Abstract
Farnesyl diphosphate (FPP) is a substrate for a diverse number of enzymes found in nature. Photoactive analogues of isoprenoid diphosphates containing either benzophenone, diazotrifluropropionate or azide groups have been useful for studying both the enzymes that synthesize FPP as well as those that employ FPP as a substrate. Here we describe the synthesis and properties of a new class of FPP analogues that links an unmodified farnesyl group to a diphosphate mimic containing a photoactive benzophenone moiety; thus, importantly, these compounds are photoactive FPP analogues that contain no modifications of the isoprenoid portion of the molecule that may interfere with substrate binding in the active site of an FPP utilizing enzyme. Two isomeric compounds containing meta- and para-substituted benzophenones were prepared. These two analogues inhibit S. cerevisiae protein farnesyltransferase (ScPFTase) with IC50 values of 5.8 (meta isomer) and 3.0 µM (para isomer); the more potent analogue, the para isomer, was shown to be a competitive inhibitor of ScPFTase with respect to FPP with a KI of 0.46 µM. Radiolabeled forms of both analogues selectively labelled the β-subunit of ScPFTase. The para isomer was also shown to label E. coli farnesyl diphosphate synthase and Drosophila melanogaster farnesyl diphosphate synthase. Finally, the para isomer was shown to be an alternative substrate for a sesquiterpene synthase from Nostoc sp. strain PCC7120, a cyanobacterial source; the compound also labeled the purified enzyme upon photolysis. Taken together, these results using a number of enzymes demonstrate that this new class of probes should be useful for a plethora of studies of FPP-utilizing enzymes.
Keywords: Benzophenone, Farnesyl Transferase, Farnesyl Diphosphate Synthase, Sesquiterpene Cyclase, Photoaffinity Labeling
1. Introduction
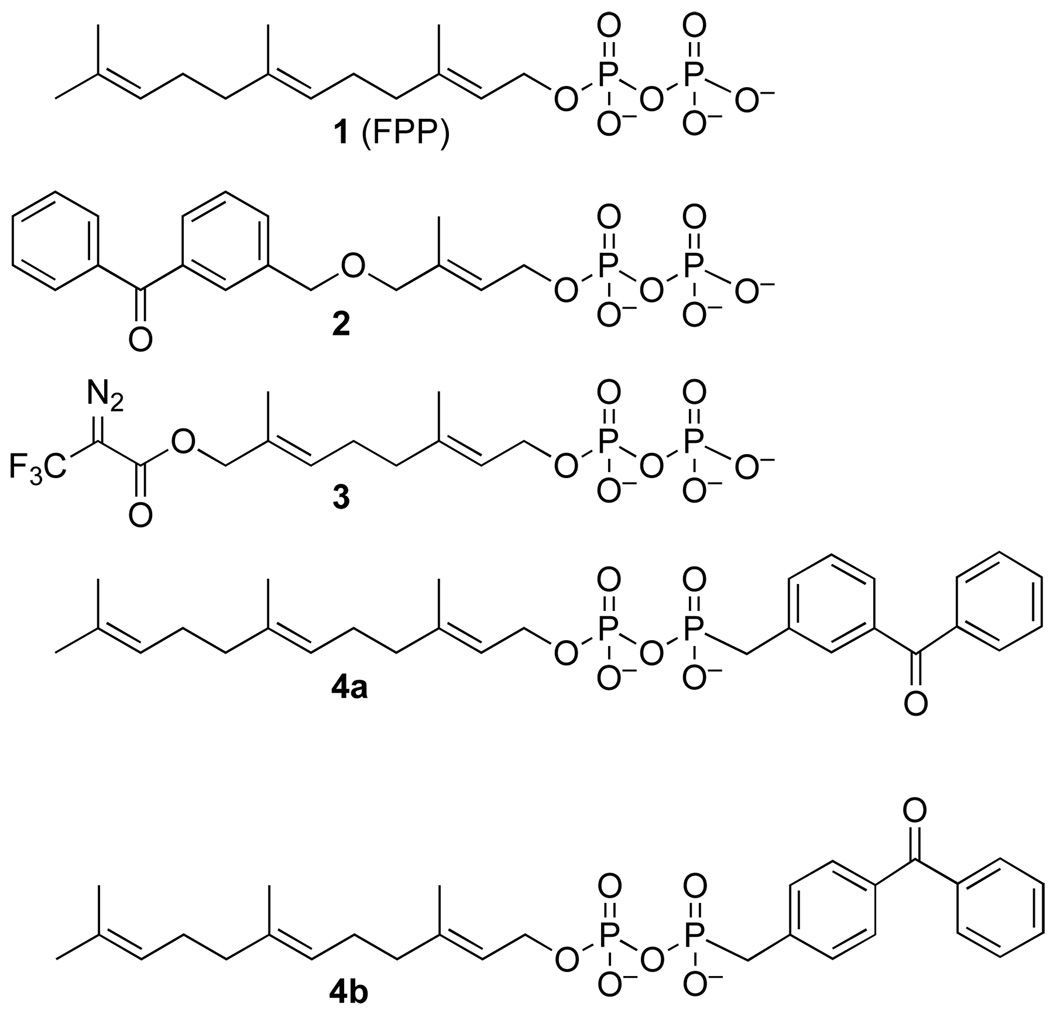
Farnesyl diphosphate (FPP, 1) is a substrate for a large number of enzymes found in nature. The products of such enzymes include sesquiterpene natural products,1, 2 side-chains of cofactors,3 biopolymers such as latex4 and lipidated proteins.5 Given the vast number of natural products derived from FPP and the medicinally important properties of many of them, the development of new probes that can be used to identify FPP-utilizing enzymes is an important area of inquiry.6 Photoactive analogues of isoprenoid diphosphates containing either benzophenone,7–16 diazotrifluropropionate17–23 or azide24, 25 groups have been useful for studying both the enzymes that synthesize FPP as well as those that employ FPP as a substrate such protein prenyltransferases. To date, most photoaffinity labeling analogues of FPP (see Figure 1, 2 and 3) incorporate the light-absorbing functionality into the isoprenoid moiety itself. While such molecules have proved to be useful, they are limited by the fact that they are only capable of labeling residues located within the isoprenoid binding site. Moreover, in cases where the overall size and shape of the active site is highly constrained, probes that incorporate bulky modifications in the prenyl group may not effectively bind. Those limitations led us to design a new type of FPP analogue (4a and 4b) that would incorporate an unmodified farnesyl group linked to a diphosphate mimic containing a photoactive benzophenone moiety. Here we describe the syntheses of these two analogues and evaluate their ability to bind and label several different FPP utilizing enzymes.
Figure 1.
Farnesyl diphosphate (FPP) and photoactive analogues
Figure 2.
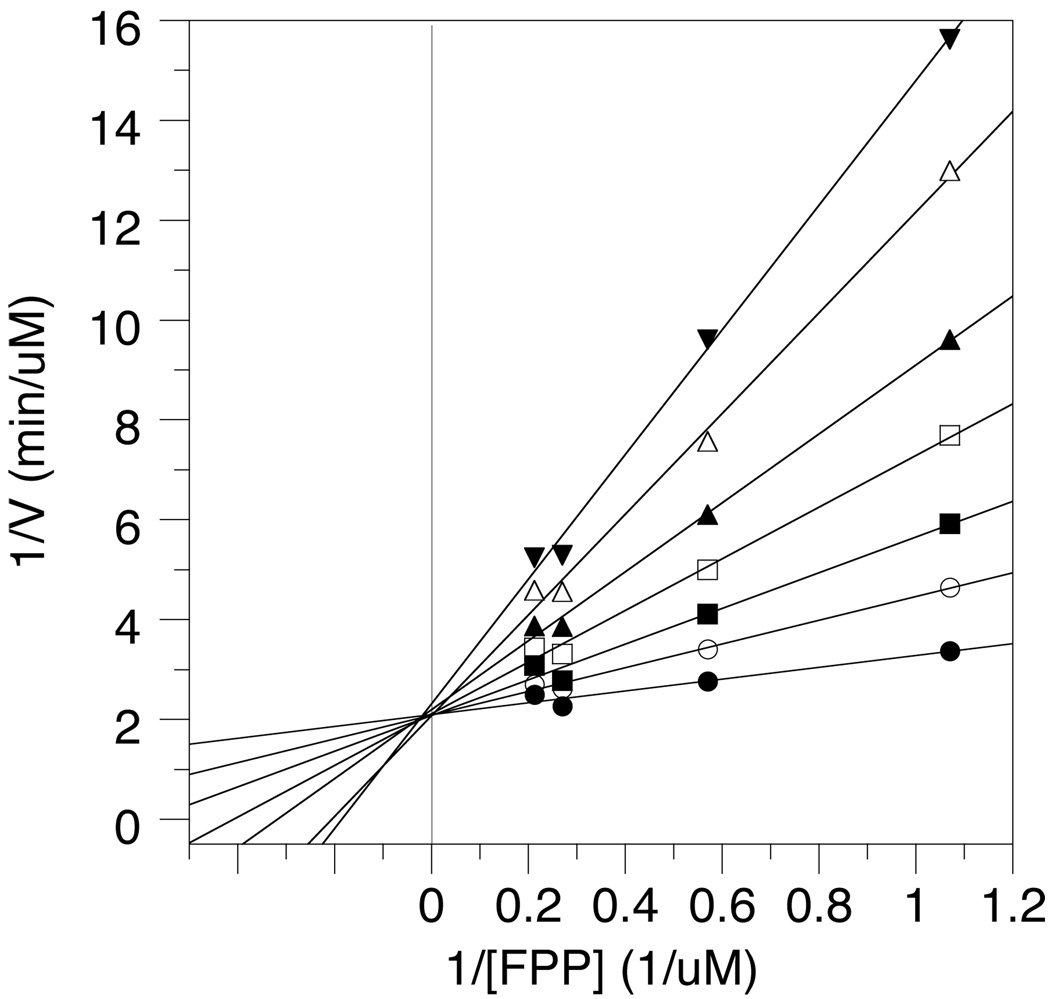
Double reciprocal plot showing the competitive inhibition of ScPFTase by 4b. The concentrations of 4b used were as follows: (unknownsymbol28) 0 µM, (O) 0.2 µM, (▪) 0.4 µM, (□) 0.7 µM, (▴) 1.0 µM, (Δ) 1.5 µM, (▾) 2.0 µM.
Figure 3.
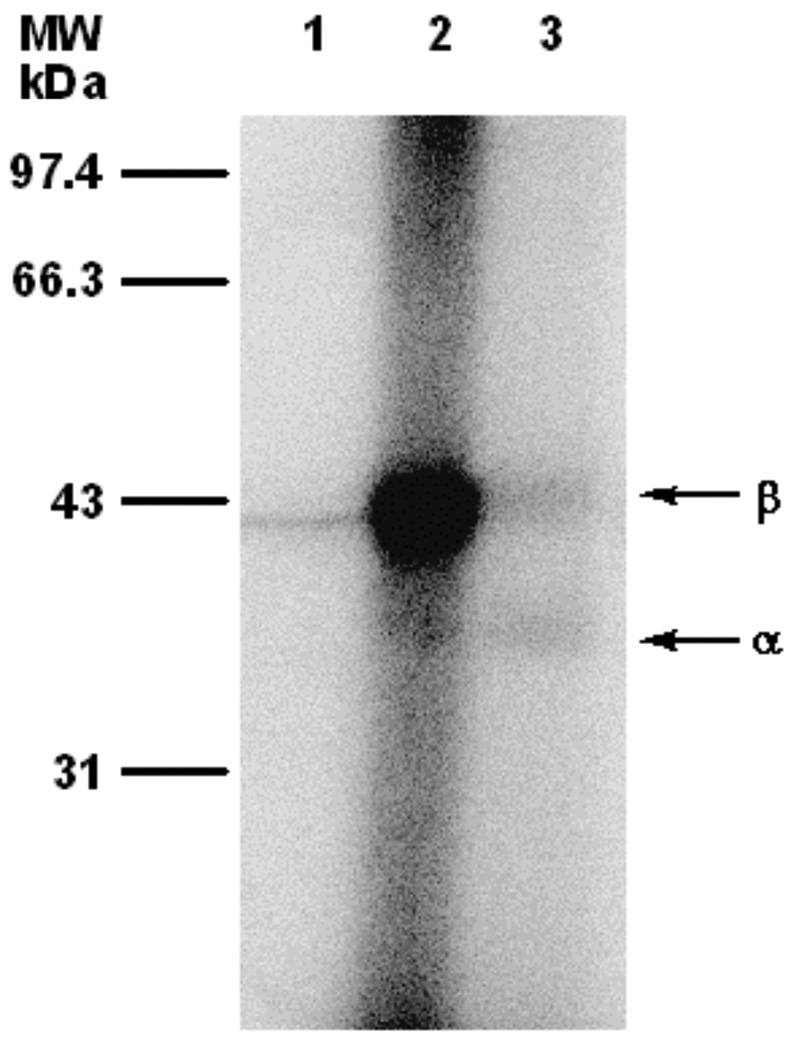
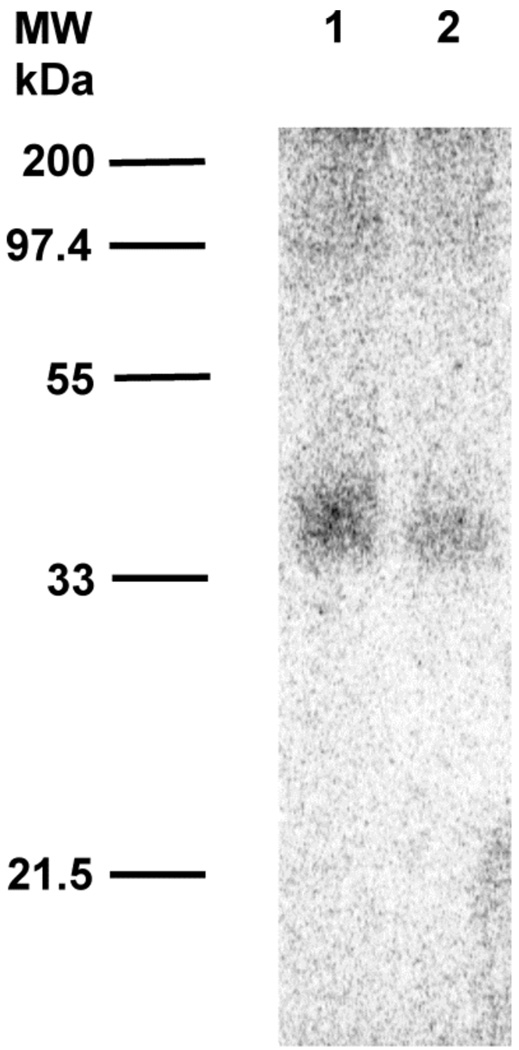
Phosphorimaging analysis of photolabeling of ScPFTase with [32P]4b after SDS-PAGE fractionation. Lane 1: ScPFTase and [32P]4b (29 µM), no UV irradiation. Lane 2: ScPFTase and [32P]4b (29 µM), with UV irradiation. Lane 3: PFTase, [32P]4b (29 µM), and FPP (100 µM), with UV irradiation.
2. Results
2.1. Synthesis of farnesyl diphosphate analogue incorporating a benzophenone photophore into the diphosphate group
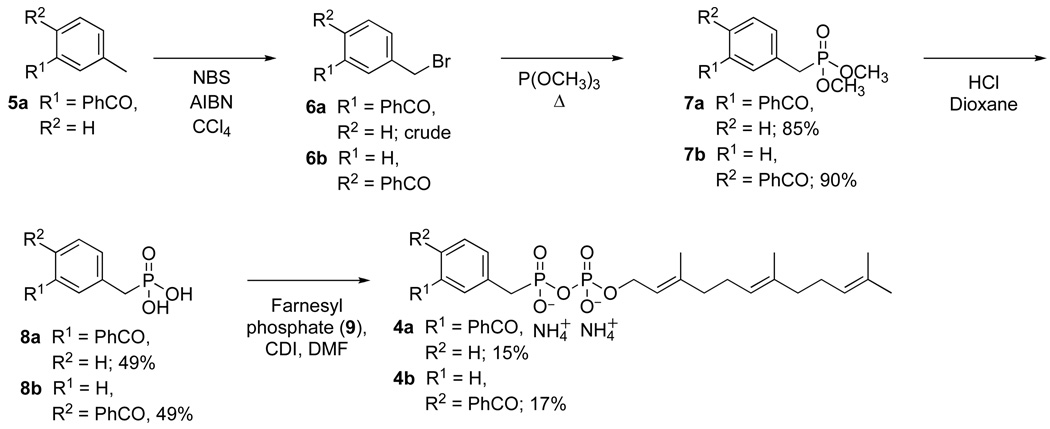
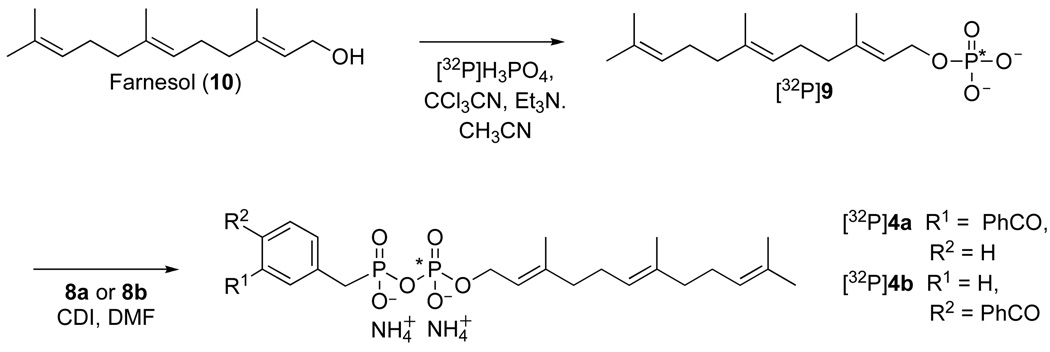
The isomeric photoactive analogues 4a and 4b were prepared using the route summarized in Scheme 1. For the preparation of the meta isomer 4a, commercially available 3-methyl- benzophenone 5a was brominated in presence of NBS in CCl4 at reflux to yield the 3-bromo-methyl-benzophenone (6a). The crude material was then treated in neat trimethylphosphite at reflux to produce the desired diester 7a in good yield (85% from 5a) followed by hydrolysis under acidic conditions to afford the unprotected phosphonate 8a in 49% yield. Coupling of 8a to farnesyl monophosphate (9) was achieved through the reaction of the two components with carbonyldiimidazole in DMF and the purified product (4a) was obtained in 15% yield after ion exchange chromatography followed by reversed-phase chromatography. Compound 4a was characterized by 1H-NMR, 31P-NMR, HR-ESI-MS and UV spectroscopy. The isomeric probe 4b was obtained from an analogous route beginning with commercially available 4-bromo-methyl-benzophenone (6b). For the photoaffinity labeling experiments described below, it was necessary to prepare 4a and 4b in radiolabeled form. In earlier work, we developed a simple method for the preparation of 32P-labeled FPP and related analogues;26, 27 a variation of that method was employed here, shown in Scheme 2, to obtain the desired labeled probes.
Scheme 1.
Synthesis of photoaffinity analogs 4a and 4b
Scheme 2.
Synthesis of 32P-labeled forms of 4a and 4b. The position of the 32P radiolabel is indicated with a (*).
First, farnesyl monophosphate ([32P]9) was synthesized through the reaction of farnesol (10) with [32P]H3PO4 using CCl3CN as the coupling agent. The radiolabeled product ([32P]9)was purified using a disposable Sep-Pak reversed-phase column and then coupled with 8a or 8b in the presence of CDI in DMF to yield 4a or 4b, respectively. The radiolabeled probes were again purified using a disposable Sep-Pak reversed-phase column and their radiochemical purities (> 80%) were assessed by TLC followed by phosphorimaging analysis.
2.2. Enzymatic studies with protein farnesyltransferase using 4a and 4b
The utility of the new probes were first evaluated in experiments with purified S. cerevisiae protein farnesyltransferase (ScPFTase) since that enzyme has been extensively characterized via structural and enzymological studies.28–32 Importantly, its substrate specificity has been thoroughly investigated including a large number of isoprenoid diphosphate analogs that have been examined as inhibitors and alternative substrates.10,11,15,17,33–38
Compounds 4a and 4b were first analyzed as substrates for ScPFTase using a continuous fluorescence assay employing the pentapeptide N-dansyl-GCVIA as the prenyl group acceptor. This assay monitors the increase in fluorescence of the dansyl group upon prenylation at the neighboring cysteine residue caused by the change in hydrophobicity as previously described.39,40 No observable change in the fluorescence of the pentapeptide was observed in reactions containing 4a or 4b in place of FPP at concentrations up to 200 µM, which suggests that these molecules are not substrates for ScPFTase.
To evaluate their potential as enzyme inhibitors, the rate of ScPFTase-catalyzed farnesylation of Ds-GCVIA was measured at a fixed concentration of FPP and varying concentrations of 4a and 4b. In those reactions, the presence of 4a or 4b attenuated the rate of peptide prenylation, indicating that these compounds are enzyme inhibitors. Plots of enzymatic rate versus concentration of inhibitor gave IC50 values (Tabulated in Table 1) of 5.8 µM for 4a and 3.0 µM for 4b. To gain additional understanding of the mechanism of ScPFTase inhibition by these compounds, the more potent inhibitor, 4b, was studied in greater detail. The rate of ScPFTase-catalyzed prenylation of Ds-GCVIA was measured in the presence of fixed concentrations of the isoprenoid substrate, FPP, and varying concentrations of 4b. Double reciprocal plots of those data gave a pattern of lines that intersect on the 1/V axis, consistent with competitive inhibition with respect to the substrate FPP (See Figure 2); this result is consistent with FPP and 4b binding at the same site. Further analysis by the method of Eadie-Hofstee gave KI value of 0.48 µM for ScPFTase.41,42
Table 1.
IC50 and KI values for inhibition of PFTase with photoactive FPP analogues.
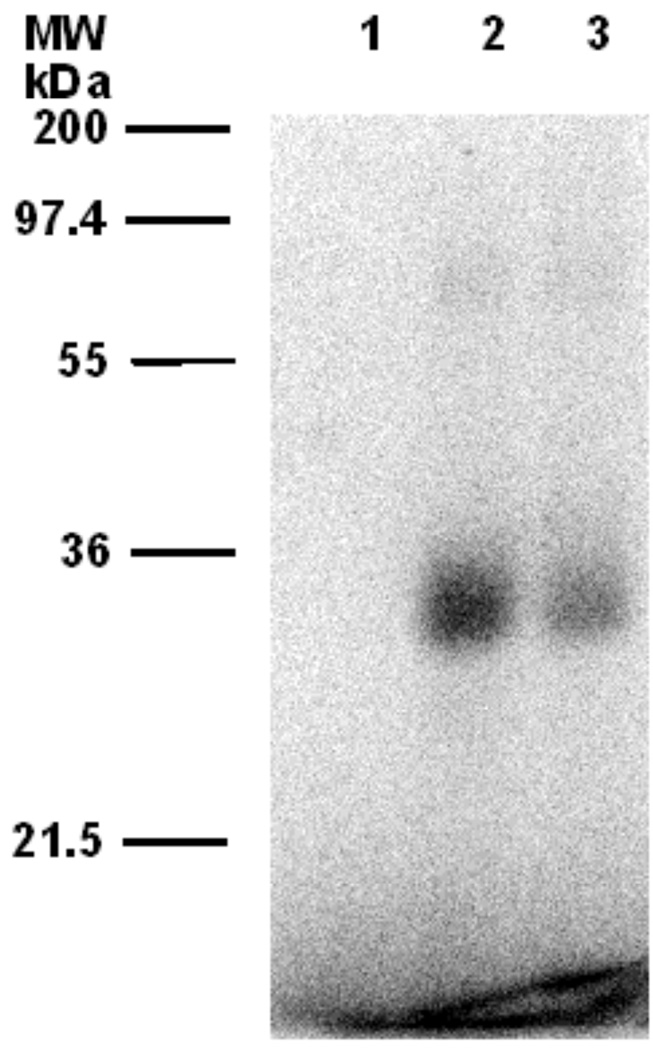
We next evaluated 32P-labelled forms of 4a and 4b for their ability to label ScPFTase upon photolysis. In those experiments, each analogue was photolyzed in the presence of purified ScPFTase and the resulting reaction mixture was then fractionated by SDS PAGE. The total protein present was visualized by staining with Coomassie Blue and the labeled protein was detected via phosphorimaging analysis. Representative photolabelling results using [32P]4b are shown in Figure 3. Photolysis of ScPFTase with [32P]4b led to strong labeling of the β-subunit of the enzyme with minimal labeling of the α-subunit (Figure 3, Lane 2). The labeling was almost completely abolished when the photolysis was performed in the presence of both [32P]4b and excess FPP (competition experiment, Figure 3, Lane 3); no labeling occurred in the absence of irradiation (Figure 3, Lane 1). Similar results were obtained in experiments with [32P]4a (Data not shown).
2.3. Labeling studies of farnesyl diphosphate synthases (FPPSase) using 4b
As noted above, a number of studies have reported the successful labeling of protein prenyltransferases using photoactive analogues. In contrast, there are relatively few reports of such probes being used to label FPPSases involved in the biosynthesis of isoprenoids. Accordingly, we examined the ability of 4b to label FPPSases from two different sources. First [32P]4b was photolyzed in the presence of E. coli FPPSase IspA (EcFPPSase)43 under conditions similar to those used for PFTase described above; results from those experiments are shown in Figure 4. Photolysis of EcFPPSase with [32P]4b led to strong labeling of the purified enzyme (Figure 4, Lane 2) which was substantially (although not completely) reduced when the photolysis was performed in the presence of both [32P]4b and FPP (competition experiment, Figure 4, Lane 3); no labeling occurred in the absence of irradiation (Figure 4, Lane 1).
Figure 4.
Phosphorimaging analysis of photolabeling of EcFPPSase with [32P]4b after SDS-PAGE separation. Lane 1: EcFPPSase and [32P]4b (29 µM), no UV irradiation. Lane 2: EcFPPSase and [32P]4b (29 µM), with UV irradiation. Lane 3: EcFPPSase, [32P]4b (29 µM), and FPP (100 µM), with UV irradiation.
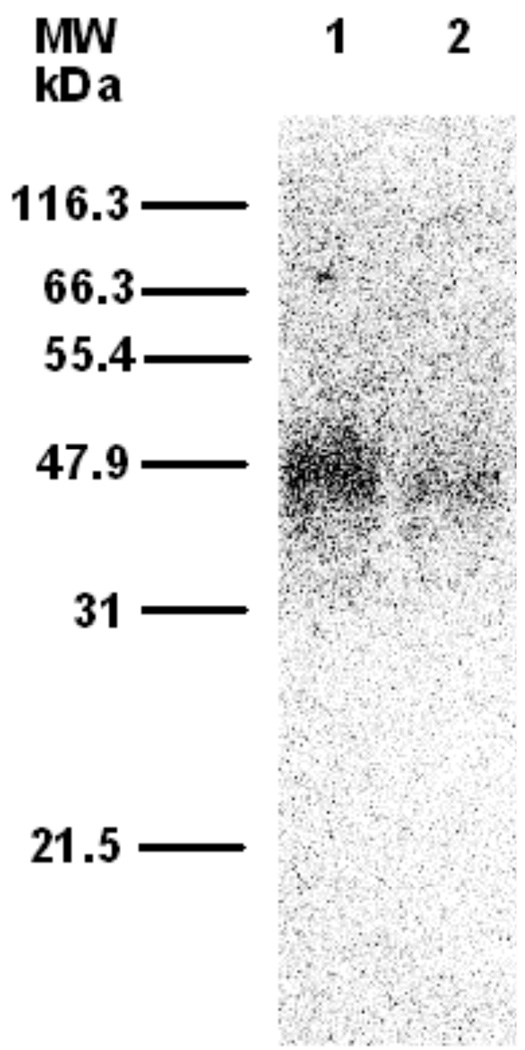
To further examine the utility of probe 4b, labeling experiments were carried out using DmFPPSase,44 an insect-derived enzyme from Drosophila melanogaster. Photolysis of purified DmFPPSase with [32P]4b resulted in labeling of the enzyme (Figure 5, Lane 1); no labeling was observed without irradiation (Data not shown). When photolysis was performed in the presence of both [32P]4b and FPP as a competitor (Figure 5, Lane 2), a significant decrease (46% of the labeling obtained in the absence of the competitor) in the amount of labeling was observed.
Figure 5.
Phosphorimaging analysis of photolabeling of DmFPPSase with [32P]4b after SDS-PAGE separation. Lane 1: DmFPPSase and [32P]4b (29 µM), with UV irradiation. Lane 2: DmFPPSase, [32P]4b (29 µM), and FPP (100 µM), with UV irradiation.
2.4. Experiments using 4b with a sesquiterpene synthase
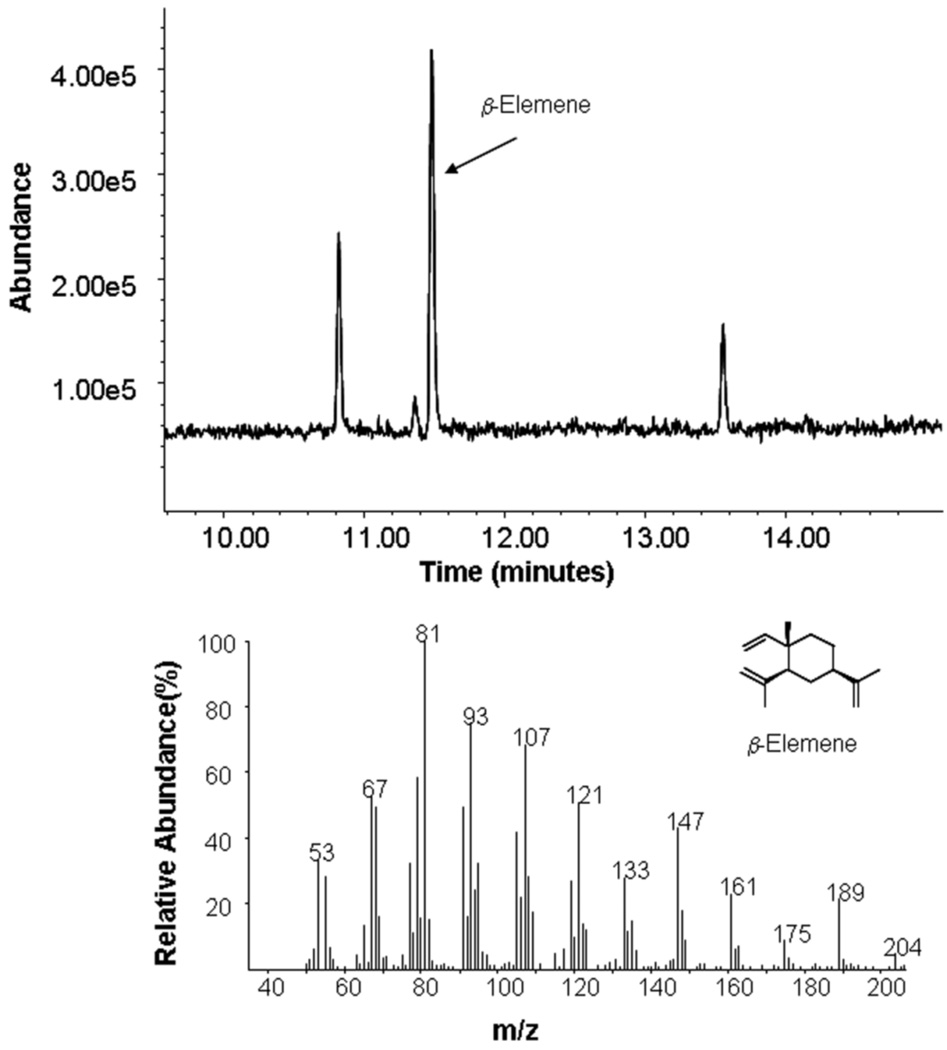
Another major class of important FPP-utilizing enzymes is the sesquiterpene synthases. To evaluate the utility of the diphosphate-directed photoprobes developed here to interact with this class of enzymes, experiments were performed using a sesquiterpene synthase from Nostoc sp. strain PCC7120, (NoSTSase);45 that particular enzyme produces germacrene A from the cyclization of FPP. Initially, it was assumed that the phosphonophosphate functionality present in 4b would not be processed by NoSTSase (as was the case with ScPFTase) and that the compound would hence serve as an inhibitor. Interestingly, addition of 4b to reactions containing purified NoSTSase and FPP did not result in a decrease in the cyclized product indicating that 4b was not an inhibitor of the enzyme. Instead, incubation of NoSTSase and 4b alone (without FPP) led to the formation of the same product (see Figure 6) obtained using FPP as a substrate, thus demonstrating that 4b is a bona fide substrate for the enzyme, although the rate of product formation was approximately 10-fold slower than with FPP. Based on these promising results, [32P]4b was also examined for its ability to label the enzyme. Photolysis of NoSTSase with [32P]4b resulted in labeling of the enzyme (See Figure 7, Lane 1) and significant protection was obtained when FPP was included in the photolysis reaction mixture (Figure 7, Lane 2) similar to that observed with the other enzymes reported here.
Figure 6.
Analysis of the product formed from incubation of 4b with NoSTSase. Above: Gas chromatographic analysis of reaction. Below: MS the of product peak (Retention Time = 11.5 min). Note: It has been previously established that the product of the cyclization reaction (germacrene A) undergoes thermal rearrangement to β-elemene at the temperature of the GC injection port.45
Figure 7.
Phosphorimaging analysis of photolabeling of NoSTSase with [32P]4b after SDS-PAGE separation. Lane 1: NoSTSase and [32P]4b (29 µM), with UV irradiation. Lane 2: NoSTSase, [32P]4b (29 µM), and FPP (100 µM), with UV irradiation.
2.4. Modeling of 4b in the active sites of FPP-utilizing enzymes
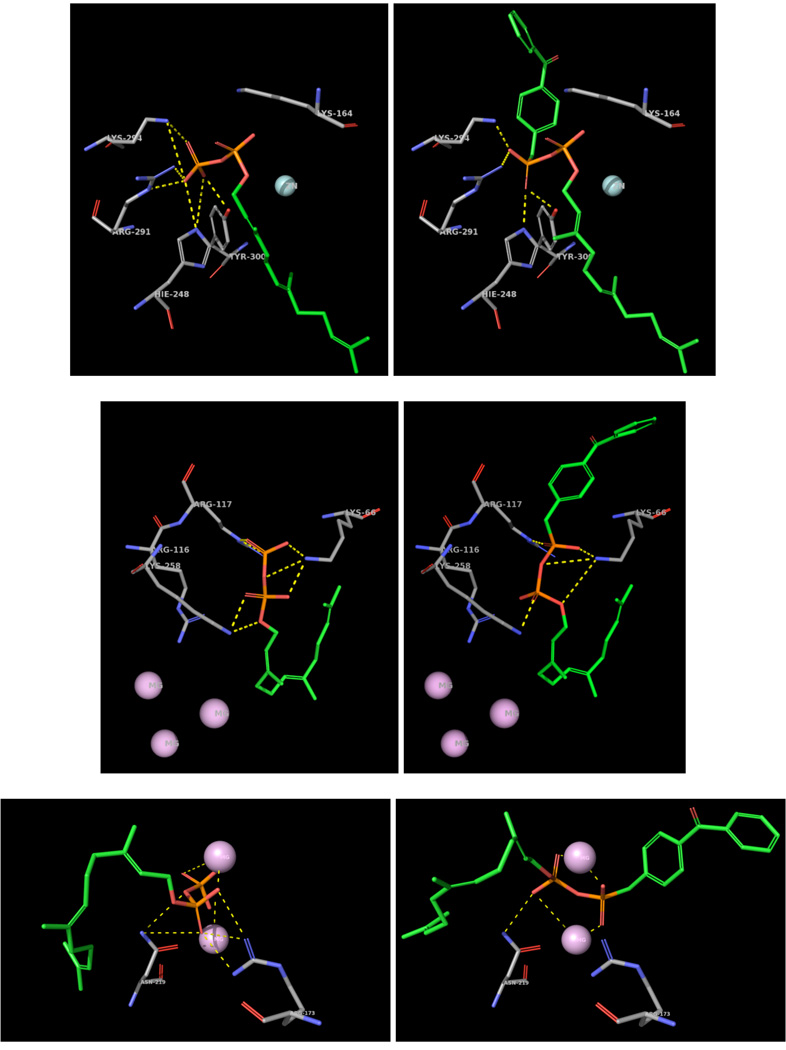
In the design of 4a and 4b, a phosphonate was used to link the benzophenone unit to the farnesyl phosphate moiety. An important result of this is the removal of one oxygen atom relative to farnesyl diphosphate. Given that a number of interactions between the diphosphate group and residues from the protein typically occur in the active sites of FPP utilizing enzymes, it was decided to model 4b in the active sites of three of the enzymes employed in this study. Since no crystal structure ScPFTase exists, the structure for RnPFTase (Rattus norvegicus) was used instead.46 For that model the benzophenone phosphonate moiety was appended onto the farnesyl diphosphate group present in the active site of the binary (RnPFTase•FPP) complex and a conformational search was performed to identify to the lowest energy conformations. The structure surrounding the diphosphate of FPP from the RnPFTase•FPP complex is shown in Figure 8 (Top Left) with the corresponding region from the RnPFTase•4b complex (Top Right). Many of the interactions between the distal phosphate (from FPP) and the protein appear to be retained in the modeled structure of the RnPFTase•4b complex; that list includes interactions with K164α, H248β and Y300β. However, interactions with R291β and K294β are absent due to the absence of the one missing oxygen atom. A similar approach was used to create a model for EcFPP synthase.47 The structure surrounding the diphosphate of FPP from the EcFPPSase•FPP complex is shown in Figure 8 (Middle Left) with the corresponding region from the EcFPPSase•4b complex (Middle Right). Residues that interact with the distal phosphate (from FPP) and the phosphonate (from 4b) include K66 R117 and K258; however, in the case of R117, it should be noted that due to the absence of one oxygen atom (due to the phosphonate substitution), one interaction between the terminal guanidino nitrogen is missing along with a longer range interaction with the internal guanidino nitrogen. Since no crystal structure of the NoSTSase employed here was available, the structure of a related enzyme, StSTSase (pentalenene synthase from Streptomyces Sp. UC5319)48 was used; mutants of that enzyme produce germacrene.49, 50 In this case, modeling was carried out using a docking algorithm. The structure surrounding the diphosphate of FPP from the StSTSase•FPP complex is shown in Figure 8 (Bottom Left) with the corresponding region from the StSTSase•4b complex (Bottom Right). In the case of StSTSase, most of the interactions between the diphosphate group and the enzyme are mediated by two Mg2+ cations; all of these interactions are preserved in the StSTSase•4b model. However, several interactions between the distal phosphate and R173 are not present in the StSTSase•4b complex.
Figure 8.
Modeling of 4b in the active sites of FPP-utilizing enzymes. Top Left: Crystal structure of NrPFTase (1JCR) with bound FPP. Shown, are residues H248β, R291β, K294β, Y300β, K164α (colored by element; carbon in grey), FPP (colored by element; carbon in green) and Zn (cyan). The yellow dashes illustrate potential hydrogen bonding or electrostatic interactions (within 4Å) of the distal phosphate. Top Right: Crystal structure of NrPFTase with 4b built on the bound FPP. The color scheme is the same as that for Top Left. Middle Left: Crystal structure of EcFPPSase (1RQ1) with FPP built by modifying the existing bound IPP. Shown, are residues K66, R116, R117, K258 (colored by element; carbon in grey), FPP (colored by element; carbon in green) and Mg (purple). The yellow dashes illustrate potential hydrogen bonding or electrostatic interactions (within 4Å) of the distal phosphate. Middle Right: Crystal structure of EcFPPSase with 4b built on the bound IPP. The color scheme is the same as that for Middle Left. Bottom Left: Crystal structure of StSTSase (Pentalenene synthase, 1PS1) with FPP docked in the active site of the enzyme. Shown, are residues R173, N219 (colored by element; carbon in grey), FPP (colored by element; carbon in green) and Mg (purple). The yellow dashes illustrate potential hydrogen bonding or electrostatic interactions (within 4Å) of the distal phosphate. Bottom Right: Crystal structure of StSTSase with 4b docked in the active site of the enzyme. The color scheme is the same as that for Bottom Left.
3. Discussion
To create a probe that would be capable of labeling residues in proximity to the diphosphate binding site of enzymes that employ FPP as a substrate, two photoactive molecules, 4a and 4b, were prepared in four (4a) or five steps (4b) from farnesyl monophosphate (9) and suitably functionalized phosphonate precursors. These compounds were competitive inhibitors of PFTase with respect to FPP with IC50 values less than 6 µM.; analogue 4b manifests a KI of 0.48 µM. In general, those values are comparable to those obtained earlier photoactive FPP analogues such as 2 and 3 (see Table 1) that incorporate the photophore into the isoprenoid moiety itself. Thus it appears that this new class of isoprenoid probes retain significant affinity for their cognate binding sites despite modification of one of the diphosphate oxygen atoms. Since the last step in the synthesis is the coupling of farnesyl monophosphate to a benzophenone phosphonate fragment, the preparation of radiolabeled forms of the probes can be easily accomplished from commercially available terpenoid precursors; accordingly, we prepared 32P-labeled forms of 4a and 4b and evaluated their ability to label PFTase. Both of these probes undergo crosslinking reactions with PFTase upon photolysis at 350 nm resulting in predominant labeling of the β-subunit of this heterodimeric enzyme; that labeling is abolished when photolysis is performed in the presence of the natural ligand, FPP. These results are consistent with the isoprenoid binding site being composed of residues primarily located on the β-subunit as evidenced from numerous x-ray crystal structures containing bound isoprenoid analogues.10, 11, 37, 51, 52 Overall, these results demonstrate that 4a and 4b bind to an FPP-utilizing enzyme in a predictable fashion similar to results obtained with other photoprobes. Moreover, the absence of crosslinking to the α-subunit indicates that these probes manifest selectivity in their photoaffinity labeling reactions. Having established this, we then examined their interactions with several other types of enzymes that use FPP as a substrate or generate it as a product. FPPSases from E. coli and D. melanogaster are both labeled by 4b upon photolysis. Since FPPSases from insect sources are involved in the biosynthesis of certain pheromones, these probes should be useful in functional studies of such proteins. Compound 4b also labels a sesquiterpene synthase from Nostoc sp. strain PCC7120 upon photolysis and interestingly, this probe is an alternative substrate for that enzyme. Given the large number of sesquiterpene synthases present in nature, these new probes should be highly useful for identifying sesquiterpene synthases from new sources. They may also be helpful in analyzing FPP binding to previously uncharacterized enzymes and for mapping their active sites. We are currently employing these molecules to assist in the identification of FPP-utilizing enzymes from a variety of sources. Finally, we note that molecular modeling of the complexes between various FPP-utilizing enzymes and 4b suggests that these enzymes are able to interact with phosphonate present in 4b in agreement with the studies reported here. However, the substitution of carbon in place of oxygen (compare the distal phosphate in FPP with the phosphonate in 4b) does eliminate some specific interactions between the probe and the various enzymes that results in decreased affinity. Significantly, despite their reduced affinity, 4a and 4b still bind with significant avidity. The KI using 4b with ScPFTase (0.48 µM) is only 6-fold higher than the KD (0.075 µM) for FPP binding to ScPFTase. Interestingly, the decrease affinity for ScPFTase caused by the phosphonate substitution is comparable to that resulting from modest modification of the isoprenoid itself (see Table 1). These results suggest that phosphonate substitutions may be useful for the design of other probes designed to target FPP-utilizing enzymes.
4. Experimental
4.1. General
All reagents were obtained from Aldrich. Analytical TLC was performed on precoated (0.25 mm) silica gel 60F-254 plates purchased from E. Merck. Flash chromatography silica gel (60–120 mesh) was obtained from E. M. Science. HPLC analysis was carried out using a Beckman Model 127/166 instrument equipped with a diode array UV detector. Analytical separations employed a Rainin Dynamax-60 Å C18 column (4.6 mm x 25 cm with a 2 cm guard column). GC/MS analysis was carried out on a Varian 3800 gas chromatograph linked to an ion-trap mass spectrometer employing a HP-1ms (Agilent Tech. Inc.) capillary column (30 m x 0.25 mm inner diameter, 1.5 µm film thickness) with He (g) as a carrier. Solid phase micro extraction (SPME) fibers (100 µm polydimethylsiloxane fiber) were obtained from Supelco. Phosphorimaging analysis was performed with a Molecular Dynamics 445 SI Phosphorimager. Dowex 50W–X8 resin was obtained from Bio-Rad and Sep-Pak columns were purchased from Waters. [32P]H3PO4 (specific activity 8500–9120 Ci/mmol) was purchased from DuPont NEN. N-dansyl-GCVIA was synthesized by Dr. Dan Mullen in the Department of Chemistry, University of Minnesota. The enzymes used in this study were purified as previously described for yeast PFTase,15 EcFPPSase,43 DmFPPSase,44 and NpSTSase.45
4.2 Chemical synthesis
4.2.1. 3-bromo-methyl-benzophenone (6a)
To a solution of 3-methyl-benzophenone (5a, 100 mg, 0.51 mmol) in CCl4 (7.0 mL) was added N-bromosuccinimide (NBS, 102 mg, 0.57 mmol) and azobisisobutyronitrile (AIBN, 0.52 mg, 0.02 mmol). The mixture was allowed to react at reflux for 24 h. The solvent was evaporated under vacuum and the crude material was used in the next reaction without purification. 1H NMR (300 MHz, CDCl3) δ = 4.52 (s, 2H), 7.42–7.52 (m, 3H), 7.57–7.63 (m, 2H), 7.69–7.73 (ddd, 1H, J = 1.5, 1.5, 7.5 Hz) 7.78–7.84 (m, 3H). 13C NMR (75 MHz, CDCl3 δ = 32.7, 128.5, 128.9, 130.1, 130.5, 132.7, 133.0, 137.3, 138.2, 138.3, 196.0. HRMS (ESI) (m/z): [M+Na]+ calcd for C14H11BrONa 296.9891 and 298.9870, found 296.9876 and 298.9861.
4.2.2 (3-Benzoyl-benzyl)-phosphonic acid dimethyl ester (7a)
A solution of crude 3-bromo-methyl-benzophenone (crude 6a, estimated to be 0.51 mmol, prepared as described above) was dissolved in P(OCH3)3 (5.0 mL) and refluxed for 12 h. Excess trimethyl P(OCH3)3 was evaporated under vacuum and the resulting crude material was purified via silica gel chromatography (Hexanes/EtOAc, 1:1, v/v) to yield 131 mg (85% from 5a). 1H NMR (300 MHz, CDCl3) δ = 3.16 (d, 2H, J = 22 Hz), 3.62 (d, 6H, J = 11 Hz), 7.32–7.40 (m, 4H) 7.46–7.51 (m, 1H) 7.66–7.71 (m, 4H). 13C NMR (75 MHz, CDCl3) δ = 32.6 (d, J = 138 Hz), 52.9 (d, J = 6.3 Hz), 128.3, 128.6 (d, J = 3.1 Hz), 128.7 (d. J = 3.7 Hz), 130.0, 131.2 (d, J = 6.8 Hz), 131.8 (d, J = 9.5 Hz), 132.5, 133.7 (d, J = 5.8 Hz), 137.4, 137.9 (d, J = 2.6 Hz), 196.2. 31P NMR (121 MHz, CDCl3) δ = 28.9.
4.2.3. (3-Benzoyl-benzyl)-phosphonic acid (8a)
A solution of dimethyl ester 7a (200 mg, 0.66 mmol) was refluxed in an aqueous solution of 4N HCl (7 mL) and dioxane (7 mL) over 12 h. The reaction mixture was extracted with EtOAc (20 mL, 4 times) and the resulting product (89 mg, 49%) was deemed to be of sufficient purity to use in the next step without additional purification. 1H NMR (300 MHz, CDCl3) δ = 3.02 (d, 2H, J = 21 Hz), 7.27–7.36 (m, 3H) 7.43–7.47 (m, 3H) 7.57 (s, 1H), 7.64 (d, 2H, J = 7.2 Hz). 13C NMR (75 MHz, CDCl3) δ = 34.1 (d, J = 134 Hz), 128.2, 130.0, 131.2 (d, J = 6.3 Hz), 132.7, 133.4 (d, J = 9.0 Hz), 134.0 (d, J = 5.9 Hz), 137.1, 137.4, 197.4. 31P NMR (121 MHz, CDCl3) δ = 24.9. HRMS (ESI) (m/z): [M-H]- calcd for C14H12O4P 275.0479, found 275.0461.
4.2.4. 3-benzoylbenzylphosphonic ((2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienyl phosphoric) anhydride (4a)
Phosphonic acid 8a (90 mg, 0.33 mmol) and CDI (79 mg, 0.39 mmol) were dissolved in DMF (1.5 mL) and allowed to react at room temperature under argon for 2 h, at which time farnesyl phosphate (9, cyclohexylamine salt) was added (130 mg, 0.326 mmol). The mixture was allowed to react for 7 days with stirring at room temperature. At that point, the reaction was quenched with water (5 mL) and extracted with EtOAc (10 mL, 4 times). The combined organic layers were dried over Na2SO4, filtered and concentrated to afford yellow oil. It was then dissolved in a solvent mixture composed of 50% CH3CN and 50% aqueous 25 mM NH4HCO3/isopropanol (50:1, v/v). This solution was applied to an ionic exchange column (Dowex 50×8–400, 2.6 × 4.5 cm), previously washed with aqueous NH4OH and equilibrated to pH 6.0 with aqueous 25 mM NH4HCO3/isopropanol (50:1, v/v). The column was eluted with additional aqueous 25 mM NH4HCO3/isopropanol (50:1, v/v) and the fractions containing the desired compound were concentrated by lyopholization. Final purification was accomplished via reversed-phase HPLC using 25 mM NH4HCO3 (solvent A) and CH3CN (solvent B) and the following elution profile performed at 5.0 mL/min: 5.0 min 0% solvent B; linear gradient from 0% to 60% solvent B over 40 min. Under these conditions the desired product eluted at 37 min. The solvent mixture was removed by lyopholization to afford 29 mg (15%) of a white solid. 1H NMR (500 MHz, CDCl3) δ = 1.51 (s, 3H), 1.52 (s, 3H), 1.55 (s, 3H), 1.63 (s, 3H), 1.92–1.98 (m, 8H), 3.1 (d, 2H, J = 21.6 Hz), 4.22 (dd, 2H, J = 5.7 Hz), 5.01–5.03 (m, 2H), 5.22 (t, 1H, J = 6.5 Hz), 7.27–7.34 (m, 2H), 7.39–7.44 (m, 2H), 7.52–7.54 (m, 2H), 7.70–7.75 (m, 3H). 31P NMR (202 MHz, CDCl3) δ = 11.5 (d, J = 23.1 Hz), −10.3 (d, J = 22.6 Hz). HRMS (ESI) (m/z): [M-H]− calcd for C29H37O7P2 559.2020, found 559.2042. UV ε = 15.7 mM−1•cm−1 at 260 nm.
4.2.5. (4-Benzoyl-benzyl)-phosphonic acid dimethyl ester (7b)
The procedure employed here was identical to that used for the preparation of 7a. Starting from 300 mg (1.1 mmol) of the bromide 6b, 298 mg (90% yield) of the desired diester product 7b was obtained. 1H NMR (300 MHz, CDCl3) δ = 3.16 (d, 2H, J = 22 Hz), 3.62 (d, 6H, J = 11 Hz), 7.32–7.40 (m, 4H) 7.46–7.51 (m, 1H) 7.66–7.71 (m, 4H). 13C NMR (75 MHz, CDCl3) δ = 32.9 (d, J = 137 Hz), 52.9 (d, J = 6.9 Hz), 128.3, 129.7 (d, J = 6.8 Hz), 129.9, 130.4 (d, J = 3.1 Hz), 132.4, 136.2 (d, J = 3.7 Hz), 136.4 (d, J = 9.5 Hz), 137.5, 196.1. 31P NMR (121 MHz, CDCl3) δ = 28.2.
4.2.6. (4-Benzoyl-benzyl)-phosphonic acid (8b)
The procedure employed here was identical to that used for the preparation of 8a. Starting from 200 mg (0.66 mmol) of the diester 7b, 90 mg (49% yield) of the desired phosphonic acid product 8b was obtained. 1H NMR (300 MHz, CDCl3) δ = 3.24 (d, 2H, J = 22.2 Hz), 7.46–7.53 (m, 4H), 7.60–7.65 (m, 1H), 7.70–7.74 (m, 4H). 13C NMR (75 MHz, CDCl3) δ = 34.9 (d, J = 134 Hz), 128.3, 129.7. 129.8, 129.9, 130.2, 130.2, 132.6, 135.7, 137.4, 138.2 197.2. 31P NMR (121 MHz, CDCl3) δ =23.6. HRMS (ESI) (m/z): [M-H]− calcd for C14H12O4P 275.0479, found 275.0466.
4.2.7. 4-benzoylbenzylphosphonic ((2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienyl phosphoric) anhydride (4b)
The procedure for the synthesis of 4b was identical to that for 4a. Starting from 90 mg (0.33 mmol) of the phosphonic acid 8b, 33 mg (17% yield) of the desired product 4b was obtained. The product eluted at 36 min (HPLC with same conditions than 4a). 1H NMR (300 MHz, CDCl3) δ = 1.34 (s, 3H), 1.35 (s, 3H), 1.39 (s, 3H), 1.43 (s, 3H), 1.71–1.82 (m, 8H), 3.0 (d, 2H, J = 22.2 Hz), 4.11 (dd, 2H, J = 6.3 Hz), 4.83–4.84 (m, 2H), 5.11 (t, 1H, J = 6.6 Hz), 7.23–7.28 (m, 4H), 7.35–7.38 (m, 1H), 7.47–7.53 (m, 4H). 31P NMR (121 MHz, CDCl3) δ =-9.6 (d, J = 25.3 Hz), 12.1 (d, J = 25.9 Hz). HRMS (ESI) (m/z): [M-H]− calcd for C29H37O7P2 559.2020, found 559.2076. UV δ = 17.8 mM−1•cm−1 at 272 nm
4.2.8. [32P]-4-benzoylbenzylphosphonic ((2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienyl phosphoric) anhydride (32P]4b)
The synthesis of [32P]4b accomplished in two steps. First, [32P]-labeled farnesyl monophosphate was prepared via phosphorylation of farnesol using a procedure similar to that we have previously employed for the preparation of isoprenyl diphosphates. Carrier free [32P]-H3PO4 (5 mCi) was diluted with unlabeled H3PO4 (1.6 mg, 20 µmol) and lyophilized. To the resulting anhydrous H3PO4 was added a solution of triethylamine (4.1 mg, 40 µmol) in CH3CN (50 µL). Farnesol (4.4 mg, 20 µmol) in 200 µL of CH3CN containing 20% of CCl3CN (v/v) was then added and the reaction was allowed to stir under N2 for 2 h after which time the volatile components were removed under a stream of N2. The crude material was purified with a Sep-Pac reversed phase C18 column using a stepwise gradient of CH3CN and 25 mM NH4HCO3 and the fractions containing [32P]9 (determined by TLC: i-PrOH/NH4OH/H2O, 6:3:1 v/v/v, Rf = 0.62, visualized by phosphoroimaging analysis) were pooled and lyophilized under vacuum. The benzophenone phosphonic acid derivative (8b, 5.5 mg, 20 µmol) was dissolved in anhydrous DMF (400µL), under Ar (g) at room temperature. CDI (3.9 mg, 24 µmol) was added and the resulting activation reaction was left stirring 2 h before it was added to the lyophilized [32P]9. The reaction was allowed to react for 2 days at which time the DMF was removed under a stream of N2 (g). The crude material was purified with a Sep-Pak reversed phase C18 column using a stepwise gradient of CH3CN and 25 mM NH4HCO3. The product eluted at 30% of CH3CN (specific activity 197 Ci/mol). The radiochemical purity was assessed by TLC (i-PrOH/NH4OH/H2O, 6:3:1 v/v/v, Rf = 0.7, 81% of product) followed by phosphoroimaging analysis.
4.3. Biological Experiments
4.3.1. Farnesyltransferase Assays
IC50 values were determined for 4a and 4b with ScPFTase by running duplicate assays with FPP and N-Ds-GCVIA maintained at 2.4 µM and 4a or 4b at concentrations of 0, 1, 2, 3, 5, 10 and 20 (µM. An enzyme concentration of 5.0 nM ScPFTase was employed. IC50 values were calculated from a direct fit of the rate versus inhibitor concentration data using KaleidaGraph software. Based on those results, a more systematic study was performed with 4b using a 4 × 6 grid of duplicate assays with FPP maintained at fixed concentrations (1, 2, 5, and 7.0 µM) and varying concentrations of 4b (0, 0.2, 0.4, 0.7, 1, 1.5 and 2 (µM for 4b). In those experiments, the concentration of ScPFTase was 5 nM, and the concentration of N-Ds-GCVIA was fixed at 2.4 (µM. The rates were determined from the initial velocity measurements and KI value calculated from an Eadie-Hofstee plot.41, 42
4.3.2. Photolabeling Reactions
All photolysis reactions were conducted at 4 °C in a UV Rayonet mini-reactor equipped with six RPR-3500 Å lamps and a circulating platform that allows up to eight samples to be irradiated simultaneously. All reactions (100 µL) were performed in silinized quartz test tubes (10 × 75 mm) and contained 52 mM Tris•HCl, pH 7.0, 5.8 mM DTT, 12 mM MgCl2, 12 (µM ZnCl2, 25 mM NH4HCO3 and the targeted enzyme at 50 µg/mL. Where appropriate, reactions contained [32P]4a (29 µM) or [32P]4b (29 µM); these concentrations were chosen to be 10 times above their experimentally measured KI values (for ScPFTase). For substrate protection experiments, FPP was added to a final concentration of 100 µM. Reactions were photolyzed for 6 h using the apparatus described above. Loading buffer [50 µL; 4.0% SDS, 12% glycerol (w/v), 50 mM Tris•HCl, 2.0% 2-mercaptoethanol (v/v), 0.01% bromophenol blue] was then added to each sample, and the samples were heated to 70°C for 20 min followed by analysis by SDS-polyacrylamide gel electrophoresis using a 12% Tris-Tricine gel. Gels were first stained with Comassie Blue or Sypro Orange to visualize the proteins followed by phosphorimaging analysis to assess the extent of crosslinking that was subsequently quantified via volume integration of the labeled protein bands (after subtraction of any background signal).
4.3.3. In Vitro Reactions with the Germacrene Producing Cyclase (NoSTSase)
Reactions were performed by incubating 10 µg of purified enzyme45 in 200 µL of reaction buffer (10 mM MgCl2 and 1.0 mM 2-mercaptoethanol in 10 mM Tris•HCl, pH 8.0) with varying concentrations of probe 4b or FPP (50, 100 or 200 µM) in a sealed glass vial. After reaction for 18 h at 25 °C, the headspace gas was sampled for 10 min by SPME using a 100 µm polydimethylsiloxane fiber. Afterwards, the full fiber was inserted into the injection port of a GC/MS for analysis. Compounds bound to SPME fibers were desorbed for 20 min in the injection port (250 °C) followed by gas chromatographic separation using a temperature program that ramped from 60 °C to 250 °C at a rate of 20 °C/min. Mass spectra were scanned in the range of 5–300 atomic mass units at 1 s intervals.
4.4. Modeling of 4b in the active sites of FPP-utilizing enzymes
For modeling of the RnPFTase•4b complex, the benzophenone moiety in 4b was built by modifying the existing FPP present in the crystal structure of the enzyme (1JCR).46 Once modified, a 5000 step mixed torsional/low-mode conformational search was conducted with MacroModel (Schrodinger, version 9.6) using the OPLS forcefield.53, 54 The rotatable bonds in the flexible tail of the ligand were specifically chosen for torsional sampling, and the protein was frozen throughout the search. The resulting conformation with the lowest potential energy was chosen for display. For EcFPPSase, FPP and the 4b were built by modifying the existing IPP present in the crystal structure of the enzyme (1RQI).47 A conformational search was performed as described for RnPFTase and the resulting conformation with the lowest potential energy was chosen for display of each ligand. For StSTSase (Pentalenene synthase),55 FPP and 4b were docked in the active site of the crystal structure of the enzyme (1PS1) using Glide (Schrodinger, version 5.0). A standard precision docking parameter was set and 10000 ligand poses per doking were run. The resulting conformation with the lowest docking score was chosen for display for each ligand.
Supplementary Material
Acknowledgment
This research was supported by the National Institutes of Health Grant GM58442 (M.D.D.), the National Science Foundation Grant DBI 0321690 (D. S.) and funds from Yulex Corp.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
1H NMR spectra for 6a, 7a, 7b, 8a, 8b, 4a and 4b, 31P NMR spectra for 6a, 7a, 7b, 8a, 8b, 4a and 4b, HR-ESI-MS spectra for 4a and 4b and HPLC chromatograms for 4a and 4b are included. Stereo images of the models shown in Figure 8 are included along with several tables that summarize important distances within the active site of each enzyme model. Pymol files for each of the enzyme models are also provided.
References
- 1.Cane DE. Comp. Nat. Prod. Chem. 1999;2:155. [Google Scholar]
- 2.Davis EM, Croteau R. Topics Curr. Chem. 2000;209:53. [Google Scholar]
- 3.Loomis WD, Croteau R. Biochem. Plants. 1980;4:363. [Google Scholar]
- 4.Cornish K. ACS Symp. Series. 1992;497:18. [Google Scholar]
- 5.Zhang FL, Casey PJ. Ann. Rev. Biochem. 1996;65:241. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 6.Fraga BM. Nat. Prod. Rep. 2008;25:1180. doi: 10.1039/b806216c. [DOI] [PubMed] [Google Scholar]
- 7.Marecak DM, Horiuchi Y, Arai H, Shimonaga M, Maki Y, Koyama T, Ogura K, Prestwich GD. Bioorg. Med. Chem. Lett. 1997;7:1973. [Google Scholar]
- 8.Zhang Y-W, Koyama T, Marecak DM, Prestwich GD, Maki Y, Ogura K. Biochemistry. 1998;37:13411. doi: 10.1021/bi972926y. [DOI] [PubMed] [Google Scholar]
- 9.Tian R, Li L, Tang W, Liu H, Ye M, Zhao ZK, Zou H. Proteomics. 2008;8:3094. doi: 10.1002/pmic.200800021. [DOI] [PubMed] [Google Scholar]
- 10.DeGraw AJ, Zhao Z, Hsieh J, Jefferies M, Distefano MD, Strickland CL, Shintani D, Nural H, McMahan C, Xie W. J. Org. Chem. 2007;72:4587. doi: 10.1021/jo0623033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turek TC, Gaon I, Distefano MD, Strickland CL. J. Org. Chem. 2001;66:3253. doi: 10.1021/jo991130x. [DOI] [PubMed] [Google Scholar]
- 12.Kale TA, Raab C, Yu N, Dean DC, Distefano MD. J. Am. Chem. Soc. 2001;123:4373. doi: 10.1021/ja0012016. [DOI] [PubMed] [Google Scholar]
- 13.Turek TC, Gaon I, Gamache D, Distefano MD. Bioorg. Med. Chem. Lett. 1997;7:2125. [Google Scholar]
- 14.Turek TC, Gaon I, Distefano MD. Tet. Lett. 1996;37:4845. [Google Scholar]
- 15.Gaon I, Turek TC, Weller VA, Edelstein RL, Singh SK, Distefano MD. J. Org. Chem. 1996;61:7738. doi: 10.1021/jo9602736. [DOI] [PubMed] [Google Scholar]
- 16.Gaon I, Turek TC, Distefano MD. Tet. Lett. 1996;37:8833. [Google Scholar]
- 17.Edelstein RL, Distefano MD. Biochem. Biophys. Res. Comm. 1997;235:377. doi: 10.1006/bbrc.1997.6792. [DOI] [PubMed] [Google Scholar]
- 18.Allen CM, Baba T. Meth. Enzymol. 1985;110:117. doi: 10.1016/s0076-6879(85)10066-2. [DOI] [PubMed] [Google Scholar]
- 19.Baba T, Allen CM. Biochemistry. 1984;23:1312. [Google Scholar]
- 20.Baba T, Muth J, Allen CM. J. Biol. Chem. 1985;260:10467. [PubMed] [Google Scholar]
- 21.Omer CA, Kral AM, Diehl RE, Prendergast GC, Powers S, Allen CM, Gibbs JB, Kohl NE. Biochemistry. 1993;32:5167. doi: 10.1021/bi00070a028. [DOI] [PubMed] [Google Scholar]
- 22.Das NP, Allen CM. Biochem. Biophys. Res. Comm. 1991;181:729. doi: 10.1016/0006-291x(91)91251-7. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama K, McGeady P, Gelb MH. Biochemistry. 1995;34:1344. doi: 10.1021/bi00004a029. [DOI] [PubMed] [Google Scholar]
- 24.Rilling HC. Meth. Enzmol. 1985;110:125. doi: 10.1016/s0076-6879(85)10067-4. [DOI] [PubMed] [Google Scholar]
- 25.Chehade KAH, Kiegiel K, Isaacs RJ, Pickett JS, Bowers KE, Fierke CA, Andres DA, Spielmann HP. J. Am. Chem. Soc. 2002;124:8206. doi: 10.1021/ja0124717. [DOI] [PubMed] [Google Scholar]
- 26.Turek TC, Gaon I, Distefano MD. J. Lab. Cmpds. Radiopharm. 1997;39:140. [Google Scholar]
- 27.Turek TC, Edelstein RL, Gaon I, Weller VA, Distefano MDIn. In: [32P]-labeled analogs of farnesyl and geranylgeranyl pyrophosphate: Synthesis and application in photoaffinity labeling experiments with protein prenyltransferases. Hays JR, Melilo DG, editors. John Wiley & Sons Ltd; 1998. p. 67. Synth. Appl. Isot. Labelled Compd. 1997, Proc. Int. Symp., 6th, 1998. [Google Scholar]
- 28.Mayer MP, Prestwich GD, Dolence JM, Bond PD, Wu H-y, Poulter CD. Gene. 1993;132:41. doi: 10.1016/0378-1119(93)90512-2. [DOI] [PubMed] [Google Scholar]
- 29.Dolence JM, Poulter CD. Proc. Nat. Acad. Sci. USA. 1995;92:5008. doi: 10.1073/pnas.92.11.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolence JM, Cassidy PB, Mathis JR, Poulter CD. Biochemistry. 1995;34:16687. doi: 10.1021/bi00051a017. [DOI] [PubMed] [Google Scholar]
- 31.Mathis JR, Poulter CD. Biochemistry. 1997;36:6367. doi: 10.1021/bi9629182. [DOI] [PubMed] [Google Scholar]
- 32.Rozema DB, Poulter CD. Biochemistry. 1999;38:13138. doi: 10.1021/bi990794y. [DOI] [PubMed] [Google Scholar]
- 33.Gibbs RA, Krishnan U, Dolence JM, Poulter CD. J. Org. Chem. 1995;60:7821. [Google Scholar]
- 34.Zahn TJ, Eilers M, Guo Z, Ksebati MB, Simon M, Scholten JD, Smith SO, Gibbs RA. J. Am. Chem. Soc. 2000;122:7153. [Google Scholar]
- 35.Reigard SA, Zahn TJ, Haworth KB, Hicks KA, Fierke CA, Gibbs RA. Biochemistry. 2005;44:11214. doi: 10.1021/bi050725l. [DOI] [PubMed] [Google Scholar]
- 36.Gibbs RA, Zahn TJ, Sebolt-Leopold JS. Curr. Med. Chem. 2001;8:1437. doi: 10.2174/0929867013372111. [DOI] [PubMed] [Google Scholar]
- 37.Turek-Etienne TC, Strickland CL, Distefano MD. Biochemistry. 2003;42:3716. doi: 10.1021/bi0266838. [DOI] [PubMed] [Google Scholar]
- 38.Kale TA, Hsieh S-aJ, Rose MW, Distefano MD. Curr. Topics Med. Chem. 2003;3:1043. doi: 10.2174/1568026033452087. [DOI] [PubMed] [Google Scholar]
- 39.Pompliano DL, Gomez RP, Anthony NJ. J. Am. Chem. Soc. 1992;114:7945. [Google Scholar]
- 40.Bond PD, Dolence JM, Poulter CD. Meth. Enzymol. 1995;250:30. doi: 10.1016/0076-6879(95)50060-x. [DOI] [PubMed] [Google Scholar]
- 41.Hofstee BHJ. Science. 1952;116:329. doi: 10.1126/science.116.3013.329. [DOI] [PubMed] [Google Scholar]
- 42.Eadie GS. J. Biol. Chem. 1942;146:85. [Google Scholar]
- 43.Lee PC, Petri R, Mijts BN, Watts KT, Schmidt-Dannert C. Metab. Eng. 2005;7:18. doi: 10.1016/j.ymben.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Sen SE, Trobaugh C, Beliveau C, Richard T, Cusson M. Insect Biochem. Mol. Biol. 2007;37:1198. doi: 10.1016/j.ibmb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. J. Bacteriol. 2008;190:6084. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long SB, Hancock PJ, Kral AM, Hellinga HW, Beese LS. Proc. Nat. Acad. Sci. USA. 2001;98:12948. doi: 10.1073/pnas.241407898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosfield DJ, Zhang Y, Dougan Douglas R, Broun A, Tari Leslie W, Swanson Ronald V, Finn J. J Biol Chem. 2004;279:8526. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 48.Cane DE, Sohng J-K, Lamberson CR, Rudnicki SM, Wu Z, Lloyd MD, Oliver JS, Hubbard BR. Biochemistry. 1994;33:5846. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 49.Seemann M, Zhai G, de Kraker J-W, Paschall CM, Christianson DW, Cane DE. J. Am. Chem. Soc. 2002;124:7681. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 50.Seemann M, Zhai G, Umezawa K, Cane D. J. Am. Chem. Soc. 1999;121:591. [Google Scholar]
- 51.Park H-W, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Science. 1997;275:1800. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 52.Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Biochemistry. 1998;37:16601. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- 53.Jorgensen WL, Tirado-Rives J. J. Am. Chem. Soc. 1988;110:1657. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 54.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J. Phys. Chem. B. 2001;105:6474. [Google Scholar]
- 55.Lesburg CA, Zhai G, Cane DE, Christianson DW. Science. 1997;277:1820. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.