Abstract
The motor protein kinesin has two heads and walks along microtubules processively using energy derived from ATP. However, how kinesin heads are coordinated to generate processive movement remains elusive. Here we created a hybrid nanomachine (DNA-kinesin) using DNA as the skeletal structure and kinesin as the functional module. Single molecule imaging of DNA-kinesin hybrid allowed us to evaluate the effects of both connect position of the heads (N, C-terminal or Mid position) and sub-nanometer changes in the distance between the two heads on motility. Our results show that although the native structure of kinesin is not essential for processive movement, it is the most efficient. Furthermore, forward bias by the power stroke of the neck linker, a 13-amino-acid chain positioned at the C-terminus of the head, and internal strain applied to the rear of the head through the neck linker are crucial for the processive movement. Results also show that the internal strain coordinates both heads to prevent simultaneous detachment from the microtubules. Thus, the inter-head coordination through the neck linker facilitates long-distance walking.
Keywords: DNA, FRET, kinesin, nanomachine, single molecule
Introduction
Conventional kinesin (kinesin-1; hereafter referred to as kinesin) is a motor protein, which drives cellular transport using the energy derived from ATP (Vale and Milligan, 2000; Hirokawa and Takemura, 2005). Kinesin's head (catalytic domain), which binds to microtubules (MTs) and hydrolyzes ATP, is located at the N-terminus of the polypeptide, whereas two identical heads are dimerized through the C-terminal coiled-coil. Kinesin walks processively along the MT over long distances (more than 1 μm) with an 8-nm step that matches the repeat distance of the MT lattice, and can generate a force of up to 7 pN (Svoboda et al, 1993; Vale et al, 1996). When walking, kinesin alternately repeats one-head and two-head bound states to move in a ‘hand-over-hand' manner (Asbury et al, 2003; Kaseda et al, 2003; Yildiz et al, 2004).
To walk efficiently over long distances, (1) the trailing head, but not the leading head, must detach from the MT at the end of the two-head bound state and (2) this detached head must bind forward, at the end of the one-head bound state. For the second condition, a bias mechanism regulated by the ‘neck linker', a 13-amino-acid chain (residues 324–336) positioned at the C-terminal part of the head, has been proposed (‘Power stroke model') (Rice et al, 1999). For detachment of the trailing head, a regulation mechanism that coordinates both heads has been proposed (for reviews, see Block, 2007; Hackney, 2007).
The power stroke model was originally proposed for the motor protein myosin (Huxley, 1969; Huxley and Simmons, 1971). In myosin, a chemical state change in the bound nucleotide generates a small mechanical conformational change in the head. This small conformational change is amplified by a rigid lever arm. The power stroke of the lever arm shifts the detached head to bias its reattachment to the next forward actin filament-binding site (Spudich, 1994, 2001; Dunn and Spudich, 2007; Shiroguchi and Kinosita, 2007). In the myosin power stroke, the length of the lever arm, which contributes to the forward displacement of the detached head, is important for directional movement (Purcell et al, 2002; Sakamoto et al, 2003, 2005). Kinesin lacks the rigid lever arm, but upon ATP binding to the head, a conformational change in the neck linker occurs during which the neck linker binds to the head (termed ‘docking'). This docking is believed to be analogous to the myosin lever arm enabling a similar walking mechanism (Rice et al, 1999; Case et al, 2000; Tomishige and Vale, 2000; Tomishige et al, 2006). By docking the neck linker to the leading head, the C-terminus of the leading head is repositioned toward the MT plus end, thus shifting the position of the detached head forward. Although the contribution of the neck linker docking is well established, there are still some arguments regarding whether this docking powers kinesin movement (Nishiyama et al, 2001, 2002; Schief and Howard, 2001; Carter and Cross, 2005; Block, 2007).
On the other hand, coordination of the heads ensures long-distance travel. To walk more than a hundred steps without dissociation, native kinesins coordinate the activities of their two heads such that one head always remains attached to the MT, while the trailing head, but not leading head, is always detaches from the MT. It is thought that there is a checkpoint (termed ‘gating'), in which the walking cycle is stalled until a specific nucleotide binding or conformational change occurs. Several types of gating mechanisms have been proposed, although two models are most popular. One is the ‘mechanical gate model' in which the power stroke of the leading head accelerates the detachment of the trailing head (Hancock and Howard, 1999), meaning the gating contributes to the velocity of the molecules (see Supplementary Discussion for details). The other is the ‘chemical gate model' according to which ATP binding of the leading head is inhibited until detachment of the trailing head occurs, meaning only the trailing head can become weak binding and subsequently detach from the MT (Rosenfeld et al, 2003; Klumpp et al, 2004). Thus simultaneous detachment from the MT by both heads is prevented by gate mechanism such that gating contributes to run length, which is a measure of processsivity. In the two-head bound state for both models, the neck linker that connects the two heads is more or less fully extended. Thus the inter-head tension (or ‘internal strain') is believed to be the origin of head–head communication.
To distinguish between these two models, two groups attempted to reduce the internal strain by extending the length of the neck linker (Hackney et al, 2003; Yildiz et al, 2008). Hackney et al (2003) inserted additional peptide residues (1–12 residues per head) between the neck linker and the coiled-coil part finding that kcat and the multi-motor sliding velocity of axonemes remain constant. However, the kinetic processivity (defined as the number of ATPs hydrolyzed per productive microtubule encounter) in these mutants is significantly less than wild type. Furthermore, another biochemical study (Rosenfeld et al, 2003) showed that the ATPases of monomers, that have no internal strain, and dimers are identical, meaning ATPase acceleration due to dimerization is negligible. These results support the chemical gate model. However, recent single molecule experiments showed conflicting results (Yildiz et al, 2008). At the single molecule level, Yildiz et al (2008) observed constructs by inserting 2–26 polyproline or seven repeats of glycine–serine residues (14GS) into each head. These extended kinesins remained processive and their run length was almost unchanged. However, the velocity significantly decreased. Interestingly, the speed recovered to near normal levels when an external tension was applied to the motor by an optical trap along the direction of movement. As this tension was applied more to the trailing head than leading head, these results were interpreted to mean that trailing head detachment was promoted by external tension, suggesting that internal force generated by the leading head's docking promotes trailing head detachment during the normal walking cycle. Thus, single molecule experiments support the mechanical gate model. To resolve this discrepancy, a novel approach is needed.
Another issue is the structural basis for the coordination. Recently, Yildiz et al (2008) showed that external tension can induce directional stepping in normally immobile kinesin constructs that lack both mechanical element (neck linker) and fuel (ATP). They proposed a hypothesis according to which the head itself can sense and respond to strain to ensure unidirectional movement. Resolving the sensing domain (or element) should provide important information regarding the coordination mechanism. However, using a classical method to construct neck linker mutants (point mutations, extensions and replacements) results in strain always being applied through the neck linker. Thus, it is difficult to conclude whether sensing tension is done by the whole head or by a specific domain.
To clarify the mechanism unequivocally, one needs to explore precisely the effect of changing the distance between the two heads and applying strain to many locations on the head. However, a construct solely based on proteins cannot fulfill all these required conditions. Therefore, we constructed a DNA-kinesin hybrid nanomachine (hereafter ‘DNA-kinesin') that connects the two monomers with DNA. Advantages using DNA are that short dsDNA can act as a rigid rod (Bustamante et al, 1994; Wang et al, 1997; Mathew-Fenn et al, 2008; see also Supplementary Results) and the DNA length can be changed incrementally 0.34 nm by changing one base, meaning one can control the distance between the two heads with sub-nanometer accuracy. In addition, by introducing a Cys residue, any surface position of the head can be labeled with DNA. Thus, both the connect position and the distance between the two heads can be fully controlled, so one can evaluate the head–head coordination mechanism precisely. Using this novel assay, we explored the origin of processive movement in kinesin.
Results
Construction and confirmation of DNA-kinesin
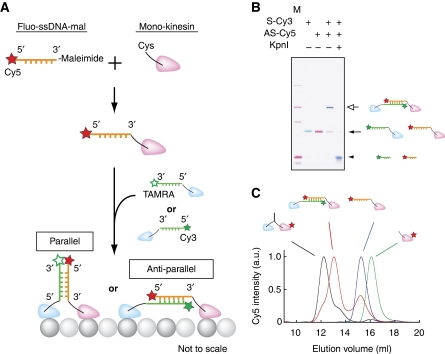
For DNA-kinesin construction, a Cys residue was introduced at a specific position in the Cys light mutant (CLM), where fluorescent dye labeling has no effect on activity (Rice et al, 1999; Tomishige et al, 2006). Then fluorescently labeled DNA–maleimide was covalently attached. DNA-kinesin dimers were obtained by mixing two hetero DNA-kinesin monomers, in which one monomer had a sense sequence and the other had an antisense sequence (Figure 1A). Attachment of the kinesin head at the 5′ or 3′ end resulted in a parallel or anti-parallel type dimer. The parallel type mimics the coiled-coil part of kinesin, whereas the anti-parallel type resembles the layout of the neck linker. Biochemical assays confirmed the dimerization of the DNA-kinesin at the bulk level (Figure 1B and C show the data of anti-parallel type DNA-kinesin connected at the C-terminal end of the neck linker (position 337; see Figure 3A). Table I lists the constructs used).
Figure 1.
Structure of DNA-kinesin. (A) A Cys residue was introduced to the surface of the kinesin Cys-Light-Mutant (CLM), after which a fluorescently labeled ssDNA was attached. By hybridizing the two DNA-kinesin monomers, we obtained ‘parallel' or ‘anti-parallel' DNA-kinesin. (B) 10% poly-acrylamide gel electrophoresis (PAGE). S-Cy3, 20 bp Cy3-labeled sense oligo nucleotide; AS-Cy5, 20 bp Cy5-labeled antisense nucleotide; M, Marker. Digestion of DNA-kinesin with restriction enzyme (KpnI) showed that the DNA was correctly hybridized (right lane). DNA was labeled at position 337 (see Figure 3A). (C) Gel filtration column experiments using a wild-type dimer (black line, K490CLM 215), DNA-kinesin heterodimer (red line, 20 bp S-Cy3+20 bp AS-Cy5), DNA-kinesin monomer (blue line, 20 bp AS-Cy5), wild-type monomer (green line, K336CLM 215). Note: we obtained similar results using 6 bp constructs (data not shown).
Table 1.
| Construct | Position | Feature (bias length) | |
|---|---|---|---|
| Figure 1 | K336CLM 337 | 337(C-terminal) | End of C-terminus (full) |
| Figure 2 | K349CLM 342 | 342(C-terminal) | End of C-terminus (full) |
| Figure 3 | K336CLM 337 | 337(C-terminal) | End of C-terminus (full) |
| Figure 4 | K336CLM 337 | 337(C-terminal) | End of C-terminus (full) |
| Figure 6 | K336CLM 2 | 2(N-terminal) | Tip of N-terminus (a,b) |
| K336CLM 7 | 7(N-terminal) | On the side of head a,b) | |
| K336CLM 23 | 23(mid) | Back part of head (none) | |
| K336CLM 43 | 43(mid) | Back part of head (none) | |
| K336CLM 101 | 101(mid) | Dorsal part of head (none) | |
| K336CLM 215 | 215(mid) | Front part of head (none) | |
| K336CLM 324 | 324(C-terminal) | Root of C-terminus (none) | |
| K336CLM 328 | 328(C-terminal) | Midpoint of C-terminus (partial) | |
| K336CLM 333 | 333(C-terminal) | End of C-terminus (almost full) | |
| Figure 7 | K336CLM 328 | 328(C-terminal) | Midpoint of C-terminus (partial) |
| K336CLM 333 | 333(C-terminal) | End of C-terminus (almost full) | |
| K336CLM 337 | 337(C-terminal) | End of C-terminus (full) | |
| Names of the constructs, points of DNA attachment and construct feature are listed. ‘Bias length' is an indicator of effective lever arm length (see Figure 5D). | |||
| aPartial or none. | |||
| bFrom the crystal structure study (PDB entry 1MKJ), the N-terminal of kinesin is known to attach to the neck linker in a docked state. Therefore, the bias length might be partial or none (the latter corresponds to the state without attachment to the neck linker). In Figures 1, 2, 3, 4, 5, 6 and 7B, EMCS, which has six carbon chains, was used as the connection linker between DNA and kinesin. To further explore the effects of the DNA-kinesin connection linker, other bi-functional linkers (AMAS: 2 carbon chains; KMUS: 11 carbon chains) were used in Figure 7D and E. For Figure 8, K336CLM 337 was heterodimerized with the constructs used in Figure 6 (K336CLM 23, 43, 101, 215, 324). | |||
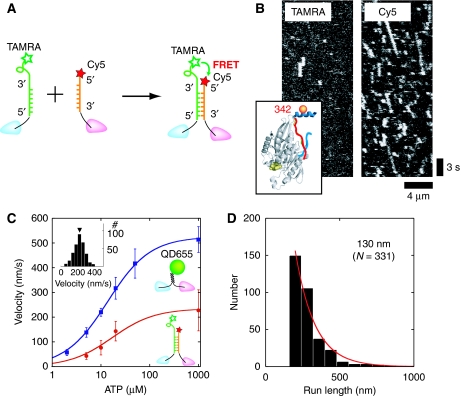
To assess the motile activity of DNA-kinesin at the single molecule level, we first observed the parallel type that replaced the coiled-coil of native kinesin with DNA at connect position 342, the C-terminal end of the neck linker (Figure 2B inset). With the parallel type, two fluorescent dyes (TAMRA and Cy5) were attached on the same side of the DNA at a short distance from each other. Thus, a high fluorescent resonance energy transfer (FRET) signal was expected (Figure 2A). On exciting these DNA-kinesin dimers using a green laser (514 nm), fluorescent spots in the TAMRA channel were rare, whereas motile fluorescent spots were only observed in the Cy5 channel (Figure 2B). The fluorescence intensity of the motile spots was the same as that of a single fluorophore. In addition, anti-correlative, simultaneous recovery of TAMRA fluorescence with photo bleaching of the Cy5 fluorophore was observed (Supplementary Figure S1B). From these results, we concluded that the motile fluorescent spots were those of single molecule DNA-kinesin. The velocity dependence on ATP concentration obeyed Michaelis–Menten kinetics, showing that the motility is an ATP-dependent process (Figure 2C). However, Vmax was 235 nm/s, half the speed of wild-type kinesin (K490CLM 416-Qdot655; 525 nm/s), and the run length (130 nm; Figure 2D) was 1/10 that of wild type (1.3 μm). A biochemical study (Hackney et al, 2003) and a recent single molecule study (Yildiz et al, 2008) have showed that the insertion of polyglycine or polyproline residues into the ‘neck linker–coiled-coil interface', which changes the distance between the two heads, lowers the velocity and decreases the run length. This may also occur in DNA-kinesin, as parallel type constructs have a carbon chain spacer between the DNA and neck linker, which acts as a soft elastic linker (see Figure 7C for details). Another possible explanation for these results is the effect of charged residues. As the charged residues of a coiled-coil (e.g. Lys) greatly contribute to run length (Thorn et al, 2000), DNA-kinesin's short run length might be the result of the construct lacking the coiled-coil region.
Figure 2.
DNA-kinesin can move processively at the single molecule level. (A) Native kinesin coiled-coil was replaced with duplex DNA. DNA was labeled at position 342 (see Figure 2B). On hybridization, a FRET signal was observed. (B) Kymograph obtained by green laser (514 nm) excitation. Owing to the high FRET condition, motile spots appeared only in the Cy5 channel (see Supplementary Figure S1 for details). (C) Velocity of the DNA-kinesin (red circle) followed Michaelis–Menten kinetics, indicating the movement was ATP hydrolysis dependent. However, the Vmax (235 nm/s) was slower than that of wild type (K490CLM 416-Qdot655; 525 nm/s, blue squares). Inset: velocity distribution of DNA-kinesin at 1 mM ATP. (D) Run length (130 nm) was shorter than that of wild type (1300 nm). See main text for details.
Effect of distance between heads
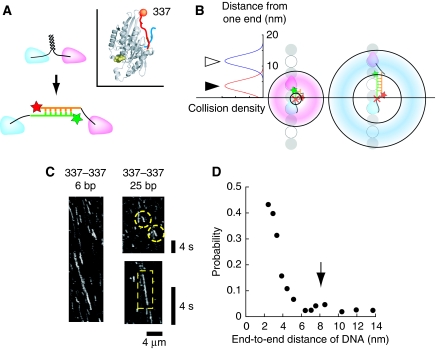
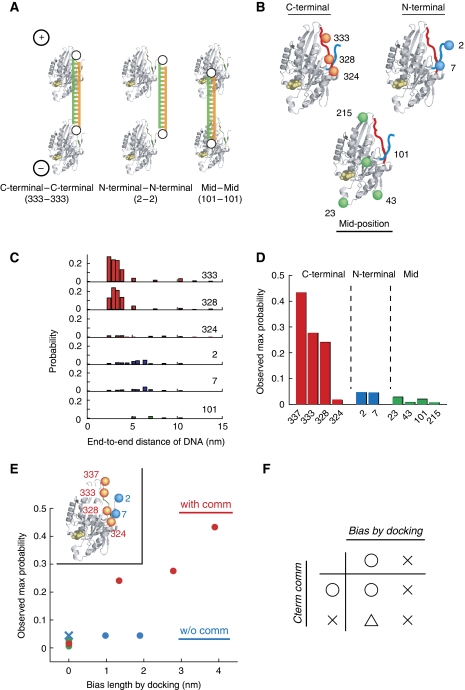
To resolve the mechanism that coordinates the two heads in kinesin walking, we measured the motile activity of anti-parallel type constructs of varying DNA lengths (6–40 bp dsDNA) at the single molecule level. As the inter-head connection linker lengthens, the tense neck linker that is more or less fully extended in native kinesin is expected to relax. Similar approaches have been used previously with contradictory results (Hackney et al, 2003; Yildiz et al, 2008). These reports, however, used protein-only constructs that had soft peptide residues (polyglycine or glycine–serine repeats; persistence length lp=0.8 nm; Sahoo et al, 2006), or semi-rigid polyproline (lp=4.4 nm; Schuler et al, 2005) insertions. These polypeptides, though, cannot be treated as rigid rod, as they behave as elastic springs (see Supplementary Results for details). Furthermore, even for semi-rigid polyproline, many free joints in the constructs exist, including those between the neck linker and inserted polypeptides and those between the polypeptides and coiled-coil. Thus, by increasing the number of inserted peptides, both the mean distance between the heads and the area on the MT the heads can access change. This might cause an increase in the number of futile steps (e.g. side steps or back steps). For less equivocal data, we observed anti-parallel type DNA-kinesin that resembles the layout of an extended neck linker (Figure 3A), but with a rod-like backbone made of DNA resulting in unique characteristics. For example, in DNA-kinesin, the mean distance between the two heads can be changed in 0.34-nm increments by changing the number of nucleotides in the rod-like DNA. As short dsDNA is a rigid rod (lp=50 nm, Bustamante et al, 1994), the area accessible on the MT by the detached head, which corresponds to the width of the doughnut in Figure 3B, is constant (see also Supplementary Figure S8C). Thus we could measure the effect of the distance between the heads more accurately using DNA-kinesin than that of a protein-only construct (see Supplementary Figures S8–S10 for details).
Figure 3.
DNA length dependence of DNA-kinesin movement. (A) Structure of anti-parallel DNA-kinesin. The overall structure is similar to that of the neck linker-extended kinesin mutants. In addition, several connect positions were feasible in DNA-kinesin (see Figure 6 for details). This is in sharp contrast to a protein-only base mutant, in which only N- or C-terminal connections can be achieved. (Inset) DNA-kinesin connected at position 337, which is at the end of the neck linker, is used for Figure 3. To simplify the results, the base construct (K336CLM) is slightly different from that of Figure 2 (K349CLM in which a short coiled-coil part (337–349) exists). (B) As short dsDNA can be treated as a rod, the area accessible on the MT by the detached head is restricted to a doughnut-shaped area (left). The width of the doughnut-shaped area is constant for various DNA lengths (right). Taken together, we can control the area accessible by the detached head. For example, with short DNA the detached head can reach the next binding site (closed arrow head); with long DNA it can reach a binding site a two-step distance away (open arrow head). Note: DNA is rigid in the longitudinal direction, but the carbon linkers between DNA and the head ensure flexibility in the rotational direction. Thus the collision probability of the binding surface of the head is not restricted to the rotational direction (see Supplementary Results). (C) A kymograph of the Cy5 channel showed that anti-parallel DNA-kinesin also walked processively for various DNA lengths. Scale bar for vertical axis=4 s, horizontal axis=4 μm. (right bottom) Enlarged kymograph of 25 bp constructs. Motile molecules are encapsulated by yellow dashes. (D) Motile probability shows dependence on DNA length. Unexpectedly, motile probability at the two-step distance (arrow) is low. Note: the peak was expected at a DNA length of 8 nm (16 nm−length of native neck linker length (8 nm)).
We measured the effect of the distance between the heads with a construct connected at position 337. Here, docking of the neck linker is not disturbed. Thus, the detached head should swing forward the length of the neck linker. Therefore, if the distance between the heads is the same as that of wild type, processive movement of DNA-kinesin should be observed. However, when the DNA length is changed, the motile probability, which is defined as ‘the number of motile molecules/number of molecules attached to MT' and is a rough indicator for run length (see Figure 4B and Supplementary Figure S4B), should decrease as the DNA-kinesin's distance between the heads becomes longer than that of wild type. This is because too long a length lowers accessibility to the next MT binding site that is only 8 nm away. However, further extending the distance between the heads to 16 nm might actually increase accessibility, as the head can now reach the second consecutive binding site (16 nm away). Therefore, a local maximum for the motile probability was expected at a DNA length of 8 nm (=16 nm−original length of the neck linker (8 nm)).
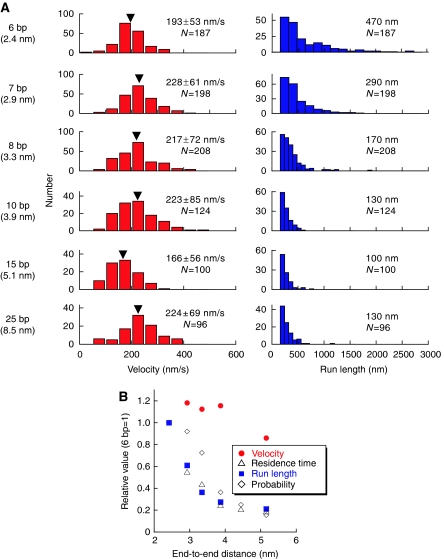
Figure 4.
Motile properties of anti-parallel type DNA-kinesin. Data of the construct connected at position 337 was analyzed more precisely. (A) Velocity (left) and run length (right) profiles show dependence on DNA length. (B) To compare, we plotted the relative values against DNA length. Data was normalized using the data from the 6 bp constructs (2.4 nm). Run length (blue squares), residence time (open triangles; original data not shown) and motile probability (open diamonds; data from Figure 3 of main text) decreased faster than velocity (red circles). To obtain the run length and residence time, data were fitted by a nonlinear least squares fitting of the cumulative probability distribution [C1*(1−exp (−t/C2))−C3 from t=0 to infinity] where C1 is a normalized parameter and C2 is the run length or the residence time. C3 was used to exclude the effect of the counting loss.
After preparation of the 337 construct, we measured the end-to-end distance of DNA. At both DNA ends, one of two fluorophores (Cy3 or Cy5) was attached. As the two fluorophores were close, the FRET signal was expected to depend on the distance between the two. The observed FRET efficiencies of many DNA lengths were similar to those values predicted from the duplex structure (Supplementary Figures S2 and S3), suggesting that we could control the end-to-end DNA distance, as expected. On measuring the motility of DNA-kinesin at variable DNA lengths (Figure 3C), the motile probability was observed to become lower as the DNA became longer, with motile molecules being negligible at a DNA length of 6 nm (=18 bp). Further extension allowed some molecules to take steps equivalent to a two-step distance, although the probability was unexpectedly low (5%, Figure 3D). At present we do not know the reason for this low probability. Recently, Yildiz et al (2008) reported that extending the neck linker length by inserting polyproline allows occasional side steps because of the linker's long reach. These steps are futile for processive movement and decrease the coordination between the heads. This may also occur upon DNA lengthening (see Supplementary Discussion for details).
To evaluate more precisely, we measured the velocity and the run length of each DNA length (Figure 4). Our DNA-kinesin results show that extending the distance between the heads by changing the length of DNA, while keeping the connect position constant (position 337), caused the run length to shorten, but the velocity to remain constant. These results are similar to those of a biochemical study (Hackney et al, 2003), but are in contrast to those from a recent single molecule study (Yildiz et al, 2008). To explain these contradicting results and better resolve the coordination mechanism, we took advantage of the unique characteristics of DNA-kinesin.
Exploring the coordination mechanism
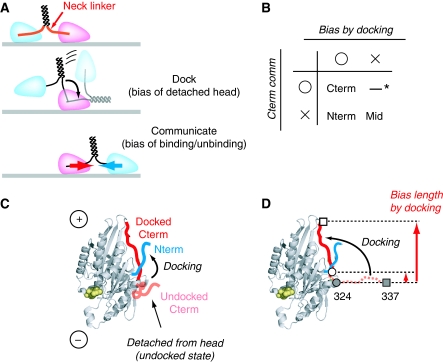
As the neck linker connects the two heads, communication between the heads should be transmitted through the neck linker. However, it is believed that the function of the neck linker (C-terminal linker) is not limited to inter-head communication (hereafter such communication is termed ‘Cterm communication'), but also involved in biasing the detached head forward by docking (Figure 5A). Thus, to evaluate the contribution of the neck linker on the head–head coordination mechanism, we needed to evaluate the power stroke and Cterm communication independently. To do this, we evaluated the effects of both by changing both the connect position and the distance between the heads using constructs with full-length neck linkers. We compared the following four conditions (Figure 5B): (1) both the power stroke and Cterm communication are active, (2) only the power stroke is active, (3) only Cterm communication is active and (4) both mechanisms are inactive. Constructs in which two monomers are connected at the C-terminus (neck linker) correspond to condition 1. A crystal structure study (PDB: 1MKJ) and molecular dynamics simulation (Hwang et al, 2008) showed that in the docked state, the N-terminus attaches to the neck linker through a β-sheet structure (Figure 5C). However, in the undocked state, a crystal structure (PDB: 1BG2) showed no sign of N-terminal residues, suggesting the N-terminus was detached. Thus, constructs connected to N-terminal residues were expected to correspond to condition 2, in which the power stroke arose from docking of the neck linker without Cterm communication. Construct connected at the root of the neck linker (position 324) corresponded to condition 3, where Cterm communication arose from the neck linker, without any forward bias of the detached head despite the power stroke (Figure 5D). Having the connection point at the midway point of the neck linker sequence (positions 333 and 328; see Figure 6A and B) allows the detached head to undergo free diffusion with the connect position acting as a pivot point. Thus, on neck linker docking, amino acids from the root of the neck linker to the connect position function as an effective lever arm, and the detached head is biased forward a distance ranging from the root to the connect position (hereafter, the distance between the root and connect position is referred to as ‘bias length'). So these constructs (333, 328) correspond to constructs that have Cterm communication but less than optimal power strokes making them intermediate constructs between conditions 1 and 3. Mid connected constructs (positions 23, 43, 101 and 215; see Figure 6B) corresponded to condition 4.
Figure 5.
Evaluating the contribution of the power stroke and communication through the neck linker. (A) Dual function of the neck linker has been proposed to facilitate processive runs. (B) To evaluate the contribution of the two functions independently, four conditions were compared. *Connection at the root of the neck linker (position 324; also see Supplementary Results). (C) The neck linker undergoes a nucleotide-dependent conformation change and so attaches to (docking) or detaches from (undocking) the head. (D) The neck linker is thought to act as a lever arm. By connecting the head to the midpoint of the neck linker, we could evaluate the effect of the effective lever arm length (bias length). For example, a small bias length is expected for connections at 324, whereas a large bias length is expected for connection at 337. Closed circle (324) and closed square (337) show the positions for the undocked state; open circle (324) and open square (337) show the positions for the docked state. A docked state crystal structure is shown (PDB entry 1MKJ).
Figure 6.
The C-terminal neck linker sequence has dual function. (A) To resolve the coordination mechanism especially with respect to uncover the contribution of the neck linker on processive movement, we compared several constructs with different connect positions. (B) Connect positions. The amino acid number is equal to that of human kinesin. N- and C-termini are colored blue and red, respectively. Positions in which fluorescent dye labeling had no effect on movement were labeled with DNA. Note: all constructs had a full-length neck linker. (C) Position dependence of movement. Note: the distance between the connect positions is different for different positions, thus the peak position changes. (D) Observed max motile probability for each connect position. Data of position 337 is from Figure 3. Note: displacement analysis showed that N-terminal connect constructs (position 2 and 7) moved unidirectionally, but Mid connect constructs (23–215) moved bi-directionally. Taken together with the mean square displacement (MSD) analysis, we concluded that N-terminal constructs move processively and Mid constructs move by diffusion (see Supplementary Figure S6). (E) Neck linker docking and C-terminal connection are critical for efficient movement. Bias length was calculated from the crystal structure (1MKJ) assuming position 324 as the starting point. ‘comm' in the figure means Cterm communication. Note: from the crystal structure study, the N-terminal of kinesin is known to attach to the neck linker in a docked state. So for the N-terminal (blue symbols), data with (•) or without (x) attachment to the neck linker is plotted. Data from C-terminal and Mid positions are plotted with red and green circles, respectively. (F) Dual functions of the neck linker are crucial for processive movement.
First, to evaluate the effect of the power stroke, we observed condition 1 and 3. If the power stroke is crucial for processive movement, then it is expected that the probability decreases as the connect position approaches the root of the neck linker, as the lever arm shortens. Thus, we observed connect positions 333, 328 and 324, in which fluorescent dye labeling had no effect on ATPase or dimer activity (Rice et al, 1999; Tomishige et al, 2006; Mori et al, 2007). As seen in Figure 6C and D, the motile probability decreased as the connection approached the root of the neck linker. Motile molecules were rarely observed for the root connection construct (connection at 324). These results show that the power stroke is critical for processive runs. Surprisingly, though, the speed of the moving molecule was higher when the connect position was nearer the root (310 nm/s for 328, 193 nm/s for 337; see below).
Having observed that forward bias by the power strokes is necessary for processivity, we then questioned whether Cterm communication was also essential. We addressed this question by observing a construct connected at the N-terminal or Mid positions (conditions 2 and 4). Surprisingly, constructs connected at the tip (2) and head (7) both showed processive movement. However, the motile probability and speed were low even at optimal DNA length. Next, we connected DNA-kinesin at the Mid positions. All observed Mid constructs failed to show processive movement but did show diffusional movement (Supplementary Figures S5 and S6). These results indicate that for efficient processive movement, both the power stroke of the neck linker and connection through the neck linker are crucial (Figure 6E and F). Therefore, taking advantage of the DNA-kinesin, we further evaluated the neck linker-connecting constructs (C-C, Figure 6A left).
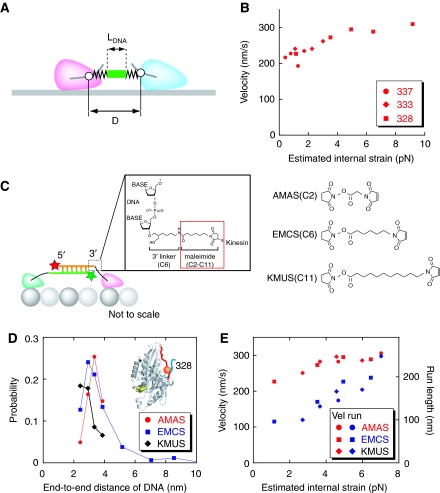
The neck linker-connecting constructs (positions 328, 333, 337) show opposite correlations for run length (motile probability) and velocity on bias length, and of these, constructs 328 and 337 had the shortest and longest run lengths, respectively, but had the fastest and slowest velocities, respectively (compare Figure 4 and Supplementary Figure S4). To understand these differences, we consider the fact that the estimated internal strain is different for each connect position. Internal strain is thought to be a key parameter for head–head coordination regardless of the mechanism (Hackney et al, 2003; Yildiz et al, 2008). Furthermore, differences in connect position mean differences in internal strain, as the number of free neck linker amino acids, which do not bind to the head and act as a spring, is different for each connect position (see Supplementary Results for details). As bias length affects run length (compare Figure 4A and Supplementary Figure S4A) making it difficult to investigate the internal strain effect on many constructs of different connection points, we first evaluated the velocity data from many connection points (position 328, 333 and 337). Results show that internal strain affects velocity slightly (Figure 7B). To compare the varying dependencies for run length and velocity on internal strain, we took advantage of DNA-kinesin, whose distance between the heads and also the spring character between heads can be independently changed by changing the DNA length and linker length, respectively. So we explored the effect of both parameters at a fixed position. We chose position 328, the halfway position of the neck linker to minimize the effect of free amino acids from the neck linker that do not bind to the head, and thus maximized the effect of the three different carbon chains: AMAS, EMCS and KMUS (Figure 7C). The collision frequency was the same for all three (Supplementary Figure S11D). This is expected as the end-to-end distance of the DNA mainly determines the collision frequency in DNA-kinesin. However, the estimated internal strain is different, and the internal strain ratio between AMAS and KMUS ranged between 1.5 and 3 (Supplementary Figure S11E). The profiles of the motile probability differ between the chains, although the peak values for motile probability were similar (Figure 7D). The shift in peak position is reasonable because a longer carbon chain spacer length compensates for the DNA length. Figure 7E shows the estimated internal force dependence of run length and velocity. The run length depends more on internal strain than velocity does. These results suggest that internal strain acts mainly on head–head coordination to prevent simultaneous detachment of both heads from MT rather than on ATPase activity (see Discussion section).
Figure 7.
Internal strain affects run length more than velocity. (A) Parameters for calculating internal strain. D, distance between two connect positions, LDNA, length of DNA (end-to-end distance). See Materials and methods section for details. (B) Velocity depends on the estimated internal strain. Data for connect positions 328, 333 and 337 were shown (carbon spacer is EMCS; see 7C). Note: the estimated value of the internal strain depends on the model, but we assume that the qualitative trend is independent of the model. (C) To compare the different dependencies of run length and velocity on internal strain, we changed the carbon chain spacers, which connect DNA and kinesin head (see text for details). We used three spacers: AMAS, EMCS and KMUS. (D) Data of the three carbon spacers are shown (connect position is 328). Motile probability of different constructs showed different peak positions. This result, namely that a short linker needs longer DNA and a long linker needs shorter DNA, is reasonable. (E) The internal strain affects run length more than velocity. Note: to simplify the interpretation, only DNA longer than the optimal motile length were plotted (data of some short DNA conditions (6 and 7 bp for AMAS, 6 bp for EMCS) were not plotted).
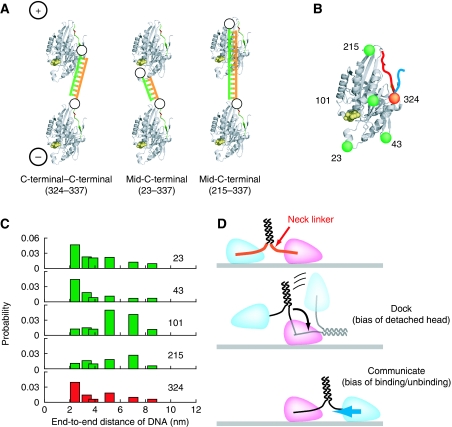
Next, we sought the structural basis for head–head coordination. Figure 6E shows the importance of Cterm communication. However, N-terminal connect constructs also moved processively, suggesting that another connect position might be able to sense internal strain to ensure processive movement. Figure 6E also shows the essential role of the forward bias produced by the docking of the neck linker. These characteristics meant that only constructs connected at the C- or N-terminal could be studied as homodimers. To evaluate the other position, we took advantage of DNA-kinesin, which allowed us to make heterodimers with various connect positions (23, 43, 101, 215, 324), whereas the other head was constantly connected at position 337 for full bias in the power stroke (Figure 8A and B). Heterodimer constructs connected at the rear of the head (23) or root of the neck linker (324) could move processively (Figure 8C). These results show that kinesin sense their internal strain through a specific domain rather than the whole head (Figure 8D).
Figure 8.
Applying strain on the back domain of the head is the origin of the head–head coordination. (A) To resolve the origin of the coordination, we compared several constructs with different connect positions. To do this, two monomers at different connect positions were dimerized: one head (head A) is connected at the location of interest while the other head (head B) is connected at the end of neck linker to achieve forward bias. (B) Connect positions. (C) Position dependence of the movement. Note: From displacement and MSD analysis, we concluded that 23 and 324 constructs move processively (see Supplementary Figure S7). (D) Dual function of the neck linker is crucial for processive movement, as internal strain applied to the rear of the leading head prevents simultaneous detachment of both heads from the MT.
Discussion
Kinesin hydrolyzes ATP to walk along MTs. Despite vast amounts of research, the coordination mechanism of the two heads to ensure processive movement has not yet been elucidated. Here we constructed a novel approach by designing DNA-kinesin to show that the power stroke of the neck linker and the head–head coordination through the neck linker are essential for efficient processive movement. Our data also suggest that communication between the two heads arises from the internal strain applied to the back domain of the head, and ensures to inhibit simultaneous detachment of both heads from the MT. Thus, the head–head coordination through the neck linker facilitates long-distance walking.
Regarding processive movement, it was not necessary to connect DNA in a manner that replicated wild-type kinesin. In other words, we could connect DNA to kinesin at points other than the C-terminal end of the neck linker, as constructs connected at the halfway residue of the neck linker and N-terminal residue also showed processive movement. These results begged the question, is it possible to walk processively without using a power stroke? It has been reported that single-headed kinesin (KIF1A) can move forward using a biased diffusion process, although very special conditions are needed (Okada and Hirokawa, 1999; Okada et al, 2003). Moreover, optical trap experiments showed that during the stepping motion of kinesin, the detached head spent most of its time diffusing (Nishiyama et al, 2001, 2002; Carter and Cross, 2005). If kinesin can walk without a power stroke (i.e. by simple biased diffusion), there should exist an optimal distance between the two heads for every construct. However, our results showed that only connecting at the C- or N-terminus could power unidirectional processive movement, although the motile probability was very low for N-terminal constructs. As for the Mid positions, we found no conditions that resulted in efficient processive movement, as only diffusional movement was observed (Figure 6C and Supplementary Figure S5). For the C-terminal (neck linker) construct, the motile probability decreased as the connection point approached the root of the neck linker in which motile molecules were rarely observed (Figure 6D and E). These results clearly show that it is impossible to walk efficiently without a power stroke and support the critical role of the power stroke in processive movement. Furthermore, C-terminal connection showed much higher motile probability compared with constructs connected at other places, suggesting that communication through the neck linker is also important for processive movement.
The reported effect of impairing inter-head coordination on kinesin motility has been controversial as a biochemical study showed a reduction in internal strain mainly affects run length, whereas a recent single molecule study showed it mainly affects velocity. Our DNA-kinesin results show that extending the distance between heads by changing the length of DNA caused the velocity to slow down and the run length to shorten. However, the effect on velocity was slight (Figure 4B and Supplementary Figure S4B). Furthermore, by changing the carbon chains between the DNA and head, we changed only the spring character between the heads while keeping the mean distance between them the same. Results also show that internal strain mainly influences run length (Figure 7E). Altogether, these results support the reported biochemical results (Hackney et al, 2003). We speculate that the discrepancies with the single molecule study (Yildiz et al, 2008) are due to differences in the constructs. Regarding run length, we and Yildiz et al (2008) used constructs derivatives from the same original construct (K560CLM), but they inserted two lysine residues per head (thus 4 lysines per dimer). As the plus charge of a lysine residue is known to increase the run length greatly (Okada and Hirokawa, 1999; Thorn et al, 2000; Tomishige et al, 2002), the effect of the lysine insertion might mask the effect of the neck linker extension when run length decreases. Indeed, our results using constructs (polyglycine extension mutants; Gn mutants in Supplementary Figure S12C) similar to their GS mutant (extension of the neck linker by inserting glycine–serine repeats) caused a decrease in run length (data not shown). Regarding velocity, the constructs used by Yildiz et al (2008) had several free joints at the junction of the coiled-coil and neck linker meaning the head can potentially bind to various sites along the MT, including rebinding to its original position or taking side steps, both of which would decrease velocity (Supplementary Figure S13E). In fact, the calculated randomness factor, which is an indicator of the tight coupling between ATP hydrolysis and the stepping motion, showed a decrease in tight coupling, suggesting futile steps. Furthermore, they also showed an increase in side steps with their neck linker extension mutants. However, in our DNA-kinesin, the two heads were connected by a rigid rod through a spring (Supplementary Figure S8B). Thus we could independently evaluate the effect of the distance between the heads and the effect of the spring (see Figure 7).
Our results suggest that the internal strain may act mainly on head–head coordination rather than on ATPase activity. This idea is supported by biochemical studies. The kcat of a monomer, which has no internal strain, is similar to that of the dimer (Jiang et al, 1997). Furthermore, ADP release does not change between monomer and dimer (Rosenfeld et al, 2003). Taken together, we hypothesize that the internal strain contributes mainly to regulating the head–head coordination, but not to the activity of the head. So then, what kind of mechanism governs the coordination mechanism? Several gating mechanisms have been proposed, with two being most widely accepted. One is the mechanical gate model, in which the power stroke of the leading head accelerates detachment of the trailing head (Hancock and Howard, 1999); the other is the chemical gate model, in which ATP binding of the leading head is inhibited until the trailing head detaches meaning only the trailing head can become weak binding and subsequently detach from MT (Rosenfeld et al, 2003; Klumpp et al, 2004). We prefer the chemical gate model for several reasons. First, if the mechanical gate model is dominant, then reducing internal strain should increase the number of futile steps resulting in a severe decrease in velocity like that observed by Yildiz et al (2008). However, the velocity of our DNA-kinesin decreased only slightly with internal strain, whereas a much greater effect was seen on run length. Second, if the mechanical gate mechanism, which assumes that the leading head's power stroke accelerates the trailing head's detachment, is true, then the trailing head remains attached to the MT (thus keeping the two-head bound state) until ATP binding and subsequent power stroke of the leading head occurs. It is known, though, that kinesin takes the one-head bound state at low ATP concentration (Hackney, 1994; Hackney et al, 2003; Alonso et al, 2007; Mori et al, 2007; Guydosh and Block, 2009), and that the detached head is very mobile (Asenjo and Sosa, 2009). Furthermore, at saturating ATP concentration, kinesin spends most of its time in the two-head bound state (Asenjo et al, 2003; Mori et al, 2007). Thus, the mechanical gate mechanism may contribute to the ATP hydrolysis process. However, our high-speed smFRET (single molecule fluorescence resonance energy transfer method; up to 1 ms time resolution) experiment found a constant dwell time (15 ms) for the two-head bound state between 4 μM and 1 mM ATP (Kikuchi, Mori and Tadakuma, in preparation), suggesting detachment is not accelerated by the power stroke of the leading head.
Our results do not, however, exclude acceleration of the trailing head's detachment in response to inter-head strain, which arises from a stretched neck linker when in the two-head bound state (without docking by the leading head's neck linker). Optical trap experiments showed directional asymmetry for ADP affinity of the kinesin head (Uemura et al, 2002; Uemura and Ishiwata, 2003), which might contribute to the trailing head's ADP hold by the forward pointing position of the neck linker during the two-head bound state. Inhibition of nucleotide release from the trailing head by internal strain might also contribute to prevent ADP release from the detached head if it rebinds to its original position. This was seen when cross-linking the neck linker to a head fixed in the docking state, in which ADP release was blocked (Hahlen et al, 2006). However, none of these studies could conclude whether leading head docking accelerated trailing head detachment. In this study, however, we presented a novel approach to explore the contribution of the connect position and of the distance between the heads. Future analysis of the stepping behavior by optical trap and FIONA (fluorescence in one nanometer accuracy) or real time observations of ATP binding and of conformational changes within the head by smFRET will help reveal the mechanism of internal strain to coordinate the heads.
A clue for the structural basis behind head–head coordination was suggested from our heterodimer results (Figure 8). Some constructs (23–337 and 324–337) showed processive movement, suggesting that internal strain may act through the back part of the head. Recently, Yildiz et al (2008) showed that external force can achieve a stepping motion with a neck linker-replaced mutant, interpreting this to mean internal strain acts on the head directly. Our results are consistent with their finding and further indicate that a specific domain senses the internal strain. Collectively, DNA-kinesin results indicate that the activity of a heterodimer can be modulated by changing the length of exogenously bound DNA. Using the power of DNA-kinesin, further studies on various heterodimers will provide insight into the coordination mechanism between heads and molecules (such as multiple kinesin (Eg5), dynein–kinesin and NCD–kinesin (Eg5)).
In this study, we used the power of DNA-kinesin to evaluate the effects of the connect position and the distance between connecting points. Our results suggest that in native kinesin, the bias length by the neck linker is maximal and, as the distance between the heads nears the contour length of the neck linker, the internal strain is also maximum. Thus, the structure of native kinesin is optimized for efficient movement indicating native kinesin is a well-designed nanomachine. Our approach using DNA as the skeletal structure and protein as the functional module, is not limited to kinesin. Furthermore, in the future, with the combination of recent DNA nanotechnology (Seeman, 2003; Rothemund, 2006), we expect studies like this will facilitate the fabrication of nanomachines.
Materials and methods
DNA cloning and protein purification
Cysteines (A2C, S7C, V23C, S43C, D101C, E215C, T324C, T328C, V333C, A337C and K342C) were introduced into a cysteine-light human ubiquitous kinesin 336-residue monomer (Figures 1 and 3, 4, 5, 6, 7, 8), 349-residue monomer (Figure 2) or a dimer of 490-residue subunits, each containing a C-terminal His6 tag (Rice et al, 1999; Tomishige et al, 2006). All constructs were verified by DNA sequencing. Monomeric and dimeric kinesins were expressed and purified as previously described (Rice et al, 1999; Tomishige et al, 2006; Mori et al, 2007). The activities of the new constructs (V23C and D101C) were confirmed by a Qdot655 -labeled (mutant/WT) hetero-dimer construct (Qdot; Invitrogen) (Tomishige et al, 2006; K490CLM V23C/WT: Vmax=510 nm/s, Km=17 μM; K490CLM D101C/WT: Vmax=496 nm/s, Km=18 μM; where control K490CLM 215/WT had Vmax=517 nm/s, Km=21 μM and control K490CLM 416/WT had Vmax=525 nm/s, Km=14 μM). The size of the streptavidin-coated Qdot was 30 nm according to the manufacturer.
Construction of DNA-kinesin
5′ and 3′ modified oligo DNAs were purchased from Sigma (Supplementary Table S1). For parallel type DNA-kinesin, sense oligo (5′ amino group (NH2) attached; 3′ TAMRA attached) and antisense oligo (5′ Cy5; 3′ NH2) were used. For anti-parallel type DNA-kinesin, sense oligo (5′ Cy3; 3′ NH2) and antisense oligo (5′ Cy5; 3′ NH2) were used. Amino groups of the oligo DNA were covalently reacted with the succinimidyl ester of the bi-functional linker (EMCS; Dojindo; for Figure 7D and E AMAS (Pierce) and KMUS (Dojindo) were also used), the opposite end of which was a thiol-reactive maleimide group. The reaction was quenched by gel filtration (NAP-5, GE) of the reaction solution. The peak fraction was used for kinesin labeling. Contamination of the unreacted bi-functional linker was low, whereas the reactivity of the obtained modified DNAs was similar to that purified by reverse-phase column chromatography (data not shown). Attachment of DNA to kinesin was performed by mixing the modified DNA and kinesins. DNA-labeled kinesins were then separated from the unreacted DNA by a gel filtration column (CHROMA SPIN+TE-30; BD). The obtained monomer DNA-kinesin was stored at −80°C until use. The dimer DNA-kinesin was obtained by mixing two DNA-kinesins monomers in Mg10 Buffer (12 mM PIPES (pH 6.8), 10 mM MgCl2 and 1 mM EGTA) for 10 min at 25°C. The reaction solution was further diluted by Mg5 Buffer (12 mM PIPES (pH 6.8), 5 mM MgCl2 and 1 mM EGTA), infused into the observation chamber and imaged by fluorescent microscopy. To confirm dimerization of DNA-kinesin, gel filtration chromatography was performed for anti-parallel type constructs (K336CLM 337C_20 bp) with a Superdex 200 10/300 GL column (GE) equipped with AKTA explore (GE) and a detection unit (FP2025 Plus, JASCO). Cy5 fluorescence (at 665 nm) was detected using HKM buffer (25 mM HEPES (pH7.4), 100 mM KCl, 5 mM MgCl2+0.1 mM ATP+BSA 0.1 mg/ml). To further confirm dimerization, 10% poly-acrylamide gel electrophoresis (PAGE) was performed with or without KpnI digestion.
Single-molecule fluorescence microscopy
Single-molecule FRET images were visualized by a total internal reflection fluorescence microscope equipped on an inverted type microscope (IX70 or IX71; Olympus), as previously described (Funatsu et al, 1995; Taguchi et al, 2001; Mori et al, 2007). Dye-labeled heterodimeric kinesins (0.5–2 nM) were attached onto the axonemes (purified from sea urchin sperm flagella) in the presence of 1 mM AMP-PNP, or moved along the axonemes in the presence of ATP and an ATP-regenerating system as described (Mori et al, 2007). Cy3 and Cy5 dyes were illuminated with an argon laser (514 nm; 35LAP321; Melles Griot) and a diode laser (635 nm; Radius 635 or Cube 635; Coherent), respectively (up to 10 mW laser power). For high FRET constructs, only the green laser was used as an excitation laser. For mid-low FRET constructs, both green and red lasers were used. Fluorescence images from Cy3 and Cy5 (or FRET) were separated by using a Dual-View (Optical Insights) and then projected side-by-side onto an electron-multiplying charge coupled device camera (iXon DV860 DCS-BV; Andor). Images were taken at a frame rate of 30 frames per s.
Data analysis
Images were analyzed using Image J (http://rsb.info.nih.gov/ij/) with custom-designed plug-in software. The position of the fluorescent spots was determined by eye, a centroid, or by a 2D Gaussian function-fitting algorithm (Press et al, 1992; Tadakuma et al, 2006). To calculate the motile probability, molecules that remained bound to the axoneme for ⩾5 frames (166 ms) in the kymograph images were analyzed. Dwell time analysis of the DNA-kinesin monomers (K336 333C_10 bp monomer, 170 ms) showed that about 40% of the attached molecules were measured under our conditions (data not shown). Furthermore, we set two pixels (=160 nm) as the threshold to judge movement. Thus, the exact calculating formula for the motile probability was as follows:
where NMove is the # of molecules that moved ⩾2 pixels, NTotal the # of molecules that remained bound to the axoneme for ⩾5 frames.
From the analysis of DNA-kinesin monomers (333C_10 bp monomer), the lower limit of the motile probability was found to be 0.0025 (=1 motile molecule/400 attached molecules). Some spots showed a sudden disruption in their trace. We removed these molecules, which jumped ⩾1 pixel within a single frame (33 ms), from the analysis.
Estimate of internal strain
We calculated the internal strain assuming the WLC (Worm Like Chain) model (Figure 7A).
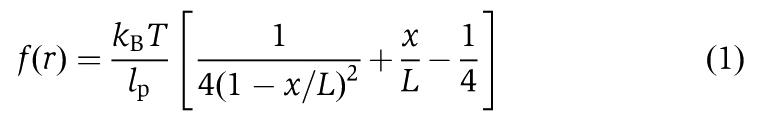 |
where, kB is the Boltzmann constant, T the temperature, lp the persistence length, D the distance between two connect positions, L the contour length, and x=D−LDNA where LDNA is length of DNA (end-to-end distance).
Equation (1) shows that the estimated internal strain depends on the value of lp. Many values of lp have been reported, all depending on the composition of the amino acids: 0.8, 1.4 and 4.4 nm for glycine–serine repeats, the neck linker and polyproline, respectively. DNA-kinesin has an amino-acid component and a carbon chain component, thus estimates should be done carefully. However in this paper, we assumed that lp is constant (0.8 nm) for all DNA-kinesin constructs to simplify the qualitative evaluation (see also Supplementary Results).
Supplementary Material
Supplementary Materials and Methods
Supplementary Figures and Table
Review Process File
Acknowledgments
We thank R Vale for permission to use the kinesin construct; M Tomishige, T Funatsu and H Taguchi for approval to use lab equipment and microscopy; M Nakajima for construction of the mutant kinesin; C Shingyoji and I Nakano for the axoneme; T Yagi and R Kamiya for the microtubule; K Kikuchi and H Tsurusawa for discussion; K Hirose for comments on the structure; S Ishiwata, K Shiroguchi, T Ueno, R Iizuka, Y Ishihama and Y Oguchi for reading the paper and for useful comments.
Author contribution: HT, YM, MH and YH designed experiments. YM and MH developed the method of constructing DNA-kinesin. YM performed the experiments and data analysis. HT wrote the paper with contribution from PK.
References
- Alonso MC, Drummond DR, Kain S, Hoeng J, Amos L, Cross RA (2007) An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science 316: 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL, Fehr AN, Block SM (2003) Kinesin moves by an asymmetric hand-overhand mechanism. Science 302: 2130–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Krohn N, Sosa H (2003) Configuration of the two kinesin motor domains during ATP hydrolysis. Nat Struct Biol 10: 836–842 [DOI] [PubMed] [Google Scholar]
- Asenjo AB, Sosa H (2009) A mobile kinesin-head intermediate during the ATP-waiting state. Proc Natl Acad Sci USA 106: 5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM (2007) Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J 92: 2986–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Marko JF, Siggia ED, Smith S (1994) Entropic elasticity of lambda-phage DNA. Science 265: 1599–1600 [DOI] [PubMed] [Google Scholar]
- Carter NJ, Cross RA (2005) Mechanics of the kinesin step. Nature 435: 308–312 [DOI] [PubMed] [Google Scholar]
- Case RB, Rice S, Hart CL, Ly B, Vale RD (2000) Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr Biol 10: 157–160 [DOI] [PubMed] [Google Scholar]
- Dunn AR, Spudich JA (2007) Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol 14: 246–248 [DOI] [PubMed] [Google Scholar]
- Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T (1995) Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature 374: 555–559 [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Block SM (2009) Direct observation of the binding sate of the kinesin head to the microtubule. Nature 461: 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA 91: 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (2007) Biochemistry. Processive motor movement. Science 316: 58–59 [DOI] [PubMed] [Google Scholar]
- Hackney DD, Stock MF, Moore J, Patterson RA (2003) Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry 42: 12011–12018 [DOI] [PubMed] [Google Scholar]
- Hahlen K, Ebbing B, Reinders J, Mergler J, Sickmann A, Woehlke G (2006) Feedback of the kinesin-1 neck-linker position on the catalytic site. J Biol Chem 281: 18868–18877 [DOI] [PubMed] [Google Scholar]
- Hancock WO, Howard J (1999) Kinesin's processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc Natl Acad Sci USA 96: 13147–13152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R (2005) Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci 6: 201–214 [DOI] [PubMed] [Google Scholar]
- Huxley AF, Simmons RM (1971) Proposed mechanism of force generation in striated muscle. Nature 233: 533–538 [DOI] [PubMed] [Google Scholar]
- Huxley HE (1969) The mechanism of muscular contraction. Science 164: 1356–1365 [PubMed] [Google Scholar]
- Hwang W, Lang MJ, Karplus M (2008) Force generation in kinesin hinges on cover-neck bundle formation. Structure 16: 62–71 [DOI] [PubMed] [Google Scholar]
- Jiang W, Stock MF, Li X, Hackney DD (1997) Influence of the kinesin neck domain on dimerization and ATPase kinetics. J Biol Chem 272: 7626–7632 [DOI] [PubMed] [Google Scholar]
- Kaseda K, Higuchi H, Hirose K (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol 5: 1079–1082 [DOI] [PubMed] [Google Scholar]
- Klumpp LM, Hoenger A, Gilbert SP (2004) Kinesin's second step. Proc Natl Acad Sci USA 101: 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew-Fenn RS, Das R, Harbury PA (2008) Remeasuring the double helix. Science 322: 446–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Vale RD, Tomishige M (2007) How kinesin waits between steps. Nature 450: 750–754 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Higuchi H, Yanagida T (2002) Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat Cell Biol 4: 790–797 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Muto E, Inoue Y, Yanagida T, Higuchi H (2001) Substeps within the 8-nm step of the ATPase cycle of single kinesin molecules. Nat Cell Biol 3: 425–428 [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N (1999) A processive single-headed motor: kinesin superfamily protein KIF1A. Science 283: 1152–1157 [DOI] [PubMed] [Google Scholar]
- Okada Y, Higuchi H, Hirokawa N (2003) Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature 424: 574–577 [DOI] [PubMed] [Google Scholar]
- Press W, Teukolsky SA, Vetterling WV, Flannery BP (1992) Numerical Recipes in C San Diego: Cambridge University Press [Google Scholar]
- Purcell TJ, Morris C, Spudich JA, Sweeney HL (2002) Role of the lever arm in the processive stepping of myosin V. Proc Natl Acad Sci USA 99: 14159–14164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD (1999) A structural change in the kinesin motor protein that drives motility. Nature 402: 778–784 [DOI] [PubMed] [Google Scholar]
- Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM (2003) How kinesin uses internal strain to walk processively. J Biol Chem 278: 18550–18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund PW (2006) Folding DNA to create nanoscale shapes and patterns. Nature 440: 297–302 [DOI] [PubMed] [Google Scholar]
- Sahoo H, Roccatano D, Zacharias M, Nau WM (2006) Distance distributions of short polypeptides recovered by fluorescence resonance energy transfer in the 10 A domain. J Am Chem Soc 128: 8118–8119 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Wang F, Schmitz S, Xu Y, Xu Q, Molloy JE, Veigel C, Sellers JR (2003) Neck length and processivity of myosin V. J Biol Chem 278: 29201–29207 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Yildez A, Selvin PR, Sellers JR (2005) Step-size is determined by neck length in myosin V. Biochemistry 44: 16203–16210 [DOI] [PubMed] [Google Scholar]
- Schief WR, Howard J (2001) Conformational changes during kinesin motility. Curr Opin Cell Biol 13: 19–28 [DOI] [PubMed] [Google Scholar]
- Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA (2005) Polyproline and the ‘spectroscopic ruler' revisited with single-molecule fluorescence. Proc Natl Acad Sci USA 102: 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC (2003) DNA in a material world. Nature 421: 427–431 [DOI] [PubMed] [Google Scholar]
- Shiroguchi K, Kinosita K Jr (2007) Myosin V walks by lever action and Brownian motion. Science 316: 1208–1212 [DOI] [PubMed] [Google Scholar]
- Spudich JA (1994) How molecular motors work. Nature 372: 515–518 [DOI] [PubMed] [Google Scholar]
- Spudich JA (2001) The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol 2: 387–392 [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365: 721–727 [DOI] [PubMed] [Google Scholar]
- Tadakuma H, Ishihama Y, Shibuya T, Tani T, Funatsu T (2006) Imaging of single mRNA molecules moving within a living cell nucleus. Biochem Biophys Res Commun 344: 772–779 [DOI] [PubMed] [Google Scholar]
- Taguchi H, Ueno T, Tadakuma H, Yoshida M, Funatsu T (2001) Single-molecule observation of protein–protein interactions in the chaperonin system. Nat Biotechnol 19: 861–865 [DOI] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD (2000) Engineering the processive run length of the kinesin motor. J Cell Biol 151: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, Vale RD (2002) Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297: 2263–2267 [DOI] [PubMed] [Google Scholar]
- Tomishige M, Stuurman N, Vale RD (2006) Single-molecule observations of neck linker conformational changes in the kinesin motor protein. Nat Struct Mol Biol 13: 887–894 [DOI] [PubMed] [Google Scholar]
- Tomishige M, Vale RD (2000) Controlling kinesin by reversible disulfide cross-linking: identifying the motility-producing conformational change. J Cell Biol 151: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Ishiwata S (2003) Loading direction regulates the affinity of ADP for kinesin. Nat Struct Biol 10: 308–311 [DOI] [PubMed] [Google Scholar]
- Uemura S, Kawaguchi K, Yajima J, Edamatsu M, Toyoshima YY, Ishiwata S (2002) Kinesin-microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci USA 99: 5977–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T (1996) Direct observation of single kinesin molecules moving along microtubules. Nature 380: 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Milligan RA (2000) The way things move: looking under the hood of molecular motor proteins. Science 288: 88–95 [DOI] [PubMed] [Google Scholar]
- Wang MD, Yin H, Landick R, Gelles J, Block SM (1997) Stretching DNA with optical tweezers. Biophys J 72: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Gennerich A, Vale RD (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR (2004) Kinesin walks hand-over-hand. Science 303: 676–678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Supplementary Figures and Table
Review Process File