Abstract
Genetic differences both between individuals and populations are studied for their evolutionary relevance and for their potential medical applications. Most of the genetic differentiation among populations are caused by random drift that should affect all loci across the genome in a similar manner. When a locus shows extraordinary high or low levels of population differentiation, this may be interpreted as evidence for natural selection. The most used measure of population differentiation was devised by Wright and is known as fixation index, or FST. We performed a genome-wide estimation of FST on about 4 millions of SNPs from HapMap project data. We demonstrated a heterogeneous distribution of FST values between autosomes and heterochromosomes. When we compared the FST values obtained in this study with another evolutionary measure obtained by comparative interspecific approach, we found that genes under positive selection appeared to show low levels of population differentiation. We applied a gene set approach, widely used for microarray data analysis, to detect functional pathways under selection. We found that one pathway related to antigen processing and presentation showed low levels of FST, while several pathways related to cell signalling, growth and morphogenesis showed high FST values. Finally, we detected a signature of selection within genes associated with human complex diseases. These results can help to identify which process occurred during human evolution and adaptation to different environments. They also support the hypothesis that common diseases could have a genetic background shaped by human evolution.
Introduction
Genetic differences are present in humans at both individual and population level. Human genetic variations are studied for their evolutionary relevance and for their potential medical applications. This studies can help scientists in understanding ancient human population migrations as well as how selective forces act on the human being [1], [2].
According to the theory of neutral variation, most of the genetic variability within species are caused by random drift of selectively neutral polymorphic alleles [3]. Genetic drift should affect all loci across the genome in a similar manner. Therefore, when a locus shows extraordinary high or low levels of variability this may be interpreted as evidence for natural selection [4]. High levels of population differentiation can suggest the acting of a positive selection of advantageous alleles in one or more populations. On the contrary, lower levels of population differentiation can be considered as the effect of balancing selection that tends to maintain a constant proportion of alleles across all populations [5].
Population differentiation is sensitive to a variety of demographic factors (including the rate of drift within populations and the extent of gene flow among them), making it difficult to rule out demographic scenarios that could account for the observed variations. Another class of tests is aimed to detect signature of natural selection by comparing data from different species. These tests explore the fact that mutations can be synonymous and non synonymous, and that non-synonymous mutations are more likely to have an effect on individual fitness. This method is also known as dN/dS. Results obtained by this comparative approach are rarely interpreted in terms of population genetics theory [6].
The human population is also not homogeneous in terms of disease susceptibility. Risks of common diseases are substantially different among ethnic groups [7]. The understanding of population genetic differentiation, especially in genes associated with diseases, can help to explain the observed variations in the prevalence of diseases. It is not difficult to forecast that, in the future, genetic structure of populations can be used in public health management [8]. Moreover, natural selection on genes that underlie human disease susceptibility has been invoked. In this framework, ancestral alleles reflect ancient adaptation. With the shift in the environment, these alleles increase the risk for common diseases [9].
Different strategies to quantify the population genetic differentiation have been elaborated [10]–[16]. One of the most used is a measure devised by Wright and known as fixation index, or FST [17], [18], which is the amount of genetic variation among groups relative to a panmictic state. As a test of selection, observed FST values are compared to those expected under neutrality. The main difficulty of this approach is to determine the distribution of FST values under neutrality [10]. Recently, however, the abundance of genetic data available allows the creation of an empirical genome-wide distribution to be used for the comparisons. Rather than statistically testing specific loci, we can use their position relative to this distribution to gain insights about their selective histories. In addition, the abundance of information about variability of many genes makes it possible to analyze not only single genes, but also sets of functionally related genes. International HapMap Project [19] by supplying data of a large number of Single Nucleotide Polymorphisms (SNPs) across many human populations, is providing an exceptional tool for studying the genetic structure of human populations.
In the present article we report the results of a genome-wide estimation of FST on 3,917,301 SNPs from the latest release of HapMap data. Our results show a heterogeneous distribution of FST values among genomic regions. Furthermore, we studied the relationship between FST and an evolutionary measure obtained by a comparative interspecific approach. We applied a gene set approach, widely used for microarray data, to detect biochemical pathways under selection. Finally, we detected a signature of selection within genes associated with complex diseases.
Results
Using FST, we estimated populations differentiation for 3,917,301 SNPs in population samples from the International HapMap Project data (Public release 27, merged II + III). To retain the largest number of SNPs broadly reflecting a continental subdivision, we used data from Yoruba (Africa), Japanese (Asia), Han Chinese (Asia) and CEPH (European descendant) individuals. Combining data from these populations we were able to compare the largest set of genotyped SNPs up to now available. We pooled Japanese and Han Chinese samples due to their geographical closeness. Furthermore, this pooling allowed us to compare our data with previous studies [20], [11]. FST was estimated according to Weir and Cockerham [18], [21].
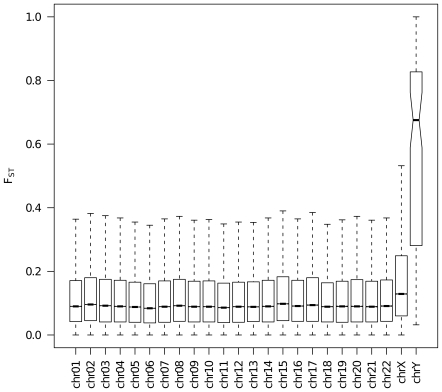
After exclusion for Minor Allele Frequency (MAF), we obtained a final SNP sample of 2,125,440 SNPs. The mean FST was 0.122 (SE = 5×10−5, median = 0.091, interquartile range = 0.131; see Supporting Information S1 for more detailed statistics). Figure 1 shows distribution of FST values for each chromosome. The median FST values of SNPs on the autosomal and sexual chromosomes were statistically different (Kruskal-Wallis test, p-value <10−16). The median FST values for X and Y chromosomes were 0.129 (mean = 0.174) and 0.676 (mean = 0.606) respectively and were notably higher than those of autosomal chromosomes. Also medians between autosomal chromosomes showed significant differences, but in a very small range of values (median range = 0.084 to 0.098).
Figure 1. Distribution of FST values across chromosomes.
For each chromosome, the box length is the interquartile range while the horizontal line inside it is the value of the median. The whiskers extend to the most extreme data point <1.5 times the interquartile range from the box. Extremes of the notches represents 95% confidence interval of the median.
For each chromosome, we computed the correlations of all pairs of FST values for neighbouring SNPs separated by a fixed number of SNPs (1 to 30). This method is commonly used to assess whether FST values are non randomly distributed across chromosomes [4], [22]. As expected, we found that correlation plots are different from those expected from a noisy signal (Figure 2). Moreover, scrambling FST values across each chromosome produced vanishing correlation values demonstrating that the distribution of data is non-random (data not shown). This result was also supported by a test for non-randomness of data (Ljung-Box test, p-value <10−16). Figure 2 shows a clear difference between correlation plots of autosomal and X-linked SNPs, the latter showing higher autocorrelation values. Chromosome Y was excluded from this analysis because of the small number of SNPs sampled.
Figure 2. Correlation between FST values.
The correlation is calculated, for each chromosome, for all pairs of SNPs separated by a fixed number of intervening SNPs. Black line shows mean value and 2σ error bars of the correlation of SNPs belonging to autosomal chromosomes. Red line shows correlation among X-linked SNPs.
To attribute FST value to genes we followed the approach by Akey et al. and Pikrell et al. [4], [16], considering FST of a gene the maximum FST value in the gene region (see Material and Methods). It is worth stressing that this approach is very conservative for genes with low FST values.
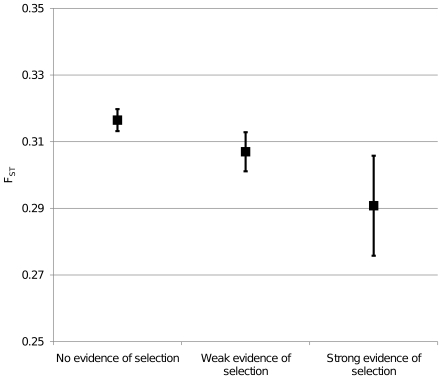
Selection affects both interspecific (between-species) and intraspecific (within-species) variability. FST is a measure of intraspecific variability. Estimation of genic dN/dS is an interspecific measure of variability [6]. We compared the gene FST values that we obtained with previously reported data from a genome-wide estimation of genic dN/dS [23]. In that article the authors divided genes into subgroups with strong, weak and no evidence of positive selection. We compared FST values of genes belonging to these subgroups. Genes with both weak and strong evidence of positive selection showed lower FST values than genes with no evidence of positive selection (ANOVA, p-value <0.001; Bonferroni post-hoc, no evidence vs. weak evidence p-value <0.02, no evidence vs. strong evidence p-value <0.005, weak evidence vs. strong evidence = N.S.; Figure 3).
Figure 3. Mean FST value of genes with and without interspecific evidence of positive selection.
Genes were grouped according to the strength of evidence of their positive selection across six species [23]. Vertical bars represent 95% confidence interval.
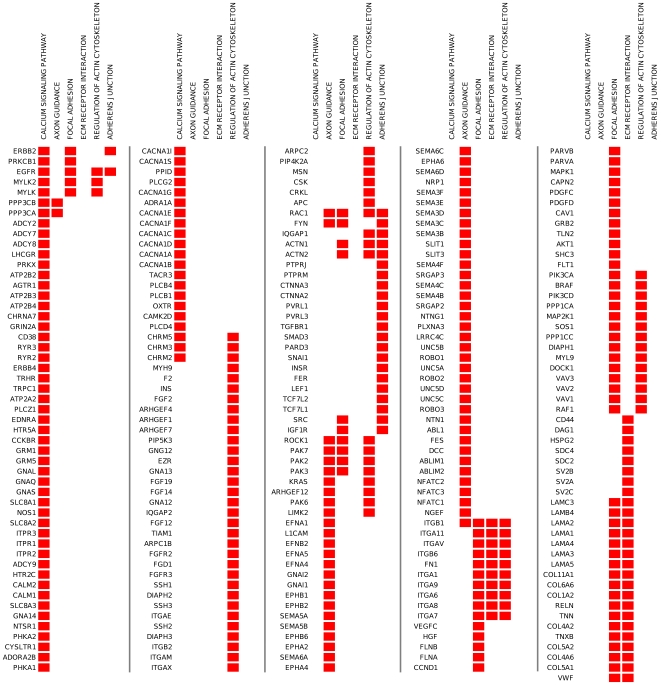
To identify functions potentially under selective pressure, we used an innovative approach, focusing on gene pathways instead of outliers. We performed this “gene set” analysis using the Gene Set Enrichment Analysis (GSEA) algorithm [24], [25]. GSEA is oriented to identify sets of functionally related genes and is currently used in the analysis of microarray data. Screening the KEGG pathway database by GSEA, we identified 6 KEGG pathways enriched by genes with high values of FST and one pathway enriched by genes with low values of FST (Table 1). In this method, the enrichment of a pathway is mainly driven by a group of genes that are called “leading edge genes” (see Material and Methods). Figure 4 shows the leading edge genes for the six pathways with high FST values. A partial overlap of genes among pathways is present.
Table 1. Enriched KEGG pathways identified by GSEA.
| Pathway | Name | KEGG ID | Size | FDR |
| Enriched by high FST genes | ||||
| Axon guidance | HS04360 | 126 | <0.001 | |
| Focal adhesion | HS04510 | 194 | 0.008 | |
| ECM receptor interaction | HS04512 | 85 | 0.009 | |
| Regulation of actin cytoskeleton | HS04810 | 199 | 0.010 | |
| Adherens junction | HS04520 | 75 | 0.010 | |
| Calcium signaling pathway | HS04020 | 168 | 0.010 | |
| Enriched by low FST genes | ||||
| Antigen processing and presentation | HS04612 | 70 | 0.001 | |
For each pathway is showed the name, the KEGG ID, the number of genes included in the pathway and the p-value after the False Discovery Rate (FDR) correction.
Figure 4. Leading edge genes of the high FST enriched KEGG pathways identified by GSEA.
Genes are indicated by gene symbols. Red box marks the presence of that gene, as leading edge gene, in that pathway.
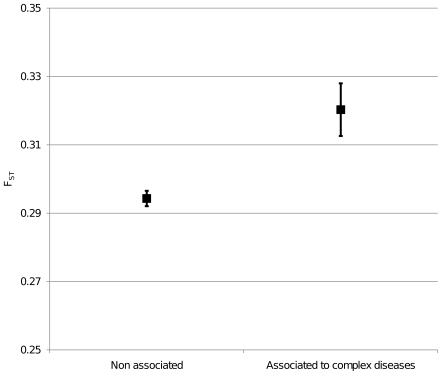
We then studied populations differentiation of genes associated with complex diseases. We used the Genetic Association Database (GAD) to select genes annotated as having positive association with complex diseases. We compared FST values of these genes with those where no association had been positively found. Genes associated with complex diseases showed a significant higher mean value of FST (t-test, p-value <0.001; Moving Block Boostrap, empirical p-value = 0.0005; Figure 5). Then, we divided diseases in subgroups according to the GAD classification of diseases. Figure 6 shows that large differences of FST values exist among disease classes, while mean FST values are usually higher than those of non associated genes.
Figure 5. Mean FST value of genes associated to complex diseases.
Genes found positively associated with complex diseases according to the Genetic Association Database are compared with the remaining ones. Vertical bars represent 95% confidence interval.
Figure 6. Mean FST values of genes in different disease classes.
Genes were grouped according to the diseases classification of Genetic Association Database. Vertical bars represent 95% confidence interval. Horizontal solid and dashed lines represent mean value and 95% confidence interval of the set of non associated genes.
Discussion
The study of the evolutionary forces acting in diseases and physiological traits is an exciting field that may drive further researches and, in the future, public health policies. The study of population genetic differentiation could help the understanding of human evolution, demographic history and disease susceptibility [26]. To study population differentiation we performed a genome-wide FST calculation using the latest available data release from the HapMap. Using this release we were able to increase both the number of SNPs and the number of individuals analysed in comparison to recent analogous studies [15]. We focused on samples from three different continents (Africa, Asia, Europe) to obtain a broad but sound measure of populations differentiation.
We found an overall mean FST value (0.122) broadly consistent with previous estimations [4], [22], [15]. The slightly higher value that we obtained could be explained by the exclusion of SNPs with MAF <0.05 and the inclusion of heterochromosomes in the calculation. Indeed, as expected [4], we observed a significantly higher median FST value of X-linked SNPs with respect to the autosomal ones. Furthermore, we found median FST value of Y-linked SNPs to be significantly higher than both the autosomal and the X-linked ones. Previous data from smaller datasets suggested a similar phenomenon [27], but, in our knowledge, this is the first observation made on Y chromosome FST in a more robust framework. The higher population differentiation for X and Y chromosomes can be due to various causes: their smaller effective population size (three-quarter and one-quarter of autosomes, respectively), the lower mutation and recombination rates and the different selective pressure between genders have been invoked [4], [6], [28].
Keinan et al. showed that there was a period of accelerated genetic drift on chromosome X associated with the human dispersal out of Africa. In particular, they estimated the autosome-to-X genetic drift ratio between North Europeans and East Asians is consistent with the expected 3/4 while it is significantly reduced between North Europeans and West Africans, and between East Asians and West Africans [29]. As possible explanations they suggested that a gender-biased process reduced the female effective population size, or that an episode of natural selection affecting chromosome X was associated with the founding of non-African populations. Our results are consistent with these finding. We computed population pair-wise FST and we found that the autosome-to-X genetic drift ratios (Q), estimated as in [29], are compatible with those reported in [29] (Asia-Europe Q = 0.72; Asia-Africa Q = 0.66; Europe-Africa Q = 0.65).
The weak but significant correlation that we found among FST values of neighbouring markers demonstrated that they are non-randomly distributed along chromosomes. This result confirms previous observations made on smaller datasets [4], [22]. We extended for the first time this observation to the X chromosome and we found that correlation was slightly stronger than that of autosomes. It has been observed that correlation between SNPs is proportional to Linkage Disequilibrium (LD) [22]. Therefore, the higher value of autocorrelation that we found can be explained by the higher value of LD in X chromosomes [22].
Population genetics approach has been largely used for studying natural selection. Other approaches include the comparative one, in which data from different species are used. The most commonly used method is to compare the ratio of nonsynonymous mutations per nonsynonymous site to the number of synonymous mutations per nonsynonymous site (dN/dS). Data from comparative studies and from population genetics are poorly connected. We found that genes with a high dN/dS ratio, indicating positive selection, showed a significantly lower FST mean value. In our knowledge this represents the first attempt to connect human population genetic data and comparative data at a genome-wide level. Our finding does not conflict with previous studies performed on a restricted number of genes [30]. It is well established that comparative data provides the most unambiguous evidence for selection, but relatively vague assertion on the type of selection and if the selection is currently acting in a population [6]. For such reasons the connection with population genetic data is needed. Further studies, mainly focused on this topic, are required to confirm and understand the relationship that we found.
We used a gene set approach to identify pathways with extraordinary levels of population genetic differentiation. The traditional approach used to perform this analysis is based on the identification of those loci outliers in a given statistic. This approach has been recently reviewed and its limits explored [10], [31]–[33]. Interestingly, similar criticisms are arising on analogous methods used in transcriptomic data analysis. In this field, alternative approaches, as the “gene set” ones, are gaining increasing interest. Among the tools implementing this approach, Gene Set Enrichment Analysis [34], [25] is one of the most used [35], [36]. The key idea underlying GSEA is to focus on gene sets, which are defined as groups of genes sharing common features (e.g. biological pathways, chromosomal position, etc.). In microarray data analysis, GSEA aims to determine whether a gene set shows statistically significant, concordant differences between two biological states or phenotypes. This method has been tailored for microarray data, however its use is being explored also in different fields [37], [38]. To the best of our knowledge, the present report is the first attempt to functionally analyse genes under selective pressure by a gene set statistical approach.
Using very conservative statistics, the GSEA analysis found differential FST values on seven KEGG pathways, one enriched by low FST genes and six enriched by high FST genes. However, it is important to note that the discrepancy between the number of low and high FST pathways is a consequence of the way by which we attributed FST values to genes rather than underlying evolutionary forces. The only pathway with decreased degree of differentiation among populations was the “antigen processing and presentation” pathway. Included in this pathway are genes involved in the antigen-presenting machinery as (i) the expression of major histocompatibility complex (MHC) molecules, (ii) the mechanism of cross-presentation, and (iii) the interaction of antigen-presenting cells. Opposing views exist concerning the evolutionary forces that shaped the innate immune system. In particular, the relative impact of purifying and balancing selection is under discussion [39], [40]. Barreiro et al demonstrated that several SNPs of genes related to the immune response to pathogens showed very high FST values [15]. On the other hand, Akey et al. reported a four times increase of proteins that perform a defense/immunity function in the group of the low FST genes [4]. Moreover, low levels of population differentiation have been previously detected at loci that are involved with host–pathogen responses (HLA class I and class II genes, beta-globin, G6PD, glycophorin A, interleukin 4 receptor-alpha and CCR5) [5]. Further evidence arises from the group of genes that we studied and that were previously described to be under positive selection. This group of genes, which we found with low FST values, was described to be enriched for several functions related to immunity and defence [23].
Among the six gene sets enriched by high FST genes, we found the “calcium signalling” pathway.
Calcium is the most abundant mineral in the body. It is also a highly versatile intracellular signal that regulates many cellular processes in response to different external stimuli, as growth factors [41]. We found very high FST values in three genes belonging to the growth factor stimulated calcium signalling pathway, namely EGFR, ERBB2, and ERBB4. It is interesting to note that a previous study from Pickrell et al. found that ERBB4 showed extreme signs of haplotype selective sweep in non-African populations [16]. The authors suggested that this gene could affect an unidentified phenotype that experienced a strong recent selection in non-African population. Our gene set approach seems to confirm this finding and expands this observation to other members of the ERBB gene family.
The other five high FST pathways are involved in the control of cell shape and mobility. Among them, four interconnected pathways (“focal adhesion”, “regulation of the actin cytoskeleton”, “adherens junction” and “extra cellular matrix receptor interaction”) govern growth-related processes and morphogenesis. Morphological traits have been demonstrated to show strong signature of positive selection [15]. These pathways were found also to be altered in a mouse model of fetal alcohol syndrome, associated with a low birth-weight phenotype [42]. Indeed, human body shape and size varies among populations showing a correlation with geographic and climate variables [43]. In addition, in the “adherens junction” pathway, one of the strongest FST values was showed by TCF7L2, the gene with largest type 2 diabetes effect size found to date [44]. This last finding is consistent with previous observations [44], [16]. Since it has been demonstrated that TCF7L2 variants also substantially influence normal birth-weight variations [45], a complex interplay between pathways that govern growth-related processes and susceptibility to type 2 diabetes could be hypothesized.
The last high FST pathway, the “axon guidance”, is involved in brain wiring during foetal development and repair throughout life. Axon guidance proteins and their relative binding partners have also an emerging role in the pathogenesis of several neurodegenerative and psychiatric diseases such as schizophrenia [46], [47]. Signature of recent positive selection inferred by identification of selective sweeps in specific populations was found in genes involved in schizophrenia [48]. Moreover, population dependent results were obtained when gene-association studies were performed using several high FST genes present in this gene set [49], [50].
It has been suggested that alleles involved in common disease could be targets of selection [51], [9], [52], [43]. The common disease/common variant (CD/CV) hypothesis proposes that common diseases are usually caused by one or a few common disease susceptibility alleles. These genetic variants represent ancestral alleles, presumably under selective pressure, that have become disadvantageous after changes in environment and of lifestyle [51], [53], [54]. We found that genes associated with complex diseases showed a significant higher mean value of FST, supporting the CD/CV hypothesis. However, several previous studies of SNPs associated with complex diseases did not find significant evidence of population differentiation [55], [56]. On the other hand, further studies observed that the distribution of maximum FST was shifted upward in regions associated with type 2 diabetes mellitus [16]. Moreover SNPs known to protect against obesity and diabetes showed very high FST values [15]. Simulation studies also provided support for the CD/CV hypothesis [57].
According to the GAD classification of diseases, we divided the overall group of the genes associated with complex diseases. Clear differences in FST means among the various classes were present. In particular, several disease classes, namely “hematological”, “infection”, and “immune”, showed an FST mean value slightly lower than the mean value of non-associated genes. Nevertheless, the majority of the classes showed FST mean values to be higher than the non-associated one. Highest FST values were detected in “pharmacogenomics” and “psychiatric” classes. GAD classifies in “pharmacogenomics” those diseases related to drug effects. It is well established that drugs effects are race/ethnic specific [58]. The GAD “psychiatric” class includes mental disorders. Why genes that confer susceptibility to mental diseases are still maintained by natural selection, is an old question which, up to now, is still unanswered. The compensatory advantage for genes associated to intermediate phenotypes has been invoked as explanation for this phenomenon, also called “psychiatric paradox” [59]. Further studies should be performed to determine if the high level of population differentiation that we found for this disease class could be related to the psychiatric paradox.
The results presented in this paper could contribute to further explorations of the ongoing selection in humans. Further studies are needed to clarify the biological pathways involved and to better elucidate the role of natural selection in human complex diseases.
Materials and Methods
Data
All analysis are based on the HapMap Public Release #27 (merged II+III) datafiles (http://www.hapmap.org). We analyzed the data from the CEPH (Utah residents with ancestry from northern and western Europe; CEU, n = 165), Yoruba in Ibadan, Nigeria (YRI, n = 167), Han Chinese in Beijing, China (CHB, n = 84) and Japanese in Tokyo, Japan (JPT, n = 86) samples. We pooled the CHB and JPT samples to form a single sample. Additional SNP information about physical positions and SNP-gene association were obtained from dbSNP build 129 (http://www.ncbi.nlm.nih.gov/projects/SNP). In particular, according to dbSNP classification, we considered all SNPs within 2 kb of a gene (locus region) as associated to that gene. Data from the International HapMap Project and dbSNP were merged in a local MySQL database by a set of script from Amigo et al. [60]. When we consider the whole Hap map dataset (autosomes and heterochromosomes) we analyzed a total of 3,917,301 SNPs.
We excluded by this analysis SNPs that were non sampled or non polymorphic in all the three samples. We excluded also SNPs with a minor allele frequency <5% in any of the 3 samples, getting a final SNP sample of 2,125,440 SNPs.
Estimates of FST
Fixation index (FST) was calculated using the unbiased estimator proposed by Weir and Cockerham [18], [21]. We implemented this calculation in a Perl script available upon request by the authors.
All analyses presented in this work were also performed by using the original FST estimator proposed by Wright [17] and results are almost identical to that obtained by the Weir and Cockerham method. This result is not surprising considering previous reports [61], [4] and the strong correlation that we found between these two measures (Spearman's ρ = 0.97, p<10−16; see Supporting Information S1).
The maximum FST values among those of the SNPs associated to the gene according to dbSNP (see above) was used to assign a FST value to each gene. This approach is consistent with previously described ones [4], [16]. We studied the correlation between FST value and gene length and we found that the former have a quite marginal effect on the latter (R2 = 0.2).
Statistical Analysis
SNPs FST values are not normally distributed across chromosomes. Thus to detect differences among medians FST values of chromosomes we used the non-parametric Kruskal-Wallis test. Conversely, FST values of genes are normally distributed (Kolmogorov-Smirnov/Lilliefor test, p<0.001,) thus comparison among these values were performed by using parametric tests (ANOVA and t-test).
All statistical analyses were performed with R ver. 2.9 (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org/). Non-randomness of data was assessed by using a Ljung-Box test (R function “Box.test”). We calculated the autocorrelation of each chromosome which can be seen as the mean correlation of all pairs of FST values separated by a fixed number of values (R function “acf”).
A list of about 4000 genes positively selected was obtained from the track “Positively Selected Genes” (database “hg18”, table “mammalPsg”) in UCSD Genome browser (http://genome.ucsc.edu). This list was produced by a genome wide scan in six mammalian genomes performed by Kosiol et al. [23]. In particular they identified (i) 400 genes with strong evidence of positive selection across species, (ii) 144 genes with strong evidence of positive selection in one or more branches, (iii) 3705 genes with weak evidence of positive selection on one or more branches, and (iv) 12280 (orthologs) genes with no significant evidence of positive selection. We pooled first and second group into a single “strong evidence of positive selection” group. Differences among groups were evaluated by ANOVA with Bonferroni post-hoc calculation.
Genes associated with complex diseases were obtained from the Genetic Association Database (GAD; October 1 2007 update; http://geneticassociationdb.nih.gov). We only kept genes with positive evidence of association, for a total of 1789 genes. According to GAD, these genes are divided into 15 classes of diseases. We excluded from the analysis four diseases classes (Other, Unknown, Mitochondrial and Normal variations) because they were not informative. Differences among groups were evaluated by a t-test and a resampling approach. In particular, we used a Moving Block Boostrap (MBB) strategy [62]. Briefly, (i) we resampled 10000 times 1789 set of adjacent SNPs {ni}j with i = 1, …,1789 and j = 1, …,10000 and with each set ni having the same number of SNPs as the i-th GAD associated gene; (ii) for each resample, we computed the FST of each set ni according to our method (the maximum FST values among those of the SNPs in the set); then, (iii) we computed the mean FST value of each resample j obtaining a distribution to which compare the mean FST value of the GAD associated genes.
Functional Analysis
We used Gene Set Enrichment Analysis (GSEA) 2.0 [63] to detect KEGG pathways enriched by genes with low or high values of FST. We provided GSEA, by its “Preranked” feature, with a list L of genes ranked according to their FST value. Given an a priori defined set of genes S representing a pathway (e.g., genes encoding products in a metabolic pathway), the goal of GSEA is to find out whether the members of S are randomly distributed throughout L or mainly found at the top or bottom (i.e. being “enriched”). Since GSEA preferably expect the values to rank for (in our case FST) to vary from negative to positive values, we linear shifted these values to get vanishing mean.
We explored the enrichment of KEGG pathways included in the software. For each pathway a False Discovery Rate (FDR) is computed, representing the statistical significance of the enrichment. For experimental conditions similar to the ours, GSEA user's guide suggests a threshold of significance FDR ≤0.05. Because of the exploratory nature of this study, we used a more conservative threshold of significance (FDR ≤0.01). Overlap among pathways was examined by the “Leading edge analysis” feature of GSEA.
Supporting Information
Additional figures and tables
(1.07 MB PDF)
Acknowledgments
We thank Michelle Kutzner for her help in the revision of the manuscript; the International HapMap Consortium for its work in creating this dataset; the authors of GSEA for making their software freely available.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: R.A. is a recipient of a fellowship by Doctorate of Computational Biology and Bioinformatics, University “Federico II”, Naples. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cavalli-Sforza LL, Menozzi P, Piazza A. Princeton: Princeton University Press; 1996. The History and Geography of Human Genes. [Google Scholar]
- 2.Barbujani G, Goldstein DB. Africans and Asians abroad: genetic diversity in Europe. Annu Rev Genomics Hum Genet. 2004;5:119–150. doi: 10.1146/annurev.genom.5.061903.180021. doi: 10.1146/annurev.genom.5.061903.180021. [DOI] [PubMed] [Google Scholar]
- 3.Kimura M. Cambridge: Cambridge University Press; 1985. The Neutral Theory of Molecular Evolution. [Google Scholar]
- 4.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis JPA, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 8.Davey Smith G, Ebrahim S, Lewis S, Hansell AL, Palmer LJ, et al. Genetic epidemiology and public health: hope, hype, and future prospects. Lancet. 2005;366:1484–1498. doi: 10.1016/S0140-6736(05)67601-5. doi: 10.1016/S0140-6736(05)67601-5. [DOI] [PubMed] [Google Scholar]
- 9.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci U S A. 2006;103:135–140. doi: 10.1073/pnas.0509691102. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, et al. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 16.Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright S. The genetic structure of populations. Annals of Eugenic. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 18.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 22.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, et al. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha VK, Lindgren CM, Eriksson K, Subramanian A, Sihag S, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavalli-Sforza LL. The Human Genome Diversity Project: past, present and future. Nat Rev Genet. 2005;6:333–340. doi: 10.1038/nrg1596. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
- 27.Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- 28.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 29.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41:66–70. doi: 10.1038/ng.303. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarazona-Santos E, Bernig T, Burdett L, Magalhaes WCS, Fabbri C, et al. CYBB, an NADPH-oxidase gene: restricted diversity in humans and evidence for differential long-term purifying selection on transmembrane and cytosolic domains. Hum Mutat. 2008;29:623–632. doi: 10.1002/humu.20667. doi: 10.1002/humu.20667. [DOI] [PubMed] [Google Scholar]
- 31.McVean G, Spencer CCA. Scanning the human genome for signals of selection. Curr Opin Genet Dev. 2006;16:624–629. doi: 10.1016/j.gde.2006.09.004. doi: 10.1016/j.gde.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 33.Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–712. doi: 10.1101/gr.5105206. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootha VK, Lindgren CM, Eriksson K, Subramanian A, Sihag S, et al. PGC-1[alpha]-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 35.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 37.Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24:2784–2785. doi: 10.1093/bioinformatics/btn516. doi: 10.1093/bioinformatics/btn516. [DOI] [PubMed] [Google Scholar]
- 38.Iorio F, Tagliaferri R, di Bernardo D. Identifying network of drug mode of action by gene expression profiling. J Comput Biol. 2009;16:241–251. doi: 10.1089/cmb.2008.10TT. doi: 10.1089/cmb.2008.10TT. [DOI] [PubMed] [Google Scholar]
- 39.Ferrer-Admetlla A, Bosch E, Sikora M, Marquès-Bonet T, Ramírez-Soriano A, et al. Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee S, Sarkar-Roy N, Wagener DK, Majumder PP. Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc Natl Acad Sci U S A. 2009;106:7073–7078. doi: 10.1073/pnas.0811357106. doi: 10.1073/pnas.0811357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 42.Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, et al. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- 43.Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, et al. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helgason A, Pálsson S, Thorleifsson G, Grant SFA, Emilsson V, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 45.Freathy RM, Weedon MN, Bennett A, Hypponen E, Relton CL, et al. Type 2 diabetes TCF7L2 risk genotypes alter birth weight: a study of 24,053 individuals. Am J Hum Genet. 2007;80:1150–1161. doi: 10.1086/518517. doi: 10.1086/518517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin L, Lesnick TG, Maraganore DM, Isacson O. Axon guidance and synaptic maintenance: preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 2009;32:142–149. doi: 10.1016/j.tins.2008.11.006. doi: 10.1016/j.tins.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allen NC, Bagade S, McQueen MB, Ioannidis JPA, Kavvoura FK, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 48.Crespi B, Summers K, Dorus S. Adaptive evolution of genes underlying schizophrenia. Proc Biol Sci. 2007;274:2801–2810. doi: 10.1098/rspb.2007.0876. doi: 10.1098/rspb.2007.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarimon J, Scholz S, Fung H, Hardy J, Eerola J, et al. Conflicting results regarding the semaphorin gene (SEMA5A) and the risk for Parkinson disease. Am J Hum Genet. 2006;78:1082–1084; author reply 1092-1094. doi: 10.1086/504727. doi: 10.1086/504727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujii T, Iijima Y, Kondo H, Shizuno T, Hori H, et al. Failure to confirm an association between the PLXNA2 gene and schizophrenia in a Japanese population. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:873–877. doi: 10.1016/j.pnpbp.2007.01.027. doi: 10.1016/j.pnpbp.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 51.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–868. doi: 10.1038/nrg2187. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe S, Kang D, Feng L, Nakagawa T, Kanellis J, et al. Uric acid, hominoid evolution, and the pathogenesis of salt-sensitivity. Hypertension. 2002;40:355–360. doi: 10.1161/01.hyp.0000028589.66335.aa. [DOI] [PubMed] [Google Scholar]
- 54.Diamond J. The double puzzle of diabetes. Nature. 2003;423:599–602. doi: 10.1038/423599a. doi: 10.1038/423599a. [DOI] [PubMed] [Google Scholar]
- 55.Lohmueller KE, Mauney MM, Reich D, Braverman JM. Variants associated with common disease are not unusually differentiated in frequency across populations. Am J Hum Genet. 2006;78:130–136. doi: 10.1086/499287. doi: 10.1086/499287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng B, Kimmel M. Simulations provide support for the common disease-common variant hypothesis. Genetics. 2007;175:763–776. doi: 10.1534/genetics.106.058164. doi: 10.1534/genetics.106.058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429:464–468. doi: 10.1038/nature02626. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 59.Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav Brain Sci. 2006;29:385–404; discussion 405-452. doi: 10.1017/S0140525X06009095. doi: 10.1017/S0140525X06009095. [DOI] [PubMed] [Google Scholar]
- 60.Amigo J, Salas A, Phillips C, Carracedo A. SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinformatics. 2008;9:428. doi: 10.1186/1471-2105-9-428. doi: 10.1186/1471-2105-9-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weicker JJ, Brumfield RT, Winker K. Estimating the unbiased estimator theta for population genetic survey data. Evolution. 2001;55:2601–2605. doi: 10.1111/j.0014-3820.2001.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 62.Lahiri S. New York: Springer; 2003. Resampling Methods for Dependent Data. [Google Scholar]
- 63.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional figures and tables
(1.07 MB PDF)