Abstract
GYPC encodes two erythrocyte surface sialoglycoproteins in humans, glycophorin C and glycophorin D (GPC and GPD), via initiation of translation at two start codons on a single transcript. The malaria-causing parasite Plasmodium falciparum uses GPC as a means of invasion into the human red blood cell. Here, we examine the molecular evolution of GYPC among the Hominoidea (Greater and Lesser Apes) and also the pattern of polymorphism at the locus in a global human sample. We find an excess of nonsynonymous divergence among species that appears to be caused solely by accelerated evolution of GYPC in the human lineage. Moreover, we find that the ability of GYPC to encode both GPC and GPD is a uniquely human trait, caused by the evolution of the GPC start codon in the human lineage. The pattern of polymorphism among humans is consistent with a hitchhiking event at the locus, suggesting that positive natural selection affected GYPC in the relatively recent past. Because GPC is exploited by P. falciparum for invasion of the red blood cell, we hypothesize that selection for evasion of P. falciparum has caused accelerated evolution of GYPC in humans (relative to other primates) and that this positive selection has continued to act in the recent evolution of our species. These data suggest that malaria has played a powerful role in shaping molecules on the surface of the human red blood cell. In addition, our examination of GYPC reveals a novel mechanism of protein evolution: co-option of untranslated region (UTR) sequence following the formation of a new start codon. In the case of human GYPC, the ancestral protein (GPD) continues to be produced through leaky translation. Because leaky translation is a widespread phenomenon among genes and organisms, we suggest that co-option of UTR sequence may be an important source of protein innovation.
Keywords: GYPC, positive selection, sialoglycoproteins, leaky translation, UTR capture
Introduction
The malaria-causing parasite Plasmodium falciparum utilizes multiple molecular pathways for invasion of human erythrocytes (Gaur et al. 2004). Underlying these diverse methods of entry is the ability of parasites to bind to the cell via numerous receptor–ligand interactions (Adams et al. 2001). Several distinct molecules comprise the human ligands targeted during invasion, including Glycophorins A, B, and C (encoded by GYPA, GYPB, and GYPC, respectively), as well as other unidentified players (Cortés 2008). Elucidating the molecular evolution of these molecules is critical to understanding the historical coevolutionary interactions between humans and P. falciparum and the origins of human alleles that provide resistance to malarial disease.
In a comparison of 280 human gene sequences with their orthologs in Old World monkeys, Wang et al. (2003) identified GYPA, GYPB, and GYPE (the latter encodes Glycophorin E, which currently has no known role in erythrocyte invasion by Plasmodium) as having among the highest rates of nonsynonymous evolution. These genes share significant homology and likely arose through duplication events in the primate lineage (Rearden et al. 1993; Onda and Fukuda 1995; Wang et al. 2003). The high rate of evolution of the glycophorins has been explained by two competing hypotheses. In their examination of GYPA, Baum et al. (2002) proposed the “decoy” hypothesis, whereby the extracellular components of erythrocytic sialoglycoproteins bind with and sequester nonerythrocytic intracellular pathogens (e.g., bacteria and viruses), thereby preventing their invasion of nucleated cells. Baum et al. (2002) suggest that species-specific pathogen pressures result in high rates of nonsynonymous divergence between species and maintenance of high levels of polymorphism within species. Wang et al. (2003) offer an alternative “evasion” hypothesis that proposes that the rapid evolution of glycophorins is in response to Plasmodium pathogens that recognize these molecules for entry into the erythrocyte. Coevolution between Plasmodium receptors and their glycophorin ligands on the human erythrocyte is proposed to drive the rapid evolution of glycophorin-encoding genes. The observation that segregating variants at P. falciparum genes encoding erythrocyte receptors have dramatically different sialoglycoprotein-binding specificities provides support for the evasion hypothesis (Mayer et al. 2002, 2004).
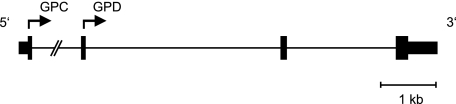
To date, analyses of the molecular evolution and population genetics of sialoglycoprotein-encoding genes have largely ignored GYPC. In part, this may be because the gene did not emerge as one of the very rapidly evolving loci identified in the Wang et al. (2003) survey. GYPC is unique among the glycophorin genes in two respects: First, its sequence is not homologous to any other gene, whereas GYPA, GYPB, and GYPE are all paralogous. Second, GYPC encodes two distinct sialoglycoproteins, Glycophorin C (GPC) and Glycophorin D (GPD), through initiation of translation at two separate start sites within a single mRNA transcript, as shown in figure 1 (Tanner et al. 1988; Le Van Kim et al. 1996). Only GPC is known to play a role in erythrocyte invasion by P. falciparum; this interaction is mediated through the parasite's EBA140 (BAEBL) receptor (Maier et al. 2003). This distinction between GPC and GPD with respect to EBA140 binding likely is due to an N-linked glycan that modifies GPC (but not GPD) and is necessary for receptor recognition (Mayer et al. 2006).
FIG. 1.—
Structure of human GYPC. The proteins GPC and GPD are encoded via initiation of translation at separate start codons in exons 1 and 2, respectively (coding exons are indicated by tall boxes, untranslated region by short boxes, and introns by lines). The gene is drawn to scale, except that exons 1 and 2 are separated by an intron that is approximately 34 kb in length.
GYPC is also of interest because an allele encoding a structural variant segregates at high frequencies among Melanesians and is responsible for the Gerbich-negative (Ge−) phenotype (Booth and McLoughlin 1972). This allele contains a large deletion that eliminates the third exon of the gene and thereby affects the extracellular components of both GPC and GPD (Serjeantson et al. 1994). Ge− is common enough to result in large numbers of homozygous individuals in affected populations; these individuals often exhibit erythrocyte ovalocytosis with no clinical symptoms (Patel et al. 2001, 2004). Importantly, P. falciparum EBA140 does not bind to Ge− cells, indicating that features of the peptide sequence in the deleted region are critical for parasite recognition (Mayer et al. 2002, 2006; Maier et al. 2003). Mayer et al. (2006) suggest that interactions between the peptide backbone and an N-linked glycan may be critical for proper exposure of the ligand recognized by EBA140. From an evolutionary perspective, the failure of EBA140 to bind with Ge− cells suggests that the Ge− phenotype may be maintained in populations through malaria selection.
Our study examines the molecular evolution of GYPC in the Hominoidea, as well as patterns of polymorphism within human populations. Our goal is to test whether an elevated rate of amino acid evolution exists at this gene. If so, we will investigate whether this signature is restricted to the human linage (consistent with recent adaptive evolution mediated by P. falciparum or another species-specific parasite) and/or restricted to extracellular regions of the protein (where interactions with P. falciparum or other parasites might occur). By examining human polymorphism in a global diversity panel, we will test whether there is a signature of recent natural selection at the GYPC locus (either in the form of positive selection increasing the frequency of a single allele or diversity-maintaining selection favoring the presence of many alleles).
Materials and Methods
Biological Materials
Nonhuman primate DNA samples were obtained from the Integrated Primate Biomaterials Resource. These included samples from five species: common chimpanzee (Pan troglodytes), bonobo (Pan paniscus), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), and white-cheeked gibbon (Nomascus leucogenys). In addition, DNA from 50 human individuals, representing a global sample of diversity (including 20 sub-Saharan Africans, 10 Europeans, 5 Middle Easterners, 10 Asians, 3 Oceanians, and 2 Native Americans) was obtained from the Coriell Cell Repository; specific sample identities are detailed in Supplementary Material online. A whole-blood sample from a single P. troglodytes individual was obtained from the Yerkes Primate Center. Whole-blood from a single human sample was obtained via venipuncture from a single subject. Human blood collection was obtained with prior informed consent and all human-subject protocols were approved by appropriate institutional review boards.
DNA Sequencing
We sequenced the nucleotide region encompassing the entire protein-coding segment of GYPC in all human and nonhuman samples (adjacent intron sequence was also examined for polymorphism in our human samples, as described in Results). Diploid sequence data were produced by direct sequencing of amplicons with overlapping primers on both strands. Primers used for amplification and sequencing are available from the authors on request. Sequences were assembled and variable sites identified using the program Sequencher v. 4.7 (GeneCodes).
Western Blotting
To investigate the protein products encoded by GYPC in humans and P. troglodytes, we used an immunoblotting procedure to stain GPC and GPD. We sampled 15 ml of whole heparinized blood from a single human and chimpanzee and prepared erythrocyte ghosts using a cell lysis procedure (Schwoch and Passow 1973; Hanahan and Ekholm 1974). Diluted membranes were run on a 15% Tris–glycine sodium dodecyl sulfate polyacrylamide gel, transferred to a nitrocellulose membrane, and treated with a 1:1,000 dilution of primary antibody. This antibody was monoclonal mouse antihuman CD236 (AbD Serotec), which recognizes a 17 amino acid motif in the fourth exon of GYPC (King et al. 1995). This motif is shared by GPC and GPD and is completely conserved between our human and P. troglodytes samples (as determined via DNA sequencing, data not shown). After primary incubation, the blot was treated with a 1:2,500 dilution of secondary antibody, rabbit F(ab′)2 antimouse IgG:HRP (AbD Serotec) and visualized using a GE Healthcare immunodetection kit.
DNA Sequence Analyses
PAML Analyses
We used the program “codeml” in the software package Phylogenetic Analysis by Maximum Likelihood v. 4.2a (PAML; Yang 2007) to fit models of sequence evolution to our Hominoidea data set (GYPC sequence from all five nonhuman primates, plus a single human sequence). This procedure requires specification of a phylogenetic tree. In our analyses, we implemented a standard unrooted Hominoidea phylogeny, shown in figure 2A. Initial branch lengths were estimated for this tree using PAML model 0 (M0), which estimates a single ratio of nonsynonymous to synonymous evolution (the dN/dS ratio, or ω) that applies to all sites in all taxa.
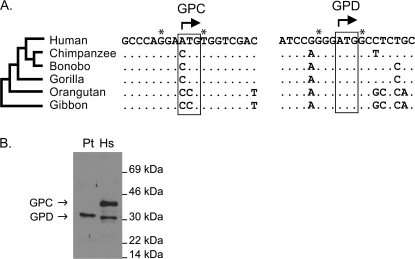
FIG. 2.—
(A) Translation initiation regions of GPC and GPD. The GPC start codon (boxed) is present only in humans, whereas the GPD start codon is conserved across all Apes. Starred sites denote loci important for efficient recognition of the start codon by the ribosome; translation is typically most efficient when there is a purine at position −3 (which is true for GPC and GPD), and a G at position +1 (which is true only for GPD). (B) Western blotting of GYPC products in human and chimpanzee. Using an antibody recognizing an epitope shared by GPC and GPD, and exactly conserved between humans and chimpanzee, we detect both proteins in human erythrocyte ghosts (Hs) but only GPD in chimpanzee (Pt).
To test for positive selection using PAML, we used two separate classes of evolutionary models. The first class includes the “site models,” where codon sites are allowed to fall into categories that differ with respect to their ω values. These values apply to all branches of the phylogeny. The second class of models are the “branch-site” models, which are similar to the site models in that different codons, can have different ω values; however, the phylogeny is split into “foreground” and “background” branches that contain categories of sites with different ω values. Of the site models, we examined several, which differ in the number of classes of sites, to test for positive selection. First, we compared the likelihood values associated with a “Nearly Neutral” model (M1a) and a “Positive Selection” model (M2a), originally described in Wong et al. (2004). Briefly, M1a assumes all codon sites fall into two classes, ω1 and w0, with a single value of ω describing each class (equal to 1 in the former, and free to vary below 1 in the latter). Model M2a adds an additional site class that is free to vary above one. In both models, the proportion of sites falling into each class is free to vary. The likelihood values associated with the data under the two models were compared using likelihood ratio tests, as described in Yang et al. (2000). In addition to M1a and M2a, we examined several additional site models, M7, M8, and M8a. Model M7 is similar to model M1a except that the sites affected by negative selection (ω < 1) approximate a beta distribution with parameters (p and q) estimated from the data. M8 is related to M7 in the same way that M2a is related to M1a—an additional class of sites is added to M8 that is affected by positive selection (ω > 1). Models M7 and M8 were initially described by Yang et al. (2000). A final model, M8a, forms an alternative null model to compare with M8 (Swanson et al. 2003). Model M8a is identical to M8, but the ω value for the positively selected class is fixed at 1. Likelihood ratio tests were used to compare M8 with M8a and M7, as described by Swanson et al. (2003) and Wong et al. (2004). The posterior probabilities associated with specific codons falling into a site class affected by positive selection (in models M8 and M2a) was calculated using the Bayes empirical Bayes (BEB) method described by Yang et al. (2005). These methods were applied to several different partitions of the data, as described in the Results section.
To examine whether humans are characterized by a different pattern of molecular evolution at GYPC compared with the rest of the Hominoidea, we used a pair of “branch-site models.” In these models, the branch of the phylogeny leading to humans was classified as the foreground branch and all others as “background branches.” We used the branch-site test of selection developed by Zhang et al. (2005) to test for positive selection acting on a subset of sites in the human lineage. This test allows for two categories of sites affected by positive selection (ω > 1) in the foreground, together with a category of sites affected by negative selection (ω < 1) and a neutrally evolving class of sites (ω = 1). The background branches contain sites in only the latter two classes. Likelihood values associated with this model are estimated and compared with a null model in which both foreground and background branches contain only sites where ω ≤ 1. In the null model, foreground branches can have a higher fraction of neutrally evolving sites (ω = 1) than background branches, making this test of selection robust to a relaxation of constraints in the foreground branches. Likelihood values for the two models were compared as described in Zhang et al. (2005). As with the site models, maximum likelihood estimates of data parameters are generated from the data. Similarly, the method generates BEB posterior probabilities associated with particular sites falling into the positively selected class.
In all PAML analyses, codons containing alignment gaps due to insertion–deletion events among species were treated as missing data and not considered in pairwise comparisons where one species contained sequence and another species did not.
Polymorphism Analyses
In our human polymorphism data set (2n=100), diploid sequence was arbitrarily phased and intron–exon boundaries annotated. We employed several measures of polymorphism and tests of neutrality using the program DNAsp v.4.50.3 (Rozas et al. 2003). These included two estimates of the population-mutation parameter (θ), including Watterson's (1975) method (θW), which is based on the number of segregating sites in a sample, and π, which is the average number of pairwise differences between sequences in a sample (Nei and Li 1979). Tests of selection included three that examine the frequency spectrum of mutations, Tajima's (1989) D (TD), Fu and Li's (1993) D (FLD), and Fay and Wu's (2000) H (FWH). Significance of these tests was assessed via coalescent simulations, implemented in DNAsp. We also performed a McDonald–Kreitman (1991) test, which examines the numbers of synonymous and nonsynonymous substitutions between species and polymorphic sites within species. For all tests requiring outgroup information, data from a single P. troglodytes were used.
Results
We sequenced the complete coding region of GYPC in six Hominoidea species. These data revealed that the translation start site associated with GPC is present in humans but not other taxa. This is due to a C to A transversion that created a novel translation start codon in humans. The translation start site used to encode GPD is well conserved among all taxa. Sequence data associated with the two start codons are shown in figure 2A. These data strongly suggest that only humans encode two protein products (GPC and GPD) at the GYPC gene. Western blotting of human and P. troglodytes erythrocyte ghosts confirms that the latter encodes only a single protein product of GYPC, conforming in size to GPD (fig. 2B).
Nonsynonymous and Synonymous Evolution
Because only humans have the ability to produce GPC, we define the shared coding region among taxa in our sample as only the portion of GYPC that encodes GPD. Limiting our analysis to this region, examination of nonsynonymous and synonymous evolution using PAML suggests that the gene has been subject to positive selection among the Hominoidea. Analyses of the entire gene (and including all six taxa) using site models suggest that models incorporating positive selection (M2a and M8) are significantly better fits to the data than models including only purifying selection and neutrally evolving sites (M1a, M7, and M8a); these results are shown in table 1 (a multispecies alignment showing all data considered in PAML analyses is included in supplementary figure 1, Supplementary Material online). When we remove from exon 3 our analysis, the portion of the gene that when deleted in humans confers resistance to malaria (the Ge− phenotype), we observe that models including positive selection no longer provide better fits to the data than nearly neutral models. Finally, when we remove humans from the data set and reperform our analysis on the full gene from the five remaining Hominoidea, we see that models incorporating positive selection are not better fits to the data than nearly neutral models. This implies that positive selection may be limited to only the human lineage. The results of the “branch-site” test bolster this inference. In this test, a model allowing positive selection in only the human lineage is a significantly better fit to the data than a model in which all branches of the phylogeny are allowed to evolve with ω ≤ 1 (table 2). The branch-site test of positive selection is not significant when any other single branch of the phylogeny is elevated to the foreground (data not shown).
Table 1.
PAML-Implemented Site Models of Positive Selection
| Modela | ln L | Parameter Estimatesb | Selected Codonsc | |
| All species, full gene | ||||
| M1a | −679.00 | |||
| M2a | −673.02* | p0 = 0.436 (ω0 = 0.032), p1 = 0.529, p2 = 0.035 (ω2 = 21.88) | 24 (0.82), 27 (0.99), 36 (0.88) | |
| M7 | −679.12 | |||
| M8A | −678.99 | |||
| M8 | −673.06* | p0 = 0.44 (p = 0.02, q = 0.01), p2 = 0.035 (ω2 = 21.39) | 24 (0.85), 27 (0.99), 36 (0.91) | |
| All species, exon 3 excluded | ||||
| M1a | −413.81 | p0 = 0.876 (ω0 = 0.209), p1 = 0.124 | ||
| M2a | −413.80 | |||
| M7 | −413.71 | p = 0.758, q = 1.756 | ||
| M8a | −413.71 | p0 = 0.999 (p = 0.747, q = 1.725), p1 = 0.001 | ||
| M8 | −413.71 | |||
| Human excluded, full gene | ||||
| M1a | −619.31 | p0 = 0.389 (ω0 = 0.0), p1 = 0.611 | ||
| M2a | −618.26 | |||
| M7 | −619.32 | p = 0.008, q = 0.005 | ||
| M8a | −619.31 | p0 = 0.389 (p = 0.005, q = 1.554), p1 = 0.611 | ||
| M8 | −618.26 | |||
*P < 0.05, comparing model including positive selection (M2a or M8) to nearly neutral model (M1a, M7, or M8a).
All models and comparisons among models are described in the text. Asterisks indicate comparisons where models that incorporate a class of sites affected by positive selection are better fits to the data than nearly neutral models (Models M2a and M8 in the “all species, full gene” comparison); in all other comparisons, models incorporating positive selection are not better fits to the data than nearly neutral models.
Parameter abbreviations are as follows: p0 = proportion of sites falling into nearly neutral site class (followed by estimate of ω0 for models M1a and M2a or shape parameters of the beta distribution (p,q) for models M7, M8a, and M8), p1 = proportion of sites falling into neutral site class (ω1 = 1); p2 = proportion of sites falling into positively selected site class (followed by estimate of ω2).
BEB estimates of sites falling into positively selected class are listed for those where the posterior probability (in parentheses) is greater than 0.8.
Table 2.
PAML-Implemented Branch-Site Tests of Positive Selection
| Model | ln L | Parameter Estimates | Selected Codons |
| Anull | −679.00 | Foreground: p0 = 0.458 (ω0 = 0.0), p1 = 0.542 | |
| Background: p0 = 0.458 (ω0 = 0.0), p1 = 0.542 | |||
| A | −673.08* | Foreground: p0 = 0.415 (ω0 = 0.0), p1 = 0.573, p2 = 0.012 (ω2 = infinite) | 27 (0.99) |
| Background: p0 = 0.420 (ω0 = 0.0), p1 = 0.580 |
NOTE.—Models are as discussed in text; parameters are as described in table 1. Foreground branch includes the branch leading to humans, background branches include all other branches of the Hominoidea phylogeny. On the foreground branch in Model A, two classes of selected sites (ω > 1) are possible (as described in Materials and Methods); however, only one class is populated in the present analysis.
*P < 0.05, comparing model A with Anull.
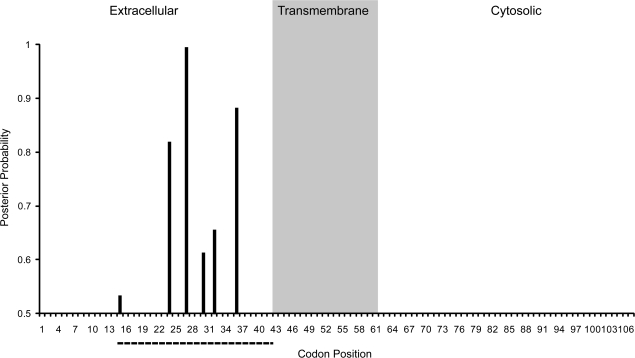
Tables 1 and 2 show that a relatively small proportion of sites within GYPC appear to be affected by positive selection. Analysis of the entire gene under the site model suggests 3.5% of codons are targeted by selection but that selection affecting this class of sites is quite strong (ω = 21.88). The branch-site models suggest an even smaller fraction of sites are targets of positive selection in the human lineage (1.2%) but that these sites are affected by very strong positive selection (a specific value of ω could not be estimated—a result that is likely affected by the small number of sites in the analysis). Results of the BEB analysis in both the site and branch-site models suggest that codons affected by positive selection all fall within the third exon of GYPC (fig. 3). In particular, all models provide strong support for the 27th codon falling into the selected site class (posterior probability > 0.99); the site models (M2a and M8) provide moderate support for the 24th and 26th codons falling into this class (posterior probability > 0.80).
FIG. 3.—
Putative selected codons within GYPC. The posterior probability of each codon having an ω > 1 is shown for those with a probability >0.5 (values taken from the BEB analysis performed under PAML model M2a for the full data set). Extracellular, transmembrane, and cytosolic regions of the encoded protein are shown. The dotted line shows the location of the exon 3 deletion, which segregates in human populations and is hypothesized to confer resistance to falciparum malaria; bases upstream of the dotted line fall within exon 2 and downstream bases fall within exon 4.
Polymorphism within Humans
To examine variation in a human sample, we resequenced 1,235 bp of GYPC from 50 individuals (2n = 100). This comprised the entire coding region of the gene (387 bp), plus noncoding DNA adjacent to each exon (exon 1 region: 50 bp 5′ untranslated region (UTR), 49 bp exon, 242 bp intron; exon 2 region: 57 bp exon, 218 bp intron; exon 3 region: 84 bp exon, 220 bp intron; exon 4: 197 bp exon, 118 bp 3′ UTR). Our survey uncovered nine segregating sites, all of which lie within or adjacent to exons 2, 3, and 4. Genotype data for the 50 samples are shown in accompanying supplementary figure 2, Supplementary Material online. Of the nine sites that vary, one is a synonymous variant within the coding region, seven are polymorphisms in noncoding DNA, and one is a nonsynonymous polymorphism. A McDonald–Kreitman test revealed no significant deviations from expected patterns under neutrality (8 silent and 1 nonsynonymous polymorphic sites, 24 silent and 12 nonsynonymous substitutions; P = 0.249 by Fisher's Exact test). In contrast, one test of neutrality based on the frequency spectrum of mutations shows significant departures from standard neutral expectations (table 3). Specifically, Fay and Wu's H (= −3.79) describes an excess of high frequency–derived alleles in the data set; Tajima's D and Fu and Li's D, in contrast, do not deviate from expected patterns under neutrality. Significance values reported in table 3 are taken from simulations of the coalescent process that do not incorporate recombination; these values are conservative relative to simulations conditioned on nonzero values of recombination.
Table 3.
Summary Statistics Describing Polymorphism at Human GYPC
| S | 9 | |
| θW (%) | 0.141 | |
| π (%) | 0.096 | |
| TD | −0.792 | P = 0.247 |
| FWH | −3.788 | P = 0.015 |
| FLD | −1.749 | P = 0.118 |
NOTE.—P values indicate probability of observing a smaller value based on neutral coalescent simulations.
Discussion
GYPC is a rapidly evolving gene that has been subject to positive natural selection in the human lineage. Our data suggest that diversifying selection has caused rapid amino acid evolution in a portion of the gene (the third exon) that is likely to affect Plasmodium invasion of the red blood cell. Our data also reveal the unexpected result that humans have evolved a novel start codon upstream of the ancestral GYPC translation initiation site. This new start codon is fixed in our global human sample and allows humans to encode two proteins, GPC and GPD, from a single transcript. DNA sequence and protein immunoblotting data confirm that all other sampled Apes encode only GPD at this locus. Our data have important implications with respect to understanding the forces driving sialoglycoprotein evolution and the coevolutionary interactions between humans and malaria-causing parasites. Moreover, our data point to a novel evolutionary pathway by which protein innovation occurs.
Positive Selection at GYPC
GYPC sequence data from the Hominoidea indicate that the locus is subject to higher rates of amino acid evolution in the human lineage than expected under neutrality. When we examine the total data set from six species, we find that site models of DNA sequence evolution that include a class of positively selected codons are significantly better fits to the data than models that do not. This result is dependent on inclusion of humans in the sequence analysis, suggesting that positive selection may be restricted to just the branch of the Hominoidea phylogeny leading to humans. We have tested this hypothesis by performing a branch-site test of accelerated evolution on the human lineage. This test confirms that models of sequence evolution in which only human GYPC harbors a class of sites affected by positive selection are better fits to the data than models where all branches of the phylogeny are evolving under purifying selection and/or neutrality. Because the null model in this latter test allows for human GYPC to have sites characterized by higher (but ≤1) ω values than other branches of the phylogeny, this test confirms that human GYPC has experienced accelerated amino acid evolution due to positive selection, not a relaxation of selective constraints.
In both the site and branch-site models of GYPC sequence evolution, the principal target of positive selection appears to be regions of the gene encoding extracellular protein components (fig. 3). All sites identified through BEB analysis as candidate targets of positive selection fall within the third exon of the gene, corresponding to the region that is deleted in the malaria-resistant Ge− allele in humans. Exclusion of the third exon of the gene from our PAML analyses eliminates any suggestion in the data that GYPC has been subject to positive selection. This result indicates that only a small portion of the gene has been targeted by positive selection and that sequence encoding intramembrane and intracellular protein regions are evolving under the constraints of purifying selection and/or neutrality.
Complementing the long-term positive selection indicated by interspecies comparisons, our human GYPC polymorphism data suggest that a recent genetic hitchhiking event has affected the locus in Homo sapiens. Specifically, our data show an excess of polymorphic sites with high frequency–derived alleles (indicated by a significantly negative value of FWH) over neutral expectations. This pattern is consistent with a selected allele being driven to very high frequency at the locus in the relatively recent past. Our failure to observe significant skews in the allele frequency spectrum using TD and FLD does not contradict our interpretation of significant FWH values. Each of these tests has limited power to detect natural selection, and their sensitivity to different aspects of the data make them complementary (Fay and Wu 2000; Przeworski 2002). Analysis of our data using a McDonald–Kreitman test also reveals no departures from neutral expectations. This latter test can lack power to detect recent positive selection because silent and replacement sites tightly linked to a target of selection will experience a reduction in variation during a selective sweep (Nielsen 2005).
The interpretation of a high frequency of derived alleles in our polymorphism survey is complicated by the fact that we sampled humans sparsely from a large number of populations. The presence of population structure in polymorphism data can affect the site frequency spectrum (Ptak and Przeworski 2002; Hammer et al. 2003). FWH, in particular, has been shown to be sensitive to population subdivision under an island model when demes are sampled unequally (Przeworski 2002; Zeng et al. 2006). Because of this, further surveys of polymorphism in individual populations will help to clarify the extent to which hitchhiking events have affected GYPC. If the observed skew in the site frequency is, in fact, caused by recent natural selection, then we predict that that the hitchhiking event must have occurred relatively recently because an excess of high frequency alleles is expected to be a highly transient signature of recent positive selection (Kim and Stephan 2000, 2002; Przeworski 2002).
What Drives Positive Selection at GYPC?
Our results suggest that GYPC has been a target of positive selection in humans over both short and long timeframes (i.e., recently enough to be evident in the pattern of polymorphism within humans and also over timescales long enough to result in an excess of amino acid substitutions between humans and related species). We propose that our data are broadly consistent with selection mediated by P. falciparum as predicted by the evasion hypothesis (Wang et al. 2003). In the case of GYPC, selection for evasion has been limited to the human lineage and is coincident with the evolution of a new protein product (GPC) at the locus. Because there is no evidence that the ancestral GYPC protein product (GPD) interacts with P. falciparum it is likely that GYPC has been subject to malaria-related selection only since the GPC start codon evolved. Subsequent to this event, rapid evolution of extracellular amino acid residues encoded by GYPC may have occurred in the human lineage to mitigate erythrocyte invasion by P. falciparum. Indeed, several studies have shown that P. falciparum EBA140 genotype variation correlates with the ability of this receptor to bind with human GPC (Mayer et al. 2002, 2006; Maier et al. 2009), consistent with rapid coevolution of GYPC and its P. falciparum receptor. Under our proposed model, the evolutionary forces favoring the initial evolution of the GPC start codon remain unclear. It may have arisen by drift in a nonmalarial ancestral population or have been favored by unknown selective forces.
Because our data suggest that accelerated amino acid evolution is restricted to the human lineage, we believe GYPC evolution is not well explained by the “decoy” hypothesis. Decoy-driven positive selection at GYPA was hypothesized on the basis of high diversity within human populations coupled with high ω values in many primate lineages (Baum et al. 2002). The broad phylogenetic distribution of rapid amino acid evolution at GYPA is potentially compatible with a generalized decoy function of Glycophorin A. In the case of human GYPC, rapid evolution mediated by interactions between GPC and P. falciparum EBA140, seems a more likely explanation for the observed evolutionary pattern.
Regardless of the specific evolutionary model invoked to explain the rapid evolution of GYPC, our results offer a possible contributing explanation to the observation that the virulence of P. falciparum is much higher in humans than other Apes (Coatney 1971). Recently, it was shown that the ability of the P. falciparum receptor EBA175 to bind to Glycophorin A is altered by species-specific sialic acid modifications to the ligand (Martin et al. 2005). Because of this, EBA175-mediated invasion of P. falciparum into P. troglodytes erythrocytes is greatly reduced. Our finding indicates that the GPC-dependent pathway (mediated by P. falciparum EBA140) is also not a possible route for P. falciparum invasion of the chimpanzee red blood cell.
GYPC Exhibits a Novel Mechanism of Protein Innovation
One of our key findings is that human GYPC recently evolved the ability to encode GPC, while retaining the ability to encode GPD. Translation of multiple protein products from a single transcript through the use of differing start codons has been termed “leaky translation,” a phenomenon that is known to affect many genes in diverse organisms (Kozak 2002). The fact that GPC and GPD are products of leaky translation is well documented and likely is caused by the stronger Kozak motif associated with the GPD start codon, despite its downstream position relative to the GPC stop codon (fig. 2; Le Van Kim et al. 1996). However, the evolutionarily derived status of this arrangement, and its restriction to humans, was not previously known.
Our study demonstrates that the evolution of human GPC occurred through co-option of 5′ UTR sequence by the protein-coding region of the gene. To our knowledge, this evolutionary pathway of protein diversification has not previously been documented. In fact, the pattern shown by GYPC suggests that leaky translation may be a general hallmark of recent protein innovation. Formation of a novel upstream codon can cause the expression of a new peptide while maintaining limited production of ancestral proteins. In some instances, leaky translation may represent a transient state as a gene shifts from encoding an ancestral to a derived protein. In other cases, selection may favor continued production of both proteins at the gene (mediated by maintenance of a stronger Kozak motif associated with downstream codons, for instance). It is unclear which of these cases applies to GYPC in humans. Regardless, we believe that the widespread occurrence of leaky translation among genes and organisms suggests that co-option of UTR sequence may represent a significant evolutionary pathway by which protein innovation occurs.
Conclusions
GYPC has been subject to positive selection in the human lineage, leading to both extensive amino acid divergence from nonhuman primates and patterns of polymorphism consistent with a recent hitchhiking event. These data are consistent with selection for evasion of P. falciparum parasites subsequent to the evolution of a novel protein product (GPC) in humans. Beyond showing an interesting pattern of adaptive evolution, GYPC also demonstrates a novel mechanism by which protein diversity can evolve: capture of UTR sequence following the formation of a novel start codon. This process leads to leaky translation of proteins from multiple initiation sites. We suggest that leaky translation may be maintained when continued production of both proteins is beneficial or may represent an evolutionarily transient state as a gene shifts from encoding an ancestral to a novel derived protein. In either case, UTR capture offers a potential means by which major protein innovation can occur through mutation of only a single nucleotide site.
Supplementary Material
Supplementary materials 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We greatly appreciate the helpful comments of two anonymous reviewers. In addition, we thank L. Banta and L. Kaplan for their help with the western blot experiment. Funding in support of this project was provided to J.A.W. by the Department of Biological Sciences at Northern Arizona University, Williams College, and the National Science Foundation (DBI-0520356).
References
- Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 2001;17:297–299. doi: 10.1016/s1471-4922(01)01948-1. [DOI] [PubMed] [Google Scholar]
- Baum J, Ward RH, Conway DJ. Natural selection on the erythrocyte surface. Mol Biol Evol. 2002;19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- Booth PB, McLoughlin K. The Gerbich blood group system, especially in Melanesians. Vox Sanguinis. 1972;22:73–84. doi: 10.1111/j.1423-0410.1972.tb03968.x. [DOI] [PubMed] [Google Scholar]
- Coatney GR. The primate malarias. Bethesda (MD): US National Institute of Allergy and Infectious Diseases; 1971. [Google Scholar]
- Cortés A. Switching Plasmodium falciparum genes on and off for erythrocyte invasion. Trends Parasitol. 2008;24:517–524. doi: 10.1016/j.pt.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur D, Mayer DC, Miller LH. Parasite ligand–host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int J Parasitol. 2004;34:1413–1429. doi: 10.1016/j.ijpara.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Blackmer F, Garrigan D, Nachman MW, Wilder JA. Human population structure and its effects on sampling Y chromosome sequence variation. Genetics. 2003;164:1495–1509. doi: 10.1093/genetics/164.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan DJ, Ekholm JE. The preparation of red cell ghosts (membranes) Meth Enzymol. 1974;31:168–172. doi: 10.1016/0076-6879(74)31018-x. [DOI] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics. 2000;155:1415–1427. doi: 10.1093/genetics/155.3.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MJ, Holmes CH, Mushens RE, Mawby W, Reid ME, Scott ML. Reactivity with erythroid and non-erythroid tissues of a murine monoclonal antibody to a synthetic peptide having amino acid sequence common to cytoplasmic domain of human glycophorins C and D. Br J Haematol. 1995;89:440–448. doi: 10.1111/j.1365-2141.1995.tb08347.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Van Kim C, Piller V, Cartron JP, Colin Y. Glycophorins C and D are generated by the use of alternative translation initiation sites. Blood. 1996;88:2364–2365. [PubMed] [Google Scholar]
- Maier AG, Baum J, Smith B, Conway DJ, Cowman AF. Polymorphisms in erythrocyte binding antigens 140 and 181 affect function and binding but not receptor specificity in Plasmodium falciparum. Infec Immun. 2009;77:1689–1699. doi: 10.1128/IAI.01331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AG, Duraisingh MT, Reeder JC, Patel SS, Kazura JW, Zimmerman PA, Cowman AF. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med. 2003;9:87–92. doi: 10.1038/nm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human–chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer DC, Jiang L, Achur RN, Kakizaki I, Gowda DC, Miller LH. The glycophorin C N-linked glycan is a critical component of the ligand for the Plasmodium falciparum erythrocyte receptor BAEBL. Proc Natl Acad Sci USA. 2006;103:2358–2362. doi: 10.1073/pnas.0510648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer DC, Mu JB, Feng X, Su XZ, Miller LH. Polymorphism in a Plasmodium falciparum erythrocyte-binding ligand changes its receptor specificity. J Exp Med. 2002;196:1523–1528. doi: 10.1084/jem.20020750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer DC, Mu JB, Kaneko O, Duan J, Su XZ, Miller LH. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc Natl Acad Sci USA. 2004;101:2518–2523. doi: 10.1073/pnas.0307318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Onda M, Fukuda M. Detailed physical mapping of the genes encoding glycophorins A, B and E, as revealed by P1 plasmids containing human genomic DNA. Gene. 1995;159:225–230. doi: 10.1016/0378-1119(95)00075-h. [DOI] [PubMed] [Google Scholar]
- Patel SS, King CL, Mgone CS, Kazura JW, Zimmerman PA. Glycophorin C (Gerbich antigen blood group) and band 3 polymorphisms in two malaria holoendemic regions of Papua New Guinea. Am J Hematol. 2004;75:1–5. doi: 10.1002/ajh.10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Mehlotra RK, Kastens W, Mgone CS, Kazura JW, Zimmerman PA. The association of the glycophorin C exon 3 deletion with ovalocytosis and malaria susceptibility in the Wosera, Papua New Guinea. Blood. 2001;98:3489–3491. doi: 10.1182/blood.v98.12.3489. [DOI] [PubMed] [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak SE, Przeworski M. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 2002;18:559–563. doi: 10.1016/s0168-9525(02)02781-6. [DOI] [PubMed] [Google Scholar]
- Rearden A, Magnet A, Kudo S, Fukuda M. Glycophorin B and glycophorin E genes arose from the glycophorin A ancestral gene via two duplications during primate evolution. J Biol Chem. 1993;268:2260–2267. [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schwoch G, Passow H. Preparation and properties of human erythrocyte ghosts. Mol Cell Biochem. 1973;2:197–218. doi: 10.1007/BF01795474. [DOI] [PubMed] [Google Scholar]
- Serjeantson SW, White BS, Bhatia K, Trent RJ. A 3.5 kb deletion in the glycophorin C gene accounts for the Gerbich-negative blood group in Melanesians. Immunol Cell Biol. 1994;72:23–27. doi: 10.1038/icb.1994.4. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner MJ, High S, Martin PG, Anstee DJ, Judson PA, Jones TJ. Genetic variants of human red-cell membrane sialoglycoprotein beta. Study of the alterations occurring in the sialoglycoprotein-beta gene. Biochem J. 1988;250:407–414. doi: 10.1042/bj2500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Tang H, Shen CK, Wu CI. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003;20:1795–1804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.