Abstract
Mitochondria, the powerhouse of eukaryotic cells, have their own translation machinery that is solely responsible for synthesis of 13 mitochondrially-encoded protein subunits of oxidative phosphorylation complexes. Phosphorylation is a well-known post-translational modification in regulation of many processes in mammalian mitochondria including oxidative phosphorylation. However, there is still very limited knowledge on phosphorylation of mitochondrial ribosomal proteins and their role(s) in ribosome function. In this study, we have identified the mitochondrial ribosomal proteins that are phosphorylated at serine, threonine or tyrosine residues. Twenty-four phosphorylated proteins were visualized by phosphorylation-specific techniques including in vitro radiolabeling, residue specific antibodies for phosphorylated residues, or ProQ phospho dye and identified by tandem mass spectrometry. Translation assays with isolated ribosomes that were phosphorylated in vitro by kinases PKA, PKCδ, or Abl Tyr showed up to 30% inhibition due to phosphorylation. Findings from this study should serve as the framework for future studies addressing the regulation mechanisms of mitochondrial translation machinery by phosphorylation and other post-translational modifications.
Keywords: ribosomal proteins, translation, phosphorylation, apoptosis, proteomics, mitochondria
Introduction
Mitochondria, according to the endosymbiotic theory, originally descended from ancient bacteria and became very specialized in producing the majority of the cell’s energy in the form of ATP, commonly referred as “powerhouse of the cell”. They have their own ribosomes to support oxidative phosphorylation through the synthesis of 13 proteins. These proteins are encoded by ~16.5 kb circular genome present in this organelle and are crucial components of the oligomeric complexes of oxidative phosphorylation in the inner membrane. Despite significant similarity to their bacterial counterparts, the 55S mitochondrial ribosomes differ mainly in composition and proteins with unknown function. They have a higher protein to RNA ratio (67% protein and 33% RNA ) compared to those of the bacterial ribosome 1-3. The small subunit (28S) is composed of a 12S rRNA and 29 proteins, while the large subunit (39S) consists of a 16S rRNA and 48 proteins 4-6. Within the last decade, all the protein components of the large and small subunits have been identified using various mass spectrometry-based proteomics approaches 5-8. However, many questions still remain on the function of many proteins and the mechanism of translational regulation.
Post-translational modifications, especially phosphorylation, are commonly known to play regulatory roles in numerous processes and likely to be involved in the mitochondrial translation through modification of its protein components. In deed, we have recently shown that the pro-apoptotic mitochondrial ribosomal protein DAP3 (MRPS29) is phosphorylated at several residues, which result in decreased cell proliferation and PARP cleavage 9. Another study has reported that mitochondrial translation elongation factor mtEF-Tu is phosphorylated, causing inhibition of protein synthesis in ischemic myocardium 10. Furthermore, components of the bacterial translational machinery have been found to be regulated by phosphorylation. The bacterial ribosomal proteins from E. coli and Streptomyces collinus were phosphorylated by a protein kinase from rabbit skeletal muscle and a protein kinase associated with the ribosomes, respectively 11-14. Our laboratory has conducted analysis of E. coli ribosomal proteins that produced additional phosphorylated proteins. Many phosphorylated sites were also mapped using tandem mass spectrometry. These recent studies clearly reveal the significant role that the phosphorylation plays in protein synthesis 15.
In this study, we conducted an extensive proteomics analyses to identity the proteins of 55S mammalian mitochondrial ribosomes that are post-translationally phosphorylated. By using 2D-gel electrophoresis, various phosphorylation-specific visualization methods, and tandem mass spectrometry, we have identified twenty-four phosphorylated proteins of which seven were small and seventeen were large subunit proteins. Interestingly, many of the bacterial (E. coli) homologs of these proteins are also phosphorylated as we reported previously 15. Furthermore, the effect of phosphorylation on translation was evaluated using potential mitochondrial kinases PKA, PKCδ, and Abl Tyr kinase in poly-U directed in vitro translation assays.
Materials and Methods
Preparation of Bovine 55S Mitochondrial Ribosome
Preparation of mitochondrial ribosomes starting from 4 kg of bovine liver was adapted from previously described methods 16, 17. To preserve protein phosphorylation, phosphatase inhibitors (2 mM imizadole, 1 mM sodium orthovanadate, 1.15 mM sodium molybdate, 1 mM sodium fluoride, and 4 mM sodium tartrate dehydrate) were added during the ribosome purification. Ribosomes separated in sucrose gradients were either pelleted by ultracentrifugation or concentrated using a Microcon Ultracel YM-10 centrifugal unit (Millipore Corp.) for further analysis.
NEPHGE Electrophoresis and Phosphoprotein Staining
Approximately 1.8 A260 units of purified ribosome sample was acetone precipitated and the pellet was resuspended in lysis buffer consisting of 9.8 M urea, 2% (w/v) NP-40, 2% ampholytes pI 3-10 and 8-10, and 100 mM DTT. The samples were loaded on NEPHGE (non-equilibrium pH gradient electrophoresis) tube gels and equilibrated in buffer containing 60 mM Tris-HCl pH 6.8, 2% SDS, 100 mM DTT, and 10% glycerol 18. The second dimension gels, usually 14%, were transferred to PVDF membranes and probed with the following primary antibodies: monoclonal anti-phosphoserine at a 1:5000 dilution (Sigma-Aldrich Inc., Product # B 7911), monoclonal anti-phosphothreonine at a 1:10000 (Sigma-Aldrich Inc., Product # B 7661) or monoclonal anti-phosphotyrosine at a 1:5000 dilution (Sigma-Aldrich Inc., Product # P 4110). The membranes were developed using the SuperSignal West Femto Max Sensitivity Substrate (Pierce Biochemicals Inc.) according to the protocol provided by the manufacturer. For further confirmation of protein phosphorylation, gels were stained with the ProQ Diamond phosphoprotein dye (Molecular Probes) and visualized by a laser scanner at an excitation wavelength of 532 nm.
In vitro Phosphorylation Assays
Mitochondrial ribosomes (1.8 A260 units) were incubated with 50 μCi [γ-32P] ATP and 200 μM cold ATP at 30 °C for 1 hr. The ribosomal proteins were separated by two-dimensional NEPHGE gels, fixed in 20% methanol/7% acetic acid, and visualized by phosphor-imaging (Amersham Biosciences Inc.). In vitro phosphorylation reactions were also performed in the absence or presence of commercial kinases. In this case, 0.4 A260 units of mitochondrial ribosomes were incubated with 5 μCi [γ-32P] ATP and 200 μM cold ATP, 2500 U of cAMP-dependent protein kinase (PKA) the catalytic subunit (New England BioLabs, Product # P6000S), 48 ng of protein kinase C delta (PKCδ) (Sigma-Aldrich Inc., Product # P 8538), and 100 U of Abl protein Tyr kinase (New England BioLabs, Product # P6050S) at 30 °C for 1 hr. The conditions for each phosphorylation assay were adapted from the protocols provided by the manufacturers. The ribosomal proteins were separated on an SDS-PAGE gel and visualized by phosphor-imaging (Amersham Biosciences Inc.) or used directly in the poly (U)-directed in vitro translation reactions.
Protein Identification by Tandem Mass Spectrometry
Phosphorylated proteins separated by two-dimensional gel electrophoresis and determined to be phosphorylated by one of three phosphorylation-specific visualization techniques were excised from the gel and in-gel digested by sequencing grade trypsin overnight. The tryptic peptides were extracted from the gel with 50% acetonitrile, 1% formic acid. The extract volume was reduced to 20 μL in autosampler inserts for analysis by capillary liquid chromatography - nanoelectrospray ionization - tandem mass spectrometry (LC-MS/MS), which consisted of a Finnigan Surveyor Micro AS autosampler, a Finnigan Surveyor MS Pump Plus quaternary gradient pump, and Finnigan LTQ linear ion trap mass spectrometer from ThermoFinnigan Electron Corporation. Samples (2-3 μL) were injected at 10 μL/min on to an LC Packings PepMapTM C18 (0.3×5mm, 5μm, 100 Å) for on-line desalting and concentration for 3 min. The trap column was put on-line with the analytical column and peptides were eluted by ramping from 2% to 40% acetonitrile (in 0.1% formic acid) over 30 or 60 minutes at ~0.3 μL/min for digests from 2D gel separations, respectively. The analytical column was a PicoFrit self-pack column with a 15 μm id spray tip packed in-house with 5μm wide pore C18 particles (Supelco Co., Bellefonte, PA) using a pressurized nitrogen. Nano electrospray ionization was achieved using the source supplied by the instrument manufacturer. Voltages applied to spray tip, tube lens, heated ion transfer tube were optimized as needed in order to maintain a stable and efficient spray. Fragmentation by Collision Induced Dissociation (CID) was triggered for the 5 most intense precursor ions from first stage MS scans performed over m/z 550 to m/z 2000 by using an intensity threshold (1000 -2000 counts) that is determined baseline of blank or standard runs. Commercial tryptic digest of BSA (Michrom Bioresources) was routinely analyzed and database searched prior to and after a batch samples to monitor instrument performance. Blank injections were employed following injection of concentrated sample to eliminate any potential carryover at the injector port.
The acquired spectra were evaluated using Xcalibur 2.0 and Bioworks 3.2 softwares. The raw CID tandem MS spectra were also converted to Mascot generic files (.mgf) using the extract msn software (ThermoFinnigan). The .dta and .mgf files were submitted to a site-licensed Sequest and Mascot (version 2.2) search engines to search against in-house generated sequences of 55S proteins, all known mitochondrial proteins, and proteins in the Swiss-Prot database. Database searches were performed with cysteine carbamidomethylation as a fixed modification. The variable modifications were methionine oxidation (+16 Da) and phosphorylation (+80 Da) of Ser, Thr, and Tyr residues and loss of water (−18 Da) from Ser and Thr due to beta elimination during loss of the phosphate moiety. Up to two missed cleavages were allowed for the protease of choice. Peptide mass tolerance and fragment mass tolerance were set to 3 and 2 Da, respectively.
Polymerization Assays
Poly (U)-directed in vitro translation reactions contained 50 mM Tris-HCl pH 7.8, 1 mM dithiothreitol, 0.1 mM spermine, 40 mM KCl, 8.5 mM MgCl2, 2.5 mM phosphoenolpyruvate, 0.18 U pyruvate kinase, 0.5 mM GTP, 50 U RNasin Plus, 12.5 μg/mL poly (U), 26.4 pmol [14C]-Phe-tRNA, 0.25 μM EF-Tumt, 1 μg EF-Gmt, and 0.4 A260 units of mitochondrial ribosomes. EF-Tumt and EF-Gmt were prepared from recombinant proteins as described previously.19, 20. The reaction mixture was incubated at 37 °C for 30 min and terminated by the addition of cold 5% trichloroacetic acid followed by incubation at 90 °C for 10 min. The precipitate was collected on nitrocellulose filter membranes and the incorporation of [14C]-Phe was quantified using a liquid scintillation counter. Results were analyzed using the two-tailed unpaired t test. Values of *P<0.05 were considered statistically significant.
Results and Discussion
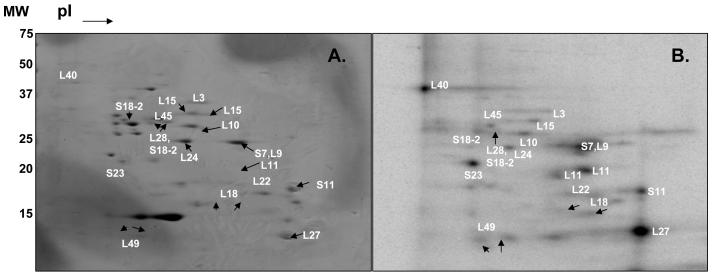
In vitro phosphorylation of mitochondrial ribosomal proteins and effects of phosphorylation on translation. To verify phosphorylation of mitochondrial ribosomal proteins, we first carried out in vitro phosphorylation of bovine 55S ribosomes. For this, we first isolated 55S ribosomes from bovine liver using a well-established sucrose gradient separation and phosphorylated the ribosomal proteins in the presence of [γ- 32P] ATP. Separation of phosphorylated ribosomal proteins by 2D-NEPHGE (non-equilibrium pH gradient electrophoresis) was shown in Figure 1. After aligning 32P-labeled proteins with the proteins in the Coomassie blue stained gel, proteins were excised from the gel and identified by tandem mass spectrometry analysis. In this analysis, a total of seventeen phosphorylated mitochondrial ribosomal proteins, MRPS7, MRPS11, MRPS18-2, MRPS23, MRPL3, MRPL9, MRPL10, MRPL11, MRPL15, MRPL18, MRPL22, MRPL24, MRPL27, MRPL28, MRPL40, MRPL45, and MRPL49, were identified by in-gel tryptic digests of the Coomassie stained 2D-gel spots corresponding to 32P -labeled phosphorylated protein spots (Figure 1 and Tables I-II). In the in vitro phosphorylation assay, the majority of proteins were possibly phosphorylated by a trace amount of endogenous kinase(s) associated with the mitochondrial ribosomes.
Figure 1.
Two-dimensional gel analysis of in vitro phosphorylated mitochondrial ribosomal proteins using [γ-32P] ATP. Bovine mitochondrial ribosomes (1.8 A260 units) were incubated in the presence of 50 μCi [γ-32P] ATP for 1 hr at 30 °C and separated on non-equilibrium pH gradient electrophoresis (NEPHGE) gels using pI 3-10 and 8-10 ampholytes. The phosphorylated proteins were excised, digested with trypsin, and analyzed by LC-MS/MS for potential phosphorylation sites using an ion trap mass spectrometer. (A) Coomassie Blue stained gel and (B) Phosphor-image of mitochondrial ribosomal proteins obtained from purified 55S ribosomes. Not all of the phosphorylated proteins identified by mass spectrometry are labeled in these 2D-gels.
TABLE I.
Characteristics of Phosphorylated Bovine Mitochondrial Ribosomal Proteins
| Protein | pI | Size (kDa) | Subunit | E. coli Phosphorylated Homologa |
|---|---|---|---|---|
| MRPS7 | 9.95 | 27.9 | 28S | Yes |
| MRPS9 | 9.35 | 45.2 | 28S | No |
| MRPS11 | 10.50 | 20.8 | 28S | Yes |
| MRPS16 | 9.69 | 15.1 | 28S | No |
| MRPS18-2 | 9.27 | 29.2 | 28S | Yes |
| MRPS23 | 8.90 | 21.6 | 28S | No |
| MRPS29 | 9.31 | 45.6 | 28S | No |
| MRPL1 | 9.03 | 36.6 | 39S | Yes |
| MRPL2 | 11.45 | 33.3 | 39S | Yes |
| MRPL3 | 9.45 | 38.6 | 39S | Yes |
| MRPL9 | 10.04 | 30.0 | 39S | No |
| MRPL10 | 9.83 | 29.3 | 39S | Yes |
| MRPL11 | 9.88 | 20.8 | 39S | No |
| MRPL13 | 9.35 | 20.6 | 39S | Yes |
| MRPL15 | 10.15 | 33.7 | 39S | No |
| MRPL18 | 9.74 | 20.4 | 39S | Yes |
| MRPL22 | 9.87 | 24.2 | 39S | Yes |
| MRPL24 | 9.47 | 24.8 | 39S | No |
| MRPL27 | 10.19 | 16.0 | 39S | No |
| MRPL28 | 9.21 | 30.0 | 39S | Yes |
| bMRPL40 | 8.75 | 48.0 | 39S | No |
| MRPL43 | 10.12 | 17.7 | 39S | No |
| MRPL45 | 9.31 | 35.2 | 39S | No |
| MRPL49 | 9.65 | 19.3 | 39S | No |
Paper accepted15.
Also known as MRPL37.
TABLE II.
Screening for Phosphorylated Proteins of Bovine Mitochondrial Ribosomes by 2D-Gel Electrophoresis coupled to LC-MS/MS Analysis
| Immunoblotting Analysis |
||||||
|---|---|---|---|---|---|---|
| Proteins | Ser | Thr | Tyr | ProQ | 32P | Phosphopeptide |
| MRPS7 | X | X | X | X | X | |
| MRPS9 | X | X | X | |||
| MRPS11 | X | X | X | X | X | |
| MRPS16 | X | X | X | |||
| MRPS18-2 | X | X | X | X | ||
| MRPS23 | X | X | X | |||
| MRPS29 | X | X | X | |||
| MRPL1 | X | X | X | |||
| MRPL2 | X | X | X | X | ||
| MRPL3 | X | X | X | X | ||
| MRPL9 | X | X | X | X | X | X |
| MRPL10 | X | X | X | X | X | X |
| MRPL11 | X | |||||
| MRPL13 | X | X | X | |||
| MRPL15 | X | X | X | X | X | |
| MRPL18 | X | X | ||||
| MRPL22 | X | X | X | X | X | |
| MRPL24 | X | X | X | X | X | X |
| MRPL27 | X | X | ||||
| MRPL28 | X | X | X | |||
| aMRPL40 | X | X | X | X | X | |
| MRPL43 | X | X | X | |||
| MRPL45 | X | X | X | X | X | |
| MRPL49 | X | X | X | X | X | X |
Also known as MRP-L37.
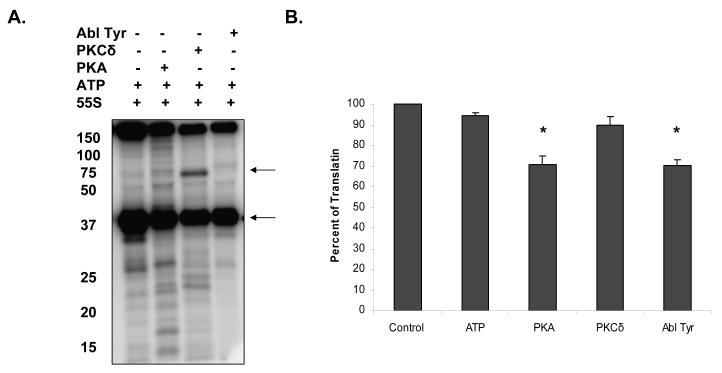
Having identified this many phosphorylated proteins in bovine mitochondria out of seventy-seven ribosomal proteins, we estimated that ~30% of the ribosomal proteins were phosphorylated. We hypothesized that post-translational modification of ribosomal proteins might affect the translation and provide a feedback inhibition for mitochondrial translation to control ATP production in oxidative phosphoryation. Phosphorylation could therefore be one of the regulatory mechanisms of protein synthesis in mammalian mitochondria. Likewise, it had already been shown that phosphorylation of the bacterial ribosomes reduced the translation significantly by ~50% 12, 13. To determine the extent of phosphorylation, mitochondrial ribosomes were incubated in the presence of [γ-32P] ATP alone and with various purified kinases known to be localized to the mitochondria such as PKA, PKCδ, and Abl protein Tyr kinase (Figure 2A). Due to the presence of endogenous kinases associated with mitochondrial ribosomes, a low level of phosphorylation of ribosomal proteins was detected under steady-state conditions. In addition to endogenous unknown kinase(s), these ribosome samples contain pyruvate dehydragenase (PDH) subunits and PDH kinase that phosphorylates the PDH E1 subunit as demonstrated by the arrow (Figure 2A). On the other hand, addition of other kinases, increased the phosphorylation of ribosomal proteins rather than the E1 subunit (Figure 2A). For this reason, these kinases may serve as candidates endogenously phosphorylating the mitochondrial ribosomes in vivo. For example, cAMP-dependent protein kinase (PKA), a Ser/Thr kinase associates with A-kinase anchoring proteins (AKAPs), which allows for PKA to localize to the mitochondrial matrix 21. Recently, a comparative analysis of wild-type and cAMP deathless S49 lymphoma cells, indicated PKA-stimulated apoptosis occurs by way of the mitochondria 22. In fact, PKA appeared to phosphorylate many of the ribosomal proteins in in vitro assays and the banding pattern resembled the control lane with only ribosomes and [γ- 32P] ATP implying the phosphorylation of the same proteins in the ribosome (Figure 2A). We have also demonstrated the phosphorylation of recombinant MRPS29 (also known as DAP3) at its highly conserved Ser and Thr residues by PKA and PKCδ in vitro9. Lastly, although Abl protein Tyr kinase has been shown to be translocated into the mitochondria upon endoplasmic reticulum stress-induced apoptosis and in response to necrotic cell death caused by oxidative stress, fewer mitochondrial ribosomal proteins were Tyr phosphorylated by this kinase compared to Ser and Thr phosphorylation by other kinases (Figure 2A) 23, 24.
Figure 2.
Mitochondrial ribosomes were phosphorylated in the presence of endogenous and mitochondrially located kinases to determine the effect on mitochondrial translation. Approximately, 0.4 A260 units of mitochondrial ribosomes were incubated with 5 μCi [γ-32P] ATP and 2500 U of cAMP-dependent protein kinase (PKA) the catalytic subunit, 48 ng of protein kinase C delta (PKCδ), and 100 U of Abl protein Tyr kinase at 30 °C for 1 hr. The ribosome in the presence of the kinase buffer served as the control. (A) The ribosomal samples were run on an SDS-PAGE gel, fixed, dried, and visualized by phosphor-imaging. The upper arrow indicates the autophosphorylation of PKCδ and lower arrow is highlighting the phosphorylation of pyruvate dehydrogenase subunit. (B) After phosphorylating the ribosome with different kinases, poly (U)-directed polymerization assays were conducted. Shown is the mean ± SD of three independent experiments. *, P < 0.05.
Having observed the phosphorylation of mitochondrial ribosomal proteins with these kinases, poly-U directed in vitro translation assays were performed to test the role of phosphorylation in protein synthesis. In these assays, prior to translation, ribosomes were phosphorylated by the addition of kinases and ATP in vitro. Upon addition of ATP and phosphorylation buffer as a control; inhibition of 6-7% was detected and this observation was in agreement with the extent of phosphorylation (Figure 2B). However, in the presence of PKA and Abl protein Tyr kinase there was a significant inhibition, ~30%, in translation (Figure 2B). Furthermore, phosphorylation by PKCδ had no significant effect on translation and was comparable to the control assay performed in the presence of ATP (Figure 2). In conclusion, modification of the Ser, Thr, and Tyr residues of mitochondrial ribosomal proteins is a mechanism by which translation can be reversibly regulated.
Phosphorylated Proteins of Bovine 55S Mitochondrial Ribosomes
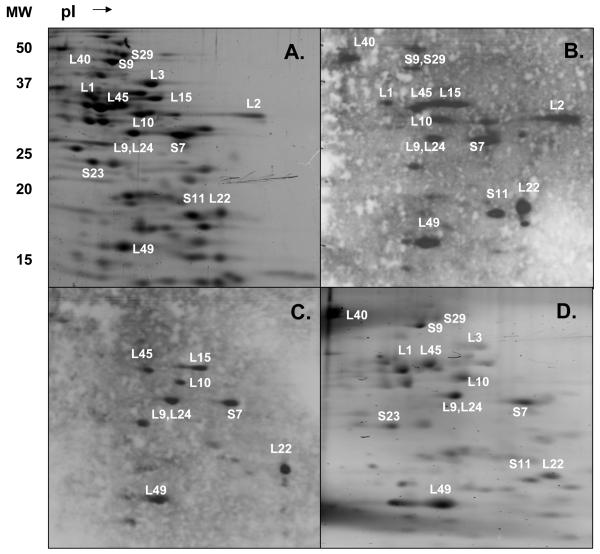
Mammalian mitochondrial 55S ribosomes are composed of two rRNA components and seventy-seven proteins 5-8. We identified twenty-four of the ribosomal proteins as phosphorylated at steady-state using 2D-gel and mass spectrometric analyses of mitochondrial ribosomes (Tables I- II). Due to basic nature of MRPs, purified 55S ribosome samples were first separated on 2D-gels and transferred to PVDF membranes for immunoblotting analysis with phospho-specific phospho-Ser, -Thr and -Tyr antibodies. Then, phosphorylated protein spots detected in blots were compared to a Sypro Ruby stained gel and the corresponding proteins were excised and digested with trypsin for protein identification by mass spectrometry using locally maintained Swiss-Prot and mitochondrial protein databases (Figure 3). In this analysis, twenty mammalian mitochondrial ribosomal proteins; MRPS7, MRPS9, MRPS11, MRPS16, MRPS18-2, MRPS29, MRPL1, MRPL2, MRPL3, MRPL9, MRPL10, MRPL13, MRPL15, MRPL22, MRPL24, MRPL28, MRPL40, MRPL43, MRPL45, and MRPL49, were phosphorylated at Ser and/or Thr residues (Figure 3B and Table II). However, only ten phosphoproteins from mitochondrial ribosomes were identified as Tyr phosphorylated including: MRPS7, MRPS18-2, MRPL2, MRPL9, MRPL10, MRPL15, MRPL22, MRPL24, MRPL45, and MRPL49 (Figure 3C and Table II). In summary, the majority of the mitochondrial ribosomal proteins were phosphorylated at Ser and Thr residues, while less than half are phosphorylated at their Tyr residues, complementing our recent phosphoproteomic analysis of E. coli ribosomal proteins 15.
Figure 3.
Two-dimensional gel analysis of phosphorylated mitochondrial ribosomal proteins. Approximately, 1.8 A260 units of bovine mitochondrial ribosomes were separated on non-equilibrium pH gradient electrophoresis (NEPHGE) gels using pI 3-10 and 8-10 ampholytes. The phosphorylated proteins were excised, digested with trypsin, and analyzed by LC-MS/MS for potential phosphorylation sites using an ion trap mass spectrometer. (A) SYPRO Ruby stained gel of the ribosomal proteins obtained from bovine 55S ribosomes. (B and C) Immunoblots of 55S ribosomes using anti-phosphoserine and anti-phosphotyrosine antibodies, respectively. Not all of the phosphorylated proteins identified by mass spectrometry are labeled in these 2D-gels for simplicity. (D) ProQ Diamond stained gel of the phosphorylated proteins from 55S ribosomes visualized at 532 nm.
To support the results obtained from immunoblotting analyses and identify additional phosphoproteins of mitochondrial ribosomes, another method was also performed. In this method, 2D-gels of ribosomal proteins were stained with a phospho-specific fluorescent dye, Pro-Q Diamond, and fourteen phosphorylated ribosomal proteins, MRPS7, MRPS9, MRPS11, MRPS23, MRPS29, MRPL1, MRPL3, MRPL9, MRPL10, MRPL22, MRPL24, MRPL40, MRPL45, and MRPL49 were identified from the tryptic digests of the 2D-gel spots with a strong fluorescent signal (Figure 1D and Table II).
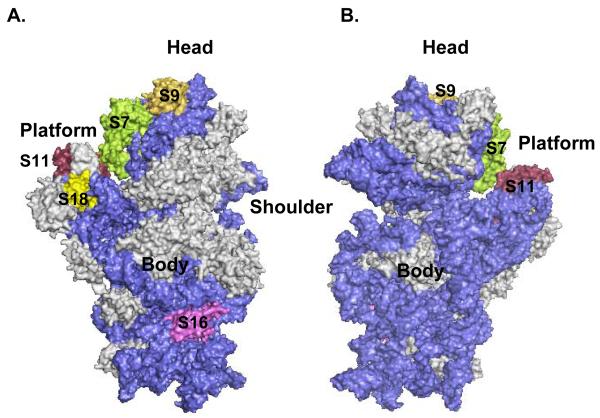
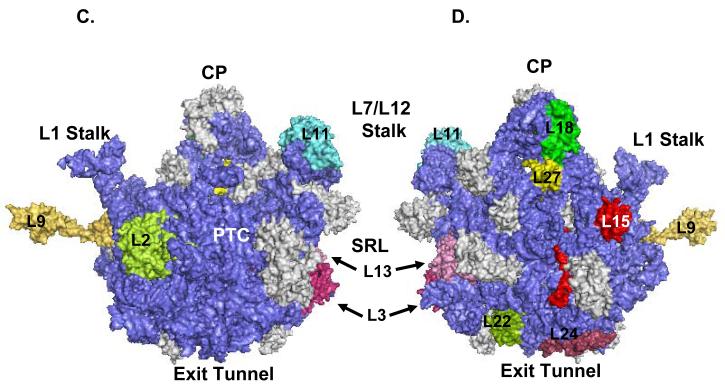
In summary, twenty-four mitochondrial ribosomal phosphoproteins were confidently detected by three different complementary approaches; specifically seven proteins from the 28S subunit and seventeen proteins from the 39S subunit were consistently identified in the LC-MS/MS analyses. Eighteen bovine mitochondrial ribosomal proteins have E. coli homologs, particularly five from the small subunit and thirteen from the large subunit, while the remaining six represent new classes of proteins. Location of the phosphorylated mitochondrial ribosomal proteins with bacterial homologs was modeled using coordinates for the crystal structure of E. coli at 3.5 Å (Figure 4) 25, 26. As seen in the model, phosphorylated proteins were mainly located in the functional sides of the ribosome such as the mRNA binding path, the peptide exit tunnel and the sarcin-ricin loop (SRL) region of the ribosome. More interestingly, based on our recent study, eleven of these proteins have phosphorylated E. coli ribosomal protein homologs implying a conserved role(s) for phosphorylation in protein synthesis 15. Moreover, as first noted for bacterial protein synthesis, our data also suggests that phosphorylation of ribosomal proteins may reduce mitochondrial translation possibly by inducing conformational or structural changes which may alter protein-protein or protein-RNA interactions during different stages of translation. Therefore, given the functional significance of phosphorylation in the bacterial and mitochondrial translation systems, phosphorylated mitochondrial ribosomal proteins will be discussed in detail emphasizing their roles in protein synthesis and newly acquired functions in apoptosis and disease.
Figure 4.
3D-Models of the E. coli ribosomal subunits displaying the location of phosphorylated mitochondrial ribosomal proteins. The phosphorylated ribosomal proteins are highlighted in different colors, while gray represents unphosphorylated ribosomal proteins, and blue is the rRNA. Coordinates of the E. coli 30S subunit and 50S subunit were obtained from the Protein Data Bank (Acc. # 2AW7 and 2AW4). (A) The 30S subunit is illustrated from the solvent side, while (B) represents a view of the small subunit from the 50S interface. (C) Representation of the 50S subunit from the 30S interface, while (D) is a view of the large subunit from the solvent side. Specific regions, peptidyl transferase center (PTC), central protuberance (CP), sarcin-ricin loop (SRL), L1 and L7/L12 stalks, exit tunnel, and the phosphorylated ribosomal proteins were labeled in the model generated by PyMOL software.
mRNA Binding Path
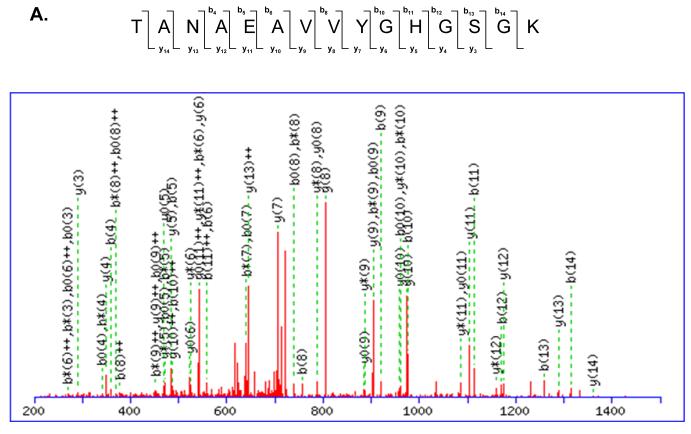
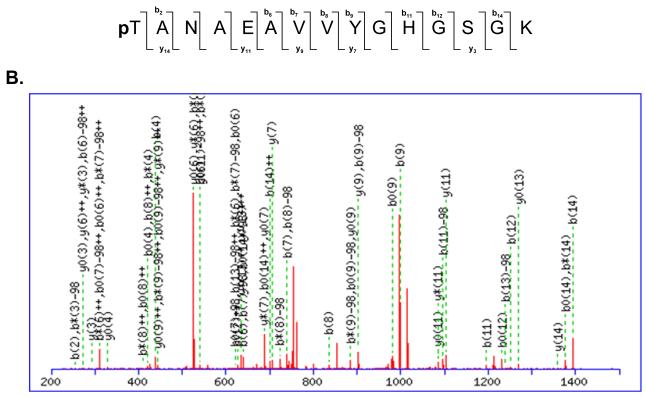
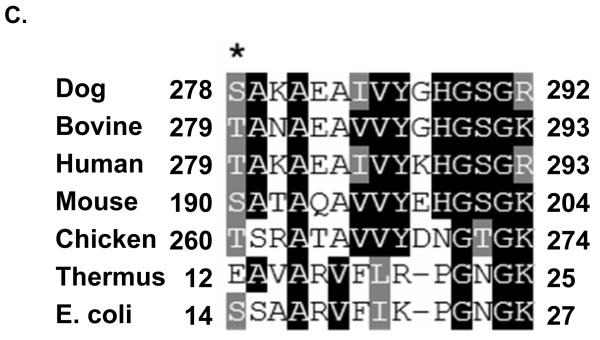
Many of these phosphorylated ribosomal proteins are located in the head region of the small subunit and interact with both mRNA and tRNA throughout the translation process 27-30. Structural studies have demonstrated that the head of the 30S subunit undergoes conformational changes during subunit association, binding of aminoacyl-tRNA or factors, and translocation 30-35. An example of MS/MS spectrum of a tryptic peptide obtained from MRPS9 and its phosphorylated form is shown in Figure 5. The molecular weight difference between the phosphorylated and nonphosphorylated form of the peptide is +80 Da, which corresponds to the addition of phosphate (HPO3) group to the peptide (Figure 5A-B). As seen in the alignment of the phosphorylated peptide from several mitochondrial and bacterial MRPS9 homologs, the homology between bacterial and mitochondrial proteins is significant and the phosphorylated Thr279 is conserved in mitochondrial and E. coli S9 (Figure 5C). E. coli ribosomal S9 was shown to be phosphorylated at Ser and Thr residues by a protein kinase from rabbit skeletal muscle, similarly we have also detected phosphorylation of MRPS9 at Ser residues by immunoblotting (Table II) 11. Bacterial S9 is located on the back of the head region of the 30S subunit and its extended C-terminal tail enters into the P site of the small subunit (Figure 4A) 28, 30, 36, 37. There are several Lys and Arg residues that make up the C-terminal tail of S9 and promote interactions with the phosphate backbone of the anticodon stem-loop tRNA and control P site specificity during initiation 37. In addition to the basic C-terminal tail, there are also highly conserved Thr and Ser residues in the C-terminal tail of the mitochondrial S9 proteins and phosphorylation of these residues would alter the charge on the S9-tail and result in modulating the interaction between this protein and the tRNA (Figure S1).
Figure 5.
Mapping of phosphorylation site(s) in mitochondrial ribosomal proteins by MS/MS. (A) A tryptic peptide (TANAEAVVYGHGSGK, m/z 730.91, 2+) obtained from MS/MS analysis of MRPS9 spot was presented. (B) The CID spectrum of the phosphorylated form of the same peptide pTANAEAVVYGHGSGK (m/z 770.93, 2+) at the first Thr residue was shown (C) The phosphopeptide obtained from bovine MRPS9 was aligned with the same region of MRPS9 homologs from dog (XP_531774), bovine (Q58DQ5), human (P82933), mouse (Q9D7N3), chicken (XP_416921), thernus (P80374), and E. coli (P0A7X3) using CLUSTALW program in Biology Workbench and the results are displayed in BOXSHADE. The asterisk indicates the site of phosphorylation mapped for MRPS9 peptide.
Another highly phosphorylated protein with a bacterial homolog is MRPS11. Bacterial S11 is located in the platform region of the small subunit and is known to interact with both mRNA and tRNA during protein synthesis (Figure 4A) 38. The platform of the 30S subunit undergoes conformational changes during translation as observed in structural studies of bacterial ribosomes, which may promote an interaction at the E site between S7 and S11 30, 33, 35, 37, 38. MRPS11 was detected to be phosphorylated by ProQ-stained, after incubation with [γ-32P] ATP, and immunoblotting with pSer- and pThr-antibodies (Figures 1 and 3, Tables I-II). However, phosphorylation of MRPS11 just by the addition of [γ-32P] ATP suggests that this protein was specifically phosphorylated by a kinase associated with the mitochondrial ribosome (Figure 1B). Furthermore, its bacterial homolog is phosphorylated primarily at Ser and Thr residues, especially Thr58 of E. coli is positioned in a loop region spanning between the N-terminal tail and helix1, where S11 and S7 interact at the mRNA binding path (Table I) 15, 38. Interestingly, Thr58 is conserved in mitochondrial (Thr127) and archaeal S11 homologs, whereby the additional negative charge may weaken the association with helix1 or trigger conformational changes in S11 hindering its interaction with S7 (Figure S1).
Sarcin-Ricin Loop Region
In our mass spectrometric analysis, MRPL3 was detected by 2D-gel immunoblotting with a phospho-Ser antibody, ProQ staining, and after incubation with [γ-32P] ATP (Tables I-II). Not surprisingly, E. coli ribosomal L3 had previously been mentioned as a phosphoprotein, whereby a protein kinase from rabbit skeletal muscle could access and phosphorylate its Ser and Thr residues (Table I) 11. Bacterial L3 is one of the ribosomal proteins responsible for arranging the sarcinricin loop (SRL) particularly domain VI of the 23S rRNA for factor binding (Figure 4C-D) 30, 39. We had mapped a few phosphorylation sites at the N-terminal domain of E. coli L3, which are partially conserved in mitochondria and might also be phosphorylated affecting protein interactions at the sarcinricin loop (Table III, Figure S1) 15. In bovine MRPL3, we have identified two phosphorylated peptides at the N-terminal domain of the protein (Table III).
TABLE III.
Phosphorylated Peptides of Bovine Mitochondrial Ribosomal Proteins Detected by LC-MS/MS Analysis
| MRP | Sequence | Phosphosite | Score | m/z | Mr(expt) | Mr(calc) | Delta |
|---|---|---|---|---|---|---|---|
| MRPS9 | |||||||
| AITYLFPSGLFEK | Thr109, Tyr110, Ser114 |
29 | 865.24 | 1728.47 | 1725.66 | 2.81 | |
| TANAEAVVYGHGSGK | Thr279 | 47 | 770.93 | 1539.85 | 1540.53 | −0.68 | |
| SSQAGAIRLAMSR | Ser339, Ser349 | 23 | 755.20 | 1508.38 | 1507.50 | 0.88 | |
| MRPS11 | |||||||
| ATGKGVTHVR | Lys145, Thr148 | 29 | 573.60 | 1145.18 | 1147.18 | −2.00 | |
| MRPS16 | |||||||
| AQEVLLAAQKTDTEATETKEN | Thr125 | 86 | 1185.97 | 2369.93 | 2370.42 | −0.51 | |
| MRPS18-2 | |||||||
| RLYQGHLR | Tyr240 | 20 | 563.49 | 1124.97 | 1122.17 | 2.80 | |
| MRPS23 | |||||||
| TQQEGSQVSR | Ser145 | 28 | 599.65 | 1197.28 | 1199.12 | −1.84 | |
| SESMGVESQTALEENPPLK | Ser153 | 114 | 1064.68 | 2127.36 | 2126.19 | 1.16 | |
| MRPL2 | |||||||
| YDPCRSADIALVAGGNR | Tyr132 | 57 | 929.93 | 1857.86 | 1857.94 | −0.08 | |
| KVNGTAIIQLPSA | Thr224, Ser231 | 59 | 738.05 | 1474.08 | 1471.49 | 2.59 | |
| QMQVLETCTATVGR | Thr242 or Thr244 | 43 | 809.80 | 1617.59 | 1616.75 | 0.84 | |
| MRPL3 | |||||||
| VLDRCTGGLGTALGSGNR | Thr19, Ser28 | 41 | 637.07 | 1908.18 | 1906.90 | 1.28 | |
| TTGGLGTALGSGNR | Ser28 | 46 | 671.32 | 1340.63 | 1341.32 | −0.69 | |
| MRPL9 | |||||||
| LSGSTPGSER | Ser37, Ser40 | 41 | 576.10 | 1150.18 | 1149.99 | 0.20 | |
| GEYWCEVTVNGLDTVR | Tyr219 | 33 | 962.31 | 1922.61 | 1920.99 | 1.63 | |
| MRPL10 | |||||||
| MIAVCQNVAMSAEDK | Ser105 | 28 | 845.95 | 1689.89 | 1689.87 | 0.02 | |
| EMVRILKSVPFLPLLGGCIDD- TILSR |
Met164, Lys169, Ser170, Thr184, Ser187 |
41 | 1062.37 | 3185.00 | 3184.45 | −0.35 | |
| ILKSVPFLPLLGGCIDDTILSR | Lys169, Ser170 | 47 | 857.99 | 2570.97 | 2572.84 | −1.87 | |
| QGFINYSKLPSLALAQGELVG- GLTLLTAR |
Tyr194, Lys196, Ser199, Thr212 |
54 | 1104.63 | 3310.87 | 3313.48 | −2.60 | |
| MRPL13 | |||||||
| QVTAAQLHR | Thr91 | 23 | 552.44 | 1102.87 | 1103.13 | −0.25 | |
| VWSPPEDYRL | Ser172 | 35 | 672.41 | 1342.78 | 1341.36 | 1.43 | |
| MRPL15 | |||||||
| DYGVQLVEEGADTFK | Tyr137 | 42 | 875.80 | 1749.59 | 1750.75 | −1.16 | |
| MRPL18 | |||||||
| HLYSTKSVVACESVGR | Ser118, Ser121, Ser127 |
34 | 988.66 | 1975.31 | 1976.90 | −0.59 | |
| MRPL24 | |||||||
| YGMSRPGSLADKKK | Tyr21, Met23 | 38 | 818.47 | 1634.93 | 1633.76 | 1.17 | |
| MRPL27 | |||||||
| TRTAVTALLSPPQAAALAVR | Ser19 | 40 | 1045.41 | 2088.81 | 2087.32 | 1.49 | |
| MRPL28 | |||||||
| VYVEELVEQLAQQALSEPAV- VQK |
Tyr230 | 82 | 885.44 | 2653.29 | 2650.87 | 2.42 | |
| MRPL40 | |||||||
| NMLSTTWKR | Met208, Ser210, Thr211 |
26 | 656.92 | 1311.82 | 1312.28 | −0.46 | |
| ESTLIQVHGSSGAQLNAK | Ser217, Thr218 | 56 | 668.12 | 2001.35 | 1999.96 | 1.39 | |
| MRPL43 | |||||||
| FLTSVLHNGLGR | Thr12 | 42 | 698.30 | 1394.58 | 1393.48 | 1.09 | |
| MRPL45 | |||||||
| CSSLVTKSNAYGQVTVR | Ser200, Ser206 | 27 | 658.62 | 1972.83 | 1973.00 | −0.17 | |
| MRPL49 | |||||||
| RLTQGTPDYPR | Thr32 | 34 | 692.76 | 1383.50 | 1383.40 | 0.10 |
MRPL13 was identified in the immunoblotting analysis of ribosomal proteins by phospho-Ser and Thr antibodies at steady-state conditions (Tables I-II). The bacterial homolog of MRPL13 was present in active sub-ribosomal particles and is primarily phosphorylated at Thr and Tyr residues (Table I) 15, 40. Bacterial L13 is oriented to interact with at least four ribosomal proteins including L3 and L6 near the factor binding site (SRL) as well as L20 and L21 at the opposite end (Figure 4C-D) 39. Therefore, phosphorylation of MRPL13 can be implicated in having a role in scaffold formation in the SRL region and factor binding to the ribosome.
L7/L12 Stalk Region
L10 and L11 in conjunction with multiple copies of L7/L12 are referred to as L7/L12 stalk in bacteria 41. Specifically, the N-terminal domain of L7/L12 binds to the C-terminal end of L10, docking this flexible protein to the large subunit of the ribosome (Figure 4C-D) 41. From an earlier report analyzing E. coli phosphorylated ribosomal proteins, L10 was Ser and Thr phosphorylated by a protein kinase from rabbit skeletal muscle (Table I) 11. Based on our 2D-gel analysis, MRPL10 was identified in each of the immunoblots with phospho-specific antibodies in addition to being detected in the ProQ phospho gel and after incubation with [γ-32P] ATP (Table II). Many of the Ser and Thr residues found in mitochondrial and bacterial homologs are conserved and phosphorylation of these residues may influence the binding ability of MRPL10 as the N-terminal domain is responsible for attaching to the large subunit (Figure S1). In addition to phosphorylation, MRPL10 is also found to be acetylated by both immunoblotting and mapping modified phosphorylated and acetylated residues by mass spectrometry (Table II and Figure S2).
Bacterial L11 is in close proximity to L10 and the L7/L12 stalk near the factor binding site on the 50S subunit (Figure 4C-D) 42. L11 is a central player in translation termination favoring UAG-dependent termination by release factor 1 (RF1), while ribosomes stripped of L11 prefer UGA-dependent termination by release factor 2 (RF2) 43. Furthermore, the N-terminal domain (residues 1-72) of L11 is required for stringent factor stimulation, when the ribosome stalls 44. This domain is very flexible and attached to a linker region (residues 73-75). Upon factor binding, the N-terminal domain swings in towards the large subunit and then moves away due to conformational changes stimulated by GTP hydrolysis 44-46. MRPL11 was only detected after incubation with [γ-32P] ATP and evolutionarily conserved Ser and Thr residues that might be involved in interactions of L11 with the translation factors and the large subunit rRNA and proteins (Tables I-II). Changes in phosphorylation of these residues could impact protein-protein and protein-rRNA interactions, and specifically act as a pivot point when the flexible N-terminal domain changes conformation upon factor binding.
Peptide Exit Tunnel
MRPL22, ~24 kDa in size, was identified in each of the 2D-gel analyses of phosphorylated mitochondrial ribosomal proteins (Tables I-II). Bacterial L22 lines the peptide exit tunnel of the large subunit and forms the narrowest portion of the channel with L4 39. L22 is a core protein associating with all six domains of the 23S rRNA 39. Furthermore, a deletion of Met82-Lys83-Arg84 in bacterial L22 leads to erythromycin resistance inhibiting peptide chain elongation 47. Several Ser, Thr, and Tyr residues are clustered in close proximity to the conserved Lys83-Arg84 in bacteria resulting in erythromycin resistance (Figure S1). Furthermore, phosphorylation of MRPL22 may be responsible for altering interactions with L4 and the 16S rRNA, acting as a mechanism to regulate the nascent polypeptide release from the exit tunnel.
MRPL24 is another ribosomal protein located near the peptide exit tunnel, and phosphorylation of this proteins was detected in the immunoblotting, ProQ phospho staining, and in vitro phosphorylation by [γ-32P] ATP (Figure 1 and 3, Tables I-II). L24 is also located at the end of the peptide exit tunnel as displayed in Figure 4D and is capable of interacting with chaperones and the signal recognition particle (SRP) in bacteria 48, 49. As an initiator protein, it functions in assembling the 50S subunit needed for protein synthesis 25, 50, 51. Therefore, phosphorylation at various residues might induce conformational changes during the assembly process or alter protein interactions with neighboring chaperones near the exit tunnel.
Apoptotic Proteins
MRPS29 and MRPL40, were identified as proapoptotic proteins. MRPS29 also known as DAP3 (Death Associated Protein 3) has been implicated in mitochondrial physiology as DAP3 deficiency is lethal in mice, while the over-expression results in mitochondrial fragmentation 52, 53. Furthermore, it is the only GTP-binding protein on the ribosome and has been localized to the base of the lower lobe of the 28S subunit by immunoelectron microscopy 54, 55. In a recent study, we have shown phosphorylation of bovine ribosomal DAP3 at several highly conserved Ser and Thr residues using a combination of 2D-gel analysis and mass spectrometry and role of these residues in induction of apoptosis 9. MRPS29 was detected in the phospho-Ser immunoblotting and has many conserved Ser residues throughout its protein sequence particularly clustering around the specific GTP-binding motifs (Tables I-II and Figure S1). In addition, two earlier studies reported the importance of DAP3 phosphorylation by Akt kinase in suppression of anoikis and by LKB1, a Ser/Thr kinase, in induction of apoptosis in cells ectopically expressing DAP3 56, 57.
Mitochondrial ribosomal protein L40 also known as MRPL37 was identified from immunoblotting with phospho-Ser and phospho-Thr antibodies as well after incubation with [γ-32P] ATP and staining with ProQ phosphoprotein dye (Tables I-II). Interestingly, this protein shows sequence similarity to another apoptotic protein MRPS30 (PDCD9) from the small subunit 58, 59. Widely expressed in multiple organs of the body, increased expression was observed in lung tumor cells compared to normal cells, as the gene may be activated in transformed cells 59. Therefore, this finding allows us to hypothesize that phosphorylation may act as a molecular switch in the stimulation or progression of apoptosis. In addition, gradually more evidence is accumulating that the mitochondrial ribosomal proteins are involved in cell death pathways, with at least two being phosphoproteins, and disease states suggesting conformational and/or functional changes of the ribosome occur as a result of this post-translational modification.
Conclusion
As we reported for bacterial ribosomes15, mitochondrial ribosomes are also phosphorylated at Ser, Thr, and Tyr residues. More importantly, we provide the first evidence for the regulatory role of phosphorylation on mitochondrial protein synthesis by in vitro phosphorylation of ribosomal proteins in the presence of mitochondrial kinases PKA, PKCδ, and Abl Tyr kinase. Therefore, findings reported in this study suggest that phosphorylation of ribosomal proteins might play a significant role in inhibition of mitochondrial protein synthesis, specifically synthesis of 13 essential components of the oxidative phosphorylation by ATP which is the final product of this process. Here, we identified the majority of the phosphorylated proteins in mitochondrial ribosomes; however, it is necessary to continue mapping phosphorylation sites by tandem mass spectrometric approaches and site-directed mutagenesis studies to specifically determine the role of each modified site in protein synthesis.
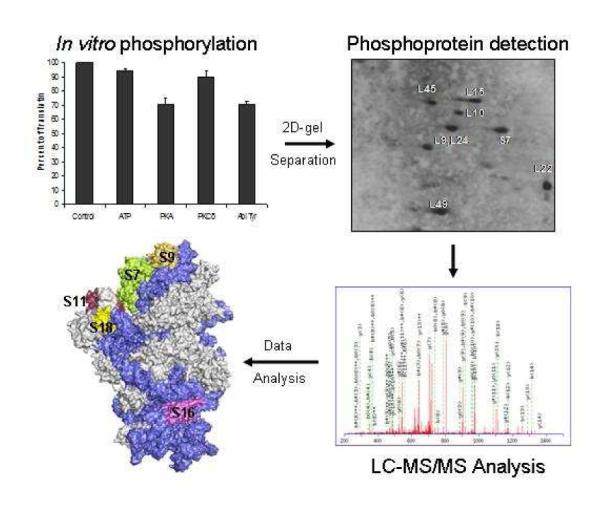
We have employed combination of 2D-gel electrophoresis and mass spectrometry to analyze the mammalian mitochondrial ribosome for phosphorylation. Moreover, we have demonstrated the phosphorylation and inhibition of protein synthesis in vitro by mitochondrial kinases PKA, PKCδ, and Abl Tyr kinase. Therefore, this study should serve as the framework for future studies addressing the regulation mechanisms of mitochondrial translation machinery responsible for the synthesis of thirteen essential proteins of oxidative phosphorylation.
Supplementary Material
Acknowledgements
We thank Seung-Kyu Lee, James L. Nasralla, and George Y. Soung for technical assistance in preparation of mitochondrial ribosomes and in-gel digestion of ribosomal proteins. This work was supported by the National Institute of Health grants GM071034 and EB005197 to E.C.K.
References
- 1.Pietromonaco S, Denslow N, O’Brien TW. Proteins of mammalian mitochondrial ribosomes. Biochimie. 1991;73:827–836. doi: 10.1016/0300-9084(91)90062-6. [DOI] [PubMed] [Google Scholar]
- 2.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115(1):97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286(1):73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton MG, O’Brien TW. Ultracentrifugal characterization of the mitochondrial ribosome and subribosomal particles of bovine liver: molecular size and composition. Biochemistry. 1974;13:5400–5403. doi: 10.1021/bi00723a024. [DOI] [PubMed] [Google Scholar]
- 5.Koc EC, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome: Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 6.Koc EC, Burkhart W, Blackburn K, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome: Analysis of the complement of ribosomal protein present. J. Biol. Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Structural compensation for deficit of rRNA with proteins in mammalian mitochondrial ribosome; systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 2001;276:21724–21736. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic Analysis of the Mammalian Mitochondrial Ribosome. Identification of Protein Components in the 28S Small Subunit. J. Biol. Chem. 2001;276(35):33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 9.Miller JL, Koc H, Koc EC. Identification of phosphorylation sites in mammalian mitochondrial ribosomal protein DAP3. Protein Sci. 2008;17(2):251–60. doi: 10.1110/ps.073185608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He H, Chen M, Scheffler NK, Gibson BW, Spremulli LL, Gottlieb RA. Phosphorylation of Mitochondrial Elongation Factor Tu in Ischemic Myocardium: Basis for Chloramphenicol-Mediated Cardioprotection. Circ. Res. 2001;89:461–467. doi: 10.1161/hh1701.096038. [DOI] [PubMed] [Google Scholar]
- 11.Traugh JA, Traut RR. Phosphorylation of ribosomal proteins of Escherichia coli by protein kinase from rabbit skeletal muscle. Biochemistry. 1972;11(13):2503–9. doi: 10.1021/bi00763a019. [DOI] [PubMed] [Google Scholar]
- 12.Mikulik K, Janda I. Protein kinase associated with ribosomes phosphorylates ribosomal proteins of Streptomyces collinus. Biochem. Biophys. Res. Commun. 1997;238(2):370–6. doi: 10.1006/bbrc.1997.7297. [DOI] [PubMed] [Google Scholar]
- 13.Mikulik K, Suchan P, Bobek J. Changes in ribosome function induced by protein kinase associated with ribosomes of Streptomyces collinus producing kirromycin. Biochem. Biophys. Res. Commun. 2001;289(2):434–43. doi: 10.1006/bbrc.2001.6017. [DOI] [PubMed] [Google Scholar]
- 14.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics. 2008;7(2):299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Soung GY, Miller JL, Koc H, Koc EC. Comprehensive analysis of phosphorylation sites in E. coli ribosomal proteins. J. Proteome Res. 2009;8(7):3390–3402. doi: 10.1021/pr900042e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews DE, Hessler RA, Denslow ND, Edwards JS, O’Brien TW. Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 1982;257:8788–8794. [PubMed] [Google Scholar]
- 17.Spremulli LL. Large-scale isolation of mitochondrial ribosomes from mammalian tissues. Methods Mol. Biol. 2007;372:265–75. doi: 10.1007/978-1-59745-365-3_19. [DOI] [PubMed] [Google Scholar]
- 18.Cahill A, Baio D, Cunningham C. Isolation and characterization of rat liver mitochondrial ribosomes. Anal. Biochem. 1995;232:47–55. doi: 10.1006/abio.1995.9962. [DOI] [PubMed] [Google Scholar]
- 19.Woriax V, Burkhart W, Spremulli LL. Cloning, sequence analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim. Biophys. Acta. 1995;1264:347–356. doi: 10.1016/0167-4781(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 20.Bhargava K, Templeton P, Spremulli LL. Expression and characterization of isoform 1 of human mitochondrial elongation factor G. Protein Expr. Purif. 2004;37(2):368–76. doi: 10.1016/j.pep.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Chen R, Fearnley IM, Peak-Chew SY, Walker JE. The phosphorylation of subunits of complex I from bovine heart mitochondria. J Biol Chem. 2004;279(25):26036–45. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Zambon AC, Vranizan K, Pothula K, Conklin BR, Insel PA. Gene expression signatures of cAMP/protein kinase A (PKA)-promoted, mitochondrial-dependent apoptosis. Comparative analysis of wild-type and cAMP-deathless S49 lymphoma cells. J. Biol. Chem. 2008;283(7):4304–13. doi: 10.1074/jbc.M708673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Bharti A, Mishra NC, Raina D, Kharbanda S, Saxena S, Kufe D. Targeting of the c-Abl tyrosine kinase to mitochondria in the necrotic cell death response to oxidative stress. J. Biol. Chem. 2001;276(20):17281–5. doi: 10.1074/jbc.M101414200. [DOI] [PubMed] [Google Scholar]
- 24.Ito Y, Pandey P, Mishra N, Kumar S, Narula N, Kharbanda S, Saxena S, Kufe D. Targeting of the c-Abl Tyrosine Kinase to Mitochondria in Endoplasmic Reticulum Stress-Induced Apoptosis. Mol. Cell. Biol. 2001;21(18):6233–6242. doi: 10.1128/MCB.21.18.6233-6242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WLT. DeLano Scientific. San Carlos; CA, U.S.A.: 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 27.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285(5436):2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 28.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407(6802):327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 29.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The Path of Messenger RNA through the Ribosome. Cell. 2001;106(2):233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 30.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal Structure of the Ribosome at 5.5 A Resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 31.Brodersen DE, Clemons WM, Jr., Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 32.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 1999;6(7):643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 33.Carter AP, Clemons J, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal Structure of an Initiation Factor Bound to the 30S Ribosomal Subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 34.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–34. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 35.Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J.Cell Biol. 2000;150(3):447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoang L, Fredrick K, Noller HF. Creating ribosomes with an all-RNA 30S subunit P site. Proc. Natl. Acad. Sci. U. S. A. 2004;101(34):12439–43. doi: 10.1073/pnas.0405227101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noller HF, Hoang L, Fredrick K. The 30S ribosomal P site: a function of 16S rRNA. FEBS Lett. 2005;579(4):855–8. doi: 10.1016/j.febslet.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 38.Robert F, Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J. Biol. Chem. 2003;278(45):44913–20. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 39.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 A Resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 40.Khaitovich P, Mankin AS, Green R, Lancaster L, Noller HF. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc. Natl. Acad. Sci.U. S. A. 1999;96(1):85–90. doi: 10.1073/pnas.96.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchiumi T, Hori K, Nomura T, Hachimori A. Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J. Biol. Chem. 1999;274(39):27578–27582. doi: 10.1074/jbc.274.39.27578. [DOI] [PubMed] [Google Scholar]
- 42.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol. Cell. 2005;20(5):723–31. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Van Dyke N, Xu W, Murgola EJ. Limitation of ribosomal protein L11 availability in vivo affects translation termination. J. Mol. Biol. 2002;319(2):329–39. doi: 10.1016/S0022-2836(02)00304-2. [DOI] [PubMed] [Google Scholar]
- 44.Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J. Mol. Biol. 2007;371(4):1047–59. doi: 10.1016/j.jmb.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 45.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Marcos A, Morreale A, Guarinos E, Briones E, Remacha M, Ortiz AR, Ballesta JP. In vivo assembling of bacterial ribosomal protein L11 into yeast ribosomes makes the particles sensitive to the prokaryotic specific antibiotic thiostrepton. Nucleic Acids Res. 2007;35(21):7109–17. doi: 10.1093/nar/gkm773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davydova N, Streltsov V, Wilce M, Liljas A, Garber M. L22 ribosomal protein and effect of its mutation on ribosome resistance to erythromycin. J. Mol. Biol. 2002;322(3):635–44. doi: 10.1016/s0022-2836(02)00772-6. [DOI] [PubMed] [Google Scholar]
- 48.Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419(6903):171–4. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 49.Ullers RS, Houben EN, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 2003;161(4):679–84. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nowotny V, Nierhaus KH. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U. S. A. 1982;79(23):7238–42. doi: 10.1073/pnas.79.23.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlunzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HA, Albrecht R, Buerger J, Wilbanks SM, Fucini P. The binding mode of the trigger factor on the ribosome: implications for protein folding and SRP interaction. Structure. 2005;13(11):1685–94. doi: 10.1016/j.str.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Mukamel Z, Kimchi A. Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. J. Biol. Chem. 2004;279(35):36732–8. doi: 10.1074/jbc.M400041200. [DOI] [PubMed] [Google Scholar]
- 53.Kim HR, Chae HJ, Thomas M, Miyazaki T, Monosov A, Monosov E, Krajewska M, Krajewski S, Reed JC. Mammalian dap3 is an essential gene required for mitochondrial homeostasis in vivo and contributing to the extrinsic pathway for apoptosis. FASEB J. 2007;21(1):188–96. doi: 10.1096/fj.06-6283com. [DOI] [PubMed] [Google Scholar]
- 54.Miyazaki T, Reed JC. A GTP-binding adapter protein couples TRAIL receptors to apoptosis-inducing proteins. Nat. Immunol. 2001;2(6):493–500. doi: 10.1038/88684. [DOI] [PubMed] [Google Scholar]
- 55.O’Brien T,W, O’Brien B,J, Norman RA. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–51. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 56.Miyazaki T, Shen M, Fujikura D, Tosa N, Kon S, Uede T, Reed JC. Functional role of death associated protein 3 (DAP3) in anoikis. J. Biol. Chem. 2004;279(43):44667–44672. doi: 10.1074/jbc.M408101200. [DOI] [PubMed] [Google Scholar]
- 57.Takeda S, Iwai A, Nakashima M, Fujikura D, Chiba S, Li HM, Uehara J, Kawaguchi S, Kaya M, Nagoya S, Wada T, Yuan J, Rayter S, Ashworth A, Reed JC, Yamashita T, Uede T, Miyazaki T. LKB1 is crucial for TRAIL-mediated apoptosis induction in osteosarcoma. Anticancer Res. 2007;27(2):761–8. [PubMed] [Google Scholar]
- 58.Koc EC, Ranasinghe A, Burkhart W, Blackburn K, Koc H, Moseley A, L.L. S. A new face on apoptosis: Death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 59.Levshenkova EV, Ukraintsev KE, Orlova VV, Alibaeva RA, Kovriga IE, Zhugdernamzhilyn O, Frolova EI. The structure and specific features of the cDNA expression of the human gene MRPL37. Bioorg. Khim. 2004;30(5):499–506. doi: 10.1023/b:rubi.0000043788.86955.ea. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.