Abstract
Computational design has been very successful in recent years: multiple novel ligand binding proteins as well as enzymes have been reported. We wanted to know in molecular detail how precise the predictions of the interactions of protein and ligands are. Therefore, we performed a structural analysis of a number of published receptors designed onto the periplasmic binding protein scaffold that were reported to bind to the new ligands with nano- to micromolar affinities. It turned out that most of these designed proteins are not suitable for structural studies due to instability and aggregation. However, we were able to solve the crystal structure of an arabinose binding protein designed to bind serotonin to 2.2 Å resolution. While crystallized in the presence of an excess of serotonin, the protein is in an open conformation with no serotonin bound, although the side-chain conformations in the empty binding pocket are very similar to the conformations predicted. During subsequent characterization using isothermal titration calorimetry, CD, and NMR spectroscopy, no indication of binding could be detected for any of the tested designed receptors, whereas wild-type proteins bound their ligands as expected. We conclude that although the computational prediction of side-chain conformations appears to be working, it does not necessarily confer binding as expected. Hence, the computational design of ligand binding is not a solved problem and needs to be revisited.
Keywords: arabinose binding protein, biosensor, serotonin receptor, glucose binding protein, ribose binding protein
Computational protein design has come a long way in recent years. It has been used to design new folds (1), binding interactions (2), and catalysts (3, 4) onto existing scaffolds. Recently, even the successful design of two nonnatural reactions have been described (5, 6). While these are great steps forward, many aspects of design leave room for improvement. For example, the substrate affinities of the new catalysts are generally only modest. This is surprising, as high affinity receptors for a number of small-molecule ligands, such as trinitrotoluene (TNT), L-lactate, serotonin (2), and the nerve agent surrogate pinacolyl methyl phosphonic acid (PMPA) (7) have been reported. These computationally designed receptors were built in the periplasmic binding protein (PBP) fold. Members of the PBP superfamily are single-chain polypeptides folding into two distinct, globular N- and C-terminal domains connected by a flexible hinge region. The ligand binding site is situated in the cleft between the two domains. Binding of the cognate ligand induces large conformational changes from an open apo state to a completely closed, ligand-bound state. Binding affinities of the designed receptors were determined indirectly by monitoring the changes in fluorescence emission intensity of a covalently attached, environmentally sensitive fluorescent probe (2, 7). We decided to structurally characterize these receptors further and thus learn about the strength and shortcomings of current computational design methods. Of the variety of published receptors, we tested five high-affinity designs: arabinose-binding protein (ABP) variants reported to bind to serotonin and L-lactate, a ribose-binding protein (RBP) variant reported to bind to TNT, and galactose/glucose-binding protein (GBP) variants reported to bind L-lactate and PMPA. No binding of the ligands to any of the designed receptors could be detected using methods that directly probe ligand binding, such as isothermal titration calorimetry (ITC) and nuclear magnetic resonance (NMR), while we were readily able to detect ligand binding and determine binding affinities of wild-type PBPs. Taken together, our findings indicate that the investigated protein designs do not bind their new ligands in the expected way. Thus, the computational design of ligand binding is not a solved problem and needs to be revisited.
Results and Discussion
Structural Analysis of Designed Receptor Proteins.
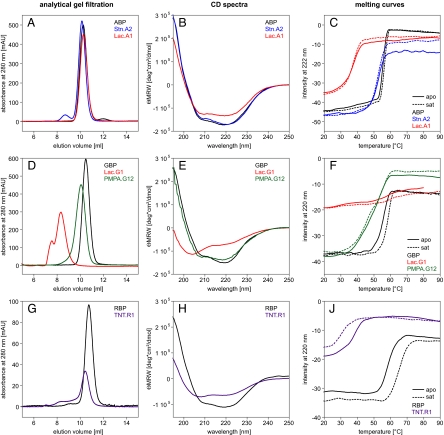
To characterize a number of PBP designs on a structural level, we expressed ABPs designed to bind L-lactate (Lac.A1) and serotonin (Stn.A2), GBPs designed to bind L-lactate (Lac.G1) and pinacolyl methyl phosphonic acid (PMPA.G12), and a RBP designed to bind trinitrotoluene (TNT.R1), as described (2, 7). For a comparative analysis, wild-type ABP, GBP, and RBP were also expressed. The His-tagged proteins were purified via affinity chromatography and subsequently subjected to analytical gel filtration. The wild-type proteins, as well as Stn.A2, eluted with a volume corresponding to the size of the monomer. In contrast, the other four designed proteins showed a mixture of oligomerization states indicating a high tendency to aggregate. Thus, these proteins were purified further via preparative gel filtration. We were able to separate monomeric Lac.A1, PMPA.G12, and TNT.R1 (Fig. 1A, D, and G). However, isolated Lac.A1 and TNT.R1 started to form dimers again overnight or at higher concentrations. In the case of Lac.G1, only higher oligomeric states could be observed (Fig. 1D).
Fig. 1.
Biophysical characteristics of designed receptor proteins: (A–C) ABP, Lac.A1, and Stn.A2, (D–F) GBP, Lac.G1, and PMPA.G12, (G–J) RBP and TNT.R1. (A) Association states measured by analytical gel filtration (0.5 mg loaded each). All three variants elute at the same volume, which corresponds to the molecular mass of monomeric protein. (B) CD spectra: the intensity of the signal of Lac.A1 is slightly reduced compared to ABP and Stn.A2. (C) Melting curves: no clear increase in stability upon addition of the respective ligand was observed for all three proteins. Thermal stability of Lac.A1 is severely reduced compared to wild-type, whereas the stability of Stn.A2 is almost the same. However, visible aggregation of Stn.A2 occurred, leading to varying signal intensity after unfolding. (D) Analytical gel filtration (0.6 mg each). Lac.G1 exists in solution only in the form of higher order oligomers. PMPA.G12 elutes in a single defined peak but with a higher apparent molecular weight than wild-type GBP. (E) CD spectra: a loss of secondary structure is observed in Lac.G1 compared to GBP and PMPA.G12. (F) Melting curves: A stability increase upon addition of ligand was only observed for wild-type GBP. No cooperative unfolding could be observed for Lac.G1. (G) Analytical gel filtration: (0.9 mg each). Monomeric TNT.R1 can be purified but aggregation increases over time. (H) CD spectra: TNT.R1 has a lower secondary structure content compared to RBP. (J) Melting curves: The thermal stability of TNT.R1 is severely reduced compared to wild type. Addition of TNT further decreases thermal stability, because of the solvent acetonitrile.
We then analyzed the secondary structure content of the designs by far-UV circular dichroism. While the CD spectra of most designs were comparable to the ones of the parent proteins, the spectrum of Lac.G1 indicates a considerable loss of secondary structure (Fig. 1 B, E, and H). Therefore, only Lac.A1, Stn.A2, PMPA.G12, and TNT.R1 were subjected to crystallization trials.
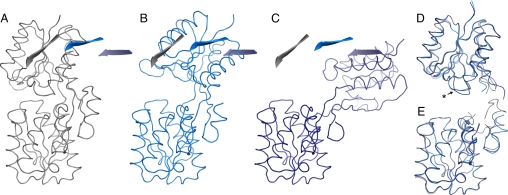
In the case of Stn.A2, we were able to grow crystals that diffracted to 2.2 Å resolution and to solve its structure by molecular replacement using the separated domains of wild-type ABP (PDB ID 5abp) (8) as search models. Two molecules (A and B) were found in the asymmetric unit, which are both in different open conformations (Fig. 2). This was surprising because the crystals were grown in the presence of 2 mM serotonin. If binding occurred in the micromolar range as reported (2), the ligand binding sites should be saturated. Furthermore, no structure of ABP in the open conformation has been reported so far. The opening angle in molecule A is 37° and, as such, is in the normal range of opening angles adopted by various wild-type PBPs in open conformation (9). Molecule B adopts a much further opened conformation with a hinge bending motion of 96°, so far not observed in any of the 135 structures of wild-type or mutant PBPs published in the protein data bank. Multiple open conformations have been described for other members of the PBP superfamily as well (10, 11). Although it is possible that wild-type ABP adopts similar conformations, the unusually high rotation angle of molecule B might be an artifact of crystal packing or a result of mutations in or near the hinge region that alter the conformational flexibility of the protein. Both the N- and C-terminal domains of molecule A superimpose with wild-type ABP (PDB ID 5abp) in closed conformation with a rmsd of 0.35 (over 116 Cα-atoms) and 0.24 (122 Cα), respectively, and of molecule B with 0.35 (111 Cα) and 0.32 (133 Cα), respectively. This clearly shows that the backbone movement is restricted to the hinge region of ABP (see Fig. 2).
Fig. 2.
Open conformations of Stn.A2 compared to wild-type ABP (5abp). (A) ABP (gray ribbon) in closed conformation. (B) Stn.A2 molecule A (light blue ribbon). (C) Stn.A2 molecule B (dark blue ribbon). All structures are aligned over their C-terminal domains. One β-strand of the N-terminal domain is highlighted as cartoon for each structure and shown in each figure to illustrate the hinge bending motion. (D) Superimposition of the N-terminal domains of ABP, and Stn.A2 molecule A and B. Loop 12–19 (includes design residues A16 and S17) adopts a different conformation in Stn.A2 molecule B (indicated by an asterisk). (E) Superimposition of the C-terminal domains of ABP, Stn.A2 molecule A and B.
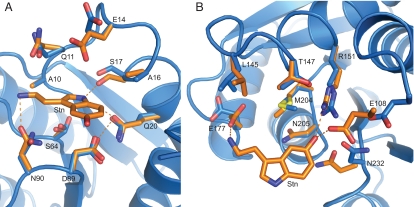
Comparison of the binding pocket in the structure and the computational model (2) shows high similarity. The binding site residues of the two domains of molecule A and B superimpose with the coordinates of the model with a rmsd over all atoms of 0.79 and 1.17 Å for the N-terminal and 1.13 and 1.06 Å for the C-terminal domain (Fig. 3 and Fig. S1). Most side chains in the binding pocket adopt a conformation that closely resembles the predictions. Some of these residues have flexible side chains, yet most side chains adopt a distinct conformation that is the same in molecules A and B in the crystal structure.
Fig. 3.
Comparison of the binding pocket residues in the Stn.A2 design prediction (orange) with the ones in the crystal structure (molecule A, blue). (A) Superimposition of the binding pocket residues in the N-terminal domain (rmsd of 0.79 over all atoms). S64 is observed in two conformations. (B) Superimposition of the binding pocket residues in the C-terminal domain (rmsd of 1.13 over all atoms).
The work of Telmer and Shilton (12) similarly illustrates the importance of structural studies of designed proteins. Although they were able to reproduce the Zn(II)-binding properties of a designed maltose binding protein (13), the crystal structure showed an unexpected Zn(II) binding mode that differed significantly from the design. As Stn.A2, this protein was also found in an open conformation, indicating that the introduced mutations interfere with the domain closure of PBPs, which might pose an additional difficulty for design in these scaffolds.
Characterization of the Binding Properties of the Designs.
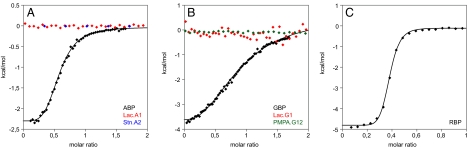
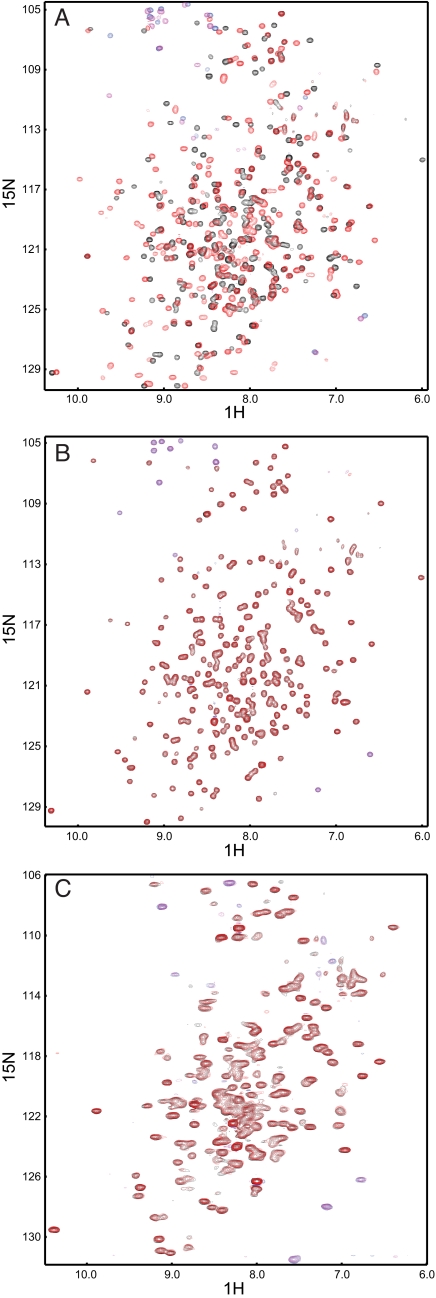
Because most of the designed proteins seemed unsuited for crystallographic characterization, we next analyzed their binding properties using numerous independent biophysical techniques. First we compared temperature-induced melting curves of wild-type and designed proteins with and without ligand. In the wild-type proteins a shift in melting temperature (Tm) can be observed upon addition of saturating concentrations of the respective ligand (Fig. 1 C, F, and J). The Tm of RBP shows a large increase in the presence of ribose: it increases by 7 °C. This result is in accordance with a previously reported shift of 6 °C (14). In ABP and GBP the Tm increases only by 0.7 °C and 2.5 °C, respectively. For GBP a 6 °C shift has been reported (15). It might be possible that in our preparation we did not remove all glucose, although we performed extensive dialysis, which might account for the smaller temperature shift. In contrast to the wild-type proteins, none of the designs showed an increase in stability upon ligand binding at the same conditions. The addition of TNT to TNT.R1 rather led to a loss in stability (see Fig. 1J), which is due to the destabilizing effect of the solvent acetonitrile. These experiments show that in general, the thermal stability of the designed receptors is strongly reduced compared to the wild-type proteins because of the numerous introduced mutations. We then tested binding by ITC. The binding constants of arabinose, glucose, and ribose to the respective wild-type proteins were determined to 3.7 ± 1.2 μM for ABP, 9.4 ± 1.1 μM for GBP, and 0.5 ± 0.1 μM for RBP (Fig. 4). The values for ABP and RBP are in accordance with previously published binding constants: 2 μM for ABP (16) and 0.13 μM for RBP (17). For GBP a binding constant of 0.2 μM has been reported (18). The difference in measurement, which is also observed in the temperature melts, might be caused by residual glucose in our preparations or the use of different solution conditions. In contrast to the wild-type proteins, the designed proteins Lac.A1, Stn.A2, Lac.G1, and PMPA.G12 did not show any change in heat above the heat of mixing upon addition of ligand (see Fig. 4). To employ a third independent technique to detect ligand binding, we performed NMR experiments in which we recorded two-dimensional 1H, 15N-correlation spectra of 15N-labeled wild-type, and designed proteins in the absence and presence of saturating ligand concentrations. Because the chemical shift of a nucleus strongly depends on its local environment, solution NMR spectroscopy is a powerful monitor of conformational changes and ligand binding, even if interactions are as weak as in the millimolar range. As expected, in the wild-type proteins we observed numerous large chemical shift changes that are caused by the protein-ligand interaction, as well as the conformational change induced by ligand binding (Fig. 5 and Fig. S2). In RBP we observed two sets of peaks in the NMR spectrum of the apo form that correspond to a mixture of open and closed conformations (see Fig. S2C). This could be because of either residual ribose in the purified protein, although it was dialyzed extensively, or an equilibrium between open and closed conformations even in the absence of ribose. However, upon addition of ribose the spectrum shifts toward a single set of peaks corresponding to the bound and closed state (see Fig. S2C). We then recorded 2D NMR spectra of the ABP-based designs Lac.A1 and Stn.A2, the RBP-based design TNT.R1, and the GBP-based design PMPA.G12. The spectra confirm that Stn.A2 is well folded. The spectral overlap observed for Lac.A1, TNT.R1, and PMPA.G12 might reflect their tendency to form higher order oligomers as observed in analytical gel filtration experiments or result from only partially folded protein (compare CD measurements). However, upon addition of a large excess of the respective ligands, we did not observe any significant chemical shift changes in the NMR spectra of the designed receptors that would indicate the expected large conformational changes upon ligand binding as observed in wild-type PBPs (see Fig. 5 and Fig. S2). The observed minor changes might be a result of unspecific binding to the open conformation or small pH effects.
Fig. 4.
Analysis of binding by ITC. Binding constants were determined from sigmoidal fits to two independent measurements. (A) Representative ITC measurements for ABP, Lac.A1, and Stn.A2. No significant change in heat upon addition of the respective ligand could be detected in the case of the designs. (B) Representative ITC measurements for GBP, Lac.G1, and PMPA.G12. No change in heat upon addition of the respective ligand could be detected in the case of the designs. (C) Representative ITC measurement for wild-type RBP. Saturation is reached for wild-type RBP already at a molar ratio of 0.5, which indicates that residual ribose remains in the purified protein sample. No ITC measurement could be performed with TNT.R1 because the ligand TNT is insoluble in aqueous buffer.
Fig. 5.
End-point chemical shift titrations of wild-type ABP and ABP-based receptors using 2D NMR spectroscopy. (A) ABP without ligand (black) and 20-fold excess of arabinose (red). Large chemical shift changes in the spectra occur due to ligand binding and ligand-induced conformational changes. (B) Stn.A2 without ligand (black) and 10-fold excess of serotonin (red). No changes upon ligand binding could be detected. (C) Lac.A1 without ligand (black) and 100-fold excess of L-lactate (red). Because of aggregation at high protein concentrations or partial folding, the spectra show unresolved areas. No significant chemical shift changes upon addition of ligand could be detected. Aliased resonances are shown in blue and magenta for protein without and with ligand, respectively.
Altogether, our combined analysis of the binding properties of the designs indicates that no specific binding of the target ligands to the respective designs occurs. In contrast to our measurements, the reported binding affinities for these receptors were based on an indirect measure via an environment-sensitive fluorophore attached to the proteins in their hinge regions. These fluorophores attached to wild-type proteins are able to sense the conformational change induced through the binding of a sugar (19). The same concept is expected to work in the designed receptors. However, this type of analysis might be susceptible to false-positive results if the ligand or its solvent directly interacts with the fluorophore or changes its environment significantly without specific ligand binding to the protein. Direct measurements, such as presented here, appear to be a better means to evaluate whether the original design strategy was successful.
Implications for the Field of Computational Biology.
Although the overall conformation of the designed binding-site residues in Stn.A2 indeed resembles the model, binding to the new ligand, serotonin, is not conferred. This should be kept in mind when judging model quality based on structures without a bound ligand. Our analysis shows the importance of experimental and structural validation to improve computational design methodologies.
We conclude that computational ligand-binding design remains an unsolved problem that needs reconsideration in its details. Do the algorithms capture all aspects important for the prediction of small molecule binding? Could the energetic cost associated with domain reorientation in PBPs (20) pose additional difficulties for the design in this particular scaffold? Boas and Harbury (21) successfully used the molecular-mechanics potential energy function to recapitulate and improve upon wild-type RBP, which points toward the fact that the general principles in receptor design hold true. Nevertheless, predicted interactions need to be verified using techniques that directly probe ligand binding.
Materials and Methods
ABP, GBP, and RBP used as wild-type reference in this study contained additional mutations to introduce a single cysteine (ABP C64A L253C, GBP S112C, RBP A234C) to allow for fluorophore labeling. Single cysteines are also present in the designed receptors (2, 7).
Protein Expression and Purification.
Proteins were expressed and purified as described previously (19). For NMR experiments, M9 minimal medium with 15NH4Cl was used for expression.
A NiNTA column (Amersham Pharmacia, HisTrap HP 5 ml) was used for the first purification step. Initially, proteins were eluted with a linear imidazole gradient. For subsequent purifications, an optimized step elution protocol was developed for each protein, thus removing the need for concentrating the sample after this step. To increase purity and to remove higher order oligomers, an additional gel filtration step (GE Healthcare 26/60 Superdex 75 HiLoad) was performed. Purity of the proteins was checked by SDS/PAGE. To remove bound sugars, the proteins were dialyzed three times 1:100 against buffer.
Analytical Methods.
Analytical gel filtration was performed using a calibrated Superdex 75 10/300 GL column (GE Healthcare). Protein concentrations were adjusted to 1.0, 1.2, and 1.7 mg/ml for ABP, GBP and RBP, and receptor mutants, respectively. Of each protein solution, 500 μl was applied and eluted with gel filtration buffer (20 mM Mops, pH 6.9, 100 mM NaCl, 1 mM DTT).
CD spectra and thermal melting curves were recorded with a JASCO J-810 spectropolarimeter in a 1-mm cuvette. Proteins were dialyzed against 10 mM Tris, pH 8.0, and concentration adjusted to 0.3 mg/ml. In the case of Lac.A1, Lac.G1, PMPA.G12, and TNT.R1, 10 mM NaCl was present to ensure that the proteins stay in solution. To investigate stability increase due to ligand binding, saturating amounts of ligand (20-fold excess) were added for thermal melting curves.
ITC was performed using a VP-ITC (Microcal). The protein concentration was adjusted to 100 μM and ligand solution was prepared with the respective concentration in dialysis buffer (20 mM Mops, pH 6.9, 100 mM NaCl, 1 mM DTT). Measurements were performed at 25 °C with a stirring speed of 300 rpm, reference power 15 μcal/s, and spacing of 300 s between injections. The data were analyzed using the microcal LLC program. Binding constants for wild-type ABP, GBP, and RBP were determined from sigmoidal fits to data from two independent experiments.
Nuclear Magnetic Resonance.
15N labeled protein was concentrated up to 500 μM, if possible, in 20 mM Mops, pH 6.9, 100 mM NaCl, 1 mM DTT. Prior to measurement 50 μl of D2O were added to 450 μl of sample. For end-point titration, saturating amounts of ligand were added in lowest possible volume to impede dilution. All NMR data were acquired at 30 °C and in the case of Lac.A1 at 20 °C on 600 or 800 MHz on Bruker Avance III spectrometers equipped with pulsed field-gradient units and triple-resonance probes (for an overview of conditions, see Table S1). Because of the relatively large molecular weight of the proteins, transverse relaxation optimized spectroscopy techniques (TROSY) were applied to record 1H, 15N-correlation spectra. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution (22). The data were processed with the NMRPipe/NMRDraw package (23) and displayed with NMRView (24).
Crystallization, Data Collection, and Refinement.
Protein crystals of Stn.A2 were obtained by standard vapor diffusion of 15.7 mg/ml protein in 20 mM Tris, pH 8.0 with 2 mM of serotonin against 20% PEG 3350, 0.2 M (NH4)citrate within 1 week. After short transfer into crystallization buffer with 10% PEG 400 (vol/vol) as cryo-protectant, crystals were flash-frozen in liquid nitrogen. X-ray data of single crystals was collected with a MarCCD 225-mm detector at the synchrotron beamline PXII (Swiss Light Source, Villigen PSI). Data were indexed and processed with XDS and converted with XDSCONV (25). Molecular replacement searches were performed with Phaser (26) using the coordinates of wild-type ABP N-terminal (residues 2–109 + 254–285 of PDB-entry 5abp) and C-terminal domains (residues 110–253 + 286–306 of PDB-entry 5abp) as search models. Model building was performed in alternating rounds of computational refinement with REFMAC 5.4.0066 (27) and manual adjustments with the program Coot (28). The quality of the final structure was judged by Molprobity (29) and WhatCheck (30). No density corresponding to the ligand serotonin was present in the refined electron density maps. Data and refinement statistics for the Stn.A2 dataset are summarized in Table S2.
Structure Analysis.
The degrees of opening of molecules A and B were determined using DynDom (31). Structural superpositions were calculated with the Pymol (32) align function.
Supplementary Material
Acknowledgments.
We thank Sooruban Shanmugaratnam for technical assistance, Andrei Lupas, Reinhard Sterner, and Steffen Schmidt for comments on the manuscript, and the Höcker group for discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of have been deposited in the Protein Data Bank, www.pdb.org (PDB ID 2wrz).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907950106/DCSupplemental.
References
- 1.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 2.Looger LL, et al. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 3.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc Natl Acad Sci USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 7.Allert M, et al. Computational design of receptors for an organophosphate surrogate of the nerve agent soman. Proc Natl Acad Sci USA. 2004;101:7907–7912. doi: 10.1073/pnas.0401309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quiocho FA, et al. Substrate specificity and affinity of a protein modulated by bound water molecules. Nature. 1989;340:404–407. doi: 10.1038/340404a0. [DOI] [PubMed] [Google Scholar]
- 9.Cuneo MJ, et al. Ligand-induced conformational changes in a thermophilic ribose-binding protein. BMC Struct Biol. 2008;8:50. doi: 10.1186/1472-6807-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjorkman AJ, Mowbray SL. Multiple open forms of ribose-binding protein trace the path of its conformational change. J Mol Biol. 1998;279:651–664. doi: 10.1006/jmbi.1998.1785. [DOI] [PubMed] [Google Scholar]
- 11.Magnusson U, et al. Hinge-bending motion of D-allose-binding protein from Escherichia coli: three open conformations. J Biol Chem. 2002;277:14077–14084. doi: 10.1074/jbc.M200514200. [DOI] [PubMed] [Google Scholar]
- 12.Telmer PG, Shilton BH. Structural studies of an engineered zinc biosensor reveal an unanticipated mode of zinc binding. J Mol Biol. 2005;354:829–840. doi: 10.1016/j.jmb.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Marvin JS, Hellinga HW. Conversion of a maltose receptor into a zinc biosensor by computational design. Proc Natl Acad Sci USA. 2001;98:4955–4960. doi: 10.1073/pnas.091083898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vercillo NC, et al. Analysis of ligand binding to a ribose biosensor using site-directed mutagenesis and fluorescence spectroscopy. Protein Sci. 2007;16:362–368. doi: 10.1110/ps.062595707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Auria S, et al. Structural and thermal stability characterization of Escherichia coli D-galactose/D-glucose-binding protein. Biotechnol Prog. 2004;20:330–337. doi: 10.1021/bp0341848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehres DG, Hogg RW. Escherichia coli K12 arabinose-binding protein mutants with altered transport properties. Protein Sci. 1992;1:1652–1660. doi: 10.1002/pro.5560011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willis RC, Furlong CE. Purification and properties of a ribose-binding protein from Escherichia coli. J Biol Chem. 1974;249:6926–6929. [PubMed] [Google Scholar]
- 18.Vyas NK, et al. Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science. 1988;242:1290–1295. doi: 10.1126/science.3057628. [DOI] [PubMed] [Google Scholar]
- 19.de Lorimier RM, et al. Construction of a fluorescent biosensor family. Protein Sci. 2002;11:2655–2675. doi: 10.1110/ps.021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millet O, et al. The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy. Proc Natl Acad Sci USA. 2003;100:12700–12705. doi: 10.1073/pnas.2134311100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boas FE, Harbury PB. Design of protein-ligand binding based on the molecular-mechanics energy model. J Mol Biol. 2008;380:415–424. doi: 10.1016/j.jmb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pervushin K, et al. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Automatic-indexing of rotation diffraction patterns. J Appl Crystallogr. 1988;21:67–71. [Google Scholar]
- 26.McCoy AJ, et al. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, et al. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooft RW, et al. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 31.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 32.DeLano WL. DeLano Scientific. CA: Palo Alto; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.