Abstract
Patients with pancreatic cancer are usually diagnosed at late stages, when the disease is incurable. Pancreatic intraepithelial neoplasia (PanIN) 3, is believed to be the immediate precursor lesion of pancreatic adenocarcinoma, and would be an ideal stage to diagnose patients, when intervention and cure are possible and patients are curable. In this study, we used quantitative proteomics to identify dysregulated proteins in PanIN 3 lesions. Altogether, over 200 dysregulated proteins were identified in the PanIN 3 tissues, with a minimum of a 1.75 fold change compared to the proteins in normal pancreas. These dysregulated PanIN 3 proteins play roles in cell motility, the inflammatory response, the blood clotting cascade, the cell cycle and its regulation, and protein degradation. Further network analysis of the proteins identified c-MYC as an important regulatory protein in PanIN 3 lesions. Finally, three of the overexpressed proteins, laminin beta-1, galectin-1, and actinin-4 were validated by IHC analysis. All three of these proteins were overexpressed in the stroma or ductal epithelial cells of advanced PanIN lesions, as well as in pancreatic cancer tissue. Our findings suggest that these three proteins may be useful as biomarkers for advanced PanIN and pancreatic cancer if further validated. The dysregulated proteins identified in this study may assist in the selection of candidates for future development of biomarkers for detecting early and curable pancreatic neoplasia.
Keywords: proteomics, pancreatic cancer, pancreatic intraepithelial neoplasia (PanIN), mass spectrometry, immunohistochemistry (IHC)
1 Introduction
Pancreatic cancer is the fourth leading cause of cancer death in the United States. Most patients diagnosed with pancreatic cancer will die within 6 months, and only 4% survive five years after diagnosis [1–3]. The high mortality of this disease is predominantly the result of the advanced stage of disease at diagnosis and the lack of effective treatments. Surgical removal of early cancers, that is less than 2 cm in size can lead to a dramatically better prognosis. Biomarkers of early pancreatic ductal neoplasia could change disease outcome and markedly improve the survival rate [4,5].
Pancreatic intraepithelial neoplasia or PanIN, represents the precursor lesion for pancreatic ductal adenocarcinoma and is graded 1–3, with PanIN 3 or high grade dysplasia representing the stage right before cancer. Advanced PanIN lesions, especially PanIN 3, would be an ideal stage to diagnose patients—right when they are on the verge of getting cancer, but at a time when intervention and cure is possible. However, the clinical diagnosis of PanIN 3 lesions is extremely difficult and the identification of biomarkers for these lesions at either RNA expression or proteomic level has been limited. Thus, a systematic quantitative proteomics study to reveal putative precancerous protein markers of pancreatic ductal neoplasia could benefit both basic research and clinical approaches to the disease.
The emerging technology of quantitative proteomics has stimulated great interest in applying the technique to investigate the proteome of diseased samples [6]. Substantial efforts have been made in the search for protein biomarkers for cancer diagnosis or prognosis [7], including pancreatic cancer [8–17]. Quantitative proteomics techniques, such as ICAT [18] and iTRAQ [19], combined with tandem mass spectrometry allow the simultaneous comparison of two or more proteomes to reveal the dysregulated proteins associated with a specific biological condition or disease. In this study, we applied both ICAT and iTRAQ approaches to systematically profile the proteome of PanIN 3 tissue in comparison to normal pancreas, pancreatitis and pancreatic cancer tissues. The differentially expressed proteins discovered in the PanIN 3 tissues are described and their implication in pancreatic tumorigenesis is discussed. Three of the overexpressed proteins identified in the PanIN 3 lesions were validated using immunohistochemistry (IHC).
2 Materials and methods
2.1 Sample preparation
Specimens
Tissue specimens were obtained from patients with histologically proven 1) PanIN 3, 2) pancreatic cancer, 3) chronic pancreatitis and 4) normal pancreas obtained from resection for benign diseases. The tissues were collected in accordance with approved Human Subject’s guidelines at the University of Washington, Virginia Mason Hospital, and the Cleveland Clinic. Pancreatic tissue specimens were collected immediately at surgery and stored in freezing media (10% DMSO) at −80 °C. Immediately adjacent tissue was routinely processed for histologic confirmation. In preparation for proteomics analysis, the tissues were placed in T-PER (Pierce, Rockford, IL) with 1 × Protease Inhibitor Cocktail (Pierce) and then lysed by homogenization followed by centrifugation at 14,000 rpm for 15 minutes. The supernatants were collected and proteins were precipitated using cold acetone (−20 °C overnight). For ICAT and iTRAQ profiling, the samples used were from a pool of tissues from each category: normal control (pool of 10), chronic pancreatitis (pool of 10), PanIN 3 (pool of 4), and pancreatic cancer (pool of 10).
ICAT
Tissue proteins were resuspended in ICAT (Applied Biosystems, Foster City, CA) denaturing buffer (50 mM Tris and 0.1% SDS). For each sample, 500 ug protein was labeled with the acid-cleavable ICAT reagents, either the isotopically light (normal pancreas) or heavy (pre-cancer) forms. The labeled normal sample and the matching labeled pre-cancerous PanIN sample were combined and digested into peptides by trypsin (Promega, Madison, WI). ICAT-labeled peptides were subsequently fractionated by cation-exchange chromatography (SCX) and purified by avidin-affinity chromatography. The resulting 40 fractions were then combined into 17 fractions. The samples were on-line separated by high-performance liquid chromatography (LC) and analyzed by electrospray ionization (ESI)/ ion-trap mass spectrometer (LTQ ThermoFinnigan, San Jose, CA).
iTRAQ
100 µg of tissue protein from each sample was resuspended in iTRAQ (Applied Biosystems, Foster City, CA) dissolution buffer (0.5 M triethylammonium bicarbonate). The cysteine groups of the proteins were reduced with 50 mM tris-(2 carboxyethyl)phosphine (TCEP) and blocked with 200 mM methyl methanethiosulfonate (MMTS). The proteins were then digested with trypsin (trypsin to protein ratio: 1/50) overnight (16–18 hours) and labeled with one channel of iTRAQ reagents according to the manufacturer’s instructions. The normal, pancreatitis, pre-cancer (PanIN 3) and cancer samples were labeled with iTRAQ reagents of 114, 115, 116 and 117, respectively, and then the samples were combined. The combined sample was separated with SCX into 13 fractions for LC MS/MS analysis using an ESI QStar tandem mass spectrometer (Applied Biosystems, Foster City, CA).
2.2 liquid chromatography and tandem mass spectrometry
For ICAT sample analysis, an LTQ linear ion trap mass spectrometer (Thermo Finnigan, San Jose, CA) was used with a microelectrospray ion source and an HP1100 solvent delivery system (Agilent, Palo Alto, CA). Samples were automatically delivered by a FAMOS autosampler (LC Packings, San Francisco, CA) to a 100 µm internal diameter fused silica capillary pre-column packed with 2 cm of 200 Å pore-size Magic C18AQ™ material (Michrom Bioresources, Auburn, CA). The samples were loaded and washed with solvent A (0.1 % formic acid, 2 % acetonitrile) on the pre-column and then eluted to an analytical column (a 75 µm × 10 cm fused silica capillary column packed with 100 Å pore-size Magic C18AQ™ material) (Michrom) with a 0–35% gradient of solvent B (100% acetonitrile) over 60 minutes at a constant column-tip flow rate of ~300 nL/min. Eluting peptides were analyzed by µLC-MS with data-dependent MS/MS acquisition, in which 1 MS scan was followed by 3 MS/MS scans.
For iTRAQ analysis, iTRAQ labeled tryptic digests were separated on an Agilent Zorbax SB-C18 column (0.075 × 150 mm) using a Proxeon easy-nLC system (Odense, Denmark) at a flow rate of 200 nL/min. Solvent A was 0.1% formic acid in deionized water and solvent B was 98% acetonitrile/0.1% formic acid in deionized water. Flow from the column was directed to a Q-STAR pulsar II workstation (Applied Biosystems, Framingham, MA) equipped with a nano-ESI source. Peptides were separated in a 60 min linear gradient (from 0% to 35% solvent B) and MS/MS spectra were acquired in positive ion mode at 2700 volts of ionization voltage and 20 units of curtain gas at a sampling rate of one spectrum per second. The top three peptides with charges ranging from 2+ to 4+ were selected for CID. Precursor ions, once selected for CID were excluded from re-selection for 60 seconds.
2.3 Proteomics data analysis
The obtained MS/MS data were processed using Trans-Proteomic Pipeline (TPP) (http://tools.proteomecenter.org). Specifically, MS/MS spectra were searched against the International Protein Index human protein database (European Bioinformatics Institute, V3.19 for ICAT experiments and V3.17 for iTRAQ experiments) using SEQUEST[20]. The ICAT data were searched using the following criteria: fixed modification of cysteine (227.13 Da) and differential modifications of cysteine (9.01 Da) and methionine (15.99 Da). For the iTRAQ data, the database search were restricted with fixed modifications of cysteine (46.01 Da), lysine (144.1 Da) and N-terminus (144.1 Da) and differential modification of methionine (15.99 Da). The database search results were validated using the PeptideProphet program[21], which uses various SEQUEST scores and a number of other parameters to calculate a probability score for each identified peptide. The identified peptides were then assigned a protein identification using the ProteinProphet software [22]. ProteinProphet allows filtering of large-scale data sets with predictable sensitivity and false-positive identification error rates, and then generates statistically validated protein identifications from the identified peptides. For both ICAT and iTRAQ experiments, cut-off probability scores were selected to ensure that the false-positive rate (error rate) for protein identification was ≤1% based on ProteinProphet. Quantification of the ratio of each protein (isotopically heavy versus light) was calculated using the ASAPRatio program [23] for ICAT and Libra [24] for iTRAQ. The quantification of ICAT data was manually verified using ASAPRatio.
2.4 Immunohistochemical analysis
IHC staining was performed on pancreatic tissue sections as previously described [13]. Briefly, staining was performed using a Discovery XT (Ventana Medical Systems/Tucson, Arizona) automated immunohistochemistry instrument with a biotin-free, multimer technology detection kit. The protocol used Proteinase K (20 µg/sml) as antigen retrieval for 4 minutes. Rabbit anti laminin polyclonal Ab from Dako (Z 0097) at a 1:100 titer was incubated for 40 minutes at room temperature. The alpha–actinin staining used a monoclonal antibody (clone number 0.T.02, Abcam Ltd., Cambridge Science Park, Cambridge, United Kingdom) at a titer of 1:400. This antibody recognizes isoforms of alpha-actinin (actinin-1 to 4). Primary rabbit anti-human galectin-1 polyclonal antibody (Research Diagnostics, Inc., Flanders, NJ) at 1:100 titer was incubated at 37 °C. The IHC staining analysis was assessed by a GI pathologists (authors MPB, ZL and JFC).
3 Results and discussion
3.1 Pancreatic intraepithelial neoplasia
PanIN lesions are thought to be the precursor lesion to pancreatic adenocarcinoma. PanIN lesions are divided into three grades, based on their cellular architecture and nuclear appearance: PanIN 1 (hyperplasia), PanIN 2 (low-grade dysplasia), and PanIN 3 (high grade dysplasia or carcinoma in-situ). PanIN 1 lesions show no nuclear abnormalities and are characterized only by the development of cytoplasmic mucin. PanIN 2 lesions demonstrate some nuclear abnormalities, such as enlarged and hyperchromatic nuclei, nuclear crowding and stratification. These lesions are found in the setting of pancreatic cancer and occasionally in the pancreas of people who have died of other causes [25–27].
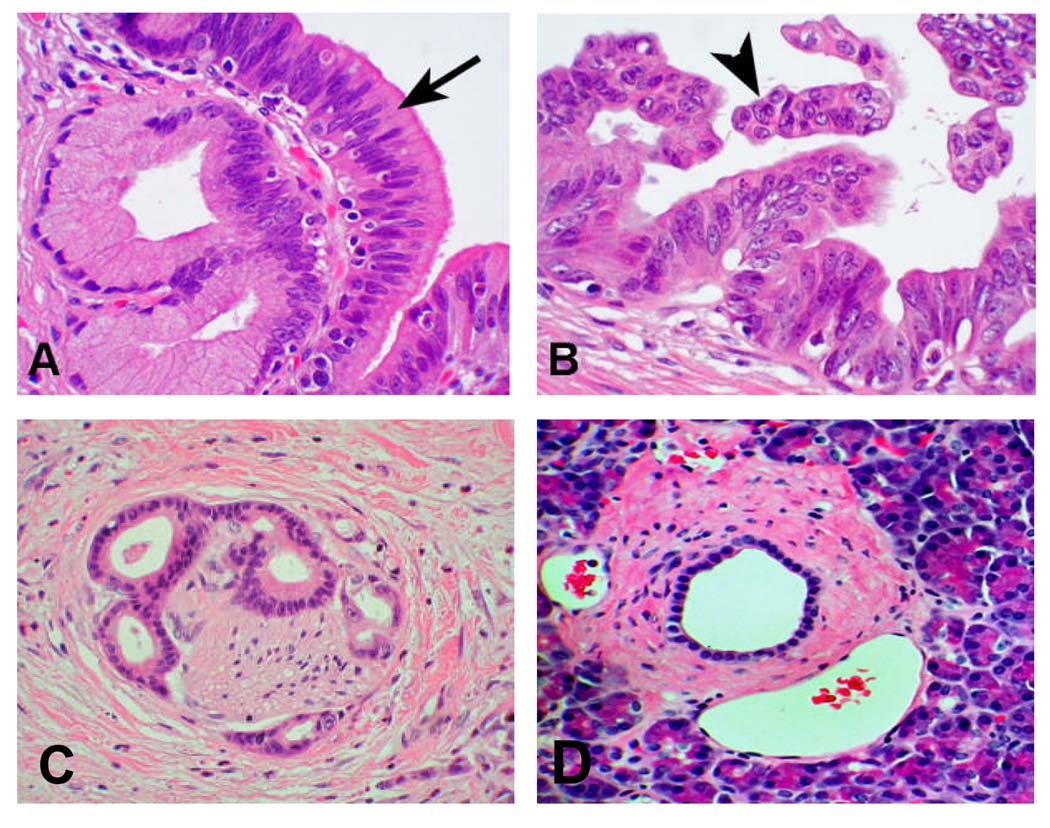
PanIN 3 nuclei are enlarged, pleomorphic, and have lost their polarity. PanIN 3 lesions are the immediate precursor of pancreatic adenocarcinoma. Autopsy studies suggest that PanIN 3 lesions are uncommon in the general population [25–27] and animal models of pancreatic tumorigenesis and case reports of PanIN 3 in humans suggest that these lesions likely progress to pancreatic cancer over time [28]. Figure 1 shows the microscopic features of a normal pancreatic duct, PanIN 2 and 3 and pancreatic adenocarcinoma. We selected the PanIN 3 lesions as the most clinically relevant target for detection of very early and curable pancreatic neoplasia.
Figure 1. Histology of PanIN and pancreatic cancer.
A. PanIN 2 or low grade ductal dysplasia; B. PanIN 3 or high grade ductal dysplasia; C. Pancreatic ductal adenocarcinoma with neural invasion; D. Normal pancreatic duct and acinar tissue.
To remove or correct for “non-specific background” in PanIN 3, the iTRAQ analysis included chronic pancreatitis tissue because the shared overlapping dysregulated proteins in pancreatitis with PanIN 3 would be of lesser interest in identifying future candidate biomarkers for pancreatic neoplasia. The comparison of the proteome of these various tissues (normal pancreas, PanIN3, chronic pancreatitis and pancreatic cancer) is expected to reveal the dysregulated proteins relevant to pancreatic tumorigenesis and its progression.
3.2 Quantitative profiling of protein expression in PanIN 3 tissue
Both ICAT and iTRAQ are established quantitative proteomics techniques and complementary to each other [29]. The ICAT method is cysteine-content biased but provides a more in-depth analysis on low abundant proteins; the iTRAQ method labels the terminal amine groups of all tryptic peptides covering a wider spectrum of peptides for identification and quantification. In addition, iTRAQ method allows multiplex comparison. A detailed comparison of ICAT and iTRAQ techniques for quantitative proteomics analysis has been reported previously [29]. Both methods were applied in this study to identify differentially expressed proteins in the whole tissue of PanIN 3 samples in comparison to normal pancreas.
ICAT
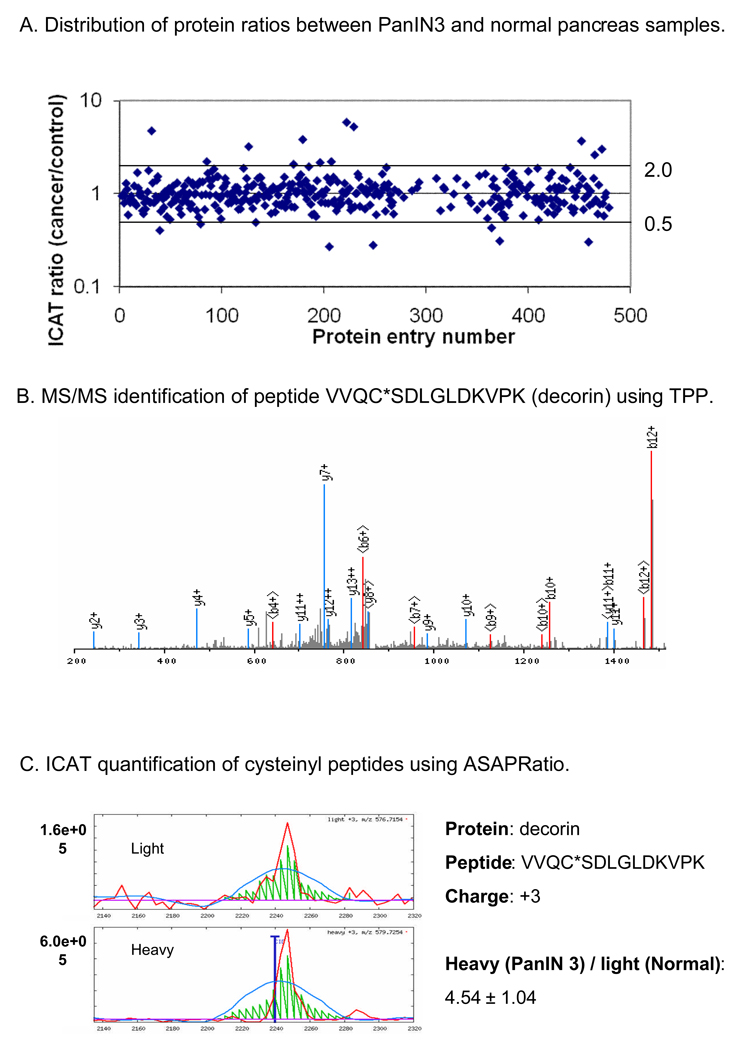
In the ICAT study, 402 proteins and protein groups, with majority of them containing cysteinyl peptides, were identified and quantified with a ProteinProphet score of 0.85, which ensures a ≤ 1% false positive rate for protein identification. Among the proteins identified, 22 and 16 proteins were over and under expressed in PanIN 3 samples, respectively, with a cut-off ratio of 1.75-fold. Figure 2A shows the distribution of protein abundance ratio between the PanIN 3 sample and the normal pancreas control. In comparison of our previous ICAT study on pancreatic cancer tissue [13], the number of the differentially expressed proteins discovered in PanIN3 tissue is significantly lower. This is likely due to the fact that at the pre-cancer stage, PanIN 3 lesions are quite small, thus, the differentially expressed proteins are less abundant and harder to detect. Figure 2B shows the MS/MS identification of tryptic peptide VVQC*SDLGLDKVPK (* indicates the labeling of ICAT reagent) derived from the protein decorin, showing that the y-ions and b-ions of the peptide are well balanced. A confident identification of a peptide provides the basis for more accurate quantification of the peptide, and subsequently, the corresponding protein. Figure 2C demonstrates the ICAT quantification of peptide VVQC*SDLGLDKVPK (decorin) using ASAPRatio software [23], which utilizes numerical and statistical methods to evaluate and calculate the abundance ratio of a peptide with identical sequences but with different isotopic tags (from different sample origins). As shown in Figure 2C, decorin was identified as one the over-expressed proteins in PanIN 3, with an overall protein ratio of 4.76±0.9, comparing its abundance in the PanIN 3 sample versus the normal pancreas control.
Figure 2. PanIN 3 protein expression profiling by ICAT.
A. Protein ratio distribution. B. An example of MS/MS spectrum for peptide identification. C. An example of peptide quantification by ICAT labeling. Note that 4.54 ± 1.04 is the ICAT ratio of a single peptide VVQC*SDLGLDKVPK, one of six peptides derived from protein decorin.
ITRAQ
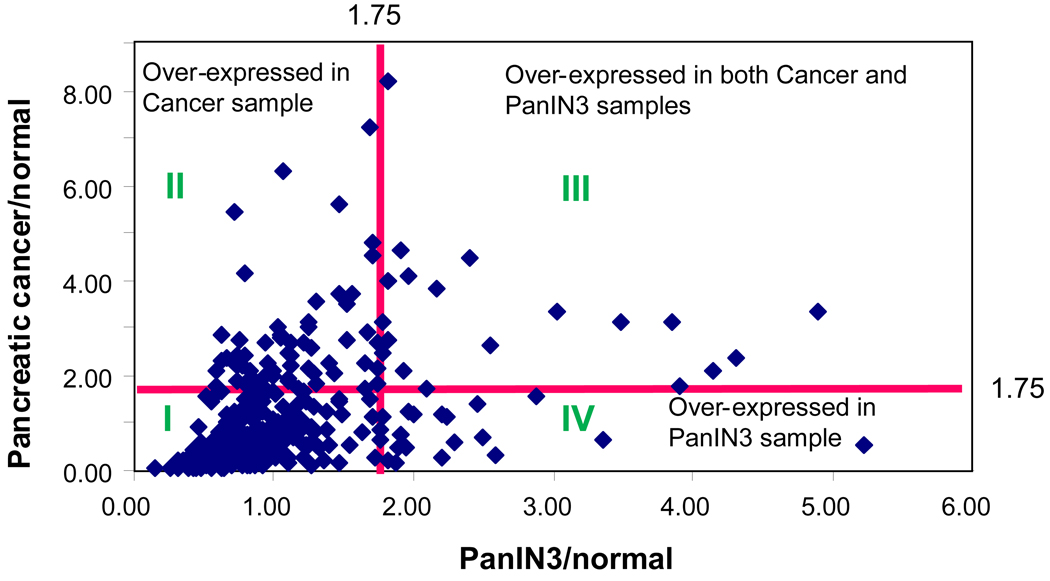
The four samples used for iTRAQ study were normal pancreas, pancreatitis, pre-cancer (PanIN 3) and cancer samples (labeled with iTRAQ reagents of 114, 115, 116 and 117 respectively). The study identified 770 proteins in total with a false positive rate of ≤ 1% based on ProteinProphet. Among these, 70 proteins were over-expressed and 133 were under-expressed in PanIN 3 using a cut-off ratio of 1.75. Figure 3 summarizes the result of one of the iTRAQ experiments, in which the tissue proteome of PanIN 3 was compared to pancreatic cancer and normal pancreas tissue. The distribution of the proteins can be divided into four groups (Figure 3) based on their abundance ratio to the normal control. The majority of the proteins are distributed in region I. These are the proteins that were similarly expressed in normal pancreas, PanIN 3 and cancer tissue samples. The proteins in region II were proteins that were over-expressed in cancer, but not in PanIN 3 tissue. The proteins in region III were over-expressed proteins in both PanIN 3 and cancer. Region IV includes proteins that were exclusively over-expressed in PanIN 3. The over-expressed proteins discovered in the PanIN 3 samples with two or more peptides for identification and quantification were summarized in Table 1. Many of these proteins were also found overexpressed in cancer or pancreatitis or both.
Figure 3. Distribution of the over-expressed proteins analyzed by iTRAQ analysis.
The distribution of the proteins can be divided into four regions based on their abundance ratio compared to the normal control; I. the proteins with expressional ratio <1.75 compared with normal control; II. the proteins overly expressed in pancreatic cancer sample only; III. the proteins overly expressed in both cancer and PanIN3 samples; IV. the proteins overly expressed in PanIN3 samples only.
Table 1.
The overexpressed (≥ 1.75) proteins identified in PanIN 3 tissues (The table only includes overexpressed proteins identified and quantified with two or more peptides)
| Protein ID | Description | Gene Symbol |
Group prob. |
Percent coverage |
No. of identified peptides |
Quantitaive Method |
Ratio (PanIN/NL) |
SD | Over expressed in cancer tissues? |
Over expressed in pancreatitis tissues? |
|---|---|---|---|---|---|---|---|---|---|---|
| IPI00412579, IPI00413986 |
25 kDa protein, 60S ribosomal protein L10a |
RPL10A;L OC137107 |
1.00 | 26.40 | 24.00 | ICA/iTRAQ | 5.88 | 1.38 | ||
| IPI00003865 | Isoform 1 of Heat shock cognate 71 kDa protein |
HSPA8 | 1.00 | 5.60 | 8.00 | ICAT | 5.26 | 0.55 | Y (ICAT) | |
| IPI00012119, IPI00219403 |
Isoform A of Decorin precursor, Isofrom D of Decorin precursor |
DCN | 1.00 | 19.20 | 16.00 | ICAT | 4.76 | 0.91 | ||
| IPI00024284, IPI00747758 |
Basement membrane-specific heparan sulfate proteoglycan core protein precursor, 365 kDa protein |
HSPG2 | 1.00 | 0.40 | 2.00 | ICAT/iTRAQ | 3.85 | 0.89 | ||
| IPI00219910 | Flavin reductase | BLVRB | 1.00 | 9.00 | 6.00 | ICAT | 3.23 | 4.27 | ||
| IPI00032179, IPI00165421 |
Antihrombin III variant, SERPINC1 protein |
SERPINC 1 | 1.00 | 5.00 | 5.00 | ICAT | 2.22 | 0.44 | Y (ICAT) | |
| IP100443909 | Isoform 1 of MIR-interacting saposin- like protein precursor |
CNPY2 | 1.00 | 34.10 | 11.00 | ICAT | 2.22 | 0.40 | ||
| IPI00302592, IPI00333541, IPI00644576 |
Filamin-A, Filamin A, alpha, Filamin A, alpha |
FLNA | 1.00 | 3.40 | 28.00 | ICAT | 2.17 | 0.66 | ||
| IPI00013991, IPI00220709, IPI00513698, IPI00646748 |
Isoform 2 of Tropomyosin beta chain, Isoform 1 of Tropomyosin beta chain, Tropomysin 2, Tropomysin 2 |
TPM2 | 1.00 | 4.90 | 4.00 | ICAT | 2.08 | 0.52 | Y (ICAT) | Y (ICAT) |
| IPI00216694 | plastin 3 | PLS3 | 1.00 | 6.50 | 5.00 | ICAT | 1.96 | 0.35 | ||
| IPI00018146 | 14-3-3 protein theta | YWHAQ | 1.00 | 11.80 | 3.00 | ICAT | 1.92 | 0.52 | ||
| IPI00013976 | Laminin beta-1 chain precursor | LAMB1 | 1.00 | 2.40 | 2.00 | ICAT | 1.89 | 0.32 | ||
| IPI00219219 | Galectin-1 | LGALS1 | 1.00 | 20.10 | 14.00 | ICAT | 1.85 | 2.95 | Y (ICAT) | |
| IPI00382804 | EEF1A protein (Fragment) | EEF1A1 | 1.00 | 24.70 | 4.00 | ICAT | 1.79 | 0.45 | ||
| IPI00294140, IPI00412253 |
Isoform 2 of Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial precursor,Isofrom 1 of Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial precursor |
MCCC2 | 1.00 | 3.80 | 5.00 | ICAT | 1.75 | 0.49 | ||
| IPI00008530, IPI00396373, IPI00556485, IPI00737483, IPI00737567, IPI00737840, IPI00739014 |
PREDICTED: similar to acidic ribosomal phosphoprotein P0 isoform 4,PREDICTED: similar to acidic ribosomal phosphoprotein P0 isoform 2,PREDICTED: similar to acidic ribosomal phosphoprotein P0 isoform 3,BLOCK 23,PREDICTED: similar to 60S acidic ribosomal protein P0 (L10E) isoform 5, RPLP0 protein,60S acidic ribosomal protein P0 |
RPLP0 | 0.99 | 10.40 | 2.00 | ICAT | 1.75 | 0.83 | ||
| IPI00382950 | Beta-globin gene from a thalassemia patient, complete cds |
HBB | 1.00 | 56.00 | 47.00 | iTRAQ | 4.00 | 0.02 | Y (ICAT/iTRAQ) | |
| IPI00026272, IPI00031562, IPI00081836, IPI00216456, IPI00216457, IPI00220855, IPI00255316, IPI00291764, IPI00303315, IPI00339274, IPI00552873, IPI00645519 |
Histone H2A type 1-B,Histone H2A type 3,Histone H2A type 1- H,Histone H2A type 1-j,Histone H2A type 1-E,Histone H2A type 1,Histone H2A type 2-C,H2A histone family, member j isoform 2,Histone H2A type 1-C,Histone H2A type 2-A,Histone H2A type 1- D,Histone H2A.c/d/i/n/p |
HIST1H2 AB;HIST1 H2AE |
1.00 | 20.50 | 5.00 | iTRAQ | 2.79 | 0.03 | Y (iCAT) | |
| IPI00473011 | Hemoglobin delta subunit | HBB;HBD | 1.00 | 58.20 | 11.00 | iTRAQ | 2.64 | 0.04 | Y (ICAT/iTRAQ) | |
| IPI00183695 | Calpactin-1 light chain | S100A10 | 0.99 | 17.10 | 4.00 | iTRAQ | 2.27 | 0.01 | Y (ICAT/iTRAQ) | Y (iTRAQ) |
| IPI00556589, IPI00719622 |
40S ribosomal protein S28,PREDICTED: similar to 40S ribosomal protein S28 |
RP528 | 0.96 | 17.40 | 2.00 | iTRAQ | 2.17 | 0.02 | ||
| IPI00008603, IPI00023006, IPI00025416 |
Actin, alpha cardiac,Actin, gamma- enteric smooth muscle,Actin, aortic smooth muscle |
ACTA2;A CTG2 |
1.00 | 51.90 | 21.00 | iTRAQ | 2.14 | 0.02 | Y (ICAT/iTRAQ) | Y (iTRAQ) |
| IPI00550315, IPI00644379 |
Ig kappa chain C region,Ig kappa chain C region |
IGKV3-20 | 0.99 | 22.10 | 2.00 | iTRAQ | 2.09 | 0.02 | Y (iTRAQ) | Y (iTRAQ) |
| IPI00386854, IPI00396378, IPI00414696, IPI00477522 |
Spice Isoform B1 of Heterogeneous nuclear ribonucleoproteins A2\B1,Splice Isoform A2 of Heterogeneous nuclear ribonucleoproteins A2\B1,Heterogeneous nuclear ribonucleoprotein A2\B1 isoform A2; Heterogeneous nuclear ribonucleoprotein A2; Heterogeneous nuclear ribonucleoprotein B1; nuclear ribonucleoprotein particle A2 protein,HNRPA2B1 protein |
HNRNPA2B1 | 1.00 | 22.10 | 6.00 | iTRAQ | 2.05 | 0.04 | ||
| IPI00477457, IPI00478348 |
46 kDa protein, Similar to Heterogeneous nuclear ribonucleoprotein H |
HNRNPF | 1.00 | 14.80 | 3.00 | iTRAQ | 1.92 | 0.02 | Y (iTRAQ) | |
| IPI00298308, IPI00739452 |
Aldehyde dehydrogenase 1 family, member L2, PREDICTED: aldehyde dehydrogenase 1 family, member L2 |
ALDH1L2 | 1.00 | 4.60 | 7.00 | iTRAQ | 1.82 | 0.07 | Y (iTRAQ) | |
| IPI00021792 | Phospholipase A2 precursor | PLA2G1B | 1.00 | 23.00 | 11.00 | iTRAQ | 5.22 | 0.03 | ||
| IPI00298547 | Protein DJ-1 | PARK7 | 1.00 | 7.90 | 2.00 | iTRAQ | 4.89 | 0.00 | Y (iTRAQ) | |
| IPI00514424, IPI00002412, IPI00514248 |
Palmitoyl-protein thioesterase 1, Palmitoyl-protein thioesterase 1 precursor,Palmitoyl-protein thioesterase 1 |
PPT1 | 0.97 | 6.30 | 2.00 | iTRAQ | 4.31 | 0.01 | Y (iTRAQ) | |
| IPI00220834 | ATP-dependent DNA helicase 2 subunit 2 |
XRCC5 | 1.00 | 2.50 | 2.00 | iTRAQ | 4.15 | 0.01 | Y (iTRAQ) | |
| IPI00748319, IPI00642800, IPI00644431, IPI00641952, IPI00552922, IPI00641926, IPI00166874, IPI00641325, IPI00643815, IPI00062206, IPI00641634, IPI00640787, IPI00641829, IPI00328343, IPI00642880 |
HLA-B associated transcript 1 (Fragment),26 kDa protein,ATP- dependent RNA helicase DDX39,HLA-B associated transcript 1 (Fragment),HLA-B associated transcript 1 (Fragment),HLA-B associated transcript 1, DDX39 protein,HLA-B associated transcript 1, HLA-B associated transcript 1, DEAD (Asp-Glu-Ala- Asp) box polypeptide 39, isoform 2,25 kDa protein,HLA-B associated transcript 1,29 kDa protein,Spliceosome RNA helicase BAT1, HLA-B associated transcript 1 |
BAT1 | 0.99 | 5.30 | 5.00 | iTRAQ | 3.48 | 0.01 | Y (iTRAQ) | |
| IPI00007752, IPI00011654, IPI00647896, IPI00645452 |
Tubulin beta-2C chain,Tubulin beta- 2 chain,Tubulin,beta polypeptide,Tubulin,beta polypeptide |
TUBB2C | iTRAQ | 3.03 | 0.01 | Y (iTRAQ) | ||||
| IPI00453473 | Histone H4 | HIST1H4A | 1.00 | 18.60 | 2.00 | iTRAQ | 2.88 | 0.03 | ||
| IPI00018553 | Caldecrin precursor | CTRC | 1.00 | 17.20 | 10.00 | iTRAQ | 2.60 | 0.12 | ||
| IPI00643041 | GTP-binding nuclear protein Ran | RAN | 0.99 | 6.50 | 4.00 | iTRAQ | 2.56 | 0.01 | Y (iTRAQ) | Y (ICAT) |
| IPI00221225, | annexin IV,ANXA4 protein | ANXA4 | 1.00 | 23.40 | 21.00 | iTRAQ | 2.45 | 0.03 | Y (ICAT) | |
| IPI00654755 | Hemoglobin beta subunit | HBB | 1.00 | 74.70 | 157.00 | iTRAQ | 2.30 | 0.01 | ||
| IPI00013860, IPI00479966 |
3-hydroxyisobutyrate dehydrogenase,mitochondrial precursor,Hypothetical protein (Fragment) |
HIBADH | 1.00 | 4.20 | 3.00 | iTRAQ | 2.25 | 0.04 | ||
| IPI00290460 | EUkaryotic translation initiation factor 3 subunit 4 |
EIF3G | 0.99 | 5.00 | 5.00 | iTRAQ | 2.21 | 0.02 | ||
| IPI00025460, IPI00022498, IPI00008752, IPI00008753, IPI00457184, IPI00008750, IPI00647104, IPI00472662, IPI00456560, IPI00413064 |
Metallothionein- 1A,Metallothionein-2,Splice Isoform 1 of Metallothionein- 1G,Metallothionein-1X,PREDICTED: similar to metallothionein 1G,Metallothionein-1H,7kDa protein,Metallothionein- 1I,PREDICTED: hypothetical protein XP_498969,Splice Isoform 2 of Mctallothoncin 1G |
MT1A | 1.00 | 13.10 | 4.00 | iTRAQ | 2.20 | 0.03 | ||
| IPI00298497 | Fibrinogen beta chain precursor | FGB | 0.97 | 2.00 | 8.00 | iTRAQ | 2.17 | 0.01 | Y (iCAT/iTRAQ) | Y (ICAT) |
| IPI00465315 | Cytochrome c | CYCS | 0.99 | 10.60 | 8.00 | iTRAQ | 2.10 | 0.01 | ||
| IPI00456969, IPI00477531 |
Dynein heavy chain, cytosolic 532 kDa protein |
DYNC1H1 | 1.00 | 1.00 | 6.00 | iTRAQ | 1.96 | 0.02 | ||
| IPI00418471 | Vimentin | VIM | 1.00 | 62.40 | 50.00 | iTRAQ | 1.96 | 0.00 | Y IICAT/iTRAQ) | Y (iTRAQ) |
| IPI00717044, IPI00001508 |
INSIGF long transport variant, Insulin precursor |
INS | 0.99 | 6.40 | 16.00 | iTRAQ | 1.94 | 0.01 | ||
| IPI00009771 | Lamin-B2 | LMNB2 | 0.99 | 1.60 | 2.00 | iTRAQ | 1.93 | 0.01 | Y (iTRAQ) | |
| IPI00736803, IPI00745291, IPI00645201, IPI00216587 |
PREDICTED: similar to ribosomal protein SB, Similar to ribosomal protein S8, Ribosomal protein S8, 405 ribosomal protein S8 |
RPS8 | 0.92 | 12.60 | 2.00 | iTRAQ | 1.91 | 0.01 | ||
| IPI00604784, IPI00306140, IPI00744153 |
glucagon preprdprotein, Glucagon precursor,21kDa protein |
GCG | 1.00 | 12.80 | 7.00 | iTRAQ | 1.91 | 0.01 | Y (iTRAQ) | |
| IPI00292709 | Phosphoenopyruvate carboxykinase, cytosolic |
PCK1 | 1.00 | 1.30 | 10.00 | iTRAQ | 1.89 | 0.02 | ||
| IPI00021447 | Alpha-amylase2B precursor | AMY2B | 1.00 | 26.60 | 4.00 | iTRAQ | 1.87 | 0.03 | ||
| IPI00745705, IPI00025476 |
Amylase,alpha 2A;pancreatic variant (Fragment),pancreatic alpha-amylase precursor |
AMY1A;1B;1C;2A | 1.00 | 30.30 | 44.00 | iTRAQ | 1.83 | 0.02 | ||
| IPI00555610 | 313 kDa protein | AHNAK | 1.00 | 10.00 | 19.00 | iTRAQ | 1.83 | 0.01 | Y (iTRAQ) | Y (iTRAQ) |
| IPI00472119, IPI00419880 |
30 kDa protein, 40S ribosomal protein S3a |
RPS3A | 1.00 | 8.00 | 9.00 | iTRAQ | 1.79 | 0.02 | ||
| IPI00329801 | Annexin AS | ANXA5 | 1.00 | 37.00 | 15.00 | iTRAQ | 1.78 | 0.01 | Y (iTRAQ) | |
| IPI00020987 | Prolargin precursor | PRELP | 1.00 | 8.60 | 4.00 | iTRAQ | 1.78 | 0.02 | Y (iTRAQ) | |
| IPI00215790 | 60S ribosomal protein L38 | RPL38 | 0.98 | 18.80 | 2.00 | iTRAQ | 1.78 | 0.03 | ||
| IPI00216085 | Cytochrome c oxidase sububit VIb isoform 1 |
COX6B1 | 0.99 | 12.90 | 6.00 | iTRAQ | 1.77 | 0.02 | ||
| IPI00744444, IPI00335168, IPI00413922 |
17 kDa protein,myosin, light polypeptide 6,alkali,smooth muscle and non-muscle isoform 1,Splice isoform Smooth muscle of Myosin light polypeptide 6 |
MYL6;MYL6B | 1.00 | 10.00 | 3.00 | iTRAQ | 1.75 | 0.00 | Y (iTRAQ) | |
| IPI00013808 | Alpha-actinin-4 | ACTN4 | 1.00 | 26.60 | 18.00 | iTRAQ | 1.75 | 0.01 | Y (iTRAQ) |
3.3 Functional roles of the dysregulated proteins in PanIN 3 tissues
To functionally annotate the dysregulated proteins identified in this study, the dysregulated proteins were entered into the GeneGO metacore program for enrichment assessment and network analysis. Among the dysregulated proteins, the top enriched category was cell motility. Eleven actin-related cytoskeleton and thirteen other cytoskeleton proteins relating to cytoskeleton remodeling were dysregulated in the PanIN 3 tissues. The cytoskeleton, especially actin cytoskeleton, plays a central role in cell motility, invasion and metastasis. Because cellular invasion has not yet occurred in PanIN lesions (these changes are a feature of cancer), dysregulation of these motility proteins in PanIN 3 lesions suggests that the dysregulation at the protein level starts early before the actual cell invasion. Other top enriched functional categories of the dysregulated proteins in PanIN 3 tissue included proteins involved in: the inflammatory response; blood clotting; cell cycle and its regulation; protein degradation.
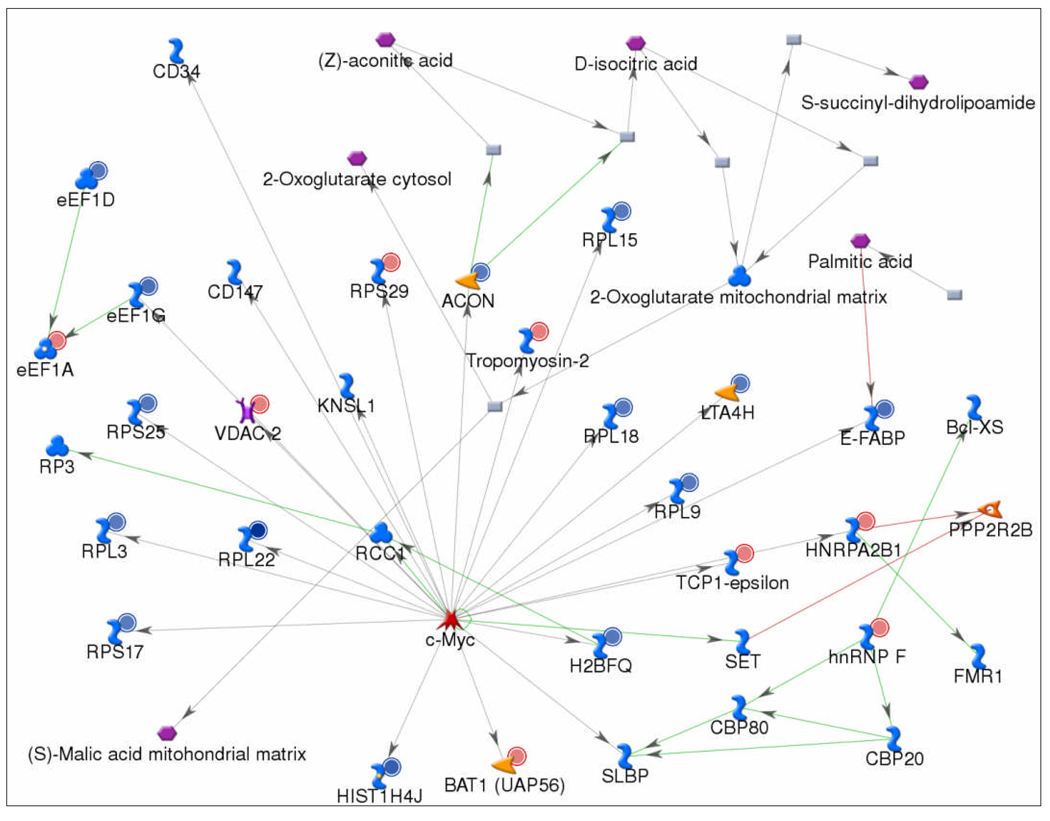
To analyze the broader interacting network among the dysregulated proteins in PanIN 3 lesions, we used the GeneGo metacore Analyze Network algorithm to construct networks using the closest 50 protein targets as a network limit. The most inclusive network, involving cell localization, cell motility, and cell morphogenesis, brought together 15 of the dysregulated proteins with one or two step interactions. The second network, as shown in Figure 4, which was inclusive of dysregulated proteins involved in RNA localization, nucleic acid transport and translation, brought together 22 of the dysregulated PanIN3 proteins with one or two step interactions. The most prominent regulatory protein in the network was c-MYC, which directly interacted with 18 of the dysregulated proteins in the network. c-MYC is an oncoprotein that is involved in over 20% of all human cancers. We previous identified c-MYC as a key regulatory protein common in pancreatic cancer and chronic pancreatitis [11]. The results in this study extend this finding to PanIN 3 tissue, identifying c-MYC as one of the common regulatory proteins present in chronic pancreatitis, PanIN 3, and pancreatic cancer.
Figure 4. Biological network analysis of the dysregulated proteins in PanIN3 tissue.
The network was generated using the Analyze Network algorithm to map the interactions between the dysregulated proteins. Nodes represent proteins; lines between the nodes indicate direct protein-protein interactions. A small red circle denotes an over-expressed protein, whereas a small blue circle denotes an under-expressed protein. The most prominent regulatory protein in the network was c-MYC, which directly interacts with 18 of the dysregulated proteins in the network.
3.4 Over-expressed proteins identified in PanIN 3 tissues
Many of the over-expressed proteins identified in PanIN 3 tissues (Table 1) were also found to be over-expressed in cancer samples in our study. The identification and quantification of these proteins were based on multiple peptides (two or more peptides) using stringent criteria. Some of the proteins listed in Table 1 are discussed below. These proteins or their isoforms have been previously related to cancer, in particular pancreatic cancer, in our studies and those of others.
Laminin beta 1 (IPI00013976)
Laminin is a glycoprotein and one of the extracellular matrix components. It consists of three different polypeptide chains (alpha, beta, gamma), which are bound to each other by disulfide bonds. Three unique peptides of laminin beta 1 were identified, including 2 cysteinyl peptides for ICAT quantification. Peptide YCIVSHLQEDK belongs to the laminin N-terminal domain. Peptide CKLNVEGEHCDVCK belongs to the EGF-like 2 domain and includes two cysteine residues which may have been derived from a potential disulfide bond. Previous studies have suggested that laminin might play a role in pancreatic cancer metastases[30], and in pancreatic cancer cell survival by inhibiting both mitochondrial dysfunction and caspase activity [31]. A previous tissue microarray study has shown that laminin beta 3 was over-expressed in high-grade PanINs and in more than 90% of pancreatic ductal carcinoma [32].
14-3-3 theta (IPI00018146)
14-3-3 theta protein is abundantly expressed in pancreas and binds to a large number of protein partners potentially implicating the signaling pathways and activity of the binding partners. Two unique cysteinyl peptides were identified for this protein in the ICAT study, including YLAEVACGDDR and AKLAEQAERYDDMATCMK. The 14-3-3 protein family, especially 14-3-3 sigma, has previously been reported as a putative biomarker for pancreatic cancer [13,33,34] and might be involved in cell cycle regulation and apoptosis [35]. 14-3-3 theta has recently been associated with lung cancer [36,37] and urothelial carcinomas [38]. Its implication with pancreatic cancer remains unclear and warrants further investigation.
Decorin (IPI00012119, IPI00219403)
Decorin is a glycoprotein and may affect fibril formation in the extracellular matrix. The ICAT study identified three cysteinyl peptides in a continuous section of the protein sequence, including DFEPSLGPVCPFR (45–57), CQCHLR (58–63) and VVQCSDLGLDKVPK (64–77), significantly increasing the identification confidence of this protein. The identified section of the protein sequence includes cysteine residues for which may have been derived from a potential disulfide crosslink, as well as a sequence variance (72–75, LDKV -> CLPS in isoform E). Decorin is a known anti-tumor factor, and has been reported to be significantly over-expressed in pancreatic cancer tissues by real time quantitative PCR and immunohistochemistry [39,40]. We report here that decorin is present in the proteome of PanIN 3 tissues at increased level compared to normal pancreas tissue. Studies suggest that pancreatic stellate cells (PSCs) might be the major source of decorin [39,40] and the expression of decorin in pancreatic cancer might represent an effort of the host to contain the growth of tumor [41]. The same study also suggested that specific post-translational modifications of decorin may be closely associated with the malignant phenotype of pancreatic cancer.
Galectin-1 (IPI00219219)
Galectin-1 is a small protein that may play a role in regulating cell apoptosis and cell differentiation. Two consecutive cysteinyl peptides of this protein were repeatedly identified in the ICAT study: DSNNLCLHFNPR (38–49) and FNAHGDANTIVCNSK (50–64), providing both identification and quantification of the protein. In concert with our findings here, previous genomic and proteomic studies reveal that galectin-1 is up-regulated in pancreatic cancer [13,42–44], and may have a role in pancreatic stellate cell activation [45,46]. The finding of galactin-1 overexpression in PanIN tissue in our study is a new finding.
Vimentin (IPI00418471)
Vimentin is a phosphoprotein and it is highly expressed in fibroblasts. Multiple peptides derived from this protein were identified in the pancreatitis, PanIN 3 and pancreatic cancer tissues of the iTRAQ study. The peptides were located at different regions of the protein sequence, including: the head region: TYSLGSALRPSTSR (phosphoserine at 39, 42 and 47); the coil 1A and linker 1 regions: FLEQQNK and ILLAELEQLK; the coil 1B region: VEVERDNLAEDIMR, DNLAEDIMR and KVESLQEEIAFLK; and the tail region: ISLPLPNFSSLNLR (phosphoserine at 412 and 420) and ETNLDSLPLVDTHSK (phosphoserine at 430). Recent studies have suggested that vimentin is associated with pancreatic cancer [13,47,48]; and vimentin is recognized as a mesenchymal marker to identify carcinoma-associated fibroblasts [47–49]. An elevated level of vimentin has also been observed in chemoresistant (gemcitabine-resistant) pancreatic tumor cells as one of the hallmarks of epithelial-to-mesenchymal transition [50].
Actinin-4 (IPI00013808)
Actinin-4 is an actin-binding protein associated with cell motility, cancer invasion and metastasis [51,52]. The iTRAQ study identified eleven peptides for alpha actinin-4 identification and quantification, including several peptides in actin-binding domain. These peptides include TFTAWCNSHLR, LMLLLEVISGER, AGTQIENIDEDFRDGLK, and DGLAFNALIHR. Previous studies have observed the over-expression of actinin-4 in colorectal cancer [53,54].
In addition to the proteins that may be implicated with pancreatic tumorgenesis, a few other over-expressed proteins that are listed in Table 1, including phospholipase A2, aldehyde dehydrogenase, alpha-amylase and a group of ribosomal proteins are worth mentioning. The abundance of these proteins has been observed to fluctuate to a significant extent in different pre-cancer and cancer samples. Some of these proteins are enzymes involving in a variety of metabolic pathways, and have been associated with pancreatic diseases in the literature. The change in abundance of these enzymes in PanIN 3 and cancer tissues did not appear to be independently related to pancreatic carcinogenesis, but rather may be a result from a variety of different factors including physiological conditions of the individual at the time the tissue was collected and the co-existence of other pancreatic disorders. For instance, phospholipase A2 is a digestion enzyme involving in calcium catalytic activity. Three peptides were identified for this protein, including peptide CCQTHDNCYDQAK, which contains a calcium binding site and cysteines for potential disulfide crosslink. Phospholipase A2 has long been associated with pancreatic diseases [55–58] and pancreatic cancer [59,60]; and the abnormal level of serum phospholipase A2 has been evaluated for its diagnostic value [61,62]. This protein was found to be under-expressed in several cancer tissues in our current and previous studies. However, the abundance of this protein in pre-cancer tissues fluctuated significantly, from over-expression to under-expression, probably due to biological variation, physiological state and the underlying pancreatic disorders that the patients involved in this study might have had. Our preliminary observations suggested that this group of proteins might not have an independent correlation with pancreatic carcinogenesis.
3.5 IHC validation
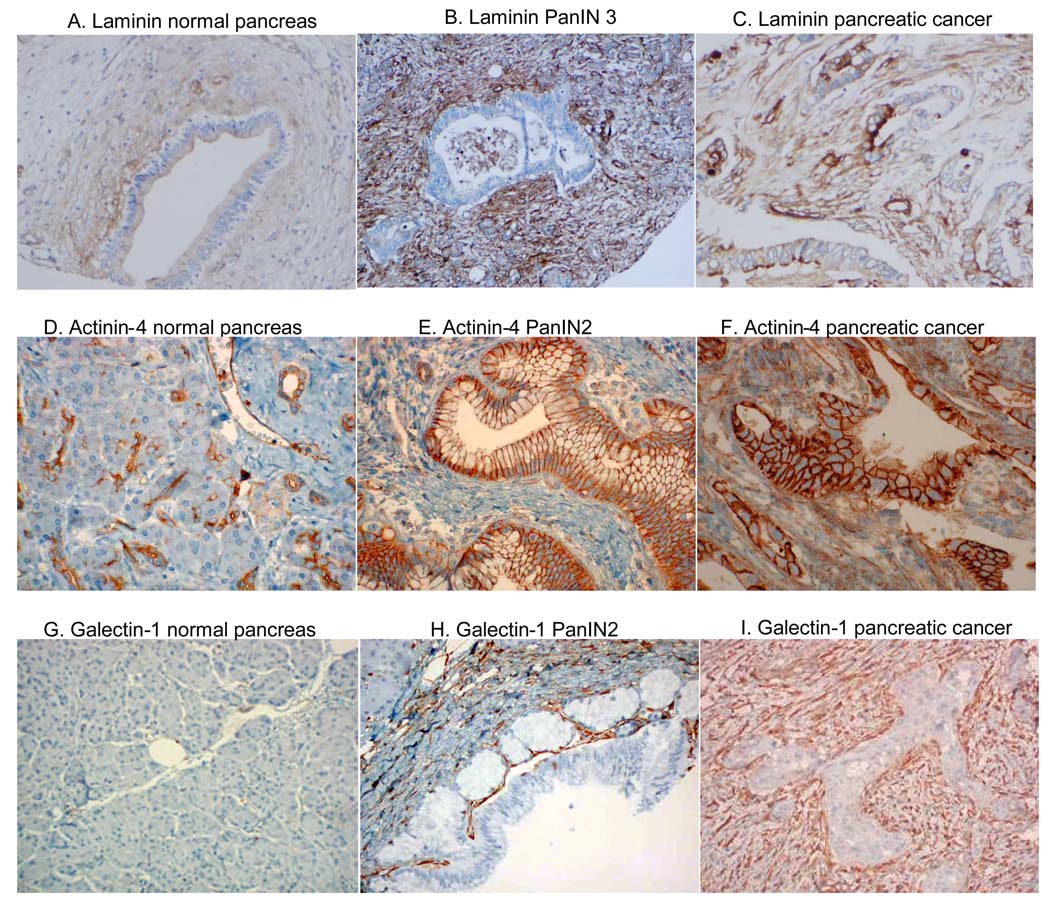
To validate and further investigate the expression of over-expressed PanIN 3 proteins, three over-expressed PanIN proteins, laminin beta-1, actinin-4 and galectin-1 were selected for IHC analysis on pancreatic tissue.
Laminin beta-1
As shown in Figure 5, laminin was marginally expressed in the stroma adjacent to pancreatic ducts in normal pancreas (Figure 5A). In contrast, laminin was highly over-expressed in the stroma adjacent to PanIN3 (Figure 5B) as well as pancreatic cancer (Figure 5C). The epithelial cells in normal pancreas and in the PanIN lesions were negative for laminin staining. However, the cell membrane and cytoplasm of adenocarcinoma cells were stained for laminin (Figure 5C). Because laminin is an extracellular matrix protein, further investigation is needed to confirm the cytoplasmic expression of laminin in pancreatic adenocarcinoma cells.
Figure 5. IHC analysis in advanced PanIN tissue and pancreatic cancer.
Laminin (A–C): note the overexpression in the stromal tissue in PanIN3 (B) and pancreatic cancer (C) compared to normal pancreas (A). Actinin-4 (D–F): note the overexpression in the ductal epithelium of PanIN2 (E), and in both the stroma and ductal epithelium of pancreatic cancer (F). Galectin-1 (G–I): note the overexpression in the stroma of PanIN2 (H) and pancreatic cancer (I) while it is absent in normal pancreas (G).
Actinin-4
Alpha actinin consists of several isoforms. The antibody used in the current study recognized isoforms 1 through 4. In normal pancreas, alpha actinin was positive in the small intralobular terminal ductules, with no staining of acinar cells (Figure 5D). PanIN 2 displayed strong staining of the dysplastic epithelium and marginal staining of the adjacent stroma (Figure 5E). In pancreatic cancer, there was marked increased and diffuse staining of both the malignant epithelium and adjacent stroma (Figure 5F). None of the 4 normal pancreas specimens displayed strong staining in ductal epithelium. In contrast, 6 out of 8 PanINs and 16 out of 21 cancer specimens had strong alpha actinin staining. Thus, actinin-4, showed increased expression in both the epithelial and stromal elements of advanced PanIN lesions and pancreatic cancer.
Galectin-1
Galectin-1 does not stain normal pancreas (Figure 5G). Increased stromal staining was observed in advanced PanIN tissue (Figure 5H). There was also a marked increased in stromal staining in pancreatic cancer (Figure 5I). The epithelial cells from the PanIN tissue and pancreatic cancer remained negative. Strong galectin-1 stromal expression was observed in none of the 33 normal pancreas controls, 70% of stroma surrounding advanced PanINs (12 out of 17 specimens), and 95% of the stroma surrounding pancreatic cancer (41 out of 43 specimens). Thus, the increased stromal expression is associated with advanced PanIN lesions and pancreatic cancer.
4 Concluding remarks
In this study, we used stable isotope labeling and mass spectrometry-based quantitative proteomics to identify the dysregulated proteins in the immediate precursor of pancreatic cancer (PanIN 3). The dysregulated proteins identified in this study may be useful in candidate protein selection for future biomarker development for early pancreatic neoplasia. These dysregulated proteins were enriched in several functional categories, including cell motility, the inflammatory response, blood clotting cascade, cell cycle and its regulation, and protein degradation. Further network analysis identified c-MYC as an important regulatory protein in PanIN 3 lesions. Finally, three of the overexpressed proteins, laminin beta-1, actinin-4 and galectin-1 were validated by IHC analysis. These three proteins were overexpressed in the stroma and/or ductal epithelial cells of PanIN 2 and 3 lesions, as well as in pancreatic ductal adenocarcinoma. The identification of tissue biomarkers present in the early stages of pancreatic cancer can provide a list of targets that could be selectively enriched for in plasma ---these tissue based proteins may be shed into the blood stream and provide the source for a blood test for early pancreatic cancer. Recent developments in proteomics technology has shown that many low abundant proteins that are shed into the circulatory system can now be quantitatively detected in blood by using targeted approaches including ELISA, immunoassays and mass spectrometry based methods. In addition, tissue biomarkers can potentially be directly detected using specimens obtained from needle biopsy such as might be obtained for clinical diagnosis. It is our hope that the information provided by this study will facilitate a better understanding of tumorigenesis of pancreatic cancer and ultimately the selection of candidate biomarkers for the earlier diagnosis of the pancreatic cancer when the disease is still treatable.
Acknowledgement
This work was supported in part by NIH/NCI grants 1R01CA107209 and K07CA116296; and funding from the Canary Foundation, Gene and Mary Ann Walters Pancreatic Cancer Foundation, the AACR-PanCAN Career Development Award for Pancreatic Cancer Research, and Federal funds from the National Heart, Lung and Blood Institute NIH, under contract # NOI-HV-28179.
Abbreviations
- PanIN
pancreatic intraepithelial neoplasia
- iTRAQ
isobaric tags for relative and absolute quantification
- IHC
immunohistochemistry
- SCX
strong cation-exchange chromatography
References
- 1.Jemal A, Thomas A, Murray T, Thun M. CA Cancer J.Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. CA Cancer J.Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.Kern S, Hruban R, Hollingsworth MA, Brand R, Adrian TE, Jaffee E, Tempero MA. Cancer Res. 2001;61:4923–4932. [PubMed] [Google Scholar]
- 4.Brand R. Cancer J. 2001;7:287–297. [PubMed] [Google Scholar]
- 5.Ariyama J, Suyama M, Satoh K, Sai J. Pancreas. 1998;16:396–401. doi: 10.1097/00006676-199804000-00030. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 7.Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. J.Proteome.Res. 2005;4:1104–1109. doi: 10.1021/pr050027n. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Pan S, Brentnall TA, Aebersold R. Mol.Cell Proteomics. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Chen R, Pan S, Yi EC, Donohoe S, Bronner MP, Potter JD, Goodlett DR, Aebersold R, Brentnall TA. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, Pan S, Cooke K, Moyes KW, Bronner MP, Goodlett DR, Aebersold R, Brentnall TA. Pancreas. 2007;34:70–79. doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, Bronner MP. Mol.Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, Pan S, Aebersold R, Brentnall TA. Proteomics-Clin.Appl. 2007;1:1582–1591. doi: 10.1002/prca.200700414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R, Yi EC, Donohoe D, Pan S, Eng J, Crispin DA, Lane Z, Goodlett DA, Bronner MP, Aebersold R, Brentnall TA. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Deng R, Lu Z, Chen Y, Zhou L, Lu X. Pancreas. 2007;34:310–317. doi: 10.1097/MPA.0b013e31802f2483. [DOI] [PubMed] [Google Scholar]
- 15.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. J.Proteome.Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 16.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Mol.Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Rosty C, Goggins M. Methods Mol.Med. 2005;103:189–197. doi: 10.1385/1-59259-780-7:189. [DOI] [PubMed] [Google Scholar]
- 18.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat.Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 19.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol.Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Eng J, McCormack AL, Yates JR. J.Am.Soc.Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 22.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal.Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 23.Li XJ, Zhang H, Ranish JA, Aebersold R. Anal.Chem. 2003;75:6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 24.Mueller LN, Brusniak MY, Mani DR, Aebersold R. J.Proteome.Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 25.Cubilla AL, Fitzgerald PJ. Cancer Res. 1976;36:2690–2698. [PubMed] [Google Scholar]
- 26.Pour PM, Sayed SE, Wolf GL. Cancer Lett. 1980;10:151–154. doi: 10.1016/0304-3835(80)90038-5. [DOI] [PubMed] [Google Scholar]
- 27.Pour PM, Sayed S, Sayed G. Am.J.Clin.Pathol. 1982;77:137–152. doi: 10.1093/ajcp/77.2.137. [DOI] [PubMed] [Google Scholar]
- 28.Brat DJ, Lillemoe KD, Yeo CJ, Warfield PB, Hruban RH. Am.J.Surg.Pathol. 1998;22:163–169. doi: 10.1097/00000478-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Wu WW, Wang G, Baek SJ, Shen RF. J.Proteome.Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 30.Grzesiak JJ, Smith KC, Burton DW, Deftos LJ, Bouvet M. Surgery. 2007;141:804–814. doi: 10.1016/j.surg.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaquero EC, Edderkaoui M, Nam KJ, Gukovsky I, Pandol SJ, Gukovskaya AS. Gastroenterology. 2003;125:1188–1202. doi: 10.1016/s0016-5085(03)01203-4. [DOI] [PubMed] [Google Scholar]
- 32.Qian J, Niu J, Li M, Chiao PJ, Tsao MS. Cancer Res. 2005;65:5045–5053. doi: 10.1158/0008-5472.CAN-04-3208. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez JA, Li M, Yao Q, Chen C, Fisher WE. World J.Surg. 2005;29:297–305. doi: 10.1007/s00268-004-7843-0. [DOI] [PubMed] [Google Scholar]
- 34.Sinha P, Hutter G, Kottgen E, Dietel M, Schadendorf D, Lage H. Electrophoresis. 1999;20:2952–2960. doi: 10.1002/(SICI)1522-2683(19991001)20:14<2952::AID-ELPS2952>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 35.Guweidhi A, Kleeff J, Giese N, El Fitori J, Ketterer K, Giese T, Buchler MW, Korc M, Friess H. Carcinogenesis. 2004;25:1575–1585. doi: 10.1093/carcin/bgh159. [DOI] [PubMed] [Google Scholar]
- 36.Pereira-Faca SR, Kuick R, Puravs E, Zhang Q, Krasnoselsky AL, Phanstiel D, Qiu J, Misek DE, Hinderer R, Tammemagi M, Landi MT, Caporaso N, Pfeiffer R, Edelstein C, Goodman G, Barnett M, Thornquist M, Brenner D, Hanash SM. Cancer Res. 2007;67:12000–12006. doi: 10.1158/0008-5472.CAN-07-2913. [DOI] [PubMed] [Google Scholar]
- 37.Qi W, Liu X, Qiao D, Martinez JD. Int.J.Cancer. 2005;113:359–363. doi: 10.1002/ijc.20492. [DOI] [PubMed] [Google Scholar]
- 38.Heidenblad M, Lindgren D, Jonson T, Liedberg F, Veerla S, Chebil G, Gudjonsson S, Borg A, Mansson W, Hoglund M. BMC.Med.Genomics. 2008;1:3. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koninger J, Giese NA, di Mola FF, Berberat P, Giese T, Esposito I, Bachem MG, Buchler MW, Friess H. Clin.Cancer Res. 2004;10:4776–4783. doi: 10.1158/1078-0432.CCR-1190-03. [DOI] [PubMed] [Google Scholar]
- 40.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Nat.Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 41.Skandalis SS, Kletsas D, Kyriakopoulou D, Stavropoulos M, Theocharis DA. Biochim.Biophys.Acta. 2006;1760:1217–1225. doi: 10.1016/j.bbagen.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 42.Berberat PO, Friess H, Wang L, Zhu Z, Bley T, Frigeri L, Zimmermann A, Buchler MW. J.Histochem.Cytochem. 2001;49:539–549. doi: 10.1177/002215540104900414. [DOI] [PubMed] [Google Scholar]
- 43.Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, Sipos B, Foerder M, Alldinger I, Jahnke B, Schackert HK, Kalthoff H, Kremer B, Kloppel G, Saeger HD. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 45.Fitzner B, Walzel H, Sparmann G, Emmrich J, Liebe S, Jaster R. Cell Signal. 2005;17:1240–-1247. doi: 10.1016/j.cellsig.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Masamune A, Satoh M, Hirabayashi J, Kasai K, Satoh K, Shimosegawa T. Am.J.Physiol Gastrointest.Liver Physiol. 2006;290:G729–G736. doi: 10.1152/ajpgi.00511.2005. [DOI] [PubMed] [Google Scholar]
- 47.Javle MM, Gibbs JF, Iwata KK, Pak Y, Rutledge P, Yu J, Black JD, Tan D, Khoury T. Ann.Surg.Oncol. 2007;14:3527–3533. doi: 10.1245/s10434-007-9540-3. [DOI] [PubMed] [Google Scholar]
- 48.Zhao S, Venkatasubbarao K, Lazor JW, Sperry J, Jin C, Cao L, Freeman JW. Cancer Res. 2008;68:4221–4228. doi: 10.1158/0008-5472.CAN-07-5123. [DOI] [PubMed] [Google Scholar]
- 49.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Cancer Biol.Ther. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- 50.Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Ann.Surg.Oncol. 2007;14:3629–3637. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 51.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. J.Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honda K, Yamada T. Seikagaku. 2007;79:643–654. [PubMed] [Google Scholar]
- 53.Hayashida Y, Honda K, Idogawa M, Ino Y, Ono M, Tsuchida A, Aoki T, Hirohashi S, Yamada T. Cancer Res. 2005;65:8836–8845. doi: 10.1158/0008-5472.CAN-05-0718. [DOI] [PubMed] [Google Scholar]
- 54.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F, Ino Y, Ono M, Hirohashi S. Gastroenterology. 2005;128:51–62. doi: 10.1053/j.gastro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Basso D, Fabris C, Panozzo MP, Meggiato T, Del Favero G, Naccarato R. Clin.Biochem. 1990;23:229–232. doi: 10.1016/0009-9120(90)90653-c. [DOI] [PubMed] [Google Scholar]
- 56.Fabris C, Basso D, Panozzo MP, Del Favero G, Meggiato T, Plebani M, Ferrara C, Fogar P, Zaninotto M, Naccarato R. Int.J.Pancreatol. 1992;11:179–184. doi: 10.1007/BF02924183. [DOI] [PubMed] [Google Scholar]
- 57.Funakoshi A, Yamada Y, Migita Y, Wakasugi H. Dig.Dis.Sci. 1993;38:502–506. doi: 10.1007/BF01316506. [DOI] [PubMed] [Google Scholar]
- 58.Miyamoto Y, Kawamata Y, Yokoigawa H, Yamamoto H, Kataoka K, Kashima K. Kaku Igaku. 1993;30:1103–1110. [PubMed] [Google Scholar]
- 59.Hanada K, Kinoshita E, Itoh M, Hirata M, Kajiyama G, Sugiyama M. FEBS Lett. 1995;373:85–87. doi: 10.1016/0014-5793(95)01005-y. [DOI] [PubMed] [Google Scholar]
- 60.Kiyohara H, Egami H, Kako H, Shibata Y, Murata K, Ohshima S, Sei K, Suko S, Kurano R, Ogawa M. Int.J.Pancreatol. 1993;13:49–57. doi: 10.1007/BF02795199. [DOI] [PubMed] [Google Scholar]
- 61.Funakoshi A, Yamada Y, Ito T, Ishikawa H, Yokota M, Shinozaki H, Wakasugi H, Misaki A, Kono M. Pancreas. 1991;6:588–594. doi: 10.1097/00006676-199109000-00013. [DOI] [PubMed] [Google Scholar]
- 62.Kitagawa M, Hayakawa T, Kondo T, Shibata T, Sakai Y, Sobajima H, Ishiguro H, Nakae Y. Gastroenterol.Jpn. 1991;26:62–68. doi: 10.1007/BF02779511. [DOI] [PubMed] [Google Scholar]