Abstract
Pseudomonas quinolone signal (PQS), 2-heptyl-3-hydroxy-4-quinolone, is an intercellular alkyl quinolone signaling molecule produced by the opportunistic pathogen Pseudomonas aeruginosa. Alkyl quinolone signaling is an atypical system that, in P. aeruginosa, controls the expression of numerous virulence factors. PQS is synthesized from the tryptophan pathway intermediate, anthranilate, which is either derived from the kynurenine pathway or from an alkyl quinolone specific anthranilate synthase encoded by phnAB. Anthranilate is converted to PQS by the enzymes encoded by the pqsABCDE operon and pqsH. PqsA forms an activated anthraniloyl-CoA thioester that shuttles anthranilate to the PqsD active site where it is transferred to Cys112 of PqsD. In the only biochemically characterized reaction, a condensation then occurs between anthraniloyl-PqsD and malonyl-CoA or malonyl-ACP, a second PqsD substrate, forming 2,4-dihydroxyquinoline (DHQ). The role PqsD plays in the biosynthesis of other alkyl quinolones, such as PQS, is unclear though it has been reported to be required for their production. No evidence however, exists that DHQ is a PQS precursor. Here we present a structural and biophysical characterization of PqsD that includes several crystal structures of the enzyme including that of the PqsD-anthranilate covalent intermediate and the inactive Cys112Ala active site mutant in complex with anthranilate. The structure reveals that PqsD is structurally similar to the FabH and chalcone synthase families of fatty acid and polyketide synthases. The crystallographic asymmetric unit contains a PqsD dimer. The PqsD monomer is composed of two nearly identical ~170 residue αβαβα domains. The structures show anthranilate-liganded Cys112 is positioned deep in the protein interior at the bottom of a ~15 Å long channel while a second anthraniloyl-CoA molecule is waiting in the cleft leading to the protein surface. Cys112, His257, and Asn287 form the FabH-like catalytic triad of PqsD. The C112A mutant is inactive although it still reversibly binds anthraniloyl-CoA. The covalent complex between anthranilate and Cys112 clearly illuminates the orientation of key elements of the PqsD catalytic machinery and represents a snapshot of a key point in the catalytic cycle.
Pseudomonas aeruginosa is a major nosocomial pathogen causing thousands of infections annually among hospitalized patients. Hospital acquired pneumonia is a particular threat to those on mechanical ventilators and P. aeruginosa is often the causative agent. Persons with AIDS and other immune disorders are also at high risk, and cystic fibrosis patients are often chronically infected, with many patients succumbing to the entrenched colonization and long term lung damage caused by P. aeruginosa (1). In addition to being resistant to all but a handful of antibiotics, P. aeruginosa produces an unusually large number of extracellular virulence factors including pyocyanin (2), pyoverdin, elastase (LasB), and ExoS (3-5). Production of many of these virulence factors is controlled by low molecular weight intercellular signaling molecules (5). In addition to the acyl homoserine lactone quorum sensing signals that many bacteria use to control various aspects of virulence, P. aeruginosa and the closely related species, Burkholderia, use an additional, and apparently far less common, alkyl quinolone based cell-to-cell communication system (6, 7). 2-heptyl-3-hydroxy-4-quinolone, also known as Pseudomonas Quinolone Signal (PQS1; Scheme 1) has been examined in the most detail, and has been shown to control the expression of multiple virulence factors as it has been demonstrated to be the ligand for PqsR, the transcriptional activator that turns on alkyl quinolone signal production (8-11).
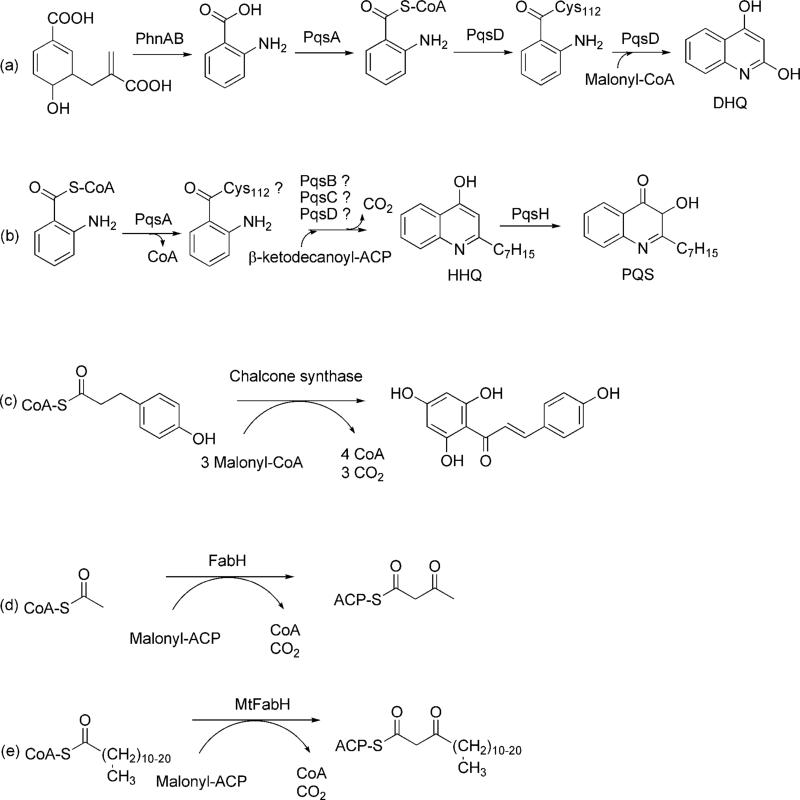
Scheme 1.
Condensation Reactions Catalyzed by PQS Biosynthetic Enzymes (a, b), Chalcone Synthase (c), FabH (d), and MtFabH (e). .
Alkyl quinolones, like pyocyanin, are derived from the key metabolic intermediate chorismic acid (Scheme 1). P. aeruginosa encodes a specific anthranilate synthase, the product of the phnAB operon, which funnels chorismate towards alkyl quinolone biosynthesis. Anthranilate is converted to PQS by the products of the pqs gene cluster which includes the pqsABCDE operon and pqsH (8). PqsA is an anthranilate-coenzyme A ligase that generates the activated thioester of anthranilate (12). Anthraniloyl-CoA (ACoA) is the first of two substrates used by PqsD. CoA delivers anthranilate to the PqsD active site where it is transferred to Cys112. In vitro, PqsD has been shown to use malonyl-CoA and malonyl-ACP as its second substrate in a reaction that forms 2,4-dihydroxyquinoline (DHQ) (13). It is unclear if PqsD has other in vivo substrates or whether it alone can catalyze an analogous reaction yielding the immediate PQS precursor 4-hydroxy-2-heptylquinoline (HHQ; Scheme 1). HHQ production has been demonstrated to involve β-ketodecanoate, supplied as a pantothenate conjugated thioester by acyl carrier protein (ACP; (8)). In principle, a reaction analogous to that proposed for DHQ biosynthesis (Scheme 2; (13)) could be catalyzed by PqsD alone, or it could involve PqsB or -C. PqsB, -C, and -D have all been annotated as condensing enzymes, in agreement with the proposed head-to-head condensation mechanism proposed for alkyl quinolone biosynthesis by Bredenbruch et al., (8). However, sequence alignments show that PqsB and -C both lack key residues associated with canonical condensation reactions suggesting they could have an alternate function. Whether DHQ is a true signaling molecule is uncertain, though some evidence points to it as having a role in cell-to-cell communication (13). It does not appear, however, that DHQ is a precursor to HHQ or PQS. PqsE is thought to be a regulatory protein involved in the modulation of cell-to-cell signaling pathways (9, 11). PqsA and PqsD have been shown to be the only proteins required for the conversion of anthranilate to DHQ (13).
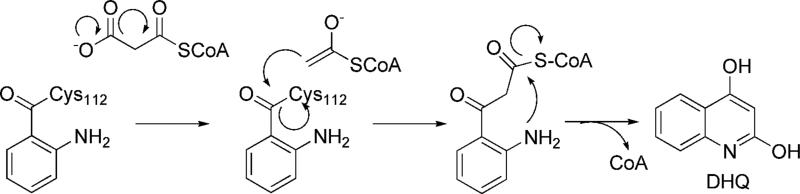
Scheme 2.
Proposed Mechanism for DHQ Formation (13).
Evaluation of the PqsD sequence indicated that it is a β-ketoacyl-ACP synthase III (FabH)-type condensing enzyme similar to authentic FabH which catalyzes the formation of acetoacetyl-ACP and is a key enzyme in bacterial fatty acid biosynthesis (Scheme 1;(14)). Indeed, a recent study has shown that PqsD catalyzes the formation DHQ from ACoA and malonyl-CoA (or malonyl-ACP (13)). There are other examples as well of FabH-like condensing enzymes that act in pathways other than fatty acid biosynthesis. The bacterial polyketide synthases (PKS) and the plant secondary metabolic enzymes chalcone synthase and stilbene synthase, for example, are FabH-type condensing enzymes (15). These enzymes differ from FabH in that they use CoA esters exclusively and they also catalyze additional chemistry, including cyclization, beyond the condensation reaction (16). PqsD exhibits functional and mechanistic attributes of both FabH and PKS-like enzymes such as chalcone synthase. For example, PqsD uses an ACP thioester substrate in a manner similar to that of FabH, but like chalcone synthase, PqsD catalyzes additional chemistry beyond a condensation reaction.
Given our ongoing interest in pyocyanin biosynthesis and the role of phenazines in virulence (2, 17, 18), we were intrigued by recent studies showing that pyocyanin production is directly regulated by alkyl quinolone signaling (11, 19). In order to evaluate the important determinants of catalysis in this crucial step in alkyl quinolone biosynthesis, to assess the features that set it apart from other condensing enzymes, and finally, to provide a framework to evaluate whether it could be a novel antimicrobial target, we have conducted a detailed structural investigation of PqsD. Here we report several crystal structures of PqsD including the 1.8 Å resolution structure of a key reaction intermediate, anthranilate modified PqsD, with anthranilate covalently bound to Cys112. The data show that PqsD has many structural similarities to a variety of condensing enzymes including E. coli FabH (20) and Mycobacterium tuberculosis FabH (MtFabH; (21). The latter has been shown to undergo a significant conformational change during catalysis in order to accommodate large (C10-C20) substrates (21). The structures presented here suggest PqsD would also need to undergo a conformational change in order to accommodate substrates such as β-ketodecanoyl thioesters that are significantly larger than its confirmed malonyl-thioester substrates.
Experimental Procedures
Protein Expression and Purification
A synthetic construct containing pqsD, optimized for overexpression in Escherichia coli, was obtained commercially (DNA 2.0) and subcloned into the expression vector pET28a (Novagen). The synthetic construct contained an N-terminal tobacco etch virus (TEV) protease cleavage site designed to allow production of fully native protein after expression of PqsD as a 6x His-tagged fusion (22). PqsD was overexpressed in E. coli strain BL21(DE3) using the autoinduction method of Studier (23). Cells were grown initially at 37°C until the culture density reached an optical density of ~0.3 at 600 nm at which time the temperature of the shaking incubator was reduced to 20°C. The bacterial cells were then harvested by centrifugation after 14-16 h and stored at −20°C until needed. PqsD was purified by cobalt ion affinity chromatography essentially as directed by the resin manufacturer (Sigma). Unfortunately, TEV protease would not cleave PqsD so instead, human α-thrombin (Haematologic Technologies) was used to remove the portion of the fusion tag encoded by the pET-28a vector. Thrombin was removed by passing the mixture over benzamidine agarose and the cleaved fusion tag was removed by a second passage over the cobalt resin. The resulting purified PqsD contained the following additional N-terminal residues GSHMASMTGGQQMGRGSENLYFQGNPIL prior to the native PqsD sequence (underlined). The N-terminal methionine was left out of the engineered expression vector coding sequence. PqsD judged pure by SDS-PAGE analysis was dialyzed against 25 mM Tris, 1 mM DTT, 1 mM EDTA (pH 7.6) and stored in 0.25 mL aliquots at −80°C.
Mutagenesis
The Cys112Ala variant of PqsD was created using the Quikchange method (Stratagene) as per the manufacturer's instructions. Cys112Ala PqsD was expressed and purified in the same manner described for the native enzyme.
Measurement of Enzymatic Activity
ACoA was synthesized from isatoic anhydride and CoA using a previously described method (24). ACoA was purified by HPLC after freeze drying the aqueous reaction mixture (~30 mL) and resuspending the dried material in ~2 mL of water. A 7.8 × 300 mm μbondapak C18 column (Waters) was used along with mobile phases of 20 mM ammonium acetate (A) and methanol (B) and a flow rate of 3.5 mL/min. The following gradient was used: 0-8 min, 0% B; 8-28 min, linear gradient 0-80% B. ACoA eluted at ~60% B. Fractions containing ACoA were pooled, the solvent was removed under vacuum and purified ACoA was stored dry at −80°C. Malonyl-CoA was purchased commercially and used without further purification. PqsD catalyzed formation of DHQ was analyzed at 37°C using a discontinuous assay with HPLC detection. Reaction mixtures typically contained 0.15 μM PqsD and variable amounts of the two substrates in 50 mM Tris, 100 mM KCl (pH 7.5). Reactions were quenched by addition of NaOH to a final concentration of 0.15M, neutralized with the same concentration of HCl, and diluted to an appropriate volume for assay with mobile phase (A). Samples were analyzed by HPLC using a Waters Acquity UPLC system equipped with photodiode array detector and electrospray ionization time-of-flight mass spectrometer. Mobile phases used for these assays were (A) 10 mM ammonium acetate (pH 5.5) and (B) acetonitrile. A BEH Shield C18 column (2.1 × 100 mm; Waters) was used to separate the reaction components using the following gradient: 0-1 min, 10 % B; 1-4 min, linear gradient to 90% B. Flow rate was 0.4 mL/min. Typical results are summarized in the supporting information.
Fluorescence spectroscopy was used to evaluate the interaction of PqsD and ACoA. PqsD or the Cys112Ala mutant (final concentration 1 μM) was added to various concentrations of ACoA in 50 mM Tris, 100 mM KCl (pH 7.5), in a 3 mL quartz cuvette. The solutions were excited at 360 nm and fluorescence emission was measured at 470 nm. Time courses were fit to equation (1), which describes a single exponential increase in fluorescence where y is the measured fluorescence intensity at a given time, y0 is the offset from zero, a is the amplitude of the fluorescence change, k is the observed rate, and t is time.
| (1) |
Isothermal Titration Calorimetry
The interaction between ACoA and Cys112Ala PqsD was evaluated using titration calorimetry. The sample cell contained ~1.4 mL of 200 μM mutant PqsD and the reference cell contained buffer only. 3 μL injections from a 3 mM stock of ACoA in the syringe were made at 300 s intervals. Analysis was complicated by the apparent tendency of PqsD to aggregate (causing large errors in the stoichiometry) so the data were analyzed by plotting total heat versus ACoA concentration and fitting to an equation describing hyperbolic binding. Plots of the data are presented in supporting information.
Molecular Weight from Laser Light Scattering
The solution molecular mass of PqsD was determined by a combination of laser light scattering and interferometric refractometry. 20 μL samples were injected onto a Shodex KW-802.5 gel filtration column attached to an Agilent 1100 series HPLC, and light scattering and concentration measurements were made using a DAWN EOS 18-angle light scattering detector (Wyatt Technology) and an OPTILAB DSP interferometric refractometer (Wyatt Technology). The column was equilibrated in 10 mM HEPES, 100 mM KCl, 0.2 mM EDTA, and 0.02% sodium azide at pH 7.50. A refractive index increment of 0.185 mL/g was used to estimate concentrations for molecular weight estimates, and bovine serum albumin was used as an isotropic scatterer for detector normalization to compensate for slight differences in the electronic gain of the 18 detectors. Molecular mass calculations were performed using ASTRA software (Wyatt Technology).
Analytical Ultracentrifugation
Sedimentation equilibrium experiments using wild-type PsqD were performed with a Beckman XL-I Optima analytical ultracentrifuge equipped with a four-hole, An-55 rotor. All experiments were performed at 25°C at rotor speeds between 15,000 and 20,000 rpm. The concentration distribution of the protein at sedimentation equilibrium was acquired in step mode as an average of 25 measurements of absorbance at 280 nm at each radial position, with nominal spacing of 0.001 cm between radial positions. Equilibrium was verified by comparing consecutive scans separated by time intervals from 40 min to 2 h. Protein samples were extensively dialyzed against 25 mM Tris, 100 mM NaCl (pH 7.6) prior to analysis. A partial specific volume (cubic centimeters per gram) of 0.7385 for PsqD was calculated from amino acid composition. In a typical experiment, equilibrium sedimentation data were collected after 16-24 h for three initial PqsD concentrations varying between 15 and 30 μM, ranging in optical density at 280 nm from 0.3 to 0.6.
The molecular weight of PqsD was calculated from a global analysis of the concentration distribution of the samples at sedimentation equilibrium in terms of a single, homogeneous species according to equation 2 where Cr is the concentration of the sample at a particular radial position, Cm is the concentration of the sample at the meniscus, M is the molecular weight, v-bar is the partial specific volume, ρ is the solvent density, ω is the angular velocity, r is the radial position in centimeters from the center of rotation, rm is the distance in centimeters from the center of rotation to the meniscus, R is the gas constant, T is the absolute (Kelvin) temperature, and B is a correction term for a nonzero baseline.
| (2) |
Crystallization
Initial crystallization screens of PqsD were performed at ambient temperature using the sitting drop vapor diffusion method with commercially available screening kits. The protein solution was concentrated to ~12 mg/mL prior to screening. Equal volumes of protein solution and reservoir solution, typically 2-4 μL total volume, were mixed for each experiment. Initial screening hits were optimized and, ultimately, a mixture containing 0.16 M MgCl2·8H2O, 0.08 M Tris HCl, 24% w/v Polyethylene Glycol 4000, and 20% v/v glycerol (pH 8.5) yielded the best results. Needle shaped (0.1x 0.2x 0.4 mm) crystals were typically obtained after ~2 days. Ligand bound structures were obtained by incubating PqsD or the Cys112Ala mutant with 7.5 mM ACoA at ambient temperature for 15 minutes prior to mixing with the crystallization solution. Optimized conditions differed slightly from that which yielded the best unliganded PqsD crystals and contained 21% PEG 3350, 0.2 M MgCl2·8H2O and 0.1 M Tris HCl (pH 8.5). Crystals reached a suitable size in ~2 days. Attempts to grow crystals in complex with malonyl-CoA or DHQ were not successful. Soaking ligands has proved successful in obtaining product complexes of related enzymes (16) however soaking native PqsD crystals in mother liquor or well solution containing DHQ did not yield a PqsD product complex.
Collection and Processing of Crystallographic Data
Native PqsD data were collected in house. Crystals were mounted in nylon loops and cooled to 100 K with a Rigaku Xtream 2000 cryo-cooler nitrogen stream without additional cryoprotection. Data were collected using a Rigaku Micro Max 007 rotating anode generator and a MAR345 CCD detector. All data were collected and processed with CrystalClear/d*Trek (25). Native PqsD crystals belonged to the P21 space group and diffracted to 1.8 Å. There were two monomers per asymmetric unit. Diffraction data for the covalent complex between anthranilate and PqsD and for the Cys112Ala mutant crystals were collected at National Synchrotron Light Source, Brookhaven National Laboratory on beamline X29 using a wavelength of 1.0809 Å. Crystals of the covalent complex exhibited the same P21 space group with slight differences in the cell dimensions and they diffracted to 1.8 Å. Cys112Ala PqsD with noncovalently bound anthranilate crystallized in the C2 space group and diffracted to 1.7 Å. These crystals were mounted in loops, cryoprotected in a solution of 25 % PEG 400, and flash cooled in liquid nitrogen prior to shipment to the synchrotron facility.
Structural Solution and Refinement
The structure of PqsD was solved by molecular replacement using the program Phaser (26). The search model was β-ketoacyl ACP synthase III from E. coli (PDB code 1HN9; (20)). The initial model was rebuilt using PHENIX auto builder (27) to match the PqsD sequence. The refinement procedure included periodic examinations of residues and subsequent refitting using the graphics program COOT (28). The model was completed and refined by using COOT for expanding the model and REFMAC5 for refining between building sessions (29). The final models are nearly complete and contain no significant disordered regions. In all structures, one chain of the crystallographic dimer exhibits clear density for a portion (7-9 residues) of the remaining affinity purification tag while the other chain begins at Gly1 of PqsD. Anthranilate modification of Cys112 was observed in both molecules of the crystallographic dimer in crystals where ACoA had been included in the crystallization mixture. An additional ACoA molecule was identified in the channel leading to each active site. Free anthranilic acid was found at the active site of Cys112Ala PqsD structure. Solvent molecules were identified and included as the refinement progressed. Statistics for data collection and refinement are shown in Table 1. The refined coordinates have been deposited in the Protein Data Bank as entries 3H76, 3H77, and 3H78.
Table 1.
Data collection and Refinement Statistics
| Native PqsD | Anthranilated-PqsD + Anthranilyl-CoA | C112A-PqsD + Anthranilic Acid | |
|---|---|---|---|
| PDB Code | 3H76 | 3H77 | 3H78 |
| space group | P21 | P21 | C2 |
| cell parameters (a, b, c) (Å) | 49.08, 90.42, 67.89 | 67.89, 59.66, 83.43 | 81.46, 59.18, 125.99 |
| cell parameters (β) (deg) | 95.38 | 103.95 | 92.30 |
| data collection statistics | |||
| resolution (Å) | 27.0–1.8 | 50.0–1.8 | 50.0–1.7 |
| mosaicity (deg) | 1.6 | 0.7 | 0.6 |
| no. of measured intensities | 153968 | 417515 | 405929 |
| no. of unique reflections | 49952 | 59980 | 62039 |
| redundancy | 3.1 | 7.0 | 6.5 |
| completeness ( %) (last shell) | 91.4 (88.3) | 99.7 (99.3) | 93.7 (84.1) |
| I/σ(I) (last shell) | 5.1 (1.6) | 7.8(6.2) | 7.8 (7.1) |
| Rmerge % (last shell)a | 9.1 (51.9) | 6.8 (40.2) | 6.1(38.7) |
| refinement statistics | |||
| resolution range (highest shell), (Å) | 25–1.8 (1.86-1.80) | 30–1.8 (1.86-1.80) | 40–1.7 (1.76-1.70) |
| no. of reflections used | 47404 | 56914 | 58860 |
| no. of protein atoms | 5007 | 5130 | 5119 |
| no. of waters | 350 | 582 | 658 |
| ligand(s) | None | anthranilic acid & anthraniloyl-CoA | anthranilic acid |
| R-factorb | 0.20 | 0.15 | 0.15 |
| Rfreec | 0.26 | 0.19 | 0.19 |
| rmsd bond lengths (Å) | 0.018 | 0.013 | 0.011 |
| rmsd bond angles (deg) | 1.9 | 1.6 | 1.3 |
| average B-factors for main chain/side chain/water (Å2) | 33.1/34.9/41.7 | 22.8/25.1/37.9 | 14.3/16.6/31.1 |
| average B-factors for ligand (Å2) | None | 33.22 | 16.67 |
Rmerge = Σ |I – (I)| /ΣI, where I is the intensity of an observed reflection and (I) is the average intensity of multiple observations.
R = Σ||Fobs| – |Fcal||/Σ|Fobs|.
Rfree = Σ||Fobs| – |Fcal||/Σ|Fobs|, where Fobs is from a test set of reflections (5 % of the total) that are not used in structural refinement.
RMS, root mean squares.
Modeling of the β-ketodecanoate-PqsD Complex
To identify plausible binding modes for ACP conjugated β-ketodecanoate, molecular docking studies were carried out using β-ketodecanoyl-CoA and each of the three structures presented here. Comparative modeling methods using chemical intuition and a viable interaction strategy have been applied to build a model that simulates the structure of PqsD in complex with β-ketodecanoyl-CoA. β-ketodecanoyl-CoA was added to the PqsD structure using the program COOT (28). The model of β-ketodecanoyl-CoA in the active site of PqsD was initially subjected to rigid body minimization with no experimental X-ray term. Residual strain in the model of the substrate plus the enzyme was relieved by conjugate gradient minimization, with no experimental X-ray term, and with inclusion of all water molecules, using CNS 1.1 (30). The optimized structure was analyzed with COOT and statistical tools therein. The RMSD values for all Cα atoms of the minimized PqsD-β-ketodecanoate models with respect to the relevant experimental model were 0.03-0.08 Å.
Results and Discussion Role of Alkyl Quinolines in Virulence
PQS, 2-heptyl-3-hydroxy-4-quinolone is both an important and unique modulator of virulence in P. aeruginosa. It is the major small molecule in alkyl quinolone signaling, one of three cell-to-cell signaling systems used by P. aeruginosa to control and coordinate production of virulence factors (31, 32). The other two systems, RhlR and LasR (so named for the transcriptional regulators involved) use the more well known and widespread homoserine lactone (HSL) signaling molecules. Specifically, the RhlR system uses N-butroyl-HSL while LasR system uses N-(3-oxo-dodecanoyl)-HSL (5). PqsR is the analogous transcriptional regulator in quinolone signaling, and the PQS-PqsR complex upregulates the genes involved in PQS production creating a positive feedback loop (10, 33).
Role of PqsD in PQS Biosynthesis
PqsD is a key biosynthetic enzyme encoded by the pqs gene cluster. It carries out the condensation and cyclization of the DHQ precursors ACoA and malonyl-ACP (13). It is unclear how the products of the pqs gene cluster produce larger alkyl quinolones such as HHQ and PQS, however, evidence indicates that β-ketodecanoic acid is involved (8), probably as a ACP thioester and that pqsB, -C, and -D are all required (13, 34). HHQ is believed to be converted to PQS via hydroxylation by PqsH, a putative aromatic hydroxylase. Interestingly, pqsH is not part of the pqs operon and, while nearby on the chromosome; it is under the apparent regulation of RhlR, not PqsR illustrating the complex regulatory systems used by P. aeruginosa to control virulence factor production. Given the interest in and development of fatty acid synthase inhibitors, including FabH inhibitors, PqsD, and possibly PqsB and -C, appear to be excellent targets for the development of antimicrobials. Disabling one of the three intertwined intercellular signaling systems in P. aeruginosa would likely have a significant impact on the production of key virulence factors.
Purification of PqsD
Using a synthetic pqsD gene optimized for expression in E. coli, approximately 100 mg of purified PqsD was obtained per liter of culture. As indicated above, TEV protease would not cut recombinant PqsD. Since the cleavage site itself was optimized, having a Gly at P1(22), we surmised that the site was physically inaccessible to TEV protease. The structure indicates that the N-terminal tail of the protein is threaded under the loop that connects the two PqsD domains (A170-G177; Figure 1). While not completely buried or occluded, it appears that this prevents TEV protease from accessing the cleavage site. The vector encoded thrombin cleavage site located approximately 20 residues toward the N-terminus of the expressed fusion protein was apparently easily accessible as cleavage occurred rapidly. Using a ~ 10 mg/mL PqsD solution and a ~1:1000 w/w ratio of thrombin to PqsD, 100 mg of PqsD was >90% cleaved in less than 1 hour at ambient temperature as judged by mass spectrometry and subsequent recovery of cleaved PqsD.
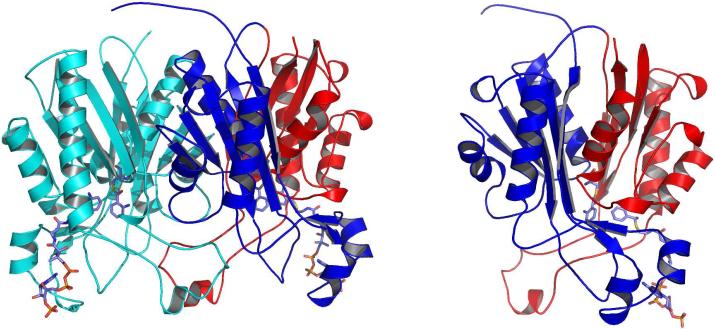
Figure 1.
Ribbon diagrams of the PqsD monomer and dimer. In the dimer representation one monomer is shown in cyan and the other is colored by domain with residues 1-174 in blue and 175-329 in red. Anthranilate-modified Cys112 and anthraniloyl-CoA molecules are shown as sticks and illustrate the locations of the active sites and CoA binding tunnels. The PqsD monomer is colored by domain as in the dimer and is rotated slightly to illustrate the structural similarity between the two domains.
Biochemical and Biophysical Characterization of PqsD
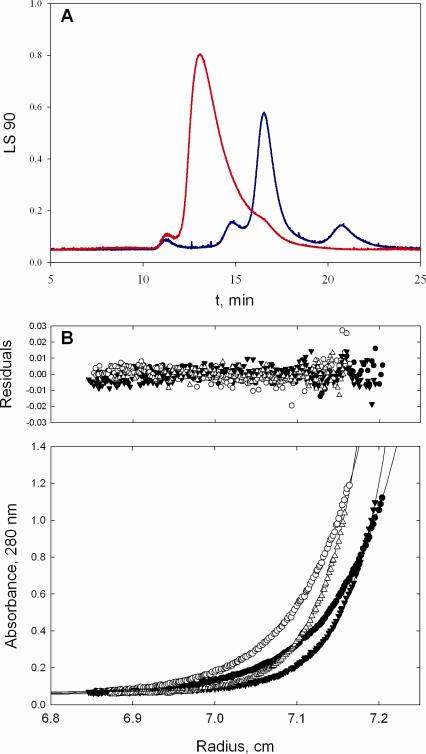
Routine laser light scattering experiments were conducted on PqsD to evaluate whether the protein was likely to crystallize or whether it was prone to aggregation, which can be an indicator of a poor crystallization candidate. Early results from laser light scattering experiments suggested that PqsD was prone to aggregation at the concentrations typically used in crystallization trials. Figure 2A shows that at higher concentrations of protein disproportionally more aggregated PqsD was present in gel filtration and laser light scattering experiments than was seen at lower concentrations. Analytical ultracentrifugation was then used to evaluate the solution molecular mass of PqsD and to ensure that aggregation was not impacting experiments such as activity measurements. Figure 2B indicates that at low to moderate concentrations, PqsD exists primarily as a dimer with a mass of ~72.8 kDa. This is consistent with other FabH-like proteins that are known to be dimers in solution. Even though light scattering clearly indicated PqsD was prone to aggregate, crystals were nevertheless obtained.
Figure 2.
Evaluation of the oligomeric state of PqsD. (A) Laser light scattering analysis indicates that PqsD begins to aggregate as it is concentrated. At 1.5 mg/mL (red trace), is predominantly a dimer with a calculated mass of ~72 kDa. At 12 mg/mL (blue trace), PqsD migrates as a species with a calculated mass of greater than 1000 kDa. (B) Analytical ultracentrifugation using sedimentation equilibrium methods confirms that PqsD is a dimer in solution at concentrations examined (0.1-0.5 mg /mL). Representative traces (bottom panel) of absorbance at 280 nm versus radial distance from the center of rotation in centimeters at 15,000 and 19,000 rpm. Distribution of residuals (top panel; Atheor – Aobs) for a single species of molecular weight 72,870.
The enzyme produced in our lab had catalytic properties consistent with the data reported by Zhang et al., (Supporting Information; (13)) and exhibited a turnover rate for DHQ formation of ~0.3 s-1 under conditions of saturating ACoA and malonyl-CoA. The Cys112Ala mutant produced no detectable DHQ. Two lines of evidence, however, suggest that the inactive mutant reversibly binds ACoA with reasonable affinity. The crystallographic results (discussed below) are fairly straightforward and show anthranilate bound in the enzyme active site. Calorimetric data also show that ACoA reversibly binds the Cys112Ala mutant with a Kd of about 57 μM (Supporting Information), a value close to the Km of 35 μM determined with the native enzyme (13).
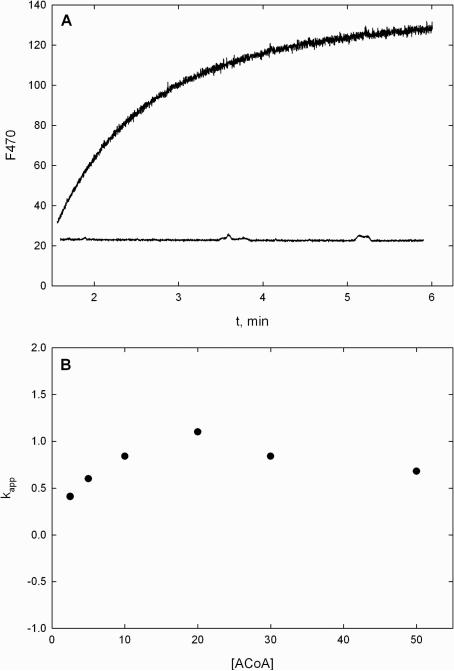
The formation of the covalent anthranilate-PqsD intermediate can readily be followed by fluorescence spectroscopy. Figure 3A illustrates the increase in fluorescence seen upon mixing PqsD and the Cys112Ala mutant with solutions of ACoA (λex= 360 nm, λem = 470 nm). An increase in fluorescence emission is seen with native PqsD but not with the mutant. Since evidence suggests that the Cys112Ala mutant is able to bind ACoA, the increase in fluorescence may result directly from the formation of the Cys112-anthranilate covalent intermediate. Alternatively, the signal change may stem from a conformational change after formation of the intermediate or a conformational change associated with CoA dissociation. Both of the latter possibilities are consistent with the relative insensitivity of the observed rate on concentration of ACoA (Figure 3B). In any event, the rate constant for the formation of the intermediate is at least slightly faster than turnover (~0.3 s−1), suggesting that another step is rate limiting in DHQ production by PqsD. We were unable to detect a change in free anthranilate fluorescence (λex= 313 nm, λem = 397 nm) using concentrations up to 2 mM when mixed with either the native or mutant PqsD enzymes.
Figure 3.
Evaluation of the interaction between PqsD and ACoA by fluorescence spectroscopy. (A) Changes in fluorescence emission at 470 nm upon mixing 1 μM PqsD or the Cys112Ala mutant with 20 μM ACoA. (B) Plot of kobs vs ACoA using native PqsD revealing that the rate is insensitive to ACoA concentration and is faster than kcat.
Overall Structure
P. aeruginosa PqsD is a 337 amino acid, 36 kDa protein that exists as a 72 kDa homodimer in solution (Figure 1). The crystallographically observed PqsD dimers in the structures presented here each bury ~ 3000 Å2 of combined surface area. The PqsD monomer is composed of two similar domains, residues 1-174 and 175-337, whose Cα atoms superimpose with an RMSD of 2.8 Å over 100 residues. Each domain consists of a five-stranded β-sheet, in which strand 4 is antiparallel, surrounded by one α-helix on the inside and two on the outside. The core of the protein is formed by the two inner helices (one from each domain) sandwiched between the two β-sheets. The most significant structural difference between the two domains involves the sequence falling between the central β-strand and the lone antiparallel β-strand of each domain. In domain 1, the 22 residue segment between Glu140 and Gly162 forms two short helices and includes key elements involved in CoA recognition and binding. In domain 2, there is only a three-residue loop (G316-A319) connecting the two β-strands. Generally, the three PqsD structures presented here are similar and can be superimposed with rms deviations ranging from 0.3-0.4 Å. Notable deviations in the positions of individual residues are discussed below.
Structure Relatives
The PqsD structure exhibits the fold first identified in the structure of thiolase (35) and it is most closely related to the E. coli β-ketoacyl-ACP Synthase III structure (FabH; (20) ). The superposition of the FabH from E. coli and PqsD structures resulted in an RMSD of 1.4 Å for 315 equivalent α-carbon atoms, indicating a very significant similarity. PqsD, not surprisingly, is similar to FabH enzymes from many species including M. tuberculosis, which encodes an unusual FabH (MtFabH) that functions in mycolic acid biosynthesis. This enzyme uses long chain (C10-20)-acyl-CoA substrates and it has been proposed that a significant conformational change permits MtFabH to bind these large molecules (21). The possibility that PqsD uses a similar mechanism should it be involved in long-chain-alkyl quinolone biosynthesis is discussed below. PqsD also bears notable similarity to chalcone synthase and stilbene synthase especially in terms of the conservation of key active site residues (15). These are plant polyketide synthases that use much the same catalytic machinery as the bacterial fatty acid synthases from which they likely evolved, yet they catalyze more diverse chemistry including, like PqsD, cyclization reactions. RMSD values of about 2.5 Å over ~300 residues are seen upon superimposing these structures on that of PqsD.
Active Site
The PqsD active site contains the Cys-His-Asn catalytic triad typical of FabH-like enzymes (Figures 4, 5). They are residues Cys112, His257, and Asn287 in PqsD. There is one, apparently independent, active site per chain. The active site is located between and under the two β-sheets and the two α-helices in between and is made up mostly of residues located on loops connecting these secondary structural elements (Figure 1). Cys112, the nucleophilic residue, captures anthranilate from ACoA in the first step of the reaction and, predictably, is required for catalysis. The Cys112Ala PqsD mutant produces no detectable DHQ. His257 and Asn287 complete the catalytic triad. The roles of His257 and Asn287 are likely more complicated than merely acting as a proton shuttle to generate a thiolate anion at Cys112. In both FabH and chalcone synthase, mutants at the corresponding histidine residue retain significant activity, and it has been shown other factors are involved in lowering the pKa of the active site cysteine residues in these proteins including a helical dipole. Therefore, alternate roles have been predicted for the His-Asn elements of the catalytic triads of these proteins. (16, 20, 36) In PqsD, these are likely involved in the decarboxylation reaction (Scheme 2) as has been demonstrated in FabH and chalcone synthase (14, 36).
Figure 4.
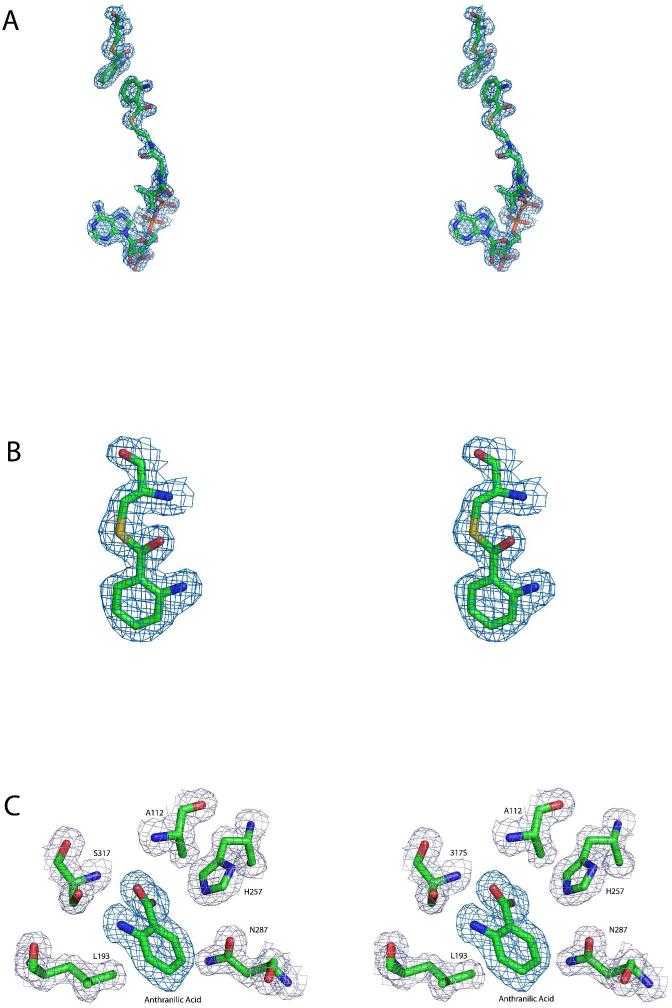
The active sites of PqsD and Cys112Ala PqsD are occupied by covalently bound and noncovalently bound ligands. (A) Stereoview of the positive difference density calculated after omitting anthranilate and anthraniloyl-CoA from a round of refinement of the liganded native structure. The map is contoured at 3σ and is depicted along with the ligands from the final refined model. (B) Magnified stereoview of the interaction between Cys112 and anthranilate illustrating the continuous density consistent with a covalent interaction. The displayed omit map was calculated as described for panel A and is shown contoured at 3σ along with the final refined model. (C) Stereoview of the positive difference density, shown in dark blue, calculated after omitting anthranilate from a round of refinement of the Cys112A PqsD structure. A noncovalently bound molecule of anthranilic acid is present in the active site. The map is contoured at 3σ and is depicted along with anthranilate from the final refined model. Nearby side chains are shown along with the final 2Fo-Fc map contoured at 1.2σ.
Figure 5.
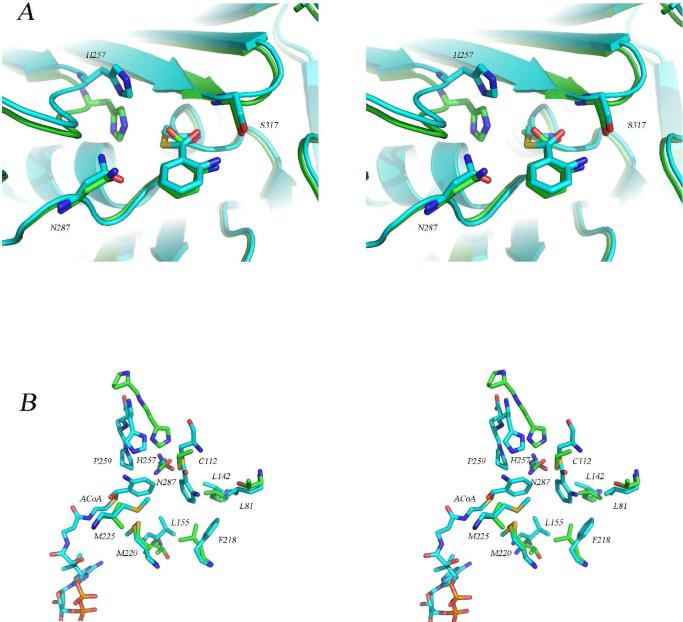
Comparison of the structure of the PqsD covalent intermediate with the structures of the Cys112Ala mutant and with E. coli FabH. (A) Stereoview of the superimposed active sites of the covalent PqsD-anthranilate complex (blue) and the Cys112Ala mutant structure in complex with anthranilate (green) illustrating the similarity in the positions of the anthraniloyl rings and the differences in the positions of His257. (B) Stereoview of the superimposed active sites of the covalent PqsD-anthranilate complex (blue) and FabH from E. coli (green) illustrating the differences in the position of the active site histidine residue and the possible role of an active site proline. In FabH, His244 is preceded by Pro243 while in PqsD, His257 is followed by Gln258 and Pro259. Also shown is the ACoA molecule occupying the active site entry tunnel and the hydrophobic pocket surrounding the reaction intermediate.
Compared to the FabH active site, the PqsD active site is somewhat larger and more hydrophobic. The approximate volumes are 650 Å3 for the FabH active site and 890 Å3 for the PqsD active site. The active site of chalcone synthase, by comparison, is 923 Å3 (16). A static evaluation of active site volumes for these proteins may be misleading however in light of recent studies on MtFabH, which has been proposed to undergo significant conformational changes during catalysis that would modulate the size of the active site cavity (21).
In the unliganded and anthranilate bound structures, the predicted catalytic triad residues are unusually far apart for residues considered to be key elements of catalysis. For example, in the unliganded structure His257 is 4.1 Å from the Cys112 sulfur and in the anthranilate-modified structure, the His257-Cys112 distance is 4.4 Å. However, in the Cys112Ala mutant, the distance between Cβ of Ala112 and the His257 ring is only 3.3, Å suggesting that sufficient flexibility exists in the active site to support a mechanism involving either His257 catalyzed deprotonation of Cys112 or a thiolate-imidazolium ion pair that serves to stabilize and orient the deprotonated cysteine((37); Figure 5A). Similar flexibility by His257 may be required in order to accommodate larger β-ketothioesters later in the reaction cycle as well.
Given the very strong similarity between PqsD and FabH, it appears that only subtle differences have evolved that permit this unique reaction to occur. An obvious difference between PqsD and FabH is that PqsD catalyzes a cyclization reaction. FabH uses acetyl- and malonyl- substrates to produce acetoacetyl-ACP while PqsD uses anthraniloyl- and malonyl- thioesters to produce the aromatic heterocycle, DHQ (Schemes 1, 2). Analysis of the active site architecture has provided some clues as to how PqsD accommodates this cyclization activity while operating with many of the same active site residues as FabH. Two Phe residues in FabH (157 and 308) are replaced by Leu159 and Ala319 in PqsD. These changes are predicted to allow binding of larger substrates and they are among the few obvious steric differences in terms of side chains lining the active site. The loop containing His257 may also be important in this regard. In both FabH and PqsD, a proline shares the loop with the active site histidine, however, in PqsD, the proline is two residues after His257 while in FabH a proline immediately precedes His244 (Figure 5B). The result is that the polypeptide chain is kinked in opposite directions in the two enzymes. As can be seen in figure 5B, the histidine residues superimpose poorly. The PqsD structure also presents some additional dilemmas. The active site, while perhaps more spacious than that of FabH, seems inadequate to handle a substrate such as β-ketodecanoyl-ACP suggesting PqsD may not be involved in the biosynthesis of the larger alkyl quinolones such as HHQ. The pocket surrounding and beyond bound anthranilate appears to be small and occluded by the covalent intermediate. The possibility that a conformational change is involved that could allow PqsD to cope with larger substrates is discussed below.
The CoA/ACP binding pocket that leads to the protein surface is about 15Å deep, measured from the protein surface to the Cys112 sulfur and the protein surface surrounding the cavity entrance is arginine rich. These residues may be involved in ACP recognition in addition to interacting with CoA (20). The CoA adenosine ring is sandwiched between the side chains of Phe32 and Arg153 near the surface of the enzyme and a number of other electrostatic interactions anchor CoA to the PqsD active site cleft. A similar scenario is seen in FabH though the residue corresponding to Phe32 of PqsD is a tryptophan. Chalcone synthase, in contrast, has no such adenosine ring sandwich feature and the CoA binds in a dissimilar fashion compared to what is seen in FabH and PqsD (16).
Structure of the Covalent Cys112-anthranilate Intermediate
Anthranilic acid is delivered to the PqsD active site as an activated CoA-bound thioester (Figure 4). Cys112, the active site nucleophile, attacks the substrate thioester bond and displaces CoA resulting in an anthranilate modified enzyme that is poised to react with either β-ketodecanoyl- or malonyl-ACP (or-CoA). The anthranilate carbonyl oxygen atom is hydrogen bonded to the backbone nitrogen atoms of Cys112 and Ser317 (an oxyanion hole) and to an active site water molecule. The 2-amino group of anthranilic acid is hydrogen bonded to the γ-oxygen of Ser317 and two active site water molecules. The anthranilate aromatic ring interacts with Phe218 and five Leu residues (81, 142, 155, 159, and 193; Figure 5B) in a very hydrophobic pocket, and the anthranilate ring of another ACoA molecule found in the entry tunnel. Density for this second substrate molecule is well defined in one subunit and less so in the other where the anthranilate ring density is strong but the CoA backbone density was marginal. The hydrophobic pocket surrounding the nonpolar portion of the anthraniloyl ring is shown in figure 5B.
Structure of the Cys112Ala Mutant in Complex with Anthranilate
We crystallized the Cys112Ala mutant in complex with ACoA. Surprisingly, we found the structure contains density consistent with free anthranilic acid, in both subunits, occupying virtually the same location as it does when covalently bound to Cys112 in native PqsD (Figures 4B, 5A). Additionally, as in the covalent intermediate structure, a second molecule of ACoA is apparently present, but in just one subunit. Only the density for the anthranilate ring portion of the molecule was modeled since the density for the CoA portion was poor. Aside from the mutation, there are only a few significant differences observed in the mutant structure. His257, as noted above however, is much closer to residue 112 in the mutant structure (Figure 5A). Interestingly, this is not due simply to rotation around the His257 Cα-Cβ bond but more so by movement of the local polypeptide chain that closes in slightly on the active site. The free anthranilic acid may be a result of crystallization being conducted at pH 8.5 since the ACoA thioester is expected to be more susceptible to hydrolysis at higher pH. Alternatively, it could suggest a thioesterase activity is present that has hydrolyzed ACoA. Such an activity would have to be present in order to liberate β-ketodecanoate (see below) if indeed, PqsD produces HHQ, however we cannot conclusively identify what residues might be involved in such an activity. This structure does illustrate, however, that other factors aside from covalent attachment to Cys112 are important in substrate recognition in PqsD. Perhaps those factors that orient bound anthranilate for reaction with the incoming second substrate are sufficient to hold the molecule in place even in the absence of a covalent interaction.
Mechanism of Alkyl Quinolone Formation
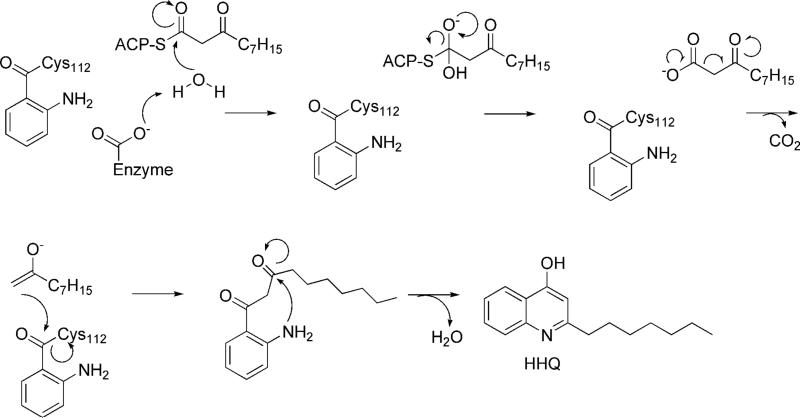
Despite exhibiting a high degree of sequence and structural homology to FabH, PqsD catalyzes an additional reaction, the cyclization forming DHQ. A plausible mechanism for this reaction has recently been proposed ( (13)Schemes 1, 2). Despite genetic data suggesting that PqsD is required for PQS production, other evidence suggests that PqsD may not be involved in a condensation reaction leading to production of longer chain alkyl quinolones. First, unlike malonyl thioesters, β-ketodecanoyl- and other long chain thioesters do not have a reactive carboxylate group exposed which, barring unforeseen chemistry, is required in order to generate the nucleophilic carbanion that attacks the thioester bond at Cys112 (Scheme 3). A mechanism for HHQ formation by PqsD would require involvement of a thioesterase activity, possibly by an activated water nucleophile (Scheme 3). The structure of HHQ dictates that, as expected, a carbon atom has to be lost during the reaction and decarboxylation of β-ketodecanoate is the most plausible scenario given the similarities between FabH and PqsD, -B, and -C. An active site base would likely have to be involved in activating a water molecule for attack, though the crystal structures do not suggest any definitive answer to this question if indeed PqsD is involved.
Scheme 3.
A Possible Mechanism for HHQ Formation.
Following the liberation of β-ketodecanoate from its ACP carrier, it would likely react in the expected fashion for a condensation of this type as depicted in Scheme 3. Cyclization is predicted to occur via a Schiff base mechanism following attack of the 2-amino group of anthranilate on the former β-keto group. The intermediate carbinolamine loses a water (the former β-keto oxygen), completing the ring closure. Scheme 3 illustrates a potential mechanism for the condensation reaction between the thioester of anthranilic acid and β-ketodecanoate.
PqsD, alternatively, could be involved in the synthesis of β-ketodecanoate itself, perhaps from malonyl-ACP and an octanoyl thioester, for use in the proposed “head-to-head” condensation reaction (8). This possibility has been noted by others (8, 13), has not been ruled out, and would be consistent with the genetic data (34, 38). The work of Bredenbruch et al., however suggests that the β-ketoacids involved in alkyl quinolone biosynthesis are derived from the rhamnolipid pathway (8). Such a scenario would suggest that PqsB or -C are involved in the putative condensation forming HHQ and as yet there is no biochemical evidence for this.
Conformational Change
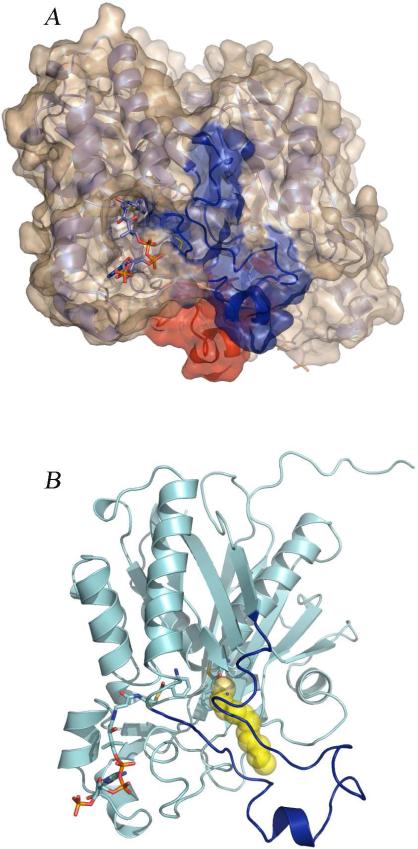
As noted above, MtFabH, which uses long-chain acyl-CoA substrates, appears to undergo a large conformational change which facilitates acyl-CoA binding. In the absence of such a change, the acyl-CoA ester would have to be threaded through both arms of the ‘L’ shaped active site of the enzyme in order for the CoA sulfur to get into position for catalysis (Figure 6). In the second step of the reaction, ACP presents malonate to the active site tethered to its pantetheine arm via the CoA binding tunnel. After completion of catalysis (in this case formation of a β-keto-ACP), the active site is proposed to again open allowing facile product release (21).
Figure 6.
Structural basis for a possible conformational change. (A) The PqsD dimer. Residues 186-222, the regions corresponding to the mobile flap in MtFabH, are colored red in subunit A and blue in subunit B. ACoA is shown in the CoA binding tunnel. (B) Close up of the PqsD monomer illustrating the ‘L’ shaped active site and a possible binding mode for β-ketodecanoate. Residues 186-222 are colored blue. The yellow spheres are decanethiol from the MtFabH structure (PDB code 2QO1). The figure was generated by superimposing PqsD and MtFabH (rmsd = 2.1 Å over 320 residues).
It is unclear whether PqsD undergoes a conformational change during catalysis, however, such a scenario could explain some of our structural observations. The first steps of the PqsD catalyzed reaction are binding of ACoA and formation of the covalent anthranilate-Cys112 intermediate. Based on the structures presented here no conformational change is required for these steps though they cannot be ruled out. The fluorescence data presented in figure 3, however, do suggest the possibility of a local conformational change but the binding of a second substrate molecule in the CoA access tunnel implies that a gross conformational change is not required. The second half of a potential reaction yielding HHQ, which is analogous to the first half of the MtFabH reaction in terms of substrate size, is difficult to envision without invoking a global conformational change since the PqsD active site in the observed conformation seems unlikely to accommodate a decanoyl chain (Figure 7). Attempts to model β-ketodecanoyl-CoA into the PqsD active site were unrevealing due to the closed conformation. However, a logical proposal based on available evidence, suggests that the nonpolar portion of β-ketodecanoate would likely bind in the hydrophobic pocket to the right (looking in the CoA binding tunnel) of enzyme bound anthranilate (Figures 6, 7). This is the location where the acyl portion of MtFabH binds and it is also the cyclization pocket in chalcone synthase (15, 21). As with MtFabH, an unfavorable threading of a long chain fatty ester would have to occur in the absence of a conformational change since in the observed (closed) conformation of PqsD, the binding pocket is both too small and blocked off by the anthranilate intermediate to accommodate β-ketodecanoate. If the flap shown in blue in figure 6B, (residues 186-222) were to open as has been proposed for the analogous residues in MtFabH, ACP-tethered β-ketodecanoate would likely have a more energetically favorable path to the catalytic site.
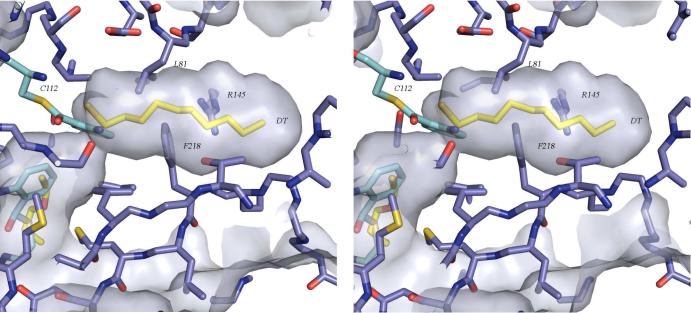
Figure 7.
Stereoview of the presumed acyl binding pocket of PqsD illustrating that a conformational change is likely required in order to accommodate β-ketodecanoate. Decanethiol from the MtFabH structure (PDB code 2QO1) was superimposed as in Figure 7 and is shown with yellow carbon atoms. In the observed conformation, Phe218, Leu81, and Arg145 are among the residues limiting access to the binding pocket.
A significant unanswered question with regard to the possible involvement of PqsD in long chain alkyl quinolone biosynthesis is that of the putative thioesterase activity. Since the proposed mechanism (Scheme 3) invokes a carbanionic intermediate that attacks the Cys112-anthranilate thioester bond, it is probable that proximity plays a key role in catalysis. After decarboxylation it is expected that the α-carbon of β-ketodecanoate will be very near the enzyme bound thioester. If PqsD catalyzes a thioesterase reaction, it therefore is predicted to occur in the immediate vicinity of the catalytic triad of PqsD. Given the predicted binding mode and the constraints imposed by the pantothenate linkage to ACP, His257 is a reasonable candidate to act as a general base catalyst.
Conclusion
The crystal structure of the PQS biosynthetic enzyme PqsD has been solved. PqsD catalyzes a key chemical step in biosynthesis of the alkyl quinolones, the condensation and cyclization of anthraniloyl and malonyl thioesters that forms DHQ (13). Structurally, PqsD most closely resembles β-ketoacyl-ACP synthase (FabH). However, in terms of catalysis, PqsD bears notable similarity to chalcone synthase which, like PqsD, catalyzes additional chemistry beyond the condensation reaction. The structure of PqsD in complex with covalently bound anthranilate reveals a hydrophobic active site that is approximately the same size as the chalcone synthase active site and ~40% larger than the FabH active site. While it remains unclear what role PqsD plays in the biosynthesis of larger alkyl quinolones, two pieces of evidence suggest that it has some role. First, it is apparently required for PQS and HHQ production (34). If true, and given that PqsD is a proven condensing enzyme, it is not unreasonable to suspect that PqsD may also catalyze HHQ formation. Additionally, the other candidates for involvement in HHQ formation, PqsB and PqsC, lack some or all of the hallmark sequence elements associated with condensing enzymes. The structures of PqsD presented here also provide a framework for designing future experiments to further analyze the enzymology of alkyl quinolone biosynthesis. Finally, these data provide a starting point for investigating whether PqsD could be a useful target for the development of antimicrobials that could potentially disable multiple downstream virulence pathways in P. aeruginosa.
Supplementary Material
Footnotes
This work was supported by NIH Grants AI067530 (J.F.P.) and AI076272 (E.C.P). Some data for this study were measured at beamline X29 at the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Abbreviations: PQS, Pseudomonas quinolone signal; TEV, tobacco etch virus; ACoA, anthraniloyl Coenzyme A; DHQ, 2,4-dihydroxyquinoline; HHQ, 4-hydroxy-2-heptylquinoline.
Supporting Information Available. LC/MS data including chromatograms, isothermal titration calorimetry data. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Wagner VE, Iglewski BH. P. aeruginosa Biofilms in CF Infection. Clin. Rev. Allergy. Immunol. 2008;35:124–134. doi: 10.1007/s12016-008-8079-9. [DOI] [PubMed] [Google Scholar]
- 2.Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel J, Balachandran P. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 2009;12:61–66. doi: 10.1016/j.mib.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Zelenitsky SA, Harding GK, Sun S, Ubhi K, Ariano RE. Treatment and outcome of Pseudomonas aeruginosa bacteraemia: an antibiotic pharmacodynamic analysis. J. Antimicrob. Chemother. 2003;52:668–674. doi: 10.1093/jac/dkg403. [DOI] [PubMed] [Google Scholar]
- 5.Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diggle SP, Lumjiaktase P, Dipilato F, Winzer K, Kunakorn M, Barrett DA, Chhabra SR, Camara M, Williams P. Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 2006;13:701–710. doi: 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Bredenbruch F, Nimtz M, Wray V, Morr M, Muller R, Haussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J. Bacteriol. 2005;187:3630–3635. doi: 10.1128/JB.187.11.3630-3635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrow JM, 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J. Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, Saucier M, Xiao G, Rahme LG. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 12.Coleman JP, Hudson LL, McKnight SL, Farrow JM, 3rd, Calfee MW, Lindsey CA, Pesci EC. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J. Bacteriol. 2008;190:1247–1255. doi: 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y-M, Frank MW, Zhu K, Mayasundarui A, Rock CO. PqsD is Responsible for the Synthesis of DHQ, an Extracellular Metabolite Produced by Pseudomonas aeruginosa. J. Biol. Chem. 2008;283:21788–21794. doi: 10.1074/jbc.M804555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies C, Heath RJ, White SW, Rock CO. The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Structure. 2000;8:185–195. doi: 10.1016/s0969-2126(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 15.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 16.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 17.Parsons JF, Song F, Parsons L, Calabrese K, Eisenstein E, Ladner JE. Structure and function of the phenazine biosynthesis protein PhzF from Pseudomonas fluorescens 2-79. Biochemistry. 2004;43:12427–12435. doi: 10.1021/bi049059z. [DOI] [PubMed] [Google Scholar]
- 18.Greenhagen BT, Shi K, Robinson H, Gamage S, Bera AK, Ladner JE, Parsons JF. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry. 2008;47:5281–5289. doi: 10.1021/bi702480t. [DOI] [PubMed] [Google Scholar]
- 19.Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 20.Qiu X, Janson CA, Smith WW, Head M, Lonsdale J, Konstantinidis AK. Refined structures of beta-ketoacyl-acyl carrier protein synthase III. J. Mol. Biol. 2001;307:341–356. doi: 10.1006/jmbi.2000.4457. [DOI] [PubMed] [Google Scholar]
- 21.Sachdeva S, Musayev FN, Alhamadsheh MM, Scarsdale JN, Wright HT, Reynolds KA. Separate entrance and exit portals for ligand traffic in Mycobacterium tuberculosis FabH. Chem. Biol. 2008;15:402–412. doi: 10.1016/j.chembiol.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 23.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Simon EJ, Shemin D. The Preparation of S-Succinyl Coenzyme A. J. Am. Chem. Soc. 1953;75:2520. [Google Scholar]
- 25.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Winn M, Isupov M, Murshudov GN. Use of TLS Parameters to Model Anisotropic Displacements in Macrololecular Refinement. Acta. Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Wagner VE, Li LL, Isabella VM, Iglewski BH. Analysis of the hierarchy of quorum-sensing regulation in Pseudomonas aeruginosa. Anal. Bioanal. Chem. 2007;387:469–479. doi: 10.1007/s00216-006-0964-6. [DOI] [PubMed] [Google Scholar]
- 32.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 33.Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. U S A. 2001;98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathieu M, Modis Y, Zeelen JP, Engel CK, Abagyan RA, Ahlberg A, Rasmussen B, Lamzin VS, Kunau WH, Wierenga RK. The 1.8 A crystal structure of the dimeric peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: implications for substrate binding and reaction mechanism. J Mol Biol. 1997;273:714–728. doi: 10.1006/jmbi.1997.1331. [DOI] [PubMed] [Google Scholar]
- 36.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 37.Jez JM, Noel JP. Mechanism of chalcone synthase. pKa of the catalytic cysteine and the role of the conserved histidine in a plant polyketide synthase. J. Biol. Chem. 2000;275:39640–39646. doi: 10.1074/jbc.M008569200. [DOI] [PubMed] [Google Scholar]
- 38.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U S A. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.