Abstract
Parkinson's disease (PD) is a complex multigenic neurodisorder frequently occurring in elderly persons. To investigate noncoding tiny microRNA mediated gene regulation, miRanda (version 1.0b) was used to predict human miRNA target sites on selected 29 genes related to PD. To verify output generated from miRanda, a similar analysis was performed only for microRNA target sites in 3′UTR using TargetScan (version 5.1). Data extracted by miRanda elucidates the mode of microRNA action based on the location of target sites in the Parkinson genes. Sites prone to action of multiple miRNAs were identified as “hot spots.” Important properties of each miRNA including multiplicity and cooperativity appear to contribute towards a complex interplay between miRNAs and their targets. Two sets of predicted results were explored for the occurrence of target sites of 112 miRNAs expressed in midbrain. Overall, convergence of results predicted by two algorithms revealed that 48 target sites for midbrain-specific miRNA occur in close proximity in 9 genes. This study will pave a way for selection of potential miRNA candidates for Parkinson's disease-related genes for quick therapeutic applications and diagnosis.
1. Introduction
Mysteries underlying the neurological disorders are as complex and bewildering as human mind itself. Aging related disorders such as Alzheimer and Parkinson's diseases are major culprits behind poor memory in elderly persons. Parkinson's disease has emerged as the second most common neurodegenerative disorder afflicting about 4 million people across the globe [1]. Steep rise in PD cases in aged population is quite distressing [2]. Though sporadic cases are more common yet in a significant fraction of western population, it can be attributed to delicate and fine tuning of gene regulation profile related to disorders [3]. PD is manifested as an outcome of interaction of copious genetic and environmental factors [4].
Parkinson's disease is characterized by motor impairments such as tremor of a limb usually restricted to one side of body. Concomitant symptoms including rigidity or stiffness of the limbs and trunk, akinesia, and impaired balance and postural instability [5–8] are often accompanied with depression to complete the clinical picture of PD. Onset of symptoms is the result of loss of neurons substantia nigra pars compacta causing a considerable decline in levels of Dopamine, a neurotransmitter. The hallmarks essential for PD diagnosis is the occurrence of eosinophilic proteinaceous inclusions, Lewy bodies in extant dopaminergic neurons. PD is incurable and various drugs prescribed for PD treatment offer merely symptomatic relief and contribute little to the halt of disease progression.
Since its description in 1817, very little was known about its etiology until recent days. The discovery of a series of genes involved in rare familial PD has instilled immense exhilaration and provided much needed impetus to research in this arena. There is mounting evidence that several genes like a-synuclein, Parkin, PTEN induced putative kinase 1 (PINK1), DJ-1, leucine-rich repeat kinase 2 (LRRK2), and ATP13A2 are misregulated in PD [9]. But whether these genes contribute in a common regulatory pathway or multiple parallel subpathways converging to same sequence in molecular pathogenesis of PD is yet to be resolved. microRNAs, generally known as negative regulators of gene expression, have attracted a lot of attention in recent times for their possible role on fine tuning of disease related genes. miRNAs are known to regulate approximately 30% of genes in human genome [10]. There is escalating evidence regarding the involvement of the abundant and endogenous 21–23 nt long RNA in various neurodegenerative disorders. Elucidation of precise biological function of these miRNAs has been the subject of many studies. miRNAs are involved in cell differentiation, development, apoptosis [11], stress resistance [12], tumor formation [13], and more importantly in neurodegenerative disorders [11, 14–16]. The establishment of role of miR-133b in mammalian midbrain dopaminergic neurons (DNs) has spurred a new interest in studies of the prospective function of these miRNAs in Parkinson's disease [17]. It appears that level of several miRNAs (miR-10a, miR-10b, miR-212, miR-132, and 495) modulates genes related to PD considerably [18]. Earlier studies provide some evidences about the involvement of miRNAs in Parkinson disease [19] but do not offer a full comprehensive view of microRNA dependent regulation of PD genes. Availability of simple, rapid, and accurate computer-based methods and development of efficient algorithms for micro RNA prediction have generated a great deal of interest [20, 21]. Here, our analysis reveals a complex interplay between microRNA and Parkinson genes for understanding the mechanism of PD pathogenesis. In practice, conventional biochemical miRNA profile is often encountered with several problems including transient, and low level of microRNA expression, tissue specificity and complex interaction with targets [20, 22]. Computational prediction of miRNA target sites can readily predict the role of miRNAs in the regulatory pathways.
The multifarious relationships shared by genes related to PD pathway, regulation of various miRNAs by other miRNAs in response to indefinite cues impose the need of an interaction map. This study aims at developing a complex interaction map between genes and microRNAs in the PD pathways, which will provide readymade clues for selection of miRNA using a comprehensive view of overall interplay between genes and microRNAs. This interaction map unravels some unexpected complexity in searching of microRNA target selection.
2. Methods
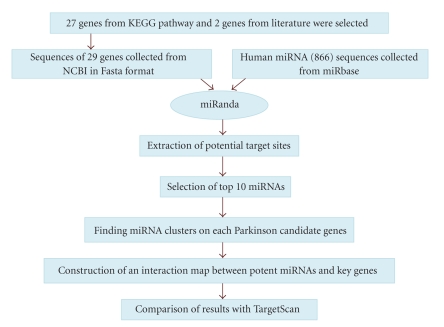
In the current study, we emphasized on identification of fraction of miRNA specific to the PD genetic pathway and propose an interaction network between these genes and targeted miRNA. Figure 1 represents stepwise workflow undertaken for the study.
Figure 1.
Schematic representation of the workflow.
2.1. Selection of Genes
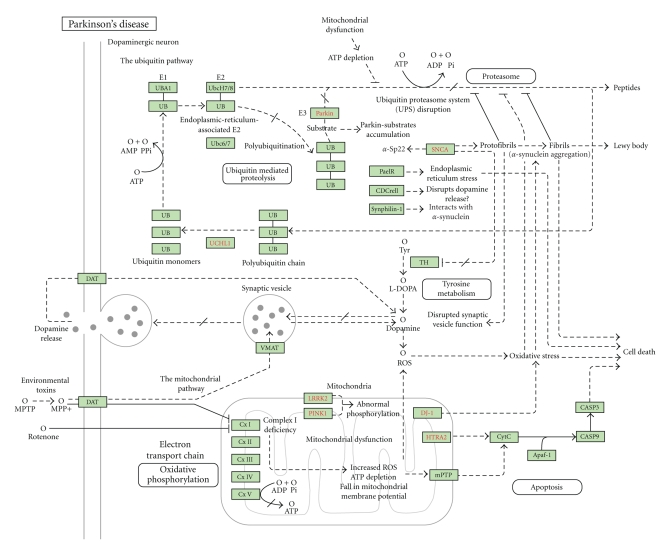
Parkinson's disease pathway (Figure 2) in Kyoto Encyclopedia of genes and Genomes (KEGG) available at http://www.genome.jp/kegg/ consists of 27 genes. Among them, genetic studies have identified few candidates such as parkin (PARK5, PARK6, PARK7, PARK8), alpha-syncline, NR4A2, synphilin-1, GBA, SNCA which, once mutated, can result Parkinson's disease like symptoms. Here, 2 more genes were also added to the PD network based on literature survey.
Figure 2.
Pathway showing genes involved in Parkinson's disease (Courtesy: KEGG pathway).
Genes selected are CASP3, CASP9, COX6B2, CYCS, GPR37, HTRA2, APAF1, UQCRFSL1, LOC100133737, LRRK2, NDUFS7, PARK2, PARK7, PTEN induced putative kinase1 (PINK1), SDHA, SEPT5, SLC6A3, SLC18A1, SLC25A4, SNCA, SNCAIP, TH, UBE1, UBE2J2, UBE2L3, ubiquitinB, UCHL1, GBA [23–25], NR4A2 [26]. Out of the 29 genes selected for this study, PINK1, PARK7, UBE2J2, GBA, and CASP9 are located on chromosome 1 while UCHL1, SNCA, CASP3, SLC25A4 are located on chromosome 4. Genes SDHA, SLC6A3, SNCAIP are present on chromosome 5. The position and location of each gene on the chromosomes is summarized (see Table S1 in supplementary Material available online at doi:10.1155/2009/363145). Sequences for the selected genes were collected in Fasta format from NCBI.
Total 866 human miRNA were downloaded in Fasta format from miRBASE (http://microrna.sanger.ac.uk/sequences/).
For prediction of miRNA targets, miRanda (version 1.0b) employs dynamic programming based on sequence complementarity, allotting higher weights to matches at the 5′ end of the mature miRNA while considering the free energy of the RNA-RNA duplex (calculated using RNAFold) [27] and the extent of conservation of the miRNA target across related genomes. miRanda [28, 29] software version 1.0b available at http://www.microrna.org/miranda was employed to predict target sites for these miRNAs. Cut-off values for prediction of target sites selected in analysis were Gap Open Penalty: 2.0, Gap Extend: 8.00 match score (S) ≥150.00, duplex free energy (ΔG) = −25.00 kcal/mol, Scaling Parameter (w) = 3.00. The selected gene sequences and human miRNA sequences were used as reference and query sequences, respectively, as input to miRanda. Conservation criterion was not used as it is reported that the nonconserved target sites may also play a part in repression.
Output generated from miRanda was used for target site identification and subsequent analysis. Top 10 microRNAs were selected based on the highest threshold match scores. Multiplicity and cooperativity were determined for all the miRNAs. Positions of the target sites for these miRNAs on 29 genes were explored and sites prone to multiple miRNAs were identified. After assembling the data for all genes, top 10 miRNAs were selected based on high multiplicity and ClustalW provided in mirBase was employed to investigate the conservation pattern in these microRNAs. A complex interaction map of interplay among 29 genes and top 10 miRNAs which showed maximum number of interaction with Parkinson disease associated genes was constructed.
2.2. Physico-Chemical Properties of Top 10 miRNAs
Important physico-chemical properties of 10 miRNAs (based on multiplicity) such as molecular weight (Kilo Dalton), free energy (δG in Kcal/mol), and composition were calculated by employing Oligo Calc: Oligonucleotide Properties Calculator [30].
Molecular weight and free energy were calculated by the following formulae:
| (1) |
where An, Un, Cn and Gn are the number of each respective nucleotide of the RNA molecule under consideration. Additionally, weight 159.0 gm/mole was added that accounts to 5′triphosphat.
| (2) |
Both (1) and (2) assume that the annealing occurs under the standard conditions of 50 nM primer, 50 mM Na+, and pH 7.0.
2.3. Validation of miRNA Target Prediction Using Multiple Programmes
In order to verify target sites of microRNA predicted solely by miRanda, TargetScan (Release 5.1) (31) was employed for prediction of target sites. miRNA targets in 3′UTR that are common in two miRNA prediction algorithms and their comparative analysis was provided.
Information regarding specific microRNAs that are expressed in midbrain was collected from WALK database (http://web.bioinformatics.ic.ac.uk/MSc07/WALK/mirna.html) and whole data was screened for target sites of midbrain-specific miRNAs.
3. Results and Discussion
Complex interplay of genes and miRNA appears to be a key factor in determining the delicate balance of disease controlling genes expression. Prediction of miRNAs using computer-based methods serves many advantages and aid in recognition of molecular hallmarks of the disease that can lead to development of effective screens for miRNA targets.
However, prediction of microRNA targets might suffer from numerous problems including tissue specific expression and lack of validation. Such weakness of in silico studies can be partially compensated by predicting targets using multiple programmes. The outputs can be effectively utilized for development of a molecular marker in diagnosis and prognosis.
3.1. miRNA Target Sites in Parkinson's Disease
Total 5501 miRNA binding target sites were predicted for 29 genes selected and the dataset of 866 human miRNA used for the analysis.
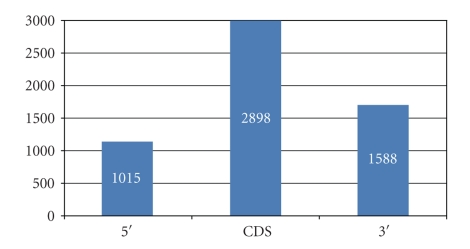
Initially, top 20 miRNA targets were selected on basis of top scores and stringent parameters for each gene. These sites were distributed diversely in 5′untranslated regions; coding region and 3′untranslated regions (UTR) (Figure 3, Figure SF1-29, and Table S2 in Supplementary Material). Surprisingly, more target sites were predicted in CDS region as compared to 3′UTR and 5′UTR. It was found that target sites predicted for CASP3, COX6B2, CYCS, SEPT5, SLC6A3, UBE2J2, UBE2L3 genes were more in 3′UTR. In contrast, no target was predicted in 3′UTR for GPR37, PINK1, SLC25A4, UQCRFSL1, LOC100133737 genes. Till recently it was believed that microRNA can target all the regions including 5′UTR, CDS, and 3′ UTR in plants and are restricted to 3′UTR in animals. But there is growing evidence that micro RNA can target the CDS [31–35] as well as 5′region [35, 36]. Location of target sites in a specified region also provides clues about mode of action of microRNAs, whether they are involved in transcriptional, posttranscriptional, and translational inhibition. Figure 4 shows the distribution pattern of miRNA target sites on the NDUFS7 genes.
Figure 3.
Distribution of predicted target sites in 5′UTR, CDS and 3′UTR as predicted using miRanda.
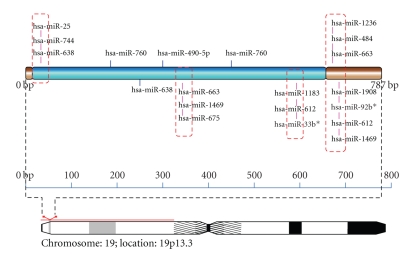
Figure 4.
Schematic representation of miRNA targets on NDUFS7.
3.2. Identification of Hotspots
Many microRNAs share either same target sites or sites located in vicinity of other microRNAs. Gathering of many miRNAs in the same site or vicinity area is commonly known as “hotspot.” Usually, miRNAs that occupy the same spot are coregulated and coexpressed [37, 38] and are involved in important biological functions [39–41]. In contrast, it is reasonable to anticipate that sequences in hotspot region might be occupied only by a predominant microRNA among the cluster members and a microRNA having affinity towards such sites may outcast other microRNAs by competitive selection. But how such selection of single microRNA occurs is still unknown. This may provide a reasonable clue regarding the variable effectiveness of microRNA pools. Researchers employ different criteria for defining cluster, mostly ranging from the presence of more than two miRNAs in a same chromosomal sites [36] occurrence in same orientation, absence of interfering transcription unit [42] or in some cases on the basis of phylogenetic relationships [37]. Keeping in view, the coannotation of these spots in important biological processes, 5501 miRNA targets were then further analyzed for identifying target prone regions in all Parkinson associated genes.
In the present study, we employed some preset criteria, which define a region as a hotspot if the region showed minimum 10 nucleotide overlap at starting position and possesses minimum overlapping five miRNA targets (see supplementary material). Total 288 regions were identified on all 29 genes spanned across 5′UTR, 3′UTR, and coding regions of the genes (Figure SF30-57 in supplement). Among all miRNA hotspots, 81, 77, and 130 regions were found in 5′UTR, 3′UTR, and coding region, respectively. Any region prone to multiple microRNA targeting could not be identified on CYCS and LOC100133737 genes. For all other genes, the number of hotspots varies between 1 and 26. It is anticipated that the genes that show the highest number of hotspots possibly undergo highest sensitivity in miRNA mediated regulation at transcriptional level because most of the sensitive sites reside in the 5′ regulatory domains of the genes. Genes such as PINK1, UBE2J2, SEPT5, and TH carried 26, 24, 23, 20 hotspots for miRNA action, respectively. The details of miRNA target prone sites against each gene are shown in Table S3. For example, in PINK1, 23 of the 26 sensitive target sites were found in the 5′UTR region, 3 regions are found in CDS only. Surprisingly, no hotspots could be identified in 3′UTR for PINK1. This revealed that PINK1 is not prone to translational inhibition. In reality, microRNA mediated transcriptional regulation is lacking broadly in animals, but frequent is plants. Abundance of predicted miRNA targets in the PINK1 regulatory regions depicts complexity of microRNA mediated target selection.
Conversely in UBE2J2, majority of such hotspots (19) were identified in 3′UTR while CDS region and 5′UTR showed only 1 and 4 hotspots, respectively. Depending on location of target sites, most likely UBE2J2 was regulated by the translational initiation. These results show that miRNAs can functionally target endogenous mRNAs in any region including coding region and 5′UTR and not restricted to the 3′UTR as described earlier. However, such dispersed distribution of miRNA target can generate more complexity in the nature of interaction, which counteracts the possibility of selection of most efficient miRNA required for functional knockdown of the PD genes.
3.3. Multiplicity and Cooperativity
In general term, one miRNA can target more than one gene (multiplicity), and one gene can be controlled by more than one miRNA (cooperativity) [29]. As reported earlier single miRNA can control hundreds of genes [43, 44]. Here, top 20 miRNAs displaying high scores were initially selected. After assembling all data, multiplicity and cooperativity were calculated. From the dataset, miRNAs that displayed maximum number of interactions with PD related genes were selected and used for constructing an interaction map. miRNAs showing maximum number of targets in PD related genes were selected as top 10 miRNA. Maximum interactions were found in 6 miRNAs, that is, hsa-miR-638, hsa-miR-1226*, hsa-miR-612, hsa-miR-612, hsa-miR1469, hsa-miR-608 and hsa-miR-939 that show high value of multiplicity. For instance, hsa-miR-638 exhibited high multiplicity showing total 119 interactions with 26 genes while no hsa-miR-638 target sites were found on three genes, namely, CASP3, PARK7, and SNCA. Defects in SNCA have been implicated in the pathogenesis of Parkinson's disease while autosomal recessive mutations in PARK7 cause early onset of Parkinson disease. CASP3 gene encodes a predominant caspase involved in cleavage of amyloid-beta 4A precursor protein, belonging to cysteine-aspartic acid protease (caspase) family which is associated with neuronal death in other neurological disorders. hsa-miR-1226* displayed 107 interactions with 25 genes but no complementary target sites could be identified for LOC100133737, PARK7, SNCA, ubiquitinB. Therefore, it can be inferred that these miRNAs may not be involved in regulation of activity of these genes.
Similarly, we analyzed the same dataset for estimating cooperativity. It was found that PINK1, SEPT5, TH exhibited high cooperativity towards top 10 miRNAs. All top 10 miRNA displayed 974 targets on the selected genes. It provides a complex picture which is difficult to comprehend the interactions. For example, PINK1 is regulated by 10 miRNAs at 81 positions, so these top 10 miRNAs demonstrate high cooperativity towards PINK1. Similarly, top 10 miRNAs exhibited 79, 78 targets in SEPT5 and TH genes. In contrast, a less degree of cooperativity was found in LOC100133737, SNCA genes suggesting a low sensitivity to, microRNA mediated regulation. Only three of 10 microRNA that is, hsa-miR-638, hsa-miR-612, and hsa-miR-661 exhibited miRNA targets on LOC10013373 (for details, see Table 1). Therefore, multiplicity and cooperativity add more complexity of microRNA selection and their sensitivity to large number of targets on PD genes.
Table 1.
Multiplicity and cooperativity for all miRNA data.
| miRNA/Genes | 1-CASP3 | 2-CASP9 | 3-COX6B2 | 4-CYCS | 5-GPR37 | 6-HTRA2 | 7-APAF1 | 8-LOC100128525 | 9-LOC100133737 | 10-LRRK2 | 11-NDUFS7 | 12-PARK2 | 13-PARK7 | 14-PINK1 | 15-SDHA | 16-SEPT5 | 17-SLC6A3 | 18-SLC18A1 | 19-SLC25A4 | 20-SNCA | 21-SNCAIP | 22-TH | 23-UBE1 | 24-UBE2J2 | 25-UBE2L3 | 26-ubiquitinB | 27-UCHL1 | 28-GBA8 | 29-NR4A2 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-638 | — | 3 | 6 | 5 | 8 | 10 | 3 | 2 | 1 | 2 | 9 | 3 | — | 4 | 1 | 12 | 5 | 1 | 4 | — | 3 | 9 | 4 | 6 | 2 | 2 | 2 | 6 | 6 | 119 |
| hsa-miR-1226* | 4 | 3 | 7 | 3 | 1 | 4 | 1 | 2 | — | 3 | 5 | 1 | — | 7 | 3 | 4 | 7 | 5 | 3 | — | 2 | 10 | 2 | 5 | 4 | — | 2 | 9 | 10 | 107 |
| hsa-miR-612 | 1 | 1 | 8 | — | 3 | 7 | — | 2 | 1 | 1 | 5 | 4 | 1 | 12 | 4 | 7 | 5 | 3 | 2 | 1 | 5 | 8 | 2 | 8 | 3 | 4 | 2 | 4 | 2 | 106 |
| hsa-miR-1469 | 1 | 2 | 3 | 1 | 6 | 12 | — | 2 | — | 1 | 6 | 3 | 1 | 9 | 4 | 14 | 5 | 1 | 2 | 1 | 1 | 8 | 2 | 7 | 2 | 3 | 2 | 3 | 4 | 106 |
| hsa-miR-939 | — | 4 | 7 | 6 | 2 | 4 | — | 3 | — | 2 | 1 | 2 | — | 5 | 2 | 1 | 7 | 5 | 3 | 1 | 8 | 10 | 6 | 3 | 5 | — | 1 | 5 | 6 | 99 |
| hsa-miR-661 | 4 | 2 | 6 | 1 | 6 | 9 | 5 | — | 1 | 1 | 3 | 2 | — | 7 | 4 | 11 | 3 | — | 2 | — | 2 | 3 | 4 | 4 | 2 | 5 | 1 | 3 | 2 | 93 |
| hsa-miR-1538 | 2 | 6 | 4 | 1 | 6 | 7 | — | 2 | — | 1 | 3 | 2 | — | 11 | 6 | 3 | 3 | 3 | 3 | 1 | 1 | 7 | 4 | 5 | 5 | — | 2 | 3 | 1 | 92 |
| hsa-miR-663 | 3 | 2 | 3 | 5 | 5 | 6 | 1 | 2 | — | 1 | 7 | 3 | — | 7 | 2 | 10 | 3 | 1 | 2 | — | 2 | 8 | 4 | 3 | 2 | — | 2 | 4 | 2 | 90 |
| hsa-miR-663b | 1 | 1 | 3 | — | 4 | — | — | 1 | — | — | 5 | 2 | 1 | 11 | 4 | 12 | 7 | — | 4 | — | 2 | 9 | 2 | 6 | 2 | — | 2 | 3 | 4 | 86 |
| hsa-miR-608 | 2 | 1 | 3 | 1 | 4 | 7 | — | — | — | 2 | 3 | 3 | 1 | 8 | 1 | 5 | 4 | 6 | 1 | — | 1 | 6 | 1 | 3 | 5 | — | — | 3 | 5 | 76 |
| Total | 18 | 25 | 50 | 23 | 45 | 66 | 10 | 16 | 3 | 14 | 47 | 25 | 4 | 81 | 31 | 79 | 49 | 25 | 26 | 4 | 27 | 78 | 31 | 50 | 32 | 14 | 16 | 43 | 42 | 974 |
Next, top 10 miRNAs were predicted based on the highest scores. hsa-miR-612 showed the maximum score 212. All the miRNAs having a score >=196 were selected and it was found that 6 miRNAs were common among both datasets, namely, miRNAs with the highest multiplicity and highest scores. Based on in silico prediction, we anticipated that these microRNAs may be master controller for regulating of PD pathway genes.
3.4. Unique miRNA Targets
microRNA targeting a single gene at single site can be termed as unique miRNA for a particular dataset and holds much importance, being specific towards a specific gene. Therefore, for targeting a specific gene these microRNAs might be useful, but single site targeting may not be sufficient to repress the activity of that gene to a desired level. It was also found that only 100 of 866 miRNAs displayed single interaction with Parkinson associated genes. Surprisingly, 3 out of 29 genes, SLC25A4, SNCAIP, and ubiquitin, did not exhibit targets for these unique miRNAs. They are not involved in single miRNA mediated regulation. One more gene UBE1 that encodes protein in the first step of ubiquitin conjugation to mark cellular proteins for degradation showed the highest no of single miRNA targets implying the need of the highest degree of specificity (Table S4 in Supplementary Material). These results predict a coherent interplay between variation of microRNA target and functional efficacy of PD genes. It is possible that simple computational prediction of microRNA targets may shed some light on the behavior of target genes involved in Parkinson disease.
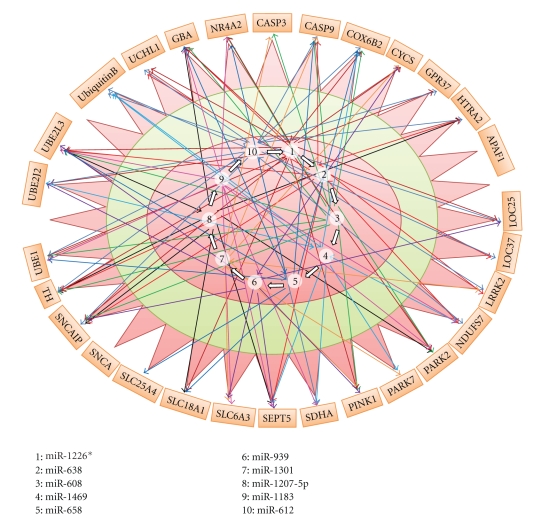
3.5. Deciphering Complexity through miRNA and Gene Interactions Map
To envisage the interrelationship of multiplicity of top 10 miRNA and their association with selected 29 genes, a gene-miRNA interaction map for Parkinson disease was constructed (Figure 5). Hsa-miR-612 showed highest number of interactions against 19 genes while four miRNAs hsa-miR-939, hsa-miR-1301, hsa-miR-1207-5p, hsa-miR-1183 showed interactions with only 9 genes. In contrast, counting the miRNA hits for individual gene, it was found that UBE2L3 exhibited 9 interactions with top scoring miRNAs whereas APAF1 and SNCA did not show any interaction with these miRNAs. These genes might not be under miRNA-based control. These findings suggest that computer-based prediction of microRNA target selection undoubtedly reduces the noise but is not sufficient to estimate efficiency of each microRNA on the multiple targets.
Figure 5.
Interaction map of miRNA and selected 29 genes. Interactions among genes and miRNAs are depicted with arrows.
Next, several important physico-chemical properties such as molecular weight (MW), free energy (δG in Kcal/mole), and the sequence composition features for the top ten miRNAs (based on multiplicity) were calculated employing Oligo Calc: Oligonucleotide tool to identify any potential specificity related to Parkinson's disease. The size of the miRNAs varied between 22 and 26 nucleotides. A narrow range of difference in properties was found among top 10 miRNAs. All microRNAs displayed a high GC content with 2 miRNAs, namely, hsa-miR-1469 and hsa-miR-663 showing exceptionally high GC composition of 91% (Table S5 in supplement). GC and AU content % of the human miRNome as taken from sequence feature statistics of 782 microRNAs from Argonaute database [45] ranges from 19.05% to 95% and 5 to 80.95%, respectively. The difference in properties is not enough for preferential selection of one miRNA from the others.
Finally, to investigate the sequence conservation of microRNA, Multiple Sequence Alignment (MSA) available at mirBase (http://microrna.sanger.ac.uk/) was used. A clear predominance of G-nucleotide site at 20th position was observed in all the top 10 miRNAs and a strong bias towards G was also found at 8th, 12th, and 15th positions.
Intersection of miRanda and TargetScan prediction —
In order to verify our above-mentioned findings depicted exclusively by miRanda, we have predicted only 3′UTR target sites using TargetScan [46] since prediction of TargetScan is limited to target sites only in 3′UTR. TargetScan relies on perfect seed complementarity and thus reduces false positive rate. Target sites predicted using miRanda and TargetScan were matched and compared (Figure 6 and Table S6).
Figure 6.
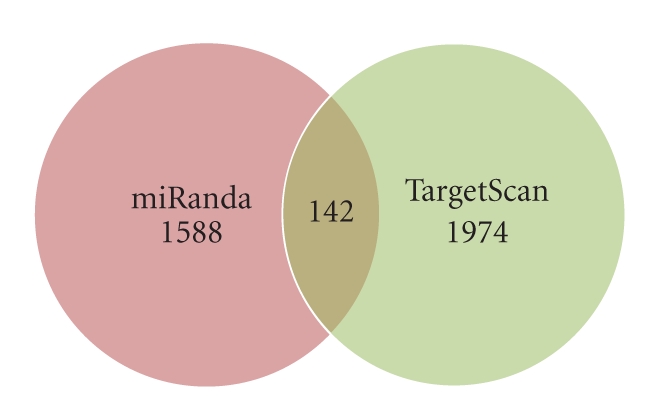
Intersection of miRanda and TargetScan.
Two genes LOC100128525 and LOC100133737 in Parkinson pathways were devoid of any predicted target sites in the 3′UTR regions as predicted using both the algorithms. Moreover, miRanda failed to predict target sites in PINK1 and SLC25A4 and resulted in prediction of target sites for only 25 genes while TargetScan predicted target sites for all remaining 27 genes. The number of targets sites predicted by two separate programmes is quite large as compared to common microRNA targets envisaged by both programmes. Target sites predicted based on only seed sequence similarity in TargetScan are greater than the 1588 sites predicted by miRanda. However, intersection includes a small number (142) of common target sites which is limited to only 12 genes (Table S7).
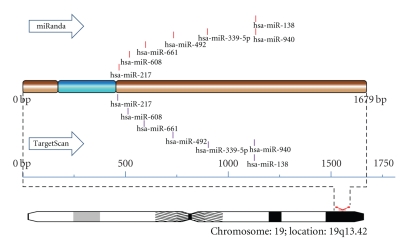
Using both algorithms, 7 microRNAs were predicted to have target sites in vicinity. Target sites for 7 microRNAs, namely, hsa-mir-138 (miRanda: 676–701, TargetScan: 693–699), hsa-miR-217 (miRanda: 6–36, TargetScan: 26–32), hsa-miR-339-5p (miRanda: 454–479,TargetScan: 471–477), hsa-miR-492 (miRanda: 274–293, TargetScan: 285–291), hsa-miR-608 (miRanda: 55–86, TargetScan: 77–83), hsa-miR-661 (miRanda: 163–187, TargetScan: 180–186), hsa-miR-940 (miRanda: 672–688, TargetScan: 681–687) were found in specified region. Similar results were found for other 8 genes and are summarized in Table 2.
Table 2.
Common microRNA target sites for various genes predicted using miRanda and TargetScan.
| Gene | microRNA | Start position of target sites predicted by miRanda | End position of target sites predicted by miRanda | Start position of target sites predicted by TargetScan | End position of target sites predicted by TargetScan |
|---|---|---|---|---|---|
| CASP9 | hsa-miR-224 | 240 | 264 | 255 | 261 |
| COX6B2 | hsa-miR-138 | 676 | 701 | 693 | 699 |
| hsa-miR-217 | 6 | 36 | 26 | 32 | |
| hsa-miR-339-5p | 454 | 479 | 471 | 477 | |
| hsa-miR-492 | 274 | 293 | 285 | 291 | |
| hsa-miR-608 | 55 | 86 | 77 | 83 | |
| hsa-miR-661 | 163 | 187 | 180 | 186 | |
| hsa-miR-940 | 672 | 688 | 681 | 687 | |
| CYCS | hsa-miR-1301 | 1812 | 1840 | 1830 | 1836 |
| hsa-miR-25 | 2191 | 2214 | 2204 | 2210 | |
| hsa-miR-510 | 2244 | 2273 | 2262 | 2268 | |
| hsa-miR-591 | 1332 | 1352 | 1341 | 1347 | |
| hsa-miR-658 | 2198 | 2232 | 2221 | 2227 | |
| hsa-miR-663 | 2283 | 2304 | 2286 | 2292 | |
| hsa-miR-769-5p | 1943 | 1974 | 1964 | 1970 | |
| hsa-miR-939 | 1990 | 2018 | 2007 | 2013 | |
| APAF1 | hsa-miR-650 | 639 | 662 | 654 | 660 |
| PARK2 | hsa-miR-361-3p | 21 | 40 | 31 | 37 |
| hsa-miR-574-5p | 96 | 118 | 110 | 116 | |
| hsa-miR-614 | 263 | 291 | 282 | 288 | |
| SEPT 5 | hsa-miR-1207-5p | 643 | 664 | 657 | 663 |
| hsa-miR-1250 | 671 | 696 | 687 | 693 | |
| hsa-miR-1825 | 600 | 622 | 614 | 620 | |
| hsa-miR-484 | 871 | 894 | 887 | 893 | |
| hsa-miR-572 | 877 | 899 | 880 | 886 | |
| hsa-miR-637 | 81 | 104 | 95 | 101 | |
| hsa-miR-663 | 160 | 183 | 176 | 182 | |
| hsa-miR-663b | 74 | 93 | 86 | 92 | |
| SLC6A3 | hsa-miR-1301 | 1613 | 1635 | 1626 | 1632 |
| hsa-miR-193b | 1518 | 1541 | 1533 | 1539 | |
| hsa-miR-331-3p | 331 | 351 | 342 | 348 | |
| hsa-miR-34a | 684 | 706 | 698 | 704 | |
| hsa-miR-449a | 686 | 706 | 698 | 704 | |
| hsa-miR-486-3p | 324 | 347 | 337 | 343 | |
| hsa-miR-601 | 406 | 432 | 423 | 429 | |
| hsa-miR-637 | 883 | 912 | 883 | 889 | |
| TH | hsa-miR-34c-5p | 211 | 240 | 230 | 236 |
| hsa-miR-658 | 217 | 245 | 236 | 242 | |
| hsa-miR-885-3p | 96 | 115 | 108 | 114 | |
| hsa-miR-920 | 107 | 125 | 116 | 122 | |
| GBA | hsa-miR-1233 | 12 | 33 | 25 | 31 |
| hsa-miR-212 | 363 | 386 | 376 | 382 | |
| hsa-miR-331-3p | 109 | 133 | 124 | 130 | |
| hsa-miR-331-3p | 159 | 178 | 169 | 175 | |
| hsa-miR-519e | 4 | 26 | 18 | 24 | |
| hsa-miR-637 | 197 | 223 | 215 | 221 | |
| hsa-miR-661 | 452 | 476 | 467 | 473 | |
| hsa-miR-766 | 160 | 183 | 175 | 181 | |
Midbrain Specific microRNAs. The predicted microRNA targets were also screened for a subset of microRNA expressed in the midbrain. It comprises of 112 microRNAs. In all, 378 target sites were predicted for 60 midbrain-specific miRNAs for miRanda across the entire length of gene (Table S8). Among them, 343 target sites were predicted for 99 midbrain-specific microRNAs using TargetScan while only 115 target sites were identified for 43 midbrain-specific microRNAs using miRanda for 3′UTR (Tables S9 and S10 in Supplementary Material). Only 39 miRNAs were found to be common in both algorithms for which 105 and 136 target sites were predicted by miRanda and TargetScan, respectively. Only target sites for hsa-miR-138 in COX6B2 (miRanda: 676–701, TargetScan: 693–699) and hsa-miR-25 in CYCS (miRanda: 2191–2214, TargetScan: 2204–2210) were predicted to occur in close proximity. The variation in results can be attributed to difference in set parameters in two independent algorithms.
4. Conclusion
Neurological disorders are complex diseases in which a wealth of information remains hidden owing to a variety of regulatory complexity. Despite of numerous studies on Parkinson diseases, existence of multiple parallel pathways or their convergence at a point is still a puzzle. The previous studies have pointed out the role of miRNAs in the etiology of PD. Absence of efficient treatment methods tends us to broaden our understanding of the candidate genes involved in disease pathway. Knowledge of putative miRNA targets on these genes achieved using fast computer-based assays will be a reasonable and powerful advancement for understanding PD. Repression of candidate genes involved in pathogenesis by a single miRNA or a group of miRNAs may aid in combating this disease. In order to offer a simplistic view of perplexing relationship of gene and miRNAs, we propose an intricate and comprehensive microRNA-gene interaction map in the PD pathway. For gaining an insight about the mechanism of these miRNA actions, distribution of these target sites at different regions was explored. Importantly, it was found that miRNA target sites are not restricted only to 3′UTR but are distributed across the entire length of gene. These hotspots may be representing favored sites for miRNA-based regulation. The miRNA and gene networks of a particular trait are poorly understood. Interaction map also provides a way for selecting important miRNAs markers required for diagnosis and therapeutics. Finally, though physiochemical properties of microRNA narrate some indicative parameters yet it is not enough to understand the complex relationship of Parkinson's disease genes and microRNAs. The weakness of the work is that as most of the computational miRNA prediction algorithms focus on 3′UTR and a number of miRNA in these search algorithms represent only a fraction of total miRNA available in microRNA repositories like mirBase, it is difficult to validate the target sites for those miRNAs and also in 5′UTR and CDS. Therefore, computational prediction may not guide optimize selection of any single microRNA for efficient knock down and level of action and further validation of these findings using experimental approaches is required. Further analysis and experimental validation of these results is mandatory for resolving complexity for the selection of microRNA targets in future.
Supplementary Material
Supplementary Materials contains Supplementary Figures showing schematic representations of miRNA targets as well as a series of Supplementary Tables.
Figure 7.
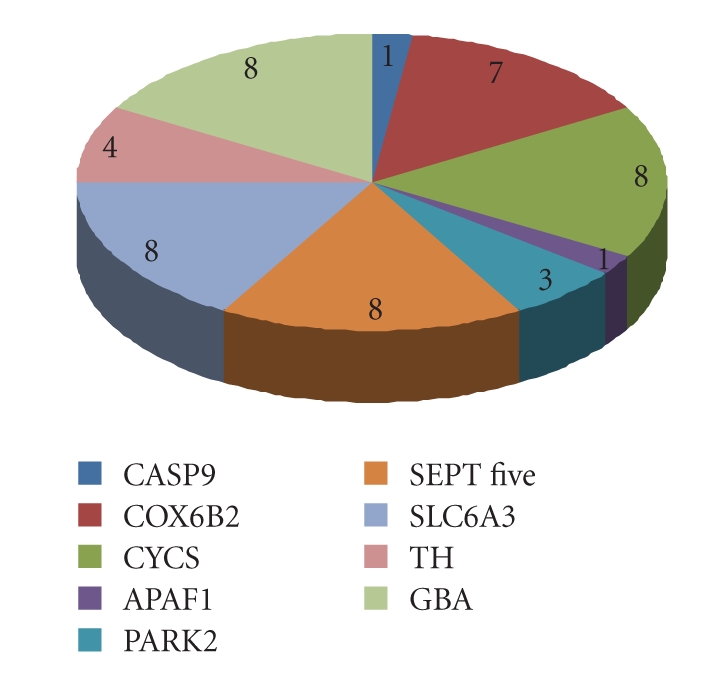
Common target sites predicted using miRanda and Targetscan, where datalabels show the number of target sites predicted for each gene.
Figure 8.
Schematic representation of miRNA targets on COX6B2.
Acknowledgments
This work was funded by grants from DBT and Wellcome Trust Senior International Fellowship to M. Pal-Bhadra and U. Bhadra, Human Frontier Science Program and DST CSIR Network (NWP 34, NWP35) projects to U. Bhadra. The authors thank anonymous reviewers for their critical and useful comments for improvement of manuscript.
References
- 1.Fitzgerald JC, Plun-Favreau H. Emerging pathways in genetic Parkinson's disease: autosomal-recessive genes in Parkinson's disease—a common pathway? FEBS Journal. 2008;275(23):5758–5766. doi: 10.1111/j.1742-4658.2008.06708.x. [DOI] [PubMed] [Google Scholar]
- 2.Wood-Kaczmar A, Gandhi S, Wood NW. Understanding the molecular causes of Parkinson's disease. Trends in Molecular Medicine. 2006;12(11):521–528. doi: 10.1016/j.molmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Davie CA. A review of Parkinson's disease. British Medical Bulletin. 2008;86(1):109–127. doi: 10.1093/bmb/ldn013. [DOI] [PubMed] [Google Scholar]
- 4.Thomas B, Beal MF. Parkinson's disease. Human Molecular Genetics. 2007;16(2):183–194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 5.Gasser T. Genetics of Parkinson's disease. Journal of Neurology. 2001;248(10):833–840. doi: 10.1007/s004150170066. [DOI] [PubMed] [Google Scholar]
- 6.Schapira AHV, Bezard E, Brotchie J, et al. Novel pharmacological targets for the treatment of Parkinson's disease. Nature Reviews Drug Discovery. 2006;5(10):845–854. doi: 10.1038/nrd2087. [DOI] [PubMed] [Google Scholar]
- 7.Cookson MR. The biochemistry of Parkinson's disease. Annual Review of Biochemistry. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 8.Jankovic J. Parkinson's disease: clinical features and diagnosis. Journal of Neurology, Neurosurgery and Psychiatry. 2008;79(4):368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 9.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annual Review of Neuroscience. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 10.Rajewsky N. microRNA target predictions in animals. Nature Genetics. 2006;38(supplement 1):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. NeuroReport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 15.Saba R, Goodman CD, Huzarewich RLCH, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS ONE. 2008;3(11, article e3652) doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushati N, Cohen SM. microRNAs in neurodegeneration. Current Opinion in Neurobiology. 2008;18(3):292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Inoue K, Ishii J, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillardon F, Mack M, Rist W, et al. MicroRNA and proteome expression profiling in early-symptomatic α-synuclein(A30P)-transgenic mice. Proteomics-Clinical Applications. 2008;2(5):697–705. doi: 10.1002/prca.200780025. [DOI] [PubMed] [Google Scholar]
- 19.Nelson PT, Wang W-X, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathology. 2008;18(1):130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Letters. 2005;579(26):5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 21.Brown JR, Sanseau P. A computational view of microRNAs and their targets. Drug Discovery Today. 2005;10(8):595–601. doi: 10.1016/S1359-6446(05)03399-4. [DOI] [PubMed] [Google Scholar]
- 22.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RHA, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 23.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson's disease in Ashkenazi Jews. The New England Journal of Medicine. 2004;351(19):1972–1977. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 24.Gan-Or Z, Giladi N, Rozovski U, et al. Genotype-phenotype correlations between GBA mutations and Parkinson's disease risk and onset. Neurology. 2008;70(24):2277–2283. doi: 10.1212/01.wnl.0000304039.11891.29. [DOI] [PubMed] [Google Scholar]
- 25.Tan E-K, Tong J, Fook-Chong S, et al. Glucocerebrosidase mutations and risk of Parkinson's disease in Chinese patients. Archives of Neurology. 2007;64(7):1056–1058. doi: 10.1001/archneur.64.7.1056. [DOI] [PubMed] [Google Scholar]
- 26.Healy DG, Abou-Sleiman PM, Ahmadi KR, et al. NR4A2 genetic variation in sporadic Parkinson's disease: a gene wide approach. Movement Disorders. 2006;21:1960–1963. doi: 10.1002/mds.21018. [DOI] [PubMed] [Google Scholar]
- 27.Wuchty S, Fontana W, Hofacker IL, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49(2):145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 28.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biology. 2003;5(1):R1–R12. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biology. 2004;2(11, article e363) doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Research. 2007;35(2):W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5′-untranslated region and coding sequence. Genetica. 2009;137(2):159–164. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 32.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lal A, Kim HH, Abdelmohsen K, et al. p16INK4a translation suppressed by miR-24. PLoS ONE. 2008;3(3, article e1864) doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 36.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 37.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. Journal of Molecular Biology. 2004;339(2):327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 38.Tanzer A, Stadler PF. Evolution of microRNAs. Methods in Molecular Biology. 2006;342:35–350. doi: 10.1385/1-59745-123-1:335. [DOI] [PubMed] [Google Scholar]
- 39.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic MicroRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Research. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 40.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a MicroRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. Journal of Virology. 2005;79(14):9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R, Wang Y-Q, Su B. Molecular evolution of a primate-specific microRNA family. Molecular Biology and Evolution. 2008;25(7):1493–1502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]
- 42.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biology. 2005;3(3, article e85) doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 45.Shahi P, Loukianiouk S, Bohne-Lang A, et al. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Research. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials contains Supplementary Figures showing schematic representations of miRNA targets as well as a series of Supplementary Tables.