Abstract
Adenine DNA glycosylase catalyzes the glycolytic removal of adenine from the promutagenic A·oxoG base pair in DNA. The general features of DNA recognition by an adenine DNA glycosylase, Bacillus stearothermophilus MutY, have previously been revealed via the X-ray structure of a catalytically inactive mutant protein bound to an A:oxoG-containing DNA duplex. Although the structure revealed the substrate adenine to be, as expected, extruded from the DNA helix and inserted into an extrahelical active site pocket on the enzyme, the substrate adenine engaged in no direct contacts with active site residues. This feature was paradoxical, because other glycosylases have been observed to engage their substrates primarily through direct contacts. The lack of direct contacts in the case of MutY suggested that either MutY uses a distinctive logic for substrate recognition or that the X-ray structure had captured a noncatalytically competent state in lesion recognition. To gain further insight into this issue, we crystallized wild-type MutY bound to DNA containing a catalytically inactive analog of 2′-deoxyadenosine in which a single 2′-H atom was replaced by fluorine. The structure of this fluorinated lesion-recognition complex (FLRC) reveals the substrate adenine buried more deeply into the active site pocket than in the prior structure and now engaged in multiple direct hydrogen bonding and hydrophobic interactions. This structure appears to capture the catalytically competent state of adenine DNA glycosylases, and it suggests a catalytic mechanism for this class of enzymes, one in which general acid-catalyzed protonation of the nucleobase promotes glycosidic bond cleavage.
Keywords: base-excision repair, general acid catalysis, substrate recognition
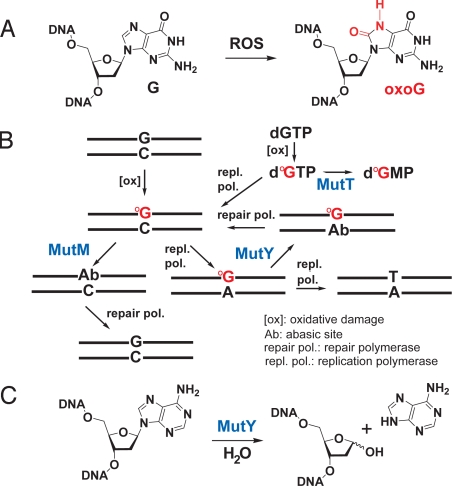
The genotoxic DNA lesion 8-oxoguanine (oxoG) arises chronically in cells through the attack of endogenous electrophilic oxidants on guanine residues (Fig. 1A). Left unrepaired, these oxoG lesions mispair with A during DNA replication, thereby giving rise to G·C to T·A transversion mutations. Most organisms possess a repair system dedicated to countering the deleterious effects of oxoG. The so-called GO system in eubacteria (1, 2) (Fig. 1B), for example, consists of proteins that directly sanitize the nucleotide precursor pool (MutT) and DNA (MutM or Fpg) of oxoG residues. Proteins that serve functions equivalent to those of MutT and MutM are found in all eukaryotic organisms (3). Failure of this first round of defense results in replicative production of oxoG:A pairs (4), which are particularly troublesome to repair, with neither nucleobase providing faithful information with which to direct correction of the other: oxoG does not belong in DNA at all, and although A is a canonical nucleobase, it is contextually aberrant in the oxoG:A pair because it replaces what should be a C. The multistep repair of oxoG:A is initiated by the highly evolutionarily conserved enzyme adenine DNA glycosylase (MutY in bacteria, hMYH in humans), which catalyzes hydrolysis of the glycosidic linkage between the oxoG-paired A and its sugar moiety (Fig. 1C) (5); the enzyme does not cleave A paired appropriately opposite T (6). The abasic site resulting from processing by MutY undergoes multistep repair to give primarily oxoG:C, which is then intercepted by MutM. The importance of the antimutagenic role played by adenine DNA glycosylases has been convincingly established upon the discovery that inherited loss-of-function mutations in hMYH are strongly associated with colorectal and gastric cancer, and in those cancers, with a dramatic increase in the incidence of G:C to T:A transversion mutations (7–9). The establishment of hMYH as a tumor suppressor protein has fueled interest in understanding how the protein functions to recognize and repair A residues paired opposite oxoG.
Fig. 1.
Generation and repair of oxoG:C and oxoG:A lesions in DNA. (A) Formation of the primary oxoG lesion by attack of reactive oxygen species (ROS) on a guanine residue. (B) The GO system, adapted from ref. 2. (C) Glycolytic reaction catalyzed by MutY.
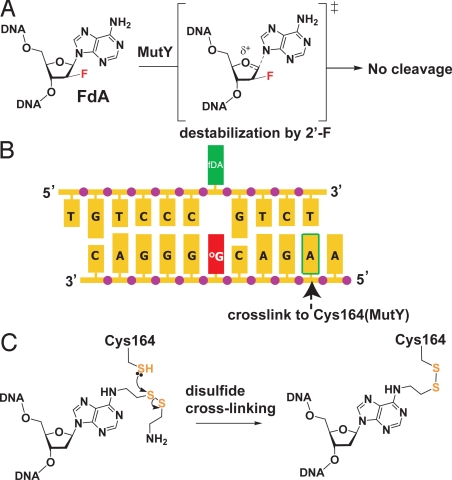
MutY, like hMYH, belongs to a large superfamily of structurally related DNA glycosylases having a signature helix-hairpin-helix motif followed by a Gly/Pro-rich loop and a key catalytic aspartic acid residue (HhH-GPD superfamily) (10, 11); members of this superfamily excise a wide variety of genotoxic lesions. The domain bearing the HhH-GPD motif lies at the N terminus of MutY and contains the active site pocket responsible for recognition and glycolytic excision of the substrate adenine (12, 13). MutY (and hMYH) also contains a C-terminal domain that is responsible for oxoG recognition and hence for discrimination in favor of oxoG:A and against T:A (14, 15). Mutation of the key catalytic Asp-144 residue in the thermophilic Bacillus stearothermophilus MutY to Asn (hereafter referred to as D144N MutY) yielded a recognition-competent but catalytically incompetent version of the protein suitable for structural studies (13). To stabilize the otherwise fleeting lesion-recognition complex (LRC) comprising D144N MutY bound to an oxoG·A-containing duplex, intermolecular disulfide cross-linking (DXL) was used. The structure thus obtained of the D144N MutY LRC revealed that, like nearly all DNA glycosylases, MutY completely extrudes the substrate adenine nucleoside from the DNA helix and inserts in into a deep extrahelical active site pocket on the N-terminal domain of the enzyme (13). The oxoG lesion is, on the other hand, fully intrahelical and makes extensive contacts to both the C- and N-terminal domains of the enzyme; residues from both domains also invade the DNA helix at the site left vacant by the extruded A. Curiously, in the D144N MutY LRC, no direct hydrogen bonding contacts were observed between the substrate A and residues of the active site pocket; instead, only water-mediated contacts were evident (see Fig. 3E). This was unexpected, as DNA glycosylases that exhibit a narrow substrate range, such as MutY, typically achieve their high specificity through direct hydrogen bonding interactions with active site residues (16–23). The absence of such contacts in the case of MutY suggested that either the protein employs a unique lesion-recognition strategy, or alternatively, that the structure captured in the D144N MutY LRC may not represent the final, catalytically competent state in the MutY-mediated base-extrusion pathway. We reasoned that a more definitive view of lesion recognition might be gained by obtaining additional structures of lesion recognition complexes. Rather than resort to active site mutations other than D144N, we elected instead to alter the DNA substrate so as to leave MutY with a fully wild-type active site. The most conservative alteration possible would consist of changing at most one atom in the substrate; such a single-atom replacement was indeed possible, because as we and others have shown, replacement of a single 2′-H in the substrate with 2′-F can completely abrogate catalysis by DNA glycosylases (24–29). As H is nearly isosteric with F, this subversion of catalysis is believed to result from destabilization of the O4′-C1′ oxocarbenium ion intermediate owing to the presence of the electron-withdrawing fluorine substituent at C2′, (Fig. 2A) (30, 31) as opposed to resulting from some debilitating structural perturbation; this made 2′-flourination an especially attractive option for the present purposes. We therefore set out to crystallize and structurally characterize a complex (hereafter designated the fluorinated lesion-recognition complex, or FLRC) comprising MutY having a fully wild-type active site bound to a DNA substrate rendered repair-incompetent by substitution of a single H atom by F on the substrate A residue (FdA residue). That structure, reported here, reveals the catalytically competent binding mode for A in the active site pocket, and suggests a catalytic mechanism featuring general acid catalysis of base-excision.
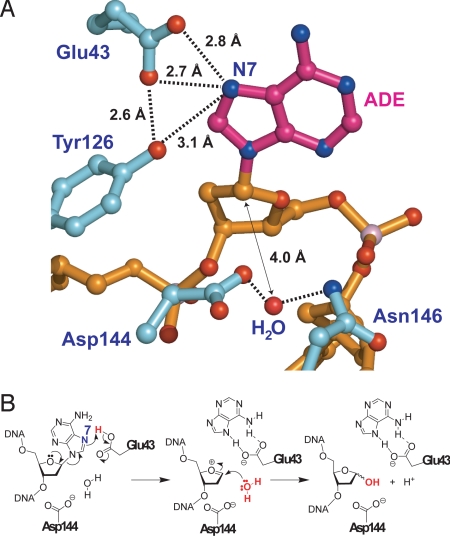
Fig. 3.
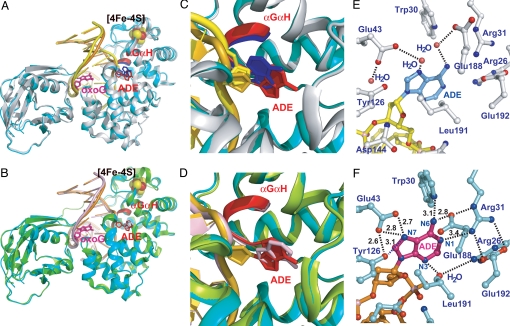
Structure of the FLRC. (A) Superposition of the FLRC (present work) and the LRC (PDB ID: 1RRQ). In the FLRC, the DNA is colored in gold, the substrate adenine in red, 8-oxoguanine in magenta, the protein backbone in cyan, the αGαH loop containing Asp-144 and Asn-146 in red, and the iron-sulfur cluster in yellow (sulfur) and red (iron) spheres. In the LRC, the coloring is the same except the DNA is yellow; substrate adenine, blue; protein backbone, gray; and the αGαH loop, blue. (B) Superposition of the FLRC and the reduced abasic complex with adenine soaked into the crystal (PDB ID: 1VRL). The color scheme is the same as in panel A, except for the reduced basic complex, for which the protein is green and the DNA and soaked adenine are pink. (C) A close-up view of the active site superposition of the FLRC and the LRC. The coloring scheme is the same as in panel A. (D) A close-up view of the active site superposition of the FLRC and the adenine-soaked complex. The coloring scheme is the same as in panel B; note the soaked adenine base in pink. (E) A view of the active site of the LRC. The substrate adenine is colored in sky blue, the phosphate backbone of the adenosine in yellow, an ordered water molecule is depicted as a red sphere, and the active site residues in gray. Hydrogen bonding interactions mediated by ordered water molecules are shown by a dotted line. (F) A view of the active site of the FLRC. The substrate adenine is colored in magenta, the phosphate backbone in gold, the active site residues in cyan, and an ordered water in red sphere. Direct hydrogen bonding interactions between the adenine and the active site residues are indicated by a dotted line with the distance on it. Numbers in panel F indicate distances in Ångstroms (Å) for direct contacts between the protein and the substrate adenine nucleobase.
Fig. 2.
Strategy for crystallization of the fluorinated lesion-recognition complex (FLRC). (A) Introduction of a fluorine atom into the substrate dA residue destabilizes the transition state (bracketed) leading to the oxocarbenium ion intermediate in glycolytic cleavage by MutY, thereby preventing the cleavage from occurring. (B) Sequence of the duplex DNA used in this work. For experiments that used intermolecular disulfide cross-linking (DXLing), the adenine residue indicated by the arrow was modified as shown in panel C, and this was cross-linked to a mutant form of Bst MutY containing a Cys residue engineered at position-164. (C) Structure of the modified adenine used in DXLing, and of the covalent linkage it forms with Cys-164 MutY.
Results and Discussion
Experimental System.
Experimental procedures are well-established for the synthesis of 2′-fluoro-2′-deoxynucleosides (FdAs) having either the 2′-ribo (α) or -arabino (β) configuration, and for their incorporation into DNA by standard phosphoramidite chemistry (16); we chose the β-epimer for the present studies, as this particular configuration leaves unmodified the face of the ribose ring expected to lie nearest the incoming nucleophilic water molecule. Nucleosides bearing a 2′-β-fluoro group are known to adopt the O4′-endo sugar pucker in DNA instead of the usual C2′-endo pucker (28, 32), but we felt it unlikely that this relatively minor difference in sugar conformation would substantially perturb the structure. In preliminary biochemical experiments, we found that wild-type Bst MutY was incapable of catalyzing base-excision on a duplex oligonucleotide containing a central FdA residue paired opposite oxoG (shown in Fig. 2B), whereas the protein could cleave the corresponding unfluorinated substrate having dA in place of FdA (data not shown). We then attempted to co-crystallize wild-type Bst MutY with the FdA·oxoG oligonucleotide, but these efforts failed to produce crystals. This was reminiscent of our experience with the original Bst MutY LRC, in which case we found it necessary to stabilize the complex and render it more homogeneous through intermolecular disulfide cross-linking (DXLing) to obtain high-quality crystals. Therefore, a DXL was introduced into the interface between MutY and the FdA:oxoG oligonucleotide at a position remote from the DNA lesion and the enzyme active site, so as to disturb neither (Fig. 2 B and C). Diffraction-quality crystals of this DXLed FLRC were obtained, and the structure was refined to 2.2 Å resolution using molecular replacement followed by cycles of refinement (see Table S1).
General Features of the MutY FLRC.
The overall architecture of the FLRC is very similar to that of the LRC (Fig. 3A), with the target A being extruded from the helical stack and inserted into the active site pocket on MutY; the oxoG is fully intrahelical, and the DNA duplex is sharply bent, with several amino acid residues penetrating the DNA at the oxoG site and the neighboring space left vacant upon departure of the substrate A (see below). The protein components of the LRC and FLRC complexes are nearly identical (heavy atom RMSD = 0.70 Å), with the only noteworthy divergence being at a small loop immediately adjacent to the extrahelical target nucleoside (αGαH loop; Fig. 3 A and C, red = FLRC, blue = LRC). The DNA in the FLRC also follows roughly the same course as in the LRC, and furthermore the features of oxoG recognition are essentially identical in the two complexes, with Tyr-88 and Gln-48 intercalating into the duplex on the 5′-side of the oxoG to create the DNA bend, and with numerous residues directly contacting both the Watson-Crick and Hoogsteen face of the oxoG nucleobase (Ser-308, Gln-48, Thr-49, and Leu-86; see ref. 13 and Fig. S1). That said, the DNA shows greater divergence than the protein component (heavy atom RMSD = 1.0 Å), especially at the extrahelical target A and 5′-to the base. All of the divergences mentioned above in the overall structure of the protein and DNA can be ascribed to a significant difference in the interaction between the target A and the MutY active site, with the extrahelical target base being plunged more deeply into the active site pocket on MutY in the FLRC than was evident in the LRC. These interactions are described in detail below. We also compared the structure of the FLRC with that of a product-like complex in which a free adenine base had been soaked into crystals of MutY bound to an abasic site analog (Fig. 3 B and D). Here, the protein and DNA in the two structures overlay very well (heavy atom RMSD = 0.53 Å), but the target A nucleobase still does not interact identically with the active site pocket.
Adenine Recognition.
As mentioned above, the extrahelical adenine in the LRC had no direct contacts with residues of the active site pocket, although two water-mediated contacts to N7 and 6-NH2 were evident (Fig. 3E). By contrast, in the FLRC, the deeper penetration of the substrate adenine nucleobase into the active site pocket enables it to make multiple hydrogen bonding contacts and also to engage in hydrophobic interactions with residues lining the pocket (Fig. 3F). Specifically, the side-chains of Glu-43 and Tyr-126 coordinately contact N7 of the substrate FdA, an interaction that would seem to indicate that the Glu-43 side-chain exists in the protonated form, rather than as the carboxylate anion. The side-chain guanidinium group of Arg-31 hydrogen bonds to N1 of FdA; the Glu-188 side-chain hydrogen bonds to N6 and also forms a salt bridge network with Arg-31, Glu-192, and Arg-26. These salt bridges make the active site pocket solvent-inaccessible by blocking the Watson-Crick face of the adenine; and Trp-30 pi-hydrogen bonds to the adenine N6. One water-mediated contact is observed, that being between the Arg-26 side-chain through an ordered water to the adenine N3. Hydrophobic active site side-chains, namely those of Ile-191, Leu-46, Val-51, Leu-28, and Trp-30 envelop the extrahelical adenine, providing further stabilization and exclusion of solvent water. In Escherichhia coli MutY, mutation of the residue corresponding to Val-51 (Val-45 in E. coli MutY) to Asn has been reported to abolish catalytic activity (12), implying that the hydrophobic interactions contributed by Val-51 and other active site residues make an important contribution to adenine recognition.
The Effect of the Asp144Asn Mutation.
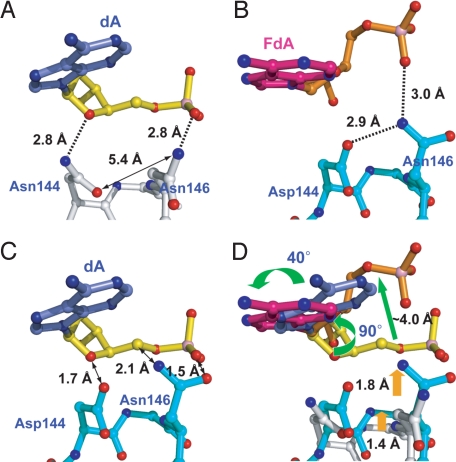
Can the differences in substrate adenine recognition between the LRC and FLRC be rationalized on the basis of the corresponding structures? In terms of chemical constitution, these structures are nearly identical, differing in just a few atoms out of some 8,000. Nevertheless, two points of difference, namely (i) the CO-NH2 group of Asn-144 (LRC) versus COO- of Asp-144 (FLRC) and (ii) the 2′-H of the substrate adenine (LRC) versus 2′-F (FLRC), are obviously associated with a substantive difference in structure. These amino acid residues are borne on the αGαH loop, which as mentioned earlier undergoes a significant positional shift in the FLRC versus the LRC. Our interpretation of the structures (Fig. 4) leads us to conclude that the key factor driving the LRC to adopt a noncatalytically-competent conformation for the substrate adenine is the presence of the mutated Asn-144 residue in place of the wild-type Asp. In the FLRC, Asp-144 hydrogen bonds to Asn-146 (Fig. 4B), which in turn hydrogen bonds with the 5′-phosphate of the substrate adenine residue (Fig. 4B). In the LRC, the hydrogen bonding contacts between Asn-144 and Asn-146 are absent (Fig. 4A). Instead, the amide group of Asn-144 hydrogen bonds with O4′ of the substrate adenine sugar moiety. To analyze how the protein conformation observed in the FLRC might be expected to interact with the DNA conformation in the LRC, and the protein conformation in the LRC with the DNA conformation in the FLRC, we superimposed these elements of the structures (Fig. 4C). Immediately evident in this superposition is the severe electronic and steric clash that would be engendered between the Asp-144/Asn-146 pair and the sugar and 5′-phosphate moiety of the substrate adenine, were the DNA in the FLRC to adopt the disposition seen in the LRC. The interaction modeled in Fig. 4C would undoubtedly be repulsive enough to force the substrate adenine sugar to move away from the conformation seen in the LRC, and this movement away is precisely what is observed in the FLRC structure (Fig. 4D). The same alternative side-chain conformation of residues 144 and 146 seen in the D144N mutant version of the Bst enzyme was also observed in the structure of the corresponding Asp-to-Asn mutant of E. coli MutY (D138N E. coli MutY, not bound to DNA, PDB ID: 1MUN; see ref. 12), whereas wild-type E. coli MutY (not bound to DNA, PDB ID: 1MUY) was found to exhibit the same 144/146 conformation as in the FLRC. These observations are consistent with the notion that the identity of the residue at position-144 controls the local conformation of the αGαH loop and the interaction between the side-chains of residues 144 and 146, with Asp-144/Asn-146 (wild-type) adopting the wild-type conformation seen in the FLRC, and Asn-144/Asn-146 adopting the alternative conformation seen in the LRC. The alternative conformation disfavors complete insertion of the adenine base into the active site pocket, while the wild-type conformation favors full insertion.
Fig. 4.
Effect of Asn versus Asp at position-144 on the interactions with Asn 146 and with DNA. (A) LRC structure, with the mutated residue Asn-144; (B) FLRC structure, with the wild-type active site Asp-144. Note the differences in the conformation of the protein with Asn-144/Asn-146 (A) versus Asp-144/Asn-146 (B); the former is referred to in the text as the alternative conformation. (C) Heavy atom superposition of the Asp-144/Asn-146-containing loop from the FLRC and substrate adenosine from the LRC. Note the steric clash indicated by a double-headed arrow. (D) Superposition of the substrate adenosine and the loop from both structures. Orange arrows indicate major positional shifts in the loop, and green arrows major shifts in DNA, upon conversion from the LRC to the FLRC.
A Mechanism for Catalysis of Glycolytic Cleavage by MutY.
The tight fit of the adenine base into the MutY lesion-recognition pocket in the FLRC structure, and the positioning of an ordered water molecule precisely positioned to intercept the oxocarbenium ion intermediate (Fig. 5A) leads us to propose that the active site structure in the FLRC is a close approximation of a catalytically competent state for this system. The positioning and inferred protonation states not only of the putative nucleophilic water molecule, and also of several key active site residues, suggest to a mechanistic proposal for catalysis of glycosidic bond scission by MutY. Biochemical and computational studies on DNA glycosylases favor a dissociative mechanism (33, 34), in which cleavage of the N-glycosidic bond and nucleophilic attack of water on the C1′ occur in discrete reaction steps (Fig. 5B). Bearing this in mind, we noted that in the FLRC, Glu-43 appears to make a bifurcated hydrogen-bonding interaction with the substrate adenine-N7, with the short distances observed between these two being energetically attainable only when Glu-43 is protonated. The proton on Glu-43 is expected to possess considerable acidity, and consequently partial or even full bonding of this proton to adenine-N7 would greatly facilitate glycosidic bond scission as depicted in Fig. 5B, by stabilizing the nucleobase leaving group in the reaction. Indeed, nonenzymatic cleavage of adenine-containing nucleosides is powerfully catalyzed by acid, and this is believed to proceed via protonation at N7 (35). In the enzyme active site, the charge relay between Glu-43 and adenine-N7 is further stabilized by interaction between the two and the side-chain hydroxyl of Tyr-126. The protonated N7 is consistent with an inverse 15N7 kinetic isotope effect seen with E. coli MutY (36). The observations that a Glu43Gln mutation in Bst MutY and a Glu37Ser mutation in E. coli MutY (equivalent to the Glu43Ser mutation in Bst MutY) abolish catalytic activity (see SI Text and Fig. S2) (12), as does use of the nonbasic analog 7-deazaadenine with the wild-type protein (37), can be rationalized in terms of general acid catalysis involving Glu-43 and adenine-N7. A transition-state analysis on E. coli MutY estimates that N7 protonation can lower the activation energy by ≈7 kcal/mol (36), which corresponds to acceleration of the excision rate by about 105 fold. The oxocarbenium ion intermediate formed by the initial dissociation step can be stabilized by Asp-144 (3.2 Å from C1′) located underneath C1′ of the sugar (33). The position of Asp-144 is physiologically relevant since, in addition to its well-recognized role as a helix cap (38, 39), Asp-144 is now optimally positioned to interact with the incipient electron-deficient oxocarbenium ion. Asp-144 is in close contact with the putative nucleophilic water (2.6 Å), which is further stabilized via hydrogen bonding to Asn-146 (2.9 Å); Asn-146 is buttressed via interactions with the 5′-phosphate (3.0 Å) and the O4′-oxygen (3.5 Å). A second-ordered water molecule is observed within hydrogen-bonding distance of the adenine-N3 (see Fig. 3D, not shown in Fig. 5A), but we ruled out this second water as the catalytic nucleophile, both because of its poor positioning but also because E. coli MutY efficiently processes the substrate analog 3-deazaadenine (40). The involvement of Glu-43 in protonating the substrate adenine and Asp-144 in deprotonating the catalytic water had been proposed earlier by Tainer and colleagues on the basis of the structure of a mutant E. coli MutY bound to adenine (corresponding residues Glu-37 and Asp-138) (12). The present structure thus supports this mechanistic proposal and clarifies the role of several other active site residues in catalysis of the base-excision repair reaction.
Fig. 5.
Adenine excision catalyzed by MutY. (A) A close-up view of the substrate adenosine interacting with catalytic residues in the FLRC. The same color scheme as in Fig. 4A. Important hydrogen bonding contacts are illustrated by dotted lines. The nucleophilic water candidate is in red sphere. Distance between C1′ of the substrate base and the water molecule is denoted by a double arrow. (B) A proposed glycolytic mechanism based on the structural information of the FLRC.
Materials and Methods
B. stearothermophilus MutY Preparation.
The Pro164Cys point mutation used for disulfide cross-linking was introduced into the B. stearothermophilus MutY gene by mega-primer mutagenesis using a parent construct wherein the gene was cloned into the pET28 (Novagen) expression vector, resulting in an ORF with an N-terminal His-tag and thrombin cleavage site (13). The protein was overexpressed at 18 °C in BL21(DE3)pLysS cells (Novagen). Cells were lysed by sonication after adding 1 mM PMSF, and the soluble lysates were subjected to Ni2+-NTA chromatography (Qiagen) followed by MonoQ column chromatography (Amersham Biosciences). After removing the His-tag by thrombin cleavage, the protein was further purified on a Superdex 75 gel-fliltration column (Amersham Pharmacia) using buffer containing 20 mM Tris, pH 7.4, 200 mM NaCl, and 5 mM βME.
Syntheses of 2′-Fluoro-2′-Deoxyadenine Phosphoramidite and Functionalized DNA.
2′-Fluoro-2′-deoxyadenine phosphoramidite was synthesized from 2′-fluoro-2′-deoxyadenosine (41) via tritylation followed by phosphitylation (24). DNA oligomers 5′-TGTCCC(FdA)GTCT-3′ (FdA, 2′-fluoro-2′-deoxyadenosinephosphoramidite) and 5′-AAGAC(oxoG)GGGAC-3′ (A, modified A with a disulfide tether for cross-linking, O6-Ph-dI and oxoG phosphoramidite purchased from Glen Research) were synthesized on an ABI 392 DNA synthesizer using standard reagents. To prepare the tether-containing DNA, the O6-Ph-dI-containing DNA was deprotected and cleaved off the resin using 1 mL concentrated aqueous NH4OH containing 0.25 mM β-ME for 6 h at 25 °C. The supernatant was collected and the resin washed with concentrated NH4OH. After concentrating the combined washes using a rotary evaporator, the oligo was treated with 1 mL 1 M aqueous cystamine containing 0.25 mM β-ME at 65 °C for 18 h. The solution was cooled to room temperature, neutralized by the addition of glacial acetic acid, desalted by a NAP column chromatography (Amersham Biosciences), and concentrated in a speed vac. The residue was purified by urea PAGE, dissolved in 10 mM Tris, pH 8.5, buffer, and annealed at 250 μM concentration.
Protein-DNA Cross-Linking Reactions and Crystallization.
The protein-DNA complex was formed by incubating 15 μM protein and 10 μM duplex DNA in a buffer containing 10 mM Tris, pH 8.5, 100 mM NaCl at 4 °C for 24 h. The complex was purified by monoQ column chromatography, and buffer exchanged into 20 mM Tris, pH 8.5, 50 mM NaCl and concentrated into 190 μM. The complexes were crystallized by the hanging-drop vapor diffusion method at room temperature using a buffer solution of 100 mM Tris, pH 8.5, 16% (wt/vol) PEG 4000, 200 μM Ca(OAc)2, 50 μM β-ME. Crystals appeared after several days and were allowed to grow for several weeks. Crystals were transferred to a cryoprotectant solution containing 100 mM Tris, pH 8.5, 16% (wt/vol) PEG 4000, 200 μM Ca(OAc)2, 50 μM β-ME, 25% glycerol and frozen in liquid nitrogen for data collection.
Data Collection and Structure Refinement.
Data were collected on beamline 24-ID at NE-CAT at the Advanced Photon Source at the Argonne National Laboratory and processed using HKL2000 (42). Crystals belong to the orthorhombic space group P212121 with unit cell dimensions of a = 38 Å, b = 86 Å, and c = 142 Å. Data collection statistics are summarized in TableS1. The coordinates of the protein from the structure of P164C/D144N B. stearothermophilus MutY bound to A·oxoG-containing duplex DNA (PDB ID: 1RRQ) were used as the initial search model in refinement using CNS. The active site residues and the duplex DNA were omitted from the initial search model. A rigid body fit and subsequent simulated annealing and energy minimizations performed in CNS provided a partial model. At this stage, electron density for the omitted active site residues and the DNA were clearly evident in a σA-weighed (43) Fo-Fc map. The model was improved by iterative rounds of simulated-annealing, energy minimization, and grouped B factor refinement in CNS (44) and model building in Coot (45) while monitoring Rfree (46). Simulated-annealing omit maps were frequently used to reduce model bias. Once the model was nearly complete, individual B-factor refinement was performed. Water molecules were added to the model using both automated methods (in CNS) and manual inspection of difference maps. Amino acid side chains of some surface residues were truncated at the α-, β-, γ-, and δ-carbon position if electron density was not visible for other atoms. The final model consists of amino acid residues 9–360 (residues 231–233 and 288–291 are missing), and 21 nucleotides of DNA, 1 calcium ion, and 44 water molecules. Refinement and model statistics are shown in TableS1. Figures were prepared using Pymol (http://pymol.sourceforge.net).
Supplementary Material
Acknowledgments.
We thank the entire staff of beamlines 24-ID of the Advanced Photon Source at Argonne National Laboratory for expert assistance with X-ray data collection and processing. This work was supported by National Institutes of Health Grant CA100742.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.A.T. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3G0Q).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902908106/DCSupplemental.
References
- 1.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 4.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 5.Williams SD, David SS. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acid Res. 1998;26:5123–5133. doi: 10.1093/nar/26.22.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manuel RC, Czerwinski EW, Lloyd RS. Identification of the structural and functional domains of MutY, an Escherichia coli DNA mismatch repair enzyme. J Biol Chem. 1996;271:16218–16226. doi: 10.1074/jbc.271.27.16218. [DOI] [PubMed] [Google Scholar]
- 7.Kim CJ, et al. Genetic alternations of the MYH gene in gastric cancer. Oncogene. 2004;23:6820–6822. doi: 10.1038/sj.onc.1207574. [DOI] [PubMed] [Google Scholar]
- 8.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 9.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash HM, et al. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein family. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 12.GuanY, et al. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 13.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 14.Noll DM, Gogos A, Granek JA, Clarke ND. The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine:adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- 15.Volk DE, et al. Structural similarities between MutT and the C-terminal domain of MutY. Biochemistry. 2000;39:7331–7336. doi: 10.1021/bi000416p. [DOI] [PubMed] [Google Scholar]
- 16.Mol CD, et al. Crystal structure and mutational analysis of human uracil-DNA glycosylase: Structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 17.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 18.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 20.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struc Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 21.Banerjee A, Yan W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 22.Lau AY, Wyatt MD, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair: Implications for DNA damage recognition, removal, and repair. DNA Repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Schärer OD, Verdine GL. A designed inhibitor of base-excision DNA repair. J Am Chem Soc. 1995;117:10781–10782. [Google Scholar]
- 25.Schärer OD, Kawate T, Gallinari P, Jiricny J, Verdine GL. Investigation of the mechanisms of DNA bindings of the human G/T glycosylase using designed inhibitors. Proc Natl Acad Sci USA. 1997;84:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chepanoske CL, Porello SL, Fujiwara T, Sugiyama H, David SS. Substrate recognition by Escherichia coli MutY using substrate analogs. Nucleic Acid Res. 1999;27:3197–3204. doi: 10.1093/nar/27.15.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doi Y, et al. Synthesis and characterization of oligonucleotides containing 2′-fluorinated thymine glycol as inhibitors of the endonuclease III reaction. Nucleic Acid Res. 2006;34:1540–1551. doi: 10.1093/nar/gkl061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman BR, Lee S, Wang S, Verdine GL. Structure of the Escherichia coli DNA glycosylase AlkA bound to the ends of duplex DNA: A system for the facile structure determination of lesion-containing DNA. Structure. 2008;16:1166–1174. doi: 10.1016/j.str.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J Am Chem Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He S, Withers SG. Assignment of sweet almond β-glucosidase as a family 1 glycosidase and identification of its active site nucleophile. J Biol Chem. 1997;272:24864–24867. doi: 10.1074/jbc.272.40.24864. [DOI] [PubMed] [Google Scholar]
- 31.Withers SG, Street IP, Bird P, Dolphin DH. 2-Deoxy-2-fluoroglucosides: A novel class of mechanism-based inhibitors. J Am Chem Soc. 1987;109:7530–7531. [Google Scholar]
- 32.Wilds CJ, Damha MJ. 2′-Deoxy-2′-fluoro-β-D-arabinonucleosides and oligonucleotides (2′F-ANA): Synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 34.Werner RM, Stivers JT. Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: Evidence for an oxocarbenium ion-uracil anion intermediate. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 35.Kampf G, Kapinos LE, Griesser R, Lippert B, Sigel H. Determination of intrinsic proton affinities of various basic sites. J Chem Soc Perkin Trans. 2002;2:1320–1327. [Google Scholar]
- 36.McCann JAB, Berti PJ. Transition-state analysis of the DNA repair enzyme MutY. J Am Chem Soc. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 37.Porello SL, Williams SD, Kuhn H, Michaels ML, David SS. Specific recognition of substrate analogs by the DNA mismatch repair enzyme MutY. J Am Chem Soc. 1996;118:10684–10692. [Google Scholar]
- 38.Eichman BF, O'Rourke EJ, Eadicella JP, Ellenberger T. Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J. 2003;22:4898–4909. doi: 10.1093/emboj/cdg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Structure and activity of a thermostable thymine-DNA glycosylase: Evidence for base twisting to remove mismatched normal DNA bases. J Mol Biol. 2002;315:373–384. doi: 10.1006/jmbi.2001.5264. [DOI] [PubMed] [Google Scholar]
- 40.Livingston AL, O'Shea VL, Kim T, Kool ET, David SS. Unnatural substrates reveal the importance of 8-oxoguanine in vivo mismatch repair by MutY. Nat Chem Biol. 2008;4:51–58. doi: 10.1038/nchembio.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma T, Lin J, Newton MG, Cheng Y, ChuCK Synthesis and anti-hepatitis B virus activity of 9-(2′-deoxy-2′-fluoro-β-L-arabinofuranosyl)purine nucleosides. J Med Chem. 1997;40:2750–2754. doi: 10.1021/jm970233+. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1999;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A. 1986;42:104–149. [Google Scholar]
- 44.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Brunger AT. Assessment of phase accuracy by cross validation: The free R value. Methods and applications. Acta Crystallogr D. 1993;54:905–921. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.