Sorbs, we were not able to show any effect of the HMGA2 locus which is currently the most consistently replicated signal associated with adult height (18). A recent study with similar sample size also failed to replicate the association, but there were still effects on bone mineral density detectable (25).
Furthermore, we showed several novel signals associated with adult height in the Sorbian population. Among them, the association was strongest for variants in GPR133 with consistent effects on height in five independent cohorts. The combined effect-size of 0.949 cm for rs1569019 was slightly higher than in the recently reported GWAS (0.2–0.6 cm) (8–10). However, in the individual populations, the SNPs showed effects up to 1.47 cm (8).
GPR133 encodes a G protein-coupled receptor (GPCR) and represents a plausible physiological candidate potentially regulating height. GPCRs recognize a variety of extracellular messenger molecules such as hormones, neurotransmitters, growth and developmental factors as well as sensory messages such as light, odors and pain (26). Two functional splice variants of GPR133 were found in fetal tissue, lung, spleen and testis (27). Interestingly, variants in another GPCR, GPR126, were previously reported to be associated with height (8,9). Moreover, associations with variants in GPR126 with trunk length were recently shown (15). It is also worth-mentioning that GPCRs are involved in osteoclast function and regulation of bone mineral density and cell growth (28–30).
In conclusion, despite certain limitations due to the small sample size, our GWAS suggests novel loci influencing height. In view of the robust replication in five different cohorts, we propose GPR133 to be a novel gene associated with adult height.
MATERIAL AND METHODS
Subjects and phenotyping
Sorbs
All subjects are part of a sample from an extensively phenotyped isolated population from Eastern Germany, the Sorbs. The Sorbs are of Slavonic origin, and lived in ethnic isolation among the Germanic majority during the past 1100 years. Today, the Sorbian-speaking, Catholic minority comprises approximately 15 000 full-blooded Sorbs resident in about 10 villages in rural Upper Lusatia (Oberlausitz), Eastern Saxony. At present, about 1000 Sorbian individuals are enrolled in the study. Sampling comprised unrelated subjects as well as families. A total of 929 subjects (544 females and 385 males) were available for the present study. Females had a mean age of 45.1 (43.6; 46.5) years, mean body mass index (BMI) of 26.4 (26.0; 26.9) kg/m2 and mean height of 163.8 (163.2; 164.4) cm (data are geometric means and 95% CI). Males had a mean age of 44.3 (42.6; 46.1) years, mean BMI of 26.8 (26.4; 27.2) kg/m2 and mean height of 177.1 (176.4; 177.8) cm.
Leipzig Cohort
A total of 1044 subjects were recruited at the University Hospital in Leipzig, Germany as a control group for diabetes studies. The subjects included 524 males and 520 females [mean age 58.2 ± 14.7 years; mean male height 176.1 (175.5; 176.7) cm; mean female height 162.8 (162.3; 163.2) cm; mean BMI 28.08 (27.66; 28.51) kg/m2].
For all log-normally distributed parameters, the data represent back-transformed geometric means and 95% CI and for all normally distributed parameters data are given as arithmetic means ± SD.
The study was approved by the ethics committee of the University of Leipzig and all subjects gave written informed consent before taking part in the study.
Berlin Cohort
A total of 1728 Caucasian individuals were included in the Metabolic Syndrome Berlin Potsdam (MesyBepo) study population from the region of Berlin/Potsdam, Germany; 577 male and 1151 females were investigated (mean male age 53.7 ± 51.9 years; mean female age 51.9 ± 13.3 years; mean male height 176 ± 8 cm; mean female height 165 ± 7 cm; mean male BMI 29 ± 5.5 kg/m2; mean female BMI 29.3 ± 6.5 kg/m2]. Data are given as arithmetic means ± SD. The study has been approved by the responsible authorities, which were the ethics committees of the University of Potsdam and the Charité-Universitätsmedizin Berlin. All subjects provided written informed consent.
DNA extraction, genotyping and SNP selection in the sorbs
Genomic DNA was extracted using QIAmp DNA Blood Midi Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer's protocol. Genotyping was performed using the 500K Affymetrix GeneChip and the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc.) by the Microarray Core Facility of the Interdisciplinary Centre for Clinical Research, University of Leipzig, Germany and by ATLAS Biolabs GmbH, Berlin, Germany. Genotypes were determined with GeneChip Genotyping Analysis Software (GTYPE) using the BRLMM algorithm for the 500K arrays and the Birdseed Algorithm for Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc.). Data underwent quality control and only SNPs fulfilling the following criteria were included: missing rate per SNP <5%, Hardy–Weinberg equilibrium (HWE) P > 0.0001, minor allele frequency (MAF) >0.01. The average genotyping rate was 99.0%. In all, 390 619 autosomal markers overlapping between the 500K Affymetrix GeneChip and the Affymetrix Genome-Wide Human SNP Array 6.0 were included in the analyses.
Statistical methods and software
Call rates per sample and cluster-plots were checked using the GeneChip Genotyping Analysis Software (GTYPE). The calculation of minor allele frequencies, HWE and missing rates per SNP was performed with PLINK (31). Pairwise IBD was calculated using–genome command in PLINK. Supplementary Material, Figure S1 provides a histogram for the distribution of pi-hat values in the pairwise comparisons. Mean IDB sharing in the Sorbian sample was 0.008 with a median < 10−6 (25th percentile < 10−6, 75th percentile 0.012). Nine hundred and seventeen out of 461 023 pairwise comparisons showed a pi_hat ≥ 0.25. We did not exclude first and second degree relatives but adjusted for genomic control. Our QQ-plot shows that we are able to correct for the inflation adequately since the observed P-values meet the observed line nearly until the top tier (Supplementary Material, Fig. S2). Genome-wide association with height was assessed by linear regression in PLINK. We corrected for age and gender, for relatedness by using genomic control (λ = 1.31) and for possible population substructure by the first four vectors of multiple dimensional scaling. Linkage disequilibrium metrics were calculated in Haploview 4.1 (32). Power calculations were carried out with Quanto (33,34).
Weighted meta-analysis for the three GWAS cohorts was performed using METAL (http://www.sph.umich.edu/csg/abecasis/metal/). Study specific P-values and effect directions were converted into a Z-statistics and weighted with sample size of each study.
Meta-analysis of rs1569019 in all five cohorts was performed in a fixed and random effect model by using the Mantel–Haenszel method based on the study specific beta estimates with STATA (version 9.0, StataCorp LP, TX, USA) (35). Other statistical analyses were performed using SPSS version 15.0.1 (SPSS, Inc.; Chicago, IL, USA).
GWAS cohorts for meta-analysis
We included data of 1496 control subjects from the DGI and additionally, we used the results of the GWA for adult height (n∼1490) performed by Dr Panos Deloukas for the Wellcome Trust Sanger Institute and published online from the British 1958 Birth Cohort DNA Collection (http://www.b58cgene.sgul.ac.uk/index.php). Both cohorts were genotyped using Affymetrix GeneChip Mapping 500K Array.
Genotyping of rs1569019 in GPR133 for replication
Genotyping of the SNPs rs1569019 in GPR133 selected for replication in subjects of two independent cohorts of German origin (Leipzig and Berlin) was performed using the TaqMan assay (Applied Biosystems, Inc.). Oligonucleotide sequences are available upon request. The TaqMan genotyping reaction was performed according to the manufacturer's protocol on an ABI PRISM 7500 sequence detector (Applied Biosystems Inc.).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by grants from the Interdisciplinary Centre for Clinical Research at the University of Leipzig (B27 to M.S., P.K. and A.T.; N06 to P.K., B.E. K.D. and D.S., Z03 to K.K.), from the German Diabetes Association (to Y.B., A.T., P.K.) and a Travel Grant from BIF (to A.T.). M.S. is supported by a grant from the DFG (KFO 152). J.S. was supported by a research group of the BMBF (Molecular Nutrition), a Heisenberg-Professorship (SP716/2-1) of the Deutsche Forschungsgemeinschaft (DFG) and a graduate school of the DFG (GK1208). The British 1958 Birth Cohort DNA collection was funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
ACKNOWLEDGEMENTS
We thank all those who participated in the studies. We especially appreciate the support by John Broxholm from the Bioinformatics Core Unit of the Wellcome Trust Centre for Human Genetics and Andre Rothe from the Coordination Centre for Clinical Trials, University of Leipzig. Furthermore, we would like to thank Claudia Vogel and Michael Babilon from the Research Laboratories and Clinic of Visceral, Transplantation, Thoracic and Vascular Surgery. We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection.
Conflict of Interest statement. The authors declare they have no conflicts of interest.
REFERENCES
- 1.Silventoinen K., Sammalisto S., Perola M., Boomsma D.I., Cornes B.K., Davis C., Dunkel L., De Lange M., Harris J.R., Hjelmborg J.V., et al. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin. Res. 2003;6:399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- 2.Visscher P.M., Medland S.E., Ferreira M.A., Morley K.I., Zhu G., Cornes B.K., Montgomery G.W., Martin N.G. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2:e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mamada M., Yorifuji T., Yorifuji J., Kurokawa K., Kawai M., Momoi T., Nakahata T. Fibrillin I gene polymorphism is associated with tall stature of normal individuals. Hum. Genet. 2007;120:733–735. doi: 10.1007/s00439-006-0263-5. [DOI] [PubMed] [Google Scholar]
- 4.Yang T.L., Xiong D.H., Guo Y., Recker R.R., Deng H.W. Comprehensive association analyses of IGF1, ESR2 and CYP17 genes with adult height in Caucasians. Eur. J. Hum. Genet. 2008;11:1380–1387. doi: 10.1038/ejhg.2008.113. [DOI] [PubMed] [Google Scholar]
- 5.Palmert M.R., Hirschhorn J.N. Genetic approaches to stature, pubertal timing, and other complex traits. Mol. Genet. Metab. 2003;80:1–10. doi: 10.1016/s1096-7192(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 6.Xiong D.H., Xu F.H., Liu P.Y., Shen H., Long J.R., Elze L., Recker R.R., Deng H.W. Vitamin D receptor gene polymorphisms are linked to and associated with adult height. J. Med. Genet. 2005;42:228–234. doi: 10.1136/jmg.2004.024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perola M., Sammalisto S., Hiekkalinna T., Martin N.G., Visscher P.M., Montgomery G.W., Benyamin B., Harris J.R., Boomsma D., Willemsen G., et al. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3:e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 9.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanna S., Jackson A.U., Nagaraja R., Willer C.J., Chen W.M., Bonnycastle L.L., Shen H., Timpson N., Lettre G., Usala G., et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weedon M.N., Lettre G., Freathy R.M., Lindgren C.M., Voight B.F., Perry J.R., Elliott K.S., Hackett R., Guiducci C., Shields B., et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weedon M.N., Frayling T.M. Reaching new heights: insights into the genetics of human stature. Trends Genet. 2008;24:595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Visscher P.M. Sizing up human height variation. Nat. Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 15.Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J.B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrada K., Krawczak M., Schreiber S., van Duijn K., Stolk L., van Meurs J.B., Liu F., Penninx B.W., Smit J.H., Vogelzangs N., et al. A genome-wide association study of northwestern Europeans involves the CNP signaling pathway in the etiology of human height variation. Hum. Mol. Genet. 2009;18:3516–3524. doi: 10.1093/hmg/ddp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson A., Marroni F., Hayward C., Franklin C.S., Kirichenko A.V., Jonasson I., Hicks A.A., Vitart V., Isaacs A., Axenovich T., et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet. 2009;18:373–380. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lettre G. Genetic regulation of adult stature. Curr. Opin. Pediatr. 2009;21:515–522. doi: 10.1097/MOP.0b013e32832c6dce. [DOI] [PubMed] [Google Scholar]
- 19.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M., et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 20.Peltonen L., Palotie A., Lange K. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 21.Wright A.F., Carothers A.D., Pirastu M. Population choice in mapping genes for complex diseases. Nat. Genet. 1999;23:397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- 22.Dempfle A., Wudy S.A., Saar K., Hagemann S., Friedel S., Scherag A., Berthold L.D., Alzen G., Gortner L., Blum W.F., et al. Evidence for involvement of the vitamin D receptor gene in idiopathic short stature via a genome-wide linkage study and subsequent association studies. Hum. Mol. Genet. 2006;15:2772–2783. doi: 10.1093/hmg/ddl218. [DOI] [PubMed] [Google Scholar]
- 23.Hirschhorn J.N., Lindgren C.M., Daly M.J., Kirby A., Schaffner S.F., Burtt N.P., Altshuler D., Parker A., Rioux J.D., Platko J., et al. Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am. J. Hum. Genet. 2001;69:106–116. doi: 10.1086/321287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sammalisto S., Hiekkalinna T., Schwander K., Kardia S., Weder A.B., Rodriguez B.L., Doria A., Kelly J.A., Bruner G.R., Harley J.B., et al. Genome-wide linkage screen for stature and body mass index in 3.032 families: evidence for sex- and population-specific genetic effects. Eur. J. Hum. Genet. 2008;17:258–266. doi: 10.1038/ejhg.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuipers A., Zhang Y., Cauley J.A., Nestlerode C.S., Chu Y., Bunker C.H., Patrick A.L., Wheeler V.W., Hoffman A.R., Orwoll E.S., et al. Association of a high mobility group gene (HMGA2) variant with bone mineral density. Bone. 2009;45:295–300. doi: 10.1016/j.bone.2009.04.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bjarnadottir T.K., Fredriksson R., Hoglund P.J., Gloriam D.E., Lagerstrom M.C., Schioth H.B. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics. 2004;84:23–33. doi: 10.1016/j.ygeno.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Bjarnadottir T.K., Geirardsdottir K., Ingemansson M., Mirza M.A., Fredriksson R., Schioth H.B. Identification of novel splice variants of Adhesion G protein-coupled receptors. Gene. 2007;387:38–48. doi: 10.1016/j.gene.2006.07.039. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao E.C., Boudignon B.M., Chang W.C., Bencsik M., Peng J., Nguyen T.D., Manalac C., Halloran B.P., Conklin B.R., Nissenson R.A. Osteoblast expression of an engineered Gs-coupled receptor dramatically increases bone mass. Proc. Natl Acad. Sci. USA. 2008;105:1209–1214. doi: 10.1073/pnas.0707457105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng J., Bencsik M., Louie A., Lu W., Millard S., Nguyen P., Burghardt A., Majumdar S., Wronski T.J., Halloran B., et al. Conditional expression of a Gi-coupled receptor in osteoblasts results in trabecular osteopenia. Endocrinology. 2008;149:1329–1337. doi: 10.1210/en.2007-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorsam R.T., Gutkind J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Gauderman W.J. Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 34.Gauderman W.J. Sample size requirements for matched case-control studies of gene-environment interaction. Stat. Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 35.Kuritz S.J., Landis J.R., Koch G.G. A general overview of Mantel-Haenszel methods: applications and recent developments. Annu. Rev. Public Health. 1988;9:123–160. doi: 10.1146/annurev.pu.09.050188.001011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.