Abstract
Proline-rich sequences (PRS) and their recognition domains have emerged as transposable protein interaction modules during eukaryotic evolution. They are especially abundant in proteins associated with pre-mRNA splicing and likely assist in the formation of the spliceosome by binding to GYF and WW domains. Here we profile PRS-mediated interactions of the CD2BP2/52K GYF domain by a site-specific peptide inhibitor and stable isotope labeling/mass spectrometry analysis. Several PRS hubs with multiple proline-rich motifs exist that can recruit GYF and/or WW domains. Saturating the PRS sites by an isolated GYF domain inhibited splicing at the level of A complex formation. The interactions mediated by PRS are therefore important to the early phases of spliceosomal assembly.
The two catalytic steps of splicing are a conceptually simple process that results in the RNA-mediated excision of introns (1). Complexity arises at the level of RNA-RNA and protein-RNA interactions that are needed for the correct spatial positioning of the critical bases, unwinding of RNA, and coupling of splicing to other processes as for example transcription (2,3), nonsense-mediated mRNA decay (4), or mRNA transport (5, 6). Pre-mRNA splicing is catalyzed by the spliceosome, a dynamic macromolecular machine, which consists of the small nuclear ribonuclear particles (snRNPs),1. U1, U2, U4/U6, and U5, and numerous non-snRNP proteins (7). The snRNPs interact with the intron in an ordered manner. First the U1 snRNP binds to the 5′ splice site, whereas the U2 snRNP stably associates with the branch site forming complex A. Subsequently the tri-snRNP U4/U6.U5 is stably integrated to form complex B. This complex undergoes large structural rearrangements that lead to the formation of the activated spliceosomal complex B*. The catalytic steps of the transesterification then occur in complex C.
Proline-rich sequences (PRS) are frequently found in spliceosomal proteins, and they are mostly assigned to unstructured regions of the corresponding full-length proteins (8). They serve as docking sites for so-called proline-rich sequence recognition domains (PRDs), and their interactions are characterized by low affinities and moderate specificities (9–13). Several of the interactions between PRD and their proline-rich binding sites within the spliceosome have been characterized in vitro in great detail, but little is known about their functional importance. One interesting spliceosomal protein that mediates PRS interactions is CD2BP2/52K (14). It is a component of the U5 snRNP (15, 16) and contains a GYF adaptor domain that recognizes PRS via a set of conserved aromatic amino acids (17–19). A likely interaction partner of the GYF domain is the core splicing protein SmB/B′, which is present in all snRNPs. The GYF domain of CD2BP2/52K comprises a second interaction surface on a site opposite to the PRS binding epitope. This site is bound by the essential splicing protein U5-15K (16, 20). Considering that the U5 snRNP protein Prp6 interacts with the N-terminal part of CD2BP2/52K (16), several independent interactions seemingly contribute to the association of the protein with the U5 snRNP. However, for the GYF domain we could identify by phage display and peptide SPOT analysis additional interaction sites in other proteins suggesting that further “moonlighting” functions of the GYF domain might exist (21, 22). This has set the basis for the current study where we performed pulldown experiments to define more generally the importance of CD2BP2/52K-GYF for the assembly of protein complexes. Utilizing the GYF domain fused to GST as bait, a large number of protein components of the U1, U2, U5, and the U4/U6.U5 tri-snRNP were co-precipitated. The association of most proteins was highly dependent on the PRS binding site in the GYF domain as mutations at this site or competition with high concentrations of a peptide inhibitor prevented their co-precipitation. Pulldown experiments with a GST-tagged U5-15K as bait in the presence and in the absence of a PRS inhibitor provided insight into PRS-dependent complex assembly of endogenous full-length CD2BP2/52K. Among the proteins identified in all experiments were SmB/B′ and NpwBP (Npw38BP/WBP-11). These proteins are known in vitro interaction partners for CD2BP2/52K-GYF, and they comprise several PRS binding motifs that are also recognized by the WW domains of FBP21 and PQBP-1 (Npw38) (21–25), respectively. We found that CD2BP2/52K-GYF and FBP21-WWs compete for the same PRS in SmB/B′ in vitro and that both are likely to interact with SmB/B′ in vivo based on fluorescence resonance energy transfer (FRET) experiments. We suggest that PRD-PRS networks within the spliceosome are important for the dynamic rearrangements occurring during spliceosomal maturation. In support of this model, addition of CD2BP2/52K GYF domain to nuclear extracts inhibited in vitro splicing of reporter pre-mRNA by blocking complex A formation.
EXPERIMENTAL PROCEDURES
Constructs
Cloning of the GST fusion construct of CD2BP2-GYF has been described previously (22). To disrupt the PRS binding site, Trp287 (GYF domain numbering, 8) was replaced by Arg, and Ala was substituted for Tyr at position 312 (GYF domain numbering, 33) using a standard PCR-based mutagenesis strategy. For the FBP21-WWs construct, a fragment encoding residues 90–219 was amplified by PCR and cloned into pGEX4T-1 via BamHI and NotI restriction sites. For the CFP- and YFP-tagged constructs, DNA fragments encoding the FBP21 residues 90–219, full-length CD2BP2, and SmB were amplified by PCR and cloned into a modified pcDNA3.1 vector using EcoRI and NotI restriction sites. A Kozak sequence was included into the 5′ primers for efficient expression. The modified pcDNA3.1 vector comprised fluorescent protein tags for either N- or C-terminal fusion (26).
Protein Preparation
GST fusion proteins of FBP21-WWs, wild-type and mutant GYF domains, and the U5-15K protein were expressed in BL21 (DE3) and harvested after 3 h of isopropyl 1-thio-β-d-galactopyranoside-induced growth at 37 °C. Pure proteins were obtained by glutathione-Sepharose and gel filtration chromatography. Proteins for the NMR experiments were purified as His- or GST-tagged constructs, and the respective tag was removed by thrombin cleavage prior to the gel filtration step. Details of the protocols were published previously (21).
NMR Spectroscopy
NMR spectroscopy was performed at 297 K on a Bruker DRX600 spectrometer equipped with a standard triple resonance probe. For the GYF epitope mapping experiments, increasing amounts of SmB peptide (SmB-2, GTPMGMPPPGMRPPPPGMRGLL) and/or full-length U5-15K were added to a solution containing 0.2 mm 15N-labeled GYF domain. Mean chemical shift differences of 15N and 1H were determined as described previously (22).
Cellular Imaging and FRET Analysis
HEK293T cells were transfected with the indicated constructs (cloned into a pcDNA3 vector backbone) using a FuGENE 6 reagent (Roche Applied Science) following the manufacturer’s recommendations. One day after transfection, live cells were imaged with an inverted confocal laser scanning microscope (LSM510-META, Carl Zeiss) and an α-Plan Fluar 100/1.45 objective. Photobleaching regions (about 0.6-μm diameter) were defined over the nucleus, and intensities were recorded in visual fields covering 7.5 × 2.5 μm2 at 25-ms intervals upon local photobleaching. FRET analyses were done with a monochromator-assisted digital epifluorescence video imaging setup (TILL-Photonics, Gräfelfing, Germany) equipped with a motorized emission filter wheel (Lambda 10/2, Sutter Instrument Co.). Determination of FRET efficiencies was performed as described earlier (27).
Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and Pulldown Experiments
SILAC was performed as described (28). In brief, cells were cultured in defined SILAC Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with lysine, arginine, and 10% (v/v) dialyzed FCS. Labeled SILAC medium was prepared by adding stable isotope-labeled l-[13C6]lysine and l-[13C6]arginine (Cambridge Isotope Laboratories, Andover, MA); for unlabeled medium normal 12C-amino acids were used. Cells were cultured for 8 days in SILAC media. After washing three times with ice-cold PBS, cells were lysed in lysis buffer (PBS with 1% Nonidet P-40, 5 mm EDTA, 1 mm DTT, 1 mm PMSF, 1 μm pepstatin A, 2 mm sodium orthovanadate, and two tablets of protease inhibitor mixture (Roche Applied Science) per 15 ml) for 30 min on ice, and cell debris were removed by centrifugation for 20 min at 16,000 × g at 4 °C. Supernatants were snap frozen and stored at −80 °C. Glutathione beads were loaded with 5 mg of GST fusion protein/ml of matrix and washed four times with ice-cold PBS. 25 μl of the protein-loaded slurry were resuspended with an equal volume of PBS and incubated with 1.2 ml of cell lysate overnight at 4 °C. For competitive inhibition, the peptide PD1 (EFGPPPGWLGR) was added to a final concentration of 2 mm. Subsequently the GST beads were centrifuged for 5 min at 500 × g at 4 °C, supernatant was removed, and the beads were washed three times with 1 ml of cold PBS. Beads were resuspended in 1 ml of cold PBS and combined by transferring 525 μl of the corresponding labeled sample to the unlabeled sample. For elution, loading dye was added, and the sample was boiled for 5 min at 95 °C. Samples were then loaded onto Tris-glycine gradient gels (4–20%, Invitrogen), and PAGE was performed under standard conditions. For MS analysis, the Coomassie-stained gel lane was cut into slices, and proteins were in-gel digested with trypsin as described (29).
Western Blot
Pulldown experiments with unlabeled lysates were performed as described above. Competitive PD1 peptide or a control peptide (FGPDLPAGD) were added to the lysates to a final concentration of 2 mm. After PAGE, proteins were transferred onto a polyvinylidene difluoride membrane. Blots were probed with an anti-SmB/B′ antibody (sc-5485, Santa Cruz Biotechnology) or PQBP-1 antibody (sc-26052, Santa Cruz Biotechnology) and a horseradish peroxidase-conjugated secondary antibody. The immunoblots were then developed with an enhanced chemiluminescence substrate (SuperSignal West Pico, Pierce) and detected by a LumiImager™ (Roche Applied Science).
Protein Identification by Mass Spectrometry
Identification and quantitation of proteins were performed on a quadrupole orthogonal acceleration time-of-flight mass spectrometer (Q-Tof Ultima, Micromass, Manchester, UK). A Micromass CapLC liquid chromatography system (Waters GmbH) was used to deliver the peptide solution to the electrospray source. 5 μl of the sample were injected and concentrated on a precolumn (PepMap C18, 5 μm, 100 Å, 5 mm × 300-μm inner diameter, Dionex, Idstein, Germany). LC separations were performed on a capillary column (Atlantis dC18, 3 μm, 100 Å, 150 mm × 75-μm inner diameter, Waters GmbH) at an eluent flow rate of 200 nl/min using a linear gradient of 3–64% B in 100 min. Mobile phase A was 0.1% formic acid (v/v) in acetonitrile-water (3:97, v/v), and B was 0.1% formic acid in acetonitrile-water (8:2, v/v). MS/MS data were acquired in a data-dependent mode (survey scanning) using one MS scan followed by MS/MS scans of the most abundant peak. To select either the [12C]Arg/Lys or the [13C]Arg/Lys signal of a tryptic peptide for MS/MS triggered by signal intensities, the adduct mass option (+6 Da) of the dynamic peak exclusion (over a time of 240 s) was used. The MS survey range was m/z 300–1550, and the MS/MS range was m/z 100–1990. Data analysis was performed by use of MassLynx version 4.0 software (Micromass-Waters).
The processed MS/MS spectra and MASCOT server (version 2.0, Matrix Science Ltd., London, UK) were used to search in-house against the UniProt/Swiss-Prot database (release 50.0 of September 21, 2006, containing 234,112 sequence entries, comprising 85,963,701 amino acids). Finally all protein identities were updated using the latest UniProtKB/Swiss-Prot release (release 56.7 of January 20, 2009, containing 408,099 sequence entries, comprising 147,085,246 amino acids). A maximum of two missed cleavages was allowed, and the mass tolerance of precursor and sequence ions was set to 100 ppm and 0.1 Da, respectively. Acrylamide modification of cysteine, methionine oxidation, [13C6]arginine, and [13C6]lysine were considered as possible modifications. A protein was accepted as identified if the total MASCOT score was greater than the significance threshold and at least two peptides appeared the first time in the report and were the first ranking peptides.
For quantitative analysis, ratios between the number of identified 13C-labeled and unlabeled forms of tryptic peptides were calculated for each protein. The proteins were categorized as labeled (more than 75% of the identified peptides were labeled), unlabeled (all identified peptides were unlabeled), and mixed (more than 25% of the identified peptides were unlabeled). Thus, the combined use of the dynamic peak exclusion and the preset 2:1 ratio (unlabeled:labeled) assured that proteins classified as labeled are highly enriched at least by a factor of 3. Unlabeled proteins showed association that was independent of the PRS recognition site or unspecific binding. For selected proteins, quantification was carried out by MSQUANT (Peter Mortensen and Matthias Mann) open source software and was based on calculations of isotope intensity ratios of at least two arginine- or lysine-containing tryptic peptides with individual MASCOT score indicating identity. An additional criterion was that no interfering mass peaks were observed. Relative protein ratios were calculated by averaging. Standard deviations of the quantification for individual proteins were obtained from MSQUANT. Analytical reproducibility was determined by multiple LC-MS measurements of selected samples showing that the experiment-to-experiment deviation of 13C:12C ratios is less than 25%.
Nuclear Extract Preparation, in Vitro Splicing and Gel Analysis
The preparation of nuclear extracts from HeLa S3 cells has been described (30). In vitro splicing of RNAs body-labeled with [32P]-UTP was performed in 50-μl reactions with either AdML-M3 or β-globin pre-mRNAs under conditions described elsewhere (30). Recombinant GYF domain was added to in vitro splicing reactions and incubated under splicing conditions for 15 minutes before the addition of exogenous pre-mRNA transcripts. After resolution on a 10% acrylamide 7 M area denaturing gel or a non-denaturing 1.5% agarose gel in 0.5× TBE at room temperature, gels were fixed in 40% methanol/10% acetic acid for 30 min. Agarose gels were sub-sequently dried at 65 °C for 1 h. Analysis of radio-labeled RNA species recovered from in vitro splicing reactions was done using a Typhoon 8600 Phosphoimager.
RESULTS
Proteomics Analysis of GYF Domain-mediated Complexes
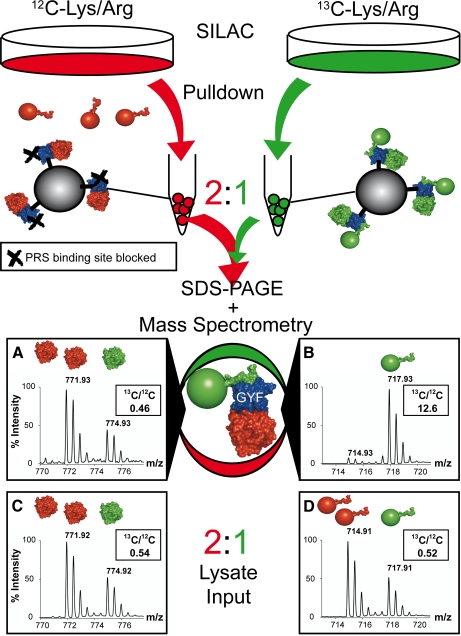
Pulldown experiments in the presence of protein interaction inhibitors provide a powerful tool to assess the contribution of individual interactions to complex formation. We combined this approach with SILAC (31). Fig. 1 summarizes the individual steps of the experimental setup. (i) Cells were cultured in growth media supplemented with either [12C]Arg/Lys (unlabeled) or [13C]Arg/Lys (labeled) and lysed. (ii) Pulldown experiments were then performed with these lysates and GST-GYF as bait. Labeled lysate was used in combination with wild-type GYF domain, whereas the unlabeled lysate was incubated either with a mutated GYF domain devoid of its PRS binding function (supplemental Fig. S1 ) or with GYF domain in the presence of a high affinity GYF domain peptide inhibitor (PD1) derived from phage display. (iii) Precipitated proteins were combined in a 2:1 (unlabeled:labeled) ratio, and (iv) the complex protein mixtures were separated by one-dimensional SDS-PAGE followed by in-gel tryptic digestion and nano-LC-MS analysis. MS/MS spectra of tryptic peptides were used to identify proteins. The relative peak intensities of the corresponding 13C- and 12C-containing peptides in the MS spectra allowed the calculation of their relative abundance as exemplified in Fig. 1. The preset 2:1 ratio (unlabeled:labeled) assured the detection of unlabeled peptides as a default (Fig. 1). Using this approach, dominance of labeled peptides was indicative of proteins that were bound by the PRS site, whereas detection of unlabeled peptides indicated an association independent of the GYF domain PRS recognition site including unspecific binders. Similar experiments were performed using GST-tagged U5-15K as bait. U5-15K is a U5 snRNP-specific protein and binds endogenous CD2BP2/52K via a second site on the GYF domain (16, 20) that is independent of the PRS binding epitope (supplemental Fig. S2). Pulldown experiments with unlabeled and labeled lysate in the presence and absence of the competing peptide, respectively, and subsequent MS analysis were carried out as described above. The use of U5-15K as bait allowed an estimation of GYF-PRS interactions in the context of full-length CD2BP2/52K bound to U5-15K.
Fig. 1.
Experimental outline for complex analysis. Separate pulldown experiments with GST-GYF fusion proteins and unlabeled (red) or 13C-labeled (green) cell lysates were performed. For the unlabeled cell lysates, the PRS binding site of the GYF domain was blocked either by mutation (W8R/Y33A mutant) or by a competing PRS peptide (2 mm). After washing, GST-GYF and associated complexes from both experiments were mixed in a ratio of 2:1 (unlabeled:labeled), separated by SDS-PAGE, and digested with trypsin. In the subsequent MS/MS analysis, protein complexes associated via the PRS binding site of the GYF domain were 13C-labeled (spectrum B, peptide 150GTIEILSDVQLIK162 of the 60 S ribosomal protein P0), whereas protein complexes bound unspecifically via the U5-15K site to the GYF domain were present in a 2:1 ratio (unlabeled:labeled; spectrum A, peptide 49GVDEVTIVNILTNR62 of annexin A2) and therefore were recognized as unlabeled in the later data analysis. Spectra C and D represent the peptides in spectra A and B, respectively, detected in a 2:1 (unlabeled:labeled) mixture of the input lysates for the pulldown experiment.
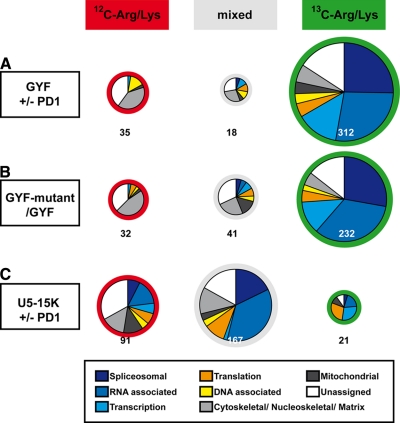
Fig. 2 gives an overview of the proteins found in the pulldown experiments. Proteins are grouped according to the isotopic state of their identified peptides. [12C]Arg/Lys “unlabeled” proteins (red) were exclusively identified by peptides containing [12C]Arg or [12C]Lys residues. For [13C]Arg/Lys “labeled” (green) proteins more than 75% of the peptides comprised [13C]Arg or [13C]Lys (see “Experimental Procedures” for details). An intermediate fraction of peptides with 13C isotopic Arg or Lys were found for “mixed” (gray) proteins. The Gene Ontology (GO) term specifications/functional annotations for the proteins of each group are presented as pie diagrams. The sets of proteins found using either competing peptide ligand (Fig. 2A) or point mutations (Fig. 2B) to block PRS binding of the GST-GYF construct compare well with each other. Spliceosomal proteins of the hPRP19 and the snRNP complexes are listed in Table I; the complete lists of proteins identified are given in supplemental Tables S1 and S2. Most of the proteins found in both experiments had the same isotopic classification (Table I and supplemental Tables S1 and S2), confirming the efficiency of the peptide-mediated inhibition of the GYF domain. The similarity of both pulldown results is also apparent in the GO term annotations (pie diagrams). More than 75% of the identified proteins were classified as labeled ([13C]Arg/Lys), which indicates PRS-dependent binding. 67% of these proteins are linked directly or indirectly to splicing (GO term annotation, spliceosomal, RNA-, or transcription-associated proteins). They encompass most of the U5 snRNP proteins and a large portion of the other snRNP components (Table I). 26% of the labeled proteins comprised potential GYF domain binding sites, including all previously identified interaction sites. In the unlabeled fraction, spliceosomal proteins did not exceed 4%, and less than 6% comprise a potential GYF domain interaction site. These results are in agreement with earlier studies describing CD2BP2/52K as a U5 snRNP component (16, 32, 33) and with our in vitro binding data (21, 22).
Fig. 2.
Classification of proteins pulldown experiments. Co-precipitated proteins from pulldown experiments were identified by mass spectrometry and classified according to their isotope label and their GO term description (as described under “Experimental Procedures”). A, results from the experiment using GST-GYF as bait in the presence or absence of inhibitory peptide PD1. B, results from pulldowns using GST-GYF and a GYF mutant, which blocks PRS binding, as bait. C, GST-U5-15K was used as bait in the absence and presence of the PD1 peptide. The size of the pie diagrams is proportional to the number (given below) of proteins they comprise.
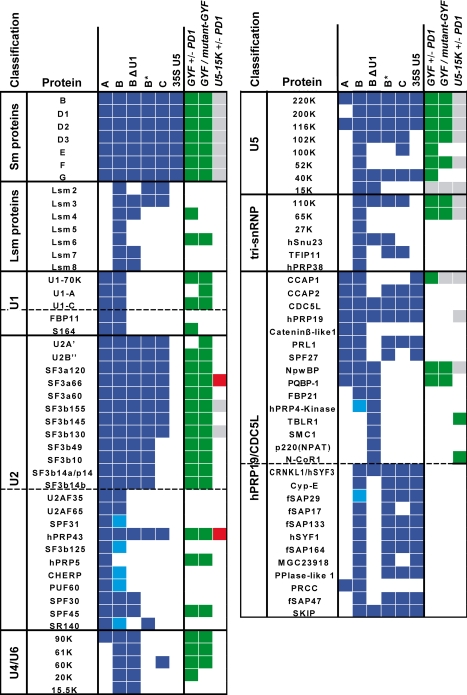
Table I. Proteins found in the pulldown experiments compared with known spliceosomal complexes.
The presence of proteins in the spliceosomal complexes A (33), B (32), BΔU1, B*, C and 35 S U5 (47) is indicated by blue squares. Light blue squares represent proteins only found once in four preparations of the spliceosomal B complex (31). Proteins identified by our pulldown experiments using GST-GYF in the presence or absence of PD1 peptide (GYF +/− PD1) using a combination of GST-tagged WT and mutant GYF domain (GYF/mutant-GYF) or using GST-tagged U5-15K in the presence or absence of PD1 peptide (U5-15K +/− PD1) are indicated in the last three columns. Green squares represent labeled proteins, red squares represent unlabeled proteins, and gray squares represent a mixed state. SmB could not be identified in pulldowns with GST-GYF according to the selection criteria. Additional evidence for selective binding from other experiments (Fig. 4C) led to the classification of SmB protein as labeled, and it is represented by green, hatched squares. Proteins are grouped according to subcomplexes. Hashed lines separate true components (upper part) from complex-related proteins (lower part).
The set of spliceosomal proteins (found to be associated in a PRS-dependent manner) could be explained by the precipitation of entire spliceosomes. Alternatively multiple distinct subcomplexes could be co-precipitated in parallel. The distribution of PRS within the spliceosome would support such a scenario (supplemental Table S1). For example, each snRNP complex, except for the U6 snRNP, comprises SmB/B′, which has multiple GYF binding motifs at its C terminus. Although SmB/B′ was identified by only a single peptide and only in the pulldown experiments with peptide inhibitor, Western blots (Fig. 4C) reconfirmed the PRS-dependent association with the isolated CD2BP2/52K GYF domain. In addition to SmB/B′, multiple in vitro interaction sites are present in the U1 snRNP proteins U1-A and U1-C; proteins of the U2 snRNP subcomplex SF3B, namely SF3a120, SF3a66, SF3b145, and SF3b49; the U5 snRNP-associated protein PSF; and other spliceosomal factors, such as NpwBP of the hPrp19-CDC5 complex (supplemental Table S3).
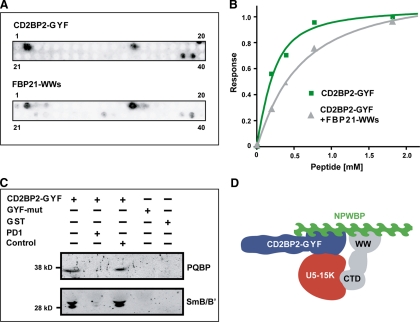
Fig. 4.
Convergence on PRS hubs by GYF and WW domain-containing proteins. A, probing GST-GYF and FBP21 WW domains for binding to a set of PRS of various origin by SPOT analysis. Complete sequences are listed in supplemental Table S3. B, NMR titration of SmB-2 peptide (NH2-GTPMGMPPPGMRPPPPGMRGLL-COOH) to 15N-labeled CD2BP2-GYF in the absence (green) or presence (gray) of FBP21 WW domains. C, purified GST fusion proteins of wild-type CD2BP2-GYF or the mutant W8R/Y33A (mut), which is devoid of PRS binding, or GST alone were incubated with cellular lysates in the presence of 2 mm peptide PD1 (EFGPPPGWLGR) or the unrelated control peptide (FGPDLPAGD) as indicated and probed for the interaction with PQBP-1 and SmB/B′ by monoclonal antibodies. D, model of the network of interactions that can be assumed from our data and the results from other laboratories. CTD denotes the C-terminal domain of PQBP-1.
Many of the proline-rich proteins that bind to the GYF domain of CD2BP2/52K have also been found to interact with one of the spliceosomal WW domains present in CA150, FBP11, and FBP21 (23). We termed proteins PRS hubs when their proline content exceeded 10% (mol/mol) and when they bound at least to one of the tested WW domains in addition to CD2BP2/52K-GYF. Except for U1-A and SmB/B′, all proteins defined as PRS hubs were found in at least two of our experiments. The spliceosomal PRS hubs are listed in Table II, and the distribution of the PRD binding sites is given in supplemental Table S3. The quantification and mass spectrometric analysis of the PRS hubs, as identified for the peptide competition and PRS binding site mutant experiments, are listed in supplemental Table S4. Proteins of the SF3B complex were recognized by most of the PRDs studied (Table II). The functional implication of such promiscuity could be a crowding effect that might facilitate the recruitment of U2 snRNP components during the assembly of the spliceosome. The large number of binding sites would provide a strong avidity effect and keep required components close to the A complex to allow for proper integration, whereas the relative low binding affinities of individual interactions would prevent a static association that would block further rearrangements during assembly. Integration into subcomplexes, on the other side, might effectively restrict possible interactions of PRS hubs and PRDs to their local counterparts. The PRS in SF1, for example, can bind to all PRDs analyzed in vitro, but for splicing in vivo, only binding to the WW domains of CA150 has been shown to be important so far (34). To determine physiologically relevant interactions between the GYF domain of endogenous CD2BP2/52K and PRS, we performed pulldown experiments using U5-15K, fused to GST, in the presence and absence of a GYF domain inhibitory peptide (Fig. 2C) that were similar to the experiments with GST-GYF as bait. The majority of proteins found in the U5-15K pulldown were classified as mixed isotopic, and they comprised fewer GYF domain recognition sites (9%). The number of specifically bound (labeled) proteins (8%) was small. However, spliceosomal and splicing-related proteins were highly abundant in the precipitate, and all components of the U5 snRNP were present. U5-15K-associated protein complexes are less dependent on GYF domain-mediated PRS recognition as judged by their mixed isotopic state. The inhibitory peptide (in the pulldown with unlabeled lysates) seems to disrupt inefficiently CD2BP2/52K-comprising complexes, such as the U5 snRNP; and consequently, unlabeled proteins are not deprived in the precipitate. The fraction of unlabeled proteins probably comprises GYF-independent interactions of U5-15K, several of which were described previously (35).
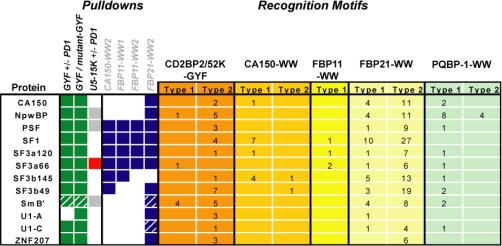
Table II. Proline-rich sequence hubs in the spliceosome.
Binding of the PRS hubs to CD2BP2/K52-GYF is compared with the interactions with spliceosomal WW domains of CA150, FBP11, and FBP21 (23). Squares represent interactions as determined by pulldown experiments with the indicated PRDs as bait. The color code for interactions with CD2BP2/K52-GYF and U5-15K is identical to that in Table I. Blue squares represent interactions with WW domains. Interactions presented by hashed blue squares are taken from another study (24). The colored part of the table indicates the number of consensus motifs as they are present in the respective proteins. The individual consensus motifs are as follows: GYF type 1, PPGX[RK] (21); GYF type 2, PPG[WFYMLIHSTAE] (22); CA150 type 1, PPPPPX[HPR] (49); CA150 type 2, PPXY (50); FBP11 type 1, PPLP (51); FBP21 type 1, [FRP]PP[GLP][MPR]X[LMP] (49); FBP21 type 2, [PG][MP][PR]X[APS] (49); PQBP-1 type 1, PPXR (39); and PQBP-1 type 2, R..PPGPPP..R (44). Square brackets enclose allowed amino acids at this position, and periods followed by the amino acid in curly brackets refer to the amino acid (arginine) following at a somewhat flexible distance from the core motif.
Proteins that were found in all pulldown experiments included the spliceosomal PRS hubs SF3a66, PSF, NpwBP, and SmB. For the latter three, we previously identified several in vitro binding sites (21). Because SmB and PSF are a component and an associated factor of the U5 snRNP, respectively (36–38), they are likely to be relevant interaction partners. NpwBP and SmB were shown to bind CD2BP2/52K by yeast two-hybrid analysis (22). Interestingly two of the four WW domain-containing proteins present in the spliceosome, PQBP-1 and CA150, were co-precipitated by CD2BP2/52K-GYF (supplemental Table S1), whereas FBP11 and FBP21 were not found. To explain this distinct precipitation pattern of the pulldown experiment we analyzed the potential interplay between GYF- and WW-containing proteins.
GYF and WW Domain-containing Proteins in the Spliceosome
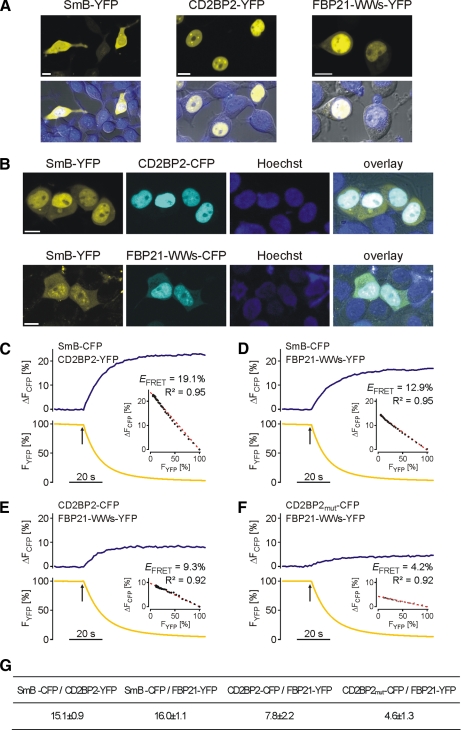
The GYF domain of CD2BP2/52K and the WW domains of FBP21 were independently shown to co-precipitate SmB/B′ (21, 24), but it is unclear whether these interactions occur under cellular conditions. We therefore studied the possible binding of CD2BP2/52K or a tandem WW domain construct of FBP21 (FBP21-WWs) to SmB by FRET analysis. Indeed all three proteins localized to the nucleus in HEK293T cells when expressed separately (Fig. 3 A). In contrast to CD2BP2/52K-YFP and YFP-tagged FBP21-WWs, however, SmB-YFP was also found in the cytosol. When SmB-YFP was co-expressed with either CD2BP2/52K-CFP or CFP-tagged FBP21-WWs, its nuclear localization became more pronounced (Fig. 3B), and a robust FRET signal was detected (Fig. 3, C and D), suggesting that binding of SmB to CD2BP2/52K or FBP21-WWs retains the protein in the nucleus.
Fig. 3.
Localization and FRET analysis of SmB-associated proteins. A, confocal laser scanning images of YFP fusion proteins of CD2BP2/52K, SmB, and FBP21. The respective proteins were transiently expressed in HEK293T cells and imaged with a confocal laser scanning microscope (LSM510-META) equipped with an α-Plan Fluar 100/1.45 objective microscope. Bar, 10 μm. B, co-expression of YFP-fused SmB with CD2BP2/52K-CFP or CFP-tagged FBP21 tandem WW domain construct. Images were recorded as described in A. The SmB-YFP was tracked to the nucleus by CD2BP2/52K-CFP or CFP-tagged FBP21-WWs. C–F, analysis of fluorescence energy transfer of a single experiment between SmB-CFP and CD2BP2-YFP (C), SmB-CFP and FBP21-YFP (D), CD2BP2-CFP and FBP21-YFP (E), and CD2BP2-CFPmut and FBP21-YFP (F). During acceptor photobleaching the donor (CFP) fluorescence signal increases whereas the acceptor (YFP) fluorescence decreases. FRET efficiency was calculated by regression analysis of the various intensities obtained during photobleaching of YFP. CD2BP2-CFPmut denotes a GYF mutant (W8R/Y33A) of CD2BP2 that is devoid of PRS binding. G, FRET efficiencies of energy transfer. Data represent mean ± S.E. of a minimum of four independent measurements.
Analysis of the recognition pattern of CD2BP2/52K GYF and FBP21 WW domains by peptide spot blots confirmed overlapping PRS selectivity. A diverse set of PRS from various proteins was probed by the GST variants of both domains and revealed a striking resemblance in their interaction profile (Fig. 4 A and supplemental Table S5) in agreement with published recognition motifs (13, 39). In spliceosomal PRS hubs, binding sites of the two PRDs are frequently overlapping, and in the described interaction partner SmB/B′, virtually all CD2BP2/52K-GYF binding sites are embedded within FBP21 recognition motifs (supplemental Table S3). NMR titrations of 15N-labeled CD2BP2/52K GYF domain with SmB peptide in the presence and absence of FBP21-WWs confirmed that the PRD competed for the same target sequence in vitro (Fig. 4B). The competition between CD2BP2/52K GYF and FBP21 WW domains might be the reason why the latter protein was not found in our pulldown experiments in which the large excess of GYF domain likely occupies all relevant FBP21 binding sites of the spliceosome.
Interestingly a lower but still significant FRET signal was found between co-expressed CD2BP2/52K-CFP and YFP-tagged FBP21-WWs (Fig. 3E). Moreover a mutant of CD2BP2 deficient in PRS binding (GYF domain mutations W8R and Y33A) showed a significant reduction in FRET signal from 7.8 ± 2.2 to 4.8 ± 1.1% (Fig. 3F), indicating that PRS-containing endogenous proteins mediate the interaction of the two proteins. Consequently the large number of binding sites in the spliceosome can prevent competitive inhibition in this experiment as observed in the in vitro experiments (Fig. 4B). Integration into different spliceosomal subcomplexes could further specify which PRD can access the PRS hubs. SmB/B′ might be the target of CD2BP2/52K in the U5 snRNP, whereas its likely interaction partner within the U2 snRNP is FBP21.
For the interplay of CD2BP2/52K-GYF and the WW domain of PQBP-1, a different scenario is likely. According to published recognition motifs, binding sites for both adaptor domains concentrate in the same PRS hubs as is the case for CD2BP2/52K-GYF and the WW domains of FBP21. NpwBP, in particular, comprises six sites for the GYF domain and 12 for the WW domain of PQBP-1 (Table II and supplemental Table S3). The motifs, however, are well separated and would allow for simultaneous binding of both adaptor domains. We reconfirmed the presence of PQBP-1 in GYF domain pulldown experiments by Western blot (Fig. 4C). The peptide inhibitor of CD2BP2/52K-GYF disrupted the association, whereas the high concentration of bait itself had no effect. Co-integration into spliceosomal complexes via the NpwBP PRS hub could be followed by a more direct and affine interplay of these proteins. Although CD2BP2/52K-GYF may initially bind to several PRS in NpwBP the additional interaction with U5-15K and the reported direct binding of PQBP-1 to U5-15K and NpwBP (40) possibly restrict accessible PRS sites. We therefore suggest the existence of a tetrameric complex consisting of CD2BP2/52K, NpwBP, PQBP-1, and U5-15K that associates in a PRS-dependent manner (Fig. 4D).
In Vitro Splicing and Complex Assembly in the Presence of the GYF Domain
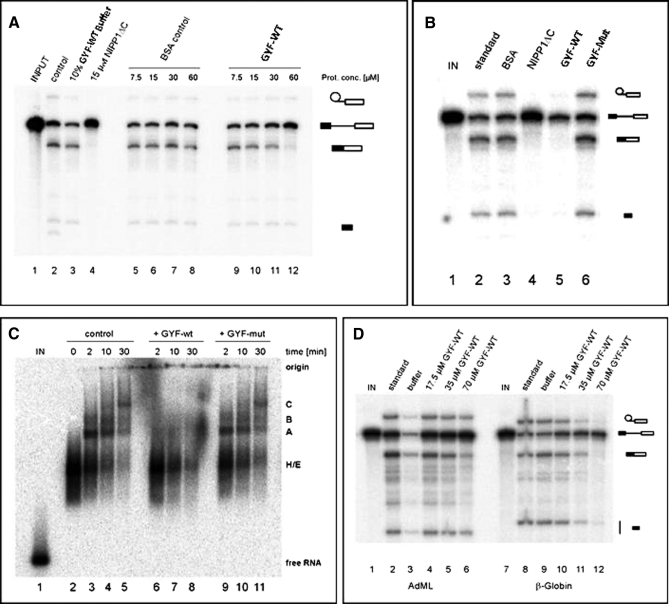
Because the isolated GYF domain was able to co-precipitate spliceosomal proteins and/or subcomplexes of different snRNPs, we were interested in its effect on splicing. Wild-type (WT) or the mutant GYF domain (devoid of PRS binding) was added to in vitro splicing reactions prior to the addition of the reporter β-globin pre-mRNA. At higher concentrations the wild-type was able to reduce the amount of spliced mRNA (Fig. 5A). The inhibitory effect depended on PRS binding because it was not seen for the mutant when added to in vitro splicing reactions (Fig. 5B). Thus, WT GYF domain is able to convey a dominant-negative effect on splicing. To clarify how the GYF domain possibly inhibits function, in vitro spliceosomal complex assembly was studied in the presence of either WT or mutant GYF domain. Separation of assembled complexes by native agarose gels (Fig. 5C) revealed that almost no A complex and subsequently no B and C complexes were formed in the presence of WT GYF. The GYF mutant displayed no inhibitory effect on spliceosome assembly. We conclude that the GYF domain, by saturating the PRS binding sites of the early spliceosome, interferes with the correct formation of spliceosomal A complexes. As a consequence, proceeding to the later complexes and splicing itself are inhibited.
Fig. 5.
Splicing inhibition by the GYF domain. In panels A, B, and D, from the top to the bottom, the position of lariat-splice intermediate, unspliced mRNA, spliced mRNA, and 5′-splice intermediate are indicated. A, inhibition of pre-mRNA splicing by WT GYF domain. BSA was used as negative control, and a C-terminal truncation mutant of the essential splicing factor NIPP1 (NIPP1ΔC) was used as positive control (44). Shown is the image of an in vitro splicing reaction containing body-labeled pre-mRNA after 45 min of incubation under splicing conditions separated on a 10% acrylamide, 7 m urea gel. B, addition of PRS binding mutant (GYF-mut) to an identical in vitro splicing reaction does not inhibit splicing. C, in vitro assembly of spliceosomal complexes. Body-labeled pre-mRNA was incubated in HeLa nuclear extract under standard splicing conditions (lanes 2–5) or in the presence of 70 μm WT GYF (lanes 6–8) and GYF mutant (lanes 9–11), respectively. Spliceosomal complexes (which appear in the order H/E, A, B, and C during spliceosome maturation) present at the indicated time points are shown separated on a 1.5% agarose gel. D, comparison of the GYF effect on the U1-independent splicing of AdML-M3 and U1-dependent splicing of β-globin pre-mRNAs. Prot. conc., protein concentration; IN, input.
In vitro, splicing of certain pre-mRNAs, e.g. a derivative of the Drosophila fushi tarazu gene and a synthetic chimera of β-globin and PIP85.A RNA sequences, is catalyzed in nuclear extracts depleted of U1 snRNP; the latter is catalyzed only when exogenous SR proteins are added to the splicing reaction (41). However, the natural β-globin pre-mRNA is strictly dependent on the presence of functional U1 snRNP in nuclear extracts that have not been supplemented with excess SR proteins, whereas splicing of AdML-M3 is U1-independent under these conditions.2To further detail possible GYF domain interactions with early spliceosomal components, e.g. the U1 snRNP, we compared its inhibitory effect on splicing of U1 snRNP-dependent β-globin and U1 snRNP-independent AdML-M3 pre-mRNA. Like β-globin pre-mRNA, in vitro splicing of AdML-M3 precursors was found to be inhibited in a concentration-dependent manner (Fig. 5D), albeit with reduced efficiency. Although splicing of β-globin pre-mRNA was almost completely blocked at concentrations of 70 μm WT GYF, splicing intermediates and spliced mRNA of AdML-M3 could be readily detected (Fig. 5D, compare lanes 6 and 12). Given the presence of PRS hubs in the U2 snRNP and splicing inhibition of both pre-mRNAs, the U2 snRNP might be the major target site of the GYF domain. Interference with the U1-U2 snRNP interactions could account for the increased sensitivity of β-globin pre-mRNA compared with AdML-M3 pre-mRNA.
DISCUSSION
In this study we define molecular assemblies that potentially depend on the GYF adaptor domain of CD2BP2/52K. We used pulldown experiments in combination with site-specific inhibition and the SILAC labeling strategy to encode the PRS dependence of co-precipitation within the isotopic composition of the proteins. The identity of co-precipitated proteins and their isotopic composition were subsequently determined by MS/MS analysis. Only a fraction of the proteins that were found to be associated in a PRS-dependent manner did themselves comprise a GYF domain binding site, suggesting the precipitation of larger complexes. The list of identified proteins comprises many of the spliceosomal components described by others (Table I and supplemental Table S1). Almost all U1, U2, and U5 and several U4/U6 snRNP proteins were found. Proteins not identified include many U6-specific Lsm proteins, U2 snRNP-related components, and most of the proteins from the hPrp19-hCDC5 complex (42, 43). It is well conceivable that different spliceosomal subcomplexes are fished out of the lysate rather than intact spliceosomes, explaining why our protein list does not reflect the composition of one particular spliceosomal complex.
The importance of PRS recognition of the endogenous CD2BP2/52K-GYF within the U5 snRNP was determined using GST-tagged U5-15K as bait. U5-15K directly binds the GYF domain of CD2BP2/52K, and NMR analysis confirmed that U5-15K binding does not compromise association of the GYF domain with PRS peptides (supplemental Fig. S2). Fewer proteins were co-precipitated in the pulldown experiment, and the dependence on PRS was reduced. However, most proteins were spliceosomal or splicing-related, and the entire U5 snRNP was identified. The increased selectivity of the endogenous GYF domain might be imposed by other parts of CD2BP2/52K and its incorporation into the U5 snRNP particle. Because full-length CD2BP2/52K interacts with U5 snRNP components such as U5-15K and Prp6 (16) independently of PRS recognition, it is tempting to speculate that PRS binding of CD2BP2/52K-GYF is more important for U5 snRNP formation or its integration into the tri-snRNP but less important for maintaining complex integrity. The reduced number of PRS-mediated binding partners of full-length CD2BP2/52K could also reflect the smaller amount of endogenous CD2BP2/52K within the U5-15K pulldown. The concentration of non-stoichiometric binding partners could drop below the detection level. Besides its role within the U5 snRNP, CD2BP2/52K-GYF could have additional functions that are more readily detected by pulldowns with isolated GYF domain. CD2BP2/52K is released from the U5 snRNP upon tri-snRNP formation (16) as are other proteins during spliceosomal maturation. NpwBP (25, 40) is not stably integrated into the hPrp19-CDC5 complex after formation of the activated complex B (32), and it represents a likely interaction partner of CD2BP2/52K. Based on the literature of the individual proteins, a complex consisting of CD2BP2/52K, U5-15K, NpwBP, and PQBP-1 could be formed (45, 46).
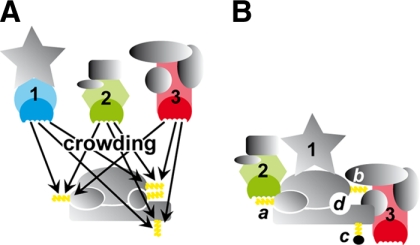
PRS hubs, proteins with numerous PRD binding sites, are present in several spliceosomal subcomplexes. The large number of docking sites for GYF and WW adaptor domains (23) could serve as integration points for PRD-associated subcomplexes (Fig. 6 A). Non-competitive, simultaneous binding is likely to occur for CD2BP2/52K-GYF and the WW domain of PQBP-1. Competition between CD2BP2/52K-GYF and FBP21-WW for SmB/B′ binding was shown in vitro, but under physiological conditions binding to different PRS hubs is more plausible. Competition for spliceosomal PRS might explain the interference of recombinant CD2BP2/52K-GYF with in vitro splicing. At the inhibitory concentrations of 60–70 μm used in our experiments, most available binding sites should be occupied by the GYF domain, which would block the access of other proteins important for spliceosomal assembly at the stage of the A complex. For example, the B complex-associated FBP21 protein was not found in our pulldowns and may well be competed out by the GYF domain.
Fig. 6.
PRS-mediated crowding of spliceosomal components. A, spliceosomal subcomplexes are kept in close proximity by multiple PRS (yellow) and PRD (blue, green, and red) interactions. Interactions are highly dynamic and allow for proper complex assembly. B, assembled complex. PRS-PRD interactions can serve to deliver components (1 and 3) or be required for their integration (2). PRS can be accessible (a) or inaccessible (b) within the complex, masked by post-translational modifications (c), or absent from the complex (d).
The inhibitory effects of CD2BP2/52K-GYF in the splicing assay (Fig. 5) indicate the importance of a correctly operating PRD-PRS interaction network within the spliceosome for productive splicing, and it can be assumed that similar effects are observable for other PRS-recognizing proteins. Major players in this network include CD2BP2/52K and the WW domain-containing proteins CA150, FBP11, FBP21, and PQBP-1 as well as their respective binding sites, which are highly abundant in the PRS hubs. Interestingly at the stage of complex B* (42), tremendous rearrangements take place, and most of the PRD-comprising proteins are released despite the continuous presence of several PRS-containing proteins. For example, SmB/B′ as a core component of the snRNPs is present in the U5 snRNP complex throughout splicing, but it appears to be available for CD2BP2/52K-GYF interactions only in the context of the isolated U5 snRNP (16) in agreement with the paucity of tri-snRNP-specific proteins in our proteomics approach. It is likely that spliceosomal proline cis-trans isomerases or post-translational modifications are modulating the accessibility or affinity of the binding sites at this point in maturation. In this context it is interesting to note that several of the proteins containing GYF domain recognition sites were recently shown to be targets of PRMT4 (47), which catalyzes the asymmetric dimethylation of arginines within PGR motifs that are neighboring or partially overlapping with the GYF domain binding motifs. Future experiments need to clarify whether proline cis-trans isomerization and post-translational modifications such as methylation or phosphorylation regulate GYF domain-mediated interactions.
In summary, we propose that PRS hubs play an important role in early spliceosomal assembly, whereas they are less important in the active spliceosome. The molecular crowding of promiscuous PRS binding sites probably leads to the required co-compartmentation of spliceosomal subcomplexes that are funneled into more stably integrated complexes (Fig. 6). Although certain PRS sites may be occupied during the various stages of splicing, many PRS sites seem to either dissociate or become inaccessible via steric hindrance or post-translational modification (Fig. 6). The premature state of loosely associated subcomplexes potentially provides a degree of flexibility that is necessary to couple splicing to other related processes such as transcription and translation. The importance of a correct PRS-PRD network for complex A and B formation is a first hint for such a peripheral but decisive role of proline-rich sequence recognition in the spliceosome.
Supplementary Material
Acknowledgments
We are thankful to Katharina Thiemke and Kathrin Motzny for excellent technical assistance. We thank Matthias Bollen und Monique Beullens, University of Leuven, Leuven, Belgium, for kindly providing us with an aliquot of NIPPΔC.
Footnotes
* This work was supported by Deutsche Forschungsgemeinschaft Grants FG806, SFB740, and SFB765 (to C. F.) and grants from the European Union (EURASNET) and the Fonds der Chemischen Industrie (to R. L.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1 and S2 and Tables S1–S8.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1 and S2 and Tables S1–S8.
2 C. Merz, unpublished data.
1 The abbreviations used are:
- snRNP
- small nuclear ribonuclear particle
- PRS
- proline-rich sequence(s)
- PRD
- proline-rich sequence recognition domain
- CD2BP2
- CD2-binding protein 2
- SILAC
- stable isotope labeling by amino acids in cell culture
- FRET
- fluorescence resonance energy transfer
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- GO
- Gene Ontology
- HEK
- human embryonic kidney
- WT
- wild-type
- GYF
- glycine-tyrosine-phenylalanine.
REFERENCES
- 1.Berget S. M., Moore C., Sharp P. A. (1977) Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A 74, 3171– 3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fong Y. W., Zhou Q. (2001) Stimulatory effect of splicing factors on transcriptional elongation. Nature 414, 929– 933 [DOI] [PubMed] [Google Scholar]
- 3.Bentley D. (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol 14, 336– 342 [DOI] [PubMed] [Google Scholar]
- 4.Maquat L. E. (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol 5, 89– 99 [DOI] [PubMed] [Google Scholar]
- 5.Le Hir H., Nott A., Moore M. J. (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci 28, 215– 220 [DOI] [PubMed] [Google Scholar]
- 6.Strässer K., Hurt E. (2001) Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413, 648– 652 [DOI] [PubMed] [Google Scholar]
- 7.Will C. L., Lührmann R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol 13, 290– 301 [DOI] [PubMed] [Google Scholar]
- 8.Pahlke D., Freund C., Leitner D., Labudde D. (2005) Statistically significant dependence of the Xaa-Pro peptide bond conformation on secondary structure and amino acid sequence. BMC Struct. Biol 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musacchio A., Wilmanns M., Saraste M. (1994) Structure and function of the SH3 domain. Prog. Biophys. Mol. Biol 61, 283– 297 [DOI] [PubMed] [Google Scholar]
- 10.Kay B. K., Williamson M. P., Sudol M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14, 231– 241 [PubMed] [Google Scholar]
- 11.Zarrinpar A., Bhattacharyya R. P., Lim W. A. (2003) The structure and function of proline recognition domains. Sci. STKE 2003, RE8. [DOI] [PubMed] [Google Scholar]
- 12.Kofler M. M., Freund C. (2006) The GYF domain. FEBS J 273, 245– 256 [DOI] [PubMed] [Google Scholar]
- 13.Kofler M., Motzny K., Freund C. (2005) GYF domain proteomics reveals interaction sites in known and novel target proteins. Mol. Cell. Proteomics 4, 1797– 1811 [DOI] [PubMed] [Google Scholar]
- 14.Nishizawa K., Freund C., Li J., Wagner G., Reinherz E. L. (1998) Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc. Natl. Acad. Sci. U.S.A 95, 14897– 14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmuth K., Urlaub H., Vornlocher H. P., Will C. L., Gentzel M., Wilm M., Lührmann R. (2002) Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. U.S.A. 99, 16719– 16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laggerbauer B., Liu S., Makarov E., Vornlocher H. P., Makarova O., Ingelfinger D., Achsel T., Lührmann R. (2005) The human U5 snRNP 52K protein (CD2BP2) interacts with U5–102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA 11, 598– 608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freund C., Dötsch V., Nishizawa K., Reinherz E. L., Wagner G. (1999) The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat. Struct. Biol 6, 656– 660 [DOI] [PubMed] [Google Scholar]
- 18.Freund C., Kühne R., Yang H., Park S., Reinherz E. L., Wagner G. (2002) Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules. EMBO J 21, 5985– 5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freund C., Kühne R., Park S., Thiemke K., Reinherz E. L., Wagner G. (2003) Structural investigations of a GYF domain covalently linked to a proline-rich peptide. J. Biomol. NMR 27, 143– 149 [DOI] [PubMed] [Google Scholar]
- 20.Nielsen T. K., Liu S., Lührmann R., Ficner R. (2007) Structural basis for the bifunctionality of the U5 snRNP 52K protein (CD2BP2). J. Mol. Biol 369, 902– 908 [DOI] [PubMed] [Google Scholar]
- 21.Kofler M., Heuer K., Zech T., Freund C. (2004) Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J. Biol. Chem 279, 28292– 28297 [DOI] [PubMed] [Google Scholar]
- 22.Kofler M., Motzny K., Beyermann M., Freund C. (2005) Novel Interaction Partners of the CD2BP2-GYF Domain. J. Biol. Chem 280, 33397– 33402 [DOI] [PubMed] [Google Scholar]
- 23.Ingham R. J., Colwill K., Howard C., Dettwiler S., Lim C. S., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G., Taylor L., Yeung B., Vassilovski G., Amin M., Chen F., Matskova L., Winberg G., Ernberg I., Linding R., O’donnell P., Starostine A., Keller W., Metalnikov P., Stark C., Pawson T. (2005) WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol 25, 7092– 7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedford M. T., Reed R., Leder P. (1998) WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc. Natl. Acad. Sci. U.S.A 95, 10602– 10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komuro A., Saeki M., Kato S. (1999) Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J. Biol. Chem 274, 36513– 36519 [DOI] [PubMed] [Google Scholar]
- 26.Schaefer M., Albrecht N., Hofmann T., Gudermann T., Schultz G. (2001) Diffusion-limited translocation mechanism of protein kinase C isotypes. FASEB J 15, 1634– 1636 [DOI] [PubMed] [Google Scholar]
- 27.Hellwig N., Albrecht N., Harteneck C., Schultz G., Schaefer M. (2005) Homo- and heteromeric assembly of TRPV channel subunits. J. Cell Sci 118, 917– 928 [DOI] [PubMed] [Google Scholar]
- 28.Blagoev B., Mann M. (2006) Quantitative proteomics to study mitogen-activated protein kinases. Methods 40, 243– 250 [DOI] [PubMed] [Google Scholar]
- 29.Klemm C., Otto S., Wolf C., Haseloff R. F., Beyermann M., Krause E. (2006) Evaluation of the titanium dioxide approach for MS analysis of phosphopeptides. J. Mass Spectrom 41, 1623– 1632 [DOI] [PubMed] [Google Scholar]
- 30.Merz C., Urlaub H., Will C. L., Lührmann R. (2007) Protein composition of human in RNPs spliced in vitro and differential regnitements for mRNP protein recruitment. RNA 13, 116– 128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376– 386 [DOI] [PubMed] [Google Scholar]
- 32.Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006) Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol 26, 5528– 5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behzadnia N., Golas M. M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C. L., Urlaub H., Stark H., Lührmann R. (2007) Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J 26, 1737– 1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin K. T., Lu R. M., Tarn W. Y. (2004) The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol 24, 9176– 9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Lindblom T., Chang A., Sudol M., Sluder A. E., Golemis E. A. (2000) Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene 257, 33– 43 [DOI] [PubMed] [Google Scholar]
- 36.Bringmann P., Rinke J., Appel B., Reuter R., Lührmann R. (1983) Purification of snRNPs U1, U2, U4, U5 and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1)RNP antisera. EMBO J 2, 1129– 1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wooley J. C., Zukerberg L. R., Chung S. Y. (1983) Polypeptide components of human small nuclear ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A 80, 5208– 5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng R., Dye B. T., Pérez I., Barnard D. C., Thompson A. B., Patton J. G. (2002) PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8, 1334– 1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otte L., Wiedemann U., Schlegel B., Pires J. R., Beyermann M., Schmieder P., Krause G., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H. (2003) WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci 12, 491– 500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waragai M., Junn E., Kajikawa M., Takeuchi S., Kanazawa I., Shibata M., Mouradian M. M., Okazawa H. (2000) PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5–15kD/dim1p via the carboxyl-terminal domain. Biochem. Biophys. Res. Commun 273, 592– 595 [DOI] [PubMed] [Google Scholar]
- 41.Crispino J. D., Mermoud J. E., Lamond A. I., Sharp P. A. (1996) Cis-acting elements distinct from the 5′ splice site promote U1-independent pre-mRNA splicing. RNA 2, 664– 673 [PMC free article] [PubMed] [Google Scholar]
- 42.Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298, 2205– 2208 [DOI] [PubMed] [Google Scholar]
- 43.Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. (2003) The Prp19p-associated complex in spliceosome activation. Science 302, 279– 282 [DOI] [PubMed] [Google Scholar]
- 44.Beullens M., Bollen M. (2002) The protein phosphatase-1 regulator NIPP1 is also a splicing factor involved in a late step of spliceosome assembly. J. Biol. Chem 277, 19855– 19860 [DOI] [PubMed] [Google Scholar]
- 45.Llorian M., Beullens M., Lesage B., Nicolaescu E., Beke L., Landuyt W., Ortiz J. M., Bollen M. (2005) Nucleocytoplasmic shuttling of the splicing factor SIPP1. J. Biol. Chem 280, 38862– 38869 [DOI] [PubMed] [Google Scholar]
- 46.Waragai M., Lammers C. H., Takeuchi S., Imafuku I., Udagawa Y., Kanazawa I., Kawabata M., Mouradian M. M., Okazawa H. (1999) PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum. Mol. Genet 8, 977– 987 [DOI] [PubMed] [Google Scholar]
- 47.Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71– 83 [DOI] [PubMed] [Google Scholar]
- 48.Makarova O. V., Makarov E. M., Urlaub H., Will C. L., Gentzel M., Wilm M., Lührmann R. (2004) A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J 23, 2381– 2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H., Columbus J., Zhang Y., Wu D., Lian L., Yang S., Goodwin J., Luczak C., Carter M., Chen L., James M., Davis R., Sudol M., Rodwell J., Herrero J. J. (2004) A map of WW domain family interactions. Proteomics 4, 643– 655 [DOI] [PubMed] [Google Scholar]
- 50.Aqeilan R. I., Palamarchuk A., Weigel R. J., Herrero J. J., Pekarsky Y., Croce C. M. (2004) Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res 64, 8256– 8261 [DOI] [PubMed] [Google Scholar]
- 51.Bedford M. T., Chan D. C., Leder P. (1997) FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J 16, 2376– 2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.