Summary
By co-opting host proteins for their replication, plus-stranded RNA viruses can support robust replication and suppress host anti-viral responses. Tomato bushy stunt tombusvirus (TBSV) recruit the cellular heat shock protein 70 (Hsp70), an abundant cytosolic chaperone, into the replicase complex. By taking advantage of yeast model host, we demonstrate that the four-member SSA subfamily of HSP70 genes is essential for TBSV replication. The constitutively-expressed SSA1 and SSA2, which are resident proteins in the viral replicase, can be complemented by the heat-inducible SSA3 and/or SSA4 for TBSV replication. Using a yeast strain carrying a temperature sensitive ssa1ts, but lacking functional SSA2/3/4, we show that inactivation of Ssa1pts led to a defect in membrane localization of the viral replication proteins, resulting in cytosolic distribution of the viral proteins and lack of replicase activity. An in vitro replicase assembly assay with Ssa1pts revealed that functional Ssa1p is required during the replicase assembly process, but not during minus- or plus-strand synthesis. Temperature shift experiments from nonpermissive to permissive in ssa1ts yeast revealed that the re-activated Ssa1pts could promote efficient TBSV replication in the absence of other SSA genes. We also demonstrate that the purified recombinant Ssa3p can facilitate the in vitro assembly of the TBSV replicase on yeast membranes, demonstrating that Ssa3p can fully complement the function of Ssa1p. Taken together, the cytosolic SSA subfamily of Hsp70 proteins play essential and multiple roles in TBSV replication.
INTRODUCTION
Due to limiting coding capacity of their genomes, plus-stranded (+)RNA viruses rely extensively on the host during their replication. These viruses hijack subcellular membranes and use the components of the host cells to make viral proteins and replicate the viral RNA. Moreover, they co-opt selected host proteins to facilitate viral genome replication (Ahlquist et al., 2003; Nagy, 2008; Noueiry and Ahlquist, 2003; Salonen, Ahola, and Kaariainen, 2005; Shi and Lai, 2005). Indeed, recent genome-wide screens with Brome mosaic virus (BMV), Tomato bushy stunt virus (TBSV), Drosophila C virus, hepatitis C virus and West Nile virus revealed that more than one hundred host proteins and many cellular pathways affected replication and infections by each (+)RNA virus (Cherry et al., 2005; Jiang et al., 2006; Krishnan et al., 2008; Kushner et al., 2003; Panavas et al., 2005b; Randall et al., 2007; Serviene et al., 2005). The functions of most of the identified host proteins during virus replication, however, are currently unknown.
TBSV, a small (+)RNA virus is used to dissect the roles of host proteins within the viral replicase, which is the key enzyme for viral genome replication. The tombusvirus replicase has been intensively characterized via proteomics approaches, in vitro replication assays and development of yeast as a model host (Li et al., 2008; Li et al., 2009; Nagy, 2008; Nagy and Pogany, 2000; Panavas and Nagy, 2003; Panaviene et al., 2004; Pogany and Nagy, 2008; Serva and Nagy, 2006; Wang and Nagy, 2008). Previous work revealed that TBSV RNA replication depends on two viral-encoded proteins, namely the RNA-dependent RNA polymerase (p92pol RdRp) and p33 replication co-factor, the key protein in recruitment of the viral RNA into replication (Monkewich et al., 2005; Pogany and Nagy, 2008; Pogany et al., 2008; Pogany, White, and Nagy, 2005). The peroxisome membrane-bound TBSV replicase also contains 6-to-10 host proteins, which are likely involved in most activities of the replicase (Nagy and Pogany, 2006; White and Nagy, 2004). The identified host proteins within the tombusvirus replicase include heat shock protein 70 (Hsp70, coded by the constitutively-expressed SSA1 and SSA2 genes in yeast) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, coded by TDH2 and TDH3 genes in yeast), which binds to the TBSV (−)RNA and affects plus-strand synthesis (Wang and Nagy, 2008). The replicase also contains translation elongation factor 1A (eEF1A), which binds to a cis-acting regulatory element in the TBSV (+)RNA as well as to p33 co-factor (Li et al., 2009). Another host-derived component is Cdc34p ubiquitin-conjugating enzyme, which ubiquitinates the p33 replication co-factor (Li et al., 2008). Down-regulation of these host factors inhibited, whereas their over-expression increased TBSV accumulation in yeast model host (Li et al., 2008; Serva and Nagy, 2006; Wang and Nagy, 2008) suggesting that they play significant roles in TBSV replication. In addition, Pex19p cytosolic transport protein binds transiently to the viral replication proteins as well as to the replicase complex, likely facilitating the transport of the replication proteins to the peroxisomal membranes, the site of replication (Pathak, Sasvari, and Nagy, 2008). The functions of the above host proteins within the viral replicase are currently under intensive investigations.
The host-coded Hsp70 chaperone family, which represents a major group among the heat shock proteins, and its co-chaperones have been suggested to promote replication of several (+)RNA and (−)RNA viruses (Brown et al., 2005; Dufresne et al., 2008; Nishikiori et al., 2006; Qanungo et al., 2004; Weeks and Miller, 2008). Based on the known cellular functions, Hsp70 and other chaperones were proposed to stimulate viral RdRp activity (Momose et al., 2002), and participate in the assembly of the viral replicase and enhance replication (Kampmueller and Miller, 2005; Tomita et al., 2003; Weeks and Miller, 2008). Plants infected by various plant viruses express cytosolic Hsp70 proteins at elevated levels, indicating that Hsp70 could play an important role during viral infections (Aparicio et al., 2005; Aranda et al., 1996; Whitham et al., 2003; Whitham, Yang, and Goodin, 2006). Moreover, a viral-encoded Hsp70-like protein is involved in the assembly of virions and cell-to-cell movement of closteroviruses (Alzhanova et al., 2001; Peremyslov, Hagiwara, and Dolja, 1999). In addition, Hsp70-like proteins are involved in nuclear localization, genome replication and cell transformation of DNA viruses, cell entry, virion assembly and disassembly, envelope protein maturation, folding of capsid proteins, and viral transcription by various viruses (Mayer, 2005). Additional cellular chaperones, such as Hsp90 proteins or the J-domain containing Hsp40 proteins have also been shown to affect virus replication, including activation of reverse transcriptase for hepadnaviruses (Hu et al., 2004; Stahl et al., 2007; Tavis, Massey, and Gong, 1998), or assembly of the BMV replicase (Tomita et al., 2003). Most of the above studies point toward Hsp70 and other cellular chaperones as major players during virus replication.
Host proteins could play different roles during tombusvirus replication. Currently, tombusvirus replication is divided into six sequential steps: RNA template selection by p33 replication protein; recruitment of the replication protein-viral RNA complex to the site of replication; assembly of the viral replicase complex; synthesis of viral progeny RNAs, including minus- and plus-strand synthesis; release of the newly made plus-strand viral RNAs from the replicase and disassembly of the viral replicase (Nagy and Pogany, 2006). Hsp70 proteins might play multiple roles/functions in TBSV replication. Accordingly, the two cytosolic Ssa1p and Ssa2p Hsp70 proteins expressed constitutively in yeast have been shown to be recruited from the cytosol to the peroxisomal membrane (the site of TBSV replication) via interaction with the p33 replication co-factor (Serva and Nagy, 2006; Wang, Stork, and Nagy, 2009). Down-regulation or over-expression of Ssa1/2p in yeast resulted in reduced and elevated level of TBSV RNA accumulation, respectively, suggesting that these Hsp70 proteins are important for TBSV RNA replication (Serva and Nagy, 2006). These Hsp70 proteins are likely involved directly in replication since Ssa1/2p have been shown as components of the highly purified tombusvirus replicase complex (Serva and Nagy, 2006). Using a HSP70 mutant yeast (ssa1ssa2), we found that the viral replication proteins remained cytosolic at an early time point, suggesting that Hsp70 is involved in subcellular localization of the viral replication proteins to intracellular membranes (Wang, Stork, and Nagy, 2009). A novel in vitro replication assay also showed that Ssa1/2p are essential for the assembly of the TBSV replicase (Pogany et al., 2008). An in vitro membrane insertion assay demonstrated that Hsp70 promoted the insertion of the viral replication proteins into the subcellular membranes (Wang, Stork, and Nagy, 2009). These functions of Hsp70 are not restricted to in vitro or yeast-based assays, since knockdown of cytosolic Hsp70 in plants or inhibition of Hsp70 with a chemical inhibitor were found to inhibit TBSV replication in a plant host (Wang, Stork, and Nagy, 2009).
The SSA subfamily of cytosolic Hsp70 consists of four genes and expression of at least one of the four SSA genes at high level is needed for yeast viability (Ahsen and Pfanner, 1997). It is currently unknown if the function of Ssa1/2p is essential for TBSV replication and whether additional cellular Hsp70 or other heat shock proteins can complement the functions of Ssa1/2p for supporting TBSV replication. To address these questions and to further dissect the roles of Hsp70 in TBSV replication, in this work, we used a yeast strain lacking functional SSA2, SSA3 and SSA4 genes, while the SSA1 gene was either wt or temperature sensitive (ts). Based on the ssa1ts strain, we demonstrate that the Ssa-subfamily of Hsp70 proteins is essential for TBSV replication in yeast. Using biochemical and temperature shift experiments, we show that Hsp70 is essential during the early steps of TBSV replication, but not during minus- and plus-strand synthesis.
RESULTS
TBSV replication can be complemented by a heat shock-inducible host factor in ssa1ssa2 mutant yeast
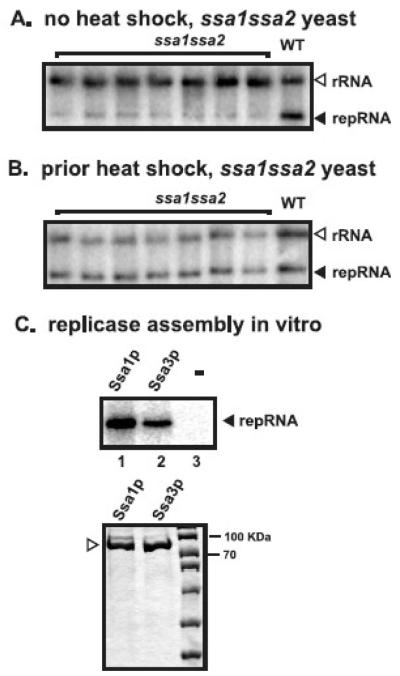
Previous work has shown that ssa1ssa2 mutant yeast can still support TBSV replication, albeit less efficiently and with significant delay when compared with the wt yeast (Wang, Stork, and Nagy, 2009). These observations indicate that either Ssa1p/Ssa2p are not essential for TBSV replication or they can be partially complemented possibly by other cellular chaperones. To test if complementation of TBSV replication can be enhanced by prior induction of heat shock proteins (Boorstein and Craig, 1990), we incubated ssa1ssa2 mutant yeast at 42°C for 30 min prior to the induction of p33/p92pol proteins and the TBSV DI-72 repRNA. In comparison with ssa1ssa2 mutant yeast grown at 23°C all the time (no heat shock), which support only 15% repRNA accumulation, the ssa1ssa2 mutant yeast treated with short heat-shock reached 80% repRNA accumulation of that measured in wt yeast (no heat shock) (Fig. 1A versus 1B). The stimulating effect of the short heat shock treatment prior to TBSV replication in ssa1ssa2 mutant yeast suggests that a heat-inducible host factor, likely a heat-shock protein, can efficiently complement the missing Ssa1/2p functions for TBSV replication.
Fig. 1.
Increased accumulation of TBSV repRNA in ssa1ssa2 mutant yeast after short heat shock treatment. (A) Northern blot analysis of TBSV repRNA accumulation level in ssa1ssa2 mutant yeast cultured continuously at 23°C. TBSV repRNA accumulates to 15% in ssa1ssa2 mutant yeast of that found in wt yeast at the 16 hour time point after induction by galactose. The ribosomal (r)RNA is used as a loading control. (B) After 30 min heat shock treatment of yeast at 42°C, TBSV repRNA replication was launched by expression of p33/p92 and DI-72(+)RNA from the GAL1/GAL10 promoters in ssa1ssa2 mutant yeast by using culture media containing galactose, followed by shaking at 23°C. Northern blot analysis shows that TBSV repRNA accumulated to ~80% in the heat-treated ssa1ssa2 mutant yeast when compared to the wt yeast at the 16 hour time point. (C) Ssa3p can facilitate the in vitro assembly of the TBSV replicase. The purified recombinant FLAG-Ssa3p (lane 2) and FLAG-Ssa1p (lane 1) were added together with purified TBSV p33 (lanes 1-3) and p92pol (lanes 1-3) to the membrane fraction of a cell-free extract prepared from untransformed yeast, which was programmed via the addition of 1 μg TBSV DI-72 (+)repRNA (lanes 1-3). The newly synthesized repRNA (marked with an arrowhead), representing mostly (+)repRNA due to a full cycle of replication, by the in vitro assembled TBSV replicase is analyzed by denaturing PAGE analysis. The bottom panel shows the coomassie-blue-stained SDS-PAGE gel with the purified recombinant FLAG-Ssa3p and FLAG-Ssa1p.
The purified heat-inducible Ssa3p Hsp70 can facilitate the assembly of the TBSV replicase in vitro
It is possible that the heat-inducible host factor supporting TBSV replication in ssa1ssa2 mutant yeast, which could also be responsible for the partial complementation of TBSV replication at the standard temperature (Wang, Stork, and Nagy, 2009), is the cytosolic, stress-inducible Ssa3p and Ssa4p Hsp70 proteins, which are highly similar to one another and show 80% sequence identity with Ssa1/Ssa2p (Becker et al., 1996; Lin et al., 2001). Accordingly, Ssa3/4p are expressed at high levels in ssa1ssa2 cells (Becker et al., 1996) and they can be efficiently induced by a short heat-shock treatment (Boorstein and Craig, 1990).
To study if Ssa3p could replace Ssa1p for supporting TBSV replication, we used the recently developed TBSV replication assay based on the addition of purified recombinant p33/p92pol and the DI-72 repRNA to a yeast cell-free extract (Pogany et al., 2008). The in vitro assembled TBSV replicase is capable of supporting authentic TBSV repRNA replication if ribonucleotides are provided (Pogany and Nagy, 2008; Pogany et al., 2008). We have shown previously that Ssa1p is required for the in vitro assembly of the TBSV replicase when only the membrane fraction of the cell-free extract is used (Pogany et al., 2008).
To test if Ssa3p can support the assembly of the TBSV replicase in vitro, we used purified recombinant Ssa3p (fused with FLAG-tag) together with purified recombinant p33/p92pol and the DI-72 repRNA and mixture of ribonucleotides in combination with the membrane fraction of the yeast cell-free extract. The in vitro replication assay revealed that the recombinant Ssa3p was capable of assembling the active TBSV replicase (Fig. 1C, lane 2), though less efficiently than Ssa1p (lane 1). Since the replicase assay required Ssa3p (Fig. 1C, lanes 2 versus 3), these data are consistent with the proposed role of Ssa3p in complementing the missing functions of Ssa1/2p in ssa1ssa2 mutant yeast (Fig. 1B).
Expression of Ssa1pts in yeast to study the role of Hsp70 in TBSV replication
There are four SSA genes in yeast coding for cytosolic Hsp70 proteins and deletion of all SSA genes makes yeast nonviable. To demonstrate if SSA1-4 genes are essential for TBSV replication, we used yeast strains with nonfunctional SSA2/3/4 genes, whereas SSA1 gene was either wt (SSA1wt ssa2 ssa3 ssa4) or ts (ssa1ts ssa2 ssa3 ssa4). We will refer to these strains as SSA1wt and ssa1ts below. The advantage of the ssa1ts is that this yeast strain can be cultured at 29°C or below, while Ssa1pts becomes nonfunctional at 35 °C or above, although the cells do not show abnormal morphology or cell cycle-specific arrest at 37 °C (Becker et al., 1996). Ssa1pts was obtained via mutagenesis and it contains P417-L mutation within the peptide-binding domain. The growth inhibition at the elevated temperature in ssa1ts cells is cytostatic (not cytocidal) since cells start growing after temperature shift to 30 °C even after kept at 37 °C for 3 days (Becker et al., 1996).
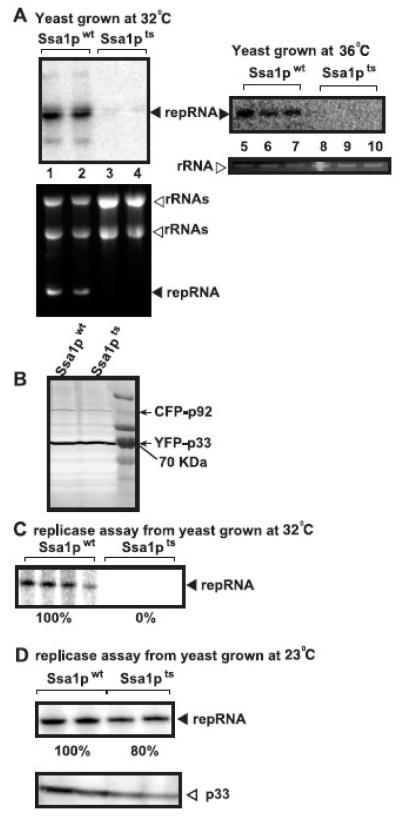
Yeast strains ssa1ts or SSA1wt were transformed with plasmids to express p33/p92pol and the repRNA from GAL1/GAL10 promoters and TBSV replication was launched by adding galactose to the culture media, followed by switching the temperature of incubation to 32°C or 36°C. Northern blot analysis of the RNA samples obtained after 18 hours of incubation at 32°C showed accumulation of TBSV repRNA to less than 1% in ssa1ts yeast when compared to that of SSA1wt (Fig. 2A, lanes 3-4 versus 1-2). The accumulation of TBSV repRNA at 36°C was below detection limit in ssa1ts yeast when compared to that of SSA1wt (Fig. 2A, lanes 8-10 versus 5-7). This low level of repRNA accumulation in ssa1ts yeast grown at 32/36 °C is likely due to plasmid-borne transcription of repRNA (Panavas and Nagy, 2003), indicating the lack of replication of the TBSV repRNA at 32 °C or at 36 °C in ssa1ts yeast. Accordingly, isolation of the membrane-bound tombusvirus replicase complexes from the above yeast strains, followed by in vitro replicase assay using the co-purified TBSV repRNA showed undetectable level of replicase activity from ssa1ts yeast grown at 32 °C, while the comparable assay from SSA1wt grown at 32 °C showed easily detectable replicase activity in vitro (Fig. 2C). In contrast, the membrane-bound tombusvirus replicase from ssa1ts yeast grown at 23 °C was almost as active as that obtained from SSA1wt grown at 23 °C (Fig. 2D). Thus, as expected, ssa1ts yeast was likely defective in assembling the tombusvirus replicase at the nonpermissive temperature.
Fig. 2.
Lack of TBSV repRNA replication in ssa1ts yeast at the nonpermissive temperature. (A) Top panels: Northern blot analysis of TBSV repRNA accumulation in ssa1ts yeast and SSA1wt yeast grown either at 32°C or 36°C, which inactivates Ssa1pts. Bottom panels: Ethidium bromide stained agarose gel shows repRNA and ribosomal rRNA levels in the total RNA samples in ssa1ts and SSA1wt yeast strains 16 hours after the induction of TBSV repRNA replication via galactose. Note that YFP-p33 and CFP-p92 each expressed from the GAL1 promoter, whereas DI-72 repRNA was expressed from the GAL10 promoter. (B) Western blot analysis to show the accumulation of YFP-p33 and CFP-p92 in ssa1ts yeast and SSA1wt yeast grown at 32°C. The yeast samples were the same as in panel A. (C) An in vitro replicase assay to test TBSV RNA synthesis in ssa1ts yeast and SSA1wt yeast grown at 32°C. We obtained the membrane fraction carrying the tombusvirus replicase from ssa1ts yeast and SSA1wt yeast grown at 32°C for 16 hours after the addition of galactose to induce TBSV replication. Note that the replicase contains the endogenous repRNA template. (D) Similar in vitro replicase assay was used to test TBSV RNA synthesis in ssa1ts yeast and SSA1wt yeast grown at 23°C. We obtained the membrane fraction carrying the tombusvirus replicase from ssa1ts yeast and SSA1wt yeast grown at 23°C for 16 hours after the addition of galactose to induce TBSV replication. The bottom panel shows a Western blot for p33 level in each membrane-bound replicase preparations.
Western blot analysis of total protein extracts form ssa1ts or SSA1wt yeast strains revealed comparable accumulation of p33 (Fig. 2B) and p92pol (not shown) replication proteins at 32°C. Thus, it is unlikely that Ssa1-4p affect the translation or degradation of the viral replication proteins. Taken together, all these results suggest that replicase activity or the assembly of the replicase is completely blocked at 32/36 °C in the yeast strain carrying ssa1ts and lacking the complementing SSA2/3/4 genes. Thus, the SSA subfamily (SSA1/2/3/4) of HSP70 genes is essential for TBSV replication and their functions cannot be complemented by other HSP70 or additional heat shock genes.
Yeast carrying ssa1ts shows defect in localization of the viral p33 replication protein when grown at 36°C
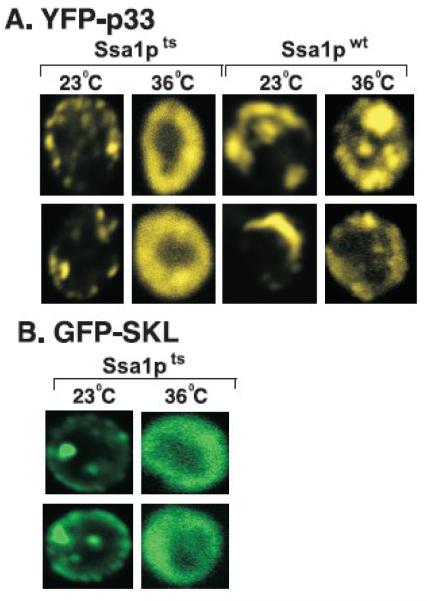
Since p33 and p92pol replication proteins become cytosolic in ssa1ssa2 mutant yeast strain at early time points instead of forming the characteristic punctate structures on the peroxisomal membranes in wt yeast (Wang, Stork, and Nagy, 2009), we tested the intracellular localization of p33 in ssa1ts yeast. The expression of the yellow fluorescent protein-tagged p33 replication protein (YFP-p33) was initiated from the GAL1 promoter by the addition of galactose to the culture media, followed by culturing at either 23°C or 36°C. Confocal laser microscopy of ssa1ts yeast cells after 18 hours of culturing at 36°C indicated cytosolic (showing “doughnut”-like distribution) localization of YFP-p33 (Fig. 3A). This is in contrast with the formation of the punctate structures by YFP-p33 when studied in ssa1ts yeast grown at 23°C or SSA1wt cultured at either 23°C or 36°C (Fig. 2A). These data suggest a defect in subcellular localization and the transport of p33 to the peroxisome membrane when none of the Ssa1-4p was functional.
Fig. 3.
Cytosolic localization of p33 replication protein in ssa1ts yeast grown at 36°C. (A) Confocal laser microscopy shows the localization of YFP-p33 in ssa1ts yeast and SSA1wt yeast grown at either 23°C or 36°C for 14 hours after induction from GAL1 promoter. The punctate structures show peroxisomal localization, while the “doughnut” -shape structure indicates cytosolic localization of p33 in yeast cells. The punctate structures formed in ssa1ts yeast grown at 23°C are reminiscent of those found in SSA1wt yeast (left side of the panel). (B) Cytosolic localization of peroxisome-targeted GFP, termed GFP-SKL, in ssa1ts yeast grown at 36°C for 14 hours after induction from GAL1 promoter. Note that GFP-SKL carries the peroxisomal targeting sequence “SKL” at the C-terminus.
Since p33, similar to many other peroxisomal membrane proteins, has been shown to be transported to the peroxisome by Pex19p cytosolic shuttle protein (Pathak, Sasvari, and Nagy, 2008), it is possible that other peroxisomal proteins might also be mislocalized in ssa1ts yeast at 36°C. To test this question, we expressed GFP-SKL, which carries the C-terminal SKL sequence known to target proteins to peroxisome in wt yeast (Kragler et al., 1993), from the GAL1 promoter. As expected, GFP-SKL was localized to the peroxisomal membrane, forming the punctate structure, in ssa1ts yeast grown at the permissive 23°C temperature (Fig. 3B). In contrast, GFP-SKL was localizeled in the cytosol in ssa1ts yeast grown at the nonpermissive 36°C (Fig. 3B). This suggests that the transportation of peroxisomal proteins as well as p33 replication protein to the peroxisome does not occur in the absence of functional Ssa1-4p proteins.
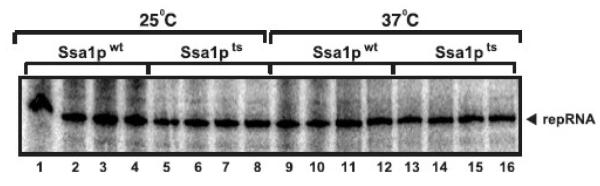
The assembled replicase from ssa1ts yeast is not sensitive to inactivation of Ssa1pts
To test if the SSA genes are required after the assembly of the viral replicase, we isolated the tombusvirus replicase bound to the membrane from ssa1ts or SSA1wt yeast cultured at 23°C, followed by in vitro replicase assay conducted at either 25°C or the nonpermissive 37 °C. These experiments revealed that the tombusvirus replicase from ssa1ts yeast was as active at the nonpermissive 37°C as at 25°C or the replicase derived from SSA1wt yeast (Fig. 4). Thus, the activity of the preassembled, membrane-bound tombusvirus replicase containing the endogenous repRNA does not depend on the presence of a functional Ssa1p.
Fig. 4.
The assembled tombusvirus replicase in the membrane fraction of ssa1ts yeast is not temperature sensitive. Denaturing PAGE analysis shows the radiolabeled repRNAs produced by tombusvirus replicase preparations in vitro obtained from ssa1ts yeast and SSA1wt yeast grown at 23°C. The in vitro replication assays were performed at either 25°C or 37°C. The membrane fraction carrying the tombusvirus replicase from ssa1ts yeast and SSA1wt yeast grown at 23°C for 16 hours after the addition of galactose to induce TBSV replication was used in the in vitro replicase assay based on the endogenous repRNA template.
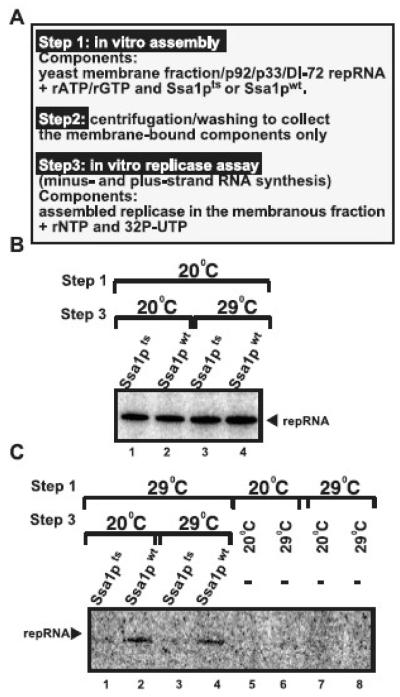
To test even more rigorously if Ssa1p is required only before/during the assembly of the tombusvirus replicase complex, but not during RNA synthesis, we used our recently developed in vitro replicase assembly assay (Pogany et al., 2008). In this assay (Fig. 5A), we added either the purified Ssa1pts or Ssa1pwt in combination with purified p33/p92pol replication proteins and DI-72(+)repRNA to the membrane fraction of yeast cell free extract. Importantly, we included only rATP and rGTP during the replicase assembly, which is required and sufficient to support in vitro assembly, but cannot support RNA synthesis that needs all four ribonucleotides. After the in vitro assembly, we collected the membrane fraction by centrifugation and removed all the soluble proteins/RNAs, including those Ssa1p/p33/p92pol molecules not bound to membranes. Then, we added the replicase buffer and all four ribonucleotides, including 32P-UTP, to the above membrane fraction to allow the RNA synthesis to occur by the preassembled TBSV replicase. Importantly, the removal of the soluble fraction before the replicase assay prevented the possibility of replicase assembly and RNA synthesis to take place simultaneously (Fig. 5A).
Fig. 5.
Ssa1p is required during the in vitro assembly of the TBSV replicase, but not during viral RNA synthesis. (A) A step-wise approach was used to separate the early steps, such as TBSV replicase assembly, from the late steps, which include viral RNA synthesis. The various components used in the procedure are listed. (B) Denaturing PAGE analysis of the radiolabeled in vitro replicase products obtained after the in vitro reconstitution of the TBSV replicase using the conditions shown during step 1 and step 3. (C) Similar analysis of the in vitro TBSV replicase assay with Ssa1pts or Ssa1pwt as in panel B, except using different conditions during step 1 and step 3 as shown. Note that samples in lanes 5-8 contained no Ssa1pts or Ssa1pwt during the replicase assembly.
When we used 20°C during the assembly step and either 20°C or 29°C during the RNA synthesis step, then we observed robust in vitro replication regardless of using Ssa1pts or Ssa1pwt (Fig. 5B). Since Ssa1pts is partially inactive at 29°C (Pogany et al., 2008) in the in vitro replication assay, we propose based on the above results that Ssa1pts is needed during the replicase assembly process, but not during RNA synthesis, which include both minus- and plus-strand synthesis in our in vitro replication assay (Pogany and Nagy, 2008; Pogany et al., 2008). Accordingly, when we used 29°C during the assembly step and 20°C during the RNA synthesis step, then we observed TBSV repRNA replication occurring only in case of the Ssa1pwt, but not with the TBSV replicase containing Ssa1pts (Fig. 5C, lanes 2, 4 versus 1 and 3). These data suggest that Ssa1pts was defective in replicase assembly at 29°C in vitro, preventing the subsequent viral RNA synthesis at 20°C or 29°C. Altogether, the obtained results are in agreement with the model that Ssa1p is required prior/during the replicase assembly, but it is not needed during minus- and plus-strand synthesis in vitro.
Downshift to 23°C restores peroxisomal localization of p33 replication protein in ssa1ts yeast
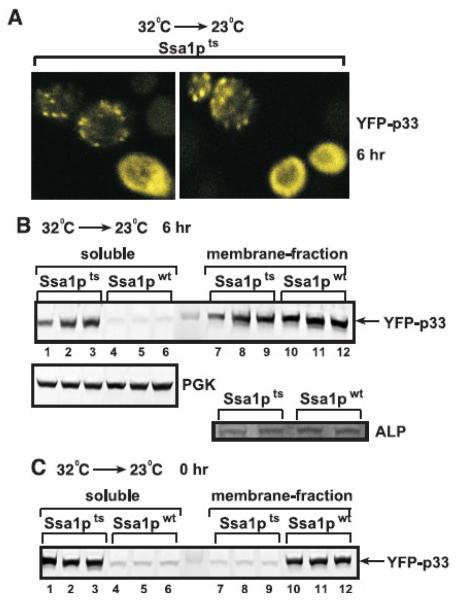
Since ssa1ts yeast cells survive short periods (up to 3 days) at the nonpermissive temperature (Becker et al., 1996), we used temperature downshift experiments to test how TBSV replication is affected by the reactivation of Ssa1pts by shifting to the permissive temperature. First, we cultured ssa1ts yeast expressing YFP-p33 at 32°C, followed by decreasing the temperature to 23°C and shutting down the production of YFP-p33 by changing the carbon-source in the media from galactose to glucose. Using confocal microscopy, we observed that about 50% of yeast cells contained the characteristic punctate structures 6 h after the downshift, suggesting peroxisomal localization of YFP-p33 (Fig. 6A). The other 50% of yeast cells still contained cytosolic YFP-p33, suggesting that the availability of the reactivated Ssa1pts is limited (Fig. 6A). Fractionation of the extracts of ssa1ts yeast cells 6h after the switch to 23°C also showed similar ~50-50% distribution of YFP-p33 between the soluble or membrane-containing fractions (Fig. 6B, lanes 1-3 versus 7-9). In contrast, YFP-p33 expressed in ssa1ts yeast at the nonpermissive temperature was found mostly in the soluble fraction (Fig. 6C, 0 hr time point). As expected, YFP-p33 expressed in SSA1wt yeast was present in the membranous fraction regardless of the growth conditions (Fig. 6B-C, 0 and 6 hr time points). The cytosolic cellular PGK protein and the membrane-bound cellular ALP protein were found in the expected fractions (Fig. 6B). Altogether, these data are consistent with the model that correct peroxisomal localization of YFP-p33 requires a functional Ssa1p and reactivation of ssa1ts by down-shifting to 23°C results in re-localization of p33 from the cytosol to the peroxisomal membrane in half of the ssa1ts yeast cells by the 6 h time point.
Fig. 6.
Re-distribution of p33 from the cytosol to the peroxisomes after shifting down the temperature from nonpermissive to permissive in ssa1ts yeast. (A) Confocal laser microscopy shows the localization of YFP-p33 in ssa1ts yeast grown at 23°C in a media containing glucose (to inhibit the synthesis of new YFP-p33 molecules) for 6 hours after culturing the yeast for 24 hours at 32°C in a media containing galactose. Note that the characteristic punctate structures indicating peroxisomal localization (Jonczyk et al., 2007; McCartney et al., 2005; Panavas et al., 2005a) are formed only in ~50% of ssa1ts yeast cells. (B) Re-localization of p33 to the membrane fraction in ssa1ts yeast after shifting down the temperature to 23°C based on fractionation experiments. Disrupted yeast cells were separated to soluble (cytosolic) and membrane fractions via differential centrifugation, followed by Western blotting to detect YFP-p33. Top panel: the samples were taken 6 hours after down-shifting to 23°C (6 hr). Bottom panel: PGK, a soluble yeast protein, and ALP membrane protein were used as internal controls in the Western-blots. See panel A for further details about the growing conditions for yeast samples. (C) Western blot analysis of localization of YFP-p33 in ssa1ts yeast cells when the samples were taken prior to down-shifting to 23°C (0 hr). Note the partial re-distribution of YFP-p33 from the soluble fraction at the 0 time point (panel C) to the membrane fraction at the 6 hour time point (panel B) in ssa1ts yeast.
To test how rapidly YFP-p33 is re-localized to the membrane, we used time course experiments based on confocal microscopy and cell fractionation. We found that 30 min after the downshift to 23°C, a small number of cells already showed several punctate structures, though the punctate structures became more visible by microscopy and detectable in the membrane fraction after 1-2 hours (Fig. 7A and D). The amounts of YFP-p33 and CFP-p92 in the membrane fraction became as abundant after 3 hours as after 6 hours (Fig. 7D, lanes 7-8 versus 11-12). Interestingly, the sizes of individual punctate structures became bigger over time after the temperature downshift in the absence of new YFP-p33 synthesis (Fig. 7A). This suggests on-going membrane rearrangement caused by the insertion of p33 into the membrane due to the presence of activated Ssa1pts. Moreover, it seems that it requires 30 min to 3 hours for the re-activated Ssa1pts to help re-localize YFP-p33 from the cytosol to the membrane. We observed the characteristic punctate structures with YFP-p33 in SSA1wt yeast shifted from 32°C to 23°C or grown continuously at 23°C over time (Fig. 7B-C), suggesting that YFP-p33 localized correctly and efficiently in the presence of functional Ssa1pwt at both temperatures.
Fig. 7.
Time-course experiments to analyze re-distribution of p33 from the cytosol to the peroxisomes after shifting down the temperature from nonpermissive to permissive in ssa1ts yeast. (A) Confocal laser microscopy shows the localization of YFP-p33 in ssa1ts yeast grown at 23°C in a media containing glucose (to shut down the production of new p33/p92/repRNA) until the shown time point after switching from 32°C. See the details about yeast culturing in the legend to Fig. 6A. Note that the sizes of the characteristic punctate structures containing YFP-p33 grew over time in ssa1ts yeast cells. (B) Localization of YFP-p33 in SSA1wt yeast grown at 23°C in a media containing glucose until the shown time point after switching from 32°C. See further details in panel A. (C) Localization of YFP-p33 in SSA1wt yeast grown continuously at 23°C in a media containing galactose. (D) Western blot analysis of fractionation experiments shows the increasing extent of re-localization of p33 and p92pol from the soluble fraction to the membrane fraction in ssa1ts yeast after shifting down the temperature to 23°C. See the details for yeast culturing in panel A. The fractions were obtained as described in Fig. 6B. (E) Western blot analysis of p33 level in total protein extracts prepared from yeast grown as described in panel A and D.
Downshift to 23°C activates the tombusvirus replicase in ssa1ts yeast
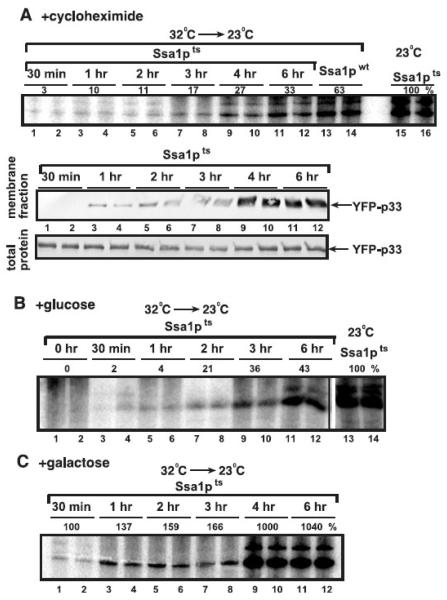
To test if the above observed changes in the subcellular localization of YFP-p33 during downshift to 23°C correlates with enhanced tombusvirus replicase activity, we measured the replicase activity on the co-purified repRNA in the membrane fraction of ssa1ts yeast. We added either cycloheximide or glucose to the growth media to inhibit the production of new YFP-p33 and CFP-p92 in ssa1ts yeast after downshifting the temperature (Fig. 8A-B). It is important to note that the cycloheximide treatment down-regulates the production of all new host and viral proteins, while glucose selectively inhibits the production of new YFP-p33 and CFP-p92, which are driven from the GAL1 promoter. These in vitro experiments revealed that the replicase activity continuously increased in samples taken from 30 min to 4-6 hours, reaching to ~30% level of the replicase activity obtained with the preparation from ssa1ts yeast cultured at 23°C (Fig. 8A). Thus, it takes from 30 min to 3-4 hours for the re-activated Ssa1pts to facilitate the assembly/activation of the tombusvirus replicase in the absence of new protein synthesis (+cyclehaximide, Fig. 8A, top panel). Similarly, the membrane-localization of YFP-p33 also needed from 60 min to 3-4 hours under the same conditions (Fig. 8A, bottom panels). The picture was somewhat similar in the first 2 hours after the downshift to 23°C in ssa1ts yeast cultured in the presence of glucose, which inhibits the production of new YFP-p33 and CFP-p92 (Fig. 8B). However, the replicase activity at 3-4 hr time points reached up to ~40% of the replicase activity obtained with the preparation from ssa1ts yeast cultured at 23°C (Fig. 8B). This moderate increase in replicase activity when compared with the cycloheximide treatment (Fig. 8A) is likely due to the newly produced active Ssa1pts after the downshift that seems to affect protein functions by the 4-hour time point. Indeed, we observed a ~6-fold increase in replicase activity from the 4-hour time point after the downshift in ssa1ts yeast cultured at 23°C in a galactose containing media, thus resulting in new production of YFP-p33 and CFP-p92 as well as Ssa1pts (Fig. 8C, lanes 9-12). Altogether, these experiments demonstrated that the re-activated Ssa1pts was capable of supporting efficient formation of the tombusvirus replicase, further confirming the essential role of cytosolic Hsp70 in tombusvirus replication.
Fig. 8.
The effect of Ssa1pts on the tombusvirus replicase activity. (A) Time-course experiments were performed to analyze the tombusvirus replicase activity in vitro in membrane fractions obtained from ssa1ts yeast after shifting down the temperature from nonpermissive to permissive. p33/p92pol replication proteins and DI-72 repRNA were expressed from the GAL1/GAL10 promoters for 16 hours at 32°C, followed by adding cycloheximide (100 μg/ml final concentration, which halted yeast growth) to stop translation and shifting the temperature to 23°C to activate Ssa1pts. Top panel: Denaturing PAGE analysis of in vitro replicase products obtained using the membrane-enriched fraction carrying the co-purified repRNA. The activity of the tombusvirus replicase (lanes 15-16) obtained from ssa1ts yeast grown continuously at 23°C in the presence of cycloheximide was taken as 100%. Middle panel: Western blotting of YFP-p33 present in the membrane fractions used for the in vitro replicase assay in the top panel. Bottom panel: Western blotting of YFP-p33 present in the total protein extract that was used to adjust the amount of replication proteins in each sample. (B) Denaturing PAGE analysis of in vitro replicase products obtained using the membrane-enriched fraction carrying the co-purified repRNA. The replicase preparations were obtained from ssa1ts yeast grown for 16 hours at 32°C in a media containing galactose, followed by changing to a media containing glucose to stop translation of p33/p92pol and shifting the temperature to 23°C to activate Ssa1pts. See further details in panel A. (C) In vitro replicase assay with tombusvirus replicase preparations obtained from ssa1ts yeast grown for 16 hours at 32°C in a media containing galactose, followed by shifting the temperature to 23°C to activate Ssa1pts. Note that p33/p92pol are produced continously under this condition. The image shows the denaturing PAGE analysis of in vitro replicase products obtained using the membrane-enriched fraction carrying the co-purified repRNA. See further details in panel A. The level of RNA synthesis by the replicase preparation obtained from yeast 30 min after the down-shift is taken as 100%.
DISCUSSION
Although host factors likely play essential and/or regulatory functions in replication of (+)RNA viruses, progress in our understanding how these factors are involved in virus replication could be difficult due to the essential roles of these factors in the viability of the host cells, functional redundancy or lack of knowledge about their functions. Therefore, yeast is a valuable model host due to amenable genetics and advanced information on many genes (Nagy, 2008). Accordingly, in this work we show the usefulness of yeast for studies on the essential Hsp70 proteins in tombusvirus replication.
Hsp70 is a highly conserved family of genes in all eukaryotic organisms showing high similarity with bacterial DnaK protein chaperones. Among the 14 HSP70 genes in yeast, SSA1-4, SSB1-2, and SSE1-2 are cytosolic, while others are localized in the ER or mitochondria (Ahsen and Pfanner, 1997; James, Pfund, and Craig, 1997). In addition, the eukaryotic cell contains a battery of other protein chaperones, such as the HSP90 family and the J-domain HSP40 co-chaperones (Brodsky and Chiosis, 2006). However, the purified tombusvirus replicase contains only Ssa1p and Ssa2p Hsp70 proteins at detectable level (Serva and Nagy, 2006), suggesting that tombusviruses recruit only this group of cytosolic chaperones. Inactivation of SSA1/2, however, only partially debilitate TBSV repRNA accumulation (Fig. 1A) (Serva and Nagy, 2006), and prior heat shock treatment enhances TBSV replication, indicating that there is a heat shock inducible factor(s), which is capable of complementing the missing Ssa1/2p function for TBSV replication in yeast (Fig. 1B). In this work, we demonstrate that complementation of SSA1/2 is due to the stress-inducible SSA3/4 genes, which are 80% homologous with SSA1/2 (Ahsen and Pfanner, 1997). This conclusion is based on (i) that recombinant Ssa3p (Fig. 1C) can facilitate the assembly of the TBSV replicase in vitro; and (ii) we did not observe complementation of TBSV replication (Fig. 2A) when we used ssa1ts yeast strain lacking functional SSA2/3/4 and carrying ssa1ts, which was inactivated at a nonpermissive temperature (Becker et al., 1996). Also, the replicase preparation from ssa1ts yeast grown at the nonpermissive temperature was inactive (Fig. 2C), arguing that the SSA1/2/3/4 subfamily of HSP70 is essential for TBSV replication, albeit individual members in the subfamily play redundant function. Thus, the numerous other heat shock proteins of yeast cannot complement the functions provided by SSA subfamily for TBSV replication.
What steps of replication are affected by Hsp70? The data presented in this and previous papers (Pogany et al., 2008; Wang, Stork, and Nagy, 2009) indicate that Ssa1p plays a role in the early steps of tombusvirus replication, including subcellular localization of the viral replication proteins, membrane insertion of the replication proteins and the assembly of the viral replicase complex. Accordingly, in vitro assembly of the TBSV replicase based on a yeast cell-free extract revealed that the constitutively expressed Ssa1p (Pogany et al., 2008) as well as the heat-induced Ssa3p (Fig. 1C) could facilitate the assembly event. Also, YFP-p33 is mislocalized to the cytosol in ssa1ts yeast cultured at the nonpermissive temperature, instead of forming puncate structures, which are characteristic for p33 associated with the peroxisome-membrane observed at the permissive temperature (Fig. 3A). Thus, functional Ssa1p is required for correct subcellular localization of the viral replication proteins. Interestingly, based on time course experiments, in which Ssa1pts was re-activated by shifting down to permissive temperature, we found correlation among accumulation of p33 in the membrane fraction (Fig. 7D), the formation of punctate structures by YFP-p33 from the diffused cytosolic stage (Fig. 7A), and the in vitro activity of the tombusvirus replicase in membrane preparations (Fig. 8A), suggesting that Ssa1p is involved in these events. Indeed, we have shown previously that Ssa1p facilitates the membrane-insertion of p33 in vitro (Wang, Stork, and Nagy, 2009). We have also shown that the targeted membranes include the peroxisome and ER membranes (Jonczyk et al., 2007; Pathak, Sasvari, and Nagy, 2008). Apparently, the membrane insertion step for p33 and p92pol is essential for the formation of active replicase complex, since the soluble, cytosolic tombusvirus replicase proteins cannot form functional replicase (Fig. 8B) (McCartney et al., 2005; Panavas et al., 2005a; Wang, Stork, and Nagy, 2009) and no accumulation of TBSV repRNA is detected in yeast cells showing cytosolic distribution of YFP-p33 (Fig. 2A and 6B).
In contrast with the multiple roles for Ssa1p in the early events of tombusvirus replication, we did not find evidence on the role of Ssa1p during viral RNA synthesis, including minus- or plus-strand synthesis. For example, the isolated replicase complex obtained from ssa1ts yeast grown at the permissive temperature was as active in viral RNA synthesis at the nonpremissive 37°C as at 25°C in vitro (Fig. 4). Moreover, an in vitro TBSV replicase assembly assay performed with Ssa1pts or Ssa1pwt at the permissive 20°C temperature, followed by RNA synthesis at a partially nonpermissive temperature for Ssa1pts showed comparable level of activity for the replicase assembled with Ssa1pts and Ssa1pwt (Fig. 5B), arguing strongly against the role of Ssa1p during viral RNA synthesis in vitro.
To gain insight into the role of Ssa1p in p33 localization and the assembly of the tombusvirus replicase in yeast, we used temperature shift experiments from nonpermissive to permissive in the absence or presence of new p33/p92pol production. It took only 30 min to detect the activity of the tombusvirus replicase after the downshift (Fig. 8A-C), which is similar to the time requirement for tombusvirus replicase assembly in vitro (Pogany and Nagy, 2008; Pogany et al., 2008). However, the activity of the tombusvirus replicase increased ~10-fold when measured 3-4 hours after the downshift, suggesting that most of the replicase assembly or localization of the replication proteins take place slowly. We speculate that this slow replicase assembly and/or subcellular localization processes are due to the limiting amount of available re-activated Ssa1pts since misfolded/mislocalized host proteins might also compete with the viral replication proteins for the service of Ssa1pts after the downshift.
This work provides evidence that the heat-inducible SSA3/4 can partially complement the functions of the constitutively-expressed SSA1/2. This is based on the finding that the stimulating effect of heat shock treatment on TBSV replication in ssa1ssa2 mutant yeast (Fig. 1) was not observed when we used Ssa1pts yeast lacking functional SSA2/3/4, suggesting that the heat-inducible host factor is likely Ssa3p and/or Ssa4p that can efficiently complement the missing Ssa1/2p functions for TBSV replication in ssa1ssa2 mutant yeast. In addition, we also observed that the recombinant Ssa3p was capable of assembling the active TBSV replicase using an in vitro replication assay (Fig. 1C).
The selective recruitment of cytosolic Hsp70 for viral TBSV replication might be more general phenomena involving numerous viruses. Co-opting cellular chaperones could be a very successful approach for viruses, since these proteins are abundant and conserved, thus facilitating their recruitment for virus infections. In addition, the cellular chaperones play significant roles in innate resistance responses by the host and their sequestration by various viruses could inhibit antiviral responses from the host cell (Mayer, 2005).
Altogether, this work firmly establishes an essential role for SSA1/2/3/4 subfamily of Hsp70 in TBSV replication. The two constitutively expressed members, Ssa1p and Ssa2p, play the major role during TBSV replication, whereas the heat-inducible Ssa3p and Ssa4p might contribute to TBSV accumulation only to a small extent at low temperature and more so at high temperatures. We show that Ssa1p is essential for TBSV accumulation in the absence of the other SSAs. The function of HSP70 is in the intracellular distribution and membrane insertion of the viral replication proteins, as well as the assembly of the viral replicase. Previous work has also shown the importance of the cytosolic Hsp70 in TBSV genomic RNA replication in plant cells and whole plant hosts based on knockdown and inhibition experiments (Wang, Stork, and Nagy, 2009). The obtained data are consistent with Hsp70 playing multiple functions during the replication of tombusvirus RNA.
Materials and Methods
Yeast strains and expression plasmids
The temperature sensitive ssa1 mutant (ssa1ts ssa2 ssa3 ssa4) strain DS10 (ssa 1-45BKD, ssa2::LEU2 ssa3::TRP1 ssa4::LYS2), the wt SSA1 strain (SSA1wt ssa2 ssa3 ssa4) and the double mutant (ssa1ssa2) strain MW123 (his3 leu2 lys2 trp1 ura3 ssa1::HIS3 ssa2::LEU2) was kindly provided by Elizabeth A. Craig (University of Wisconsin) (Becker et al., 1996). We used pESC-YFP-p33-DI-RNA dual construct (Jonczyk et al., 2007) to express YFP-p33 and DI-72 (+)RNA from GAL1 and GAL10 promoters, respectively.
To express GFP-SKL carrying the three-amino acid SKL peroxisomal targeting signal (Kragler et al., 1993), we PCR amplified the GFP sequence in combination with the C-terminally fused SKL sequence from pYES-GFP (Panavas et al., 2005a) as the template using the combination of primers #1292 (5′-CGGCAAGCTTACCATGGTGAGCAAGGGCGAGGAGCTGT), and #2410 (5′-CGCGGGATCCTTATAATTTAGACTTGTACAGCTCGTCCATGC) as well as #1292 (5′-CGGCAAGCTTACCATGGTGAGCAAGGGCGAGGAGCTGT), and #2411 (5′-CGCGGGATCCTTACAACTTGGACTTGTACAGCTCGTCCATGC). The two separate PCR products for GFP-SKL were treated with HindIII and BamHI, followed by their separate ligations into pYES-NT/C. The obtained two GFP-SKL constructs behaved the same way in our experiments (not shown).
Subcellular fractionation
Yeast cells were grown to an optical density (OD600) between 0.8 and 1.0. One hundred mg of cells were broken by using a cell-homogenizer (Fast prep) in 600 μl of yeast lysis buffer (200 mM sorbitol, 50 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma) (Wang, Stork, and Nagy, 2009). The obtained extracts were centrifuged in a microcentrifuge for 5 min at 100 × g to pellet cell debris. The supernatant was subsequently centrifuged at 21,000 × g to separate cytosolic and membrane-associated proteins into supernatant and pellet fractions, respectively (Wang, Stork, and Nagy, 2009). Pellets were re-suspended in the lysis buffer and aliquots corresponding to equal OD600 units of the original cell culture were analyzed by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting procedures as described previously (Jonczyk et al., 2007; Panaviene et al., 2004).
RNA analysis
Total RNA isolation and Northern blot analysis were performed as described previously (Panavas and Nagy, 2003; Panaviene et al., 2004). Briefly, for extraction of total RNA, yeast cells were broken by a cell-homogenizer (Genogrinder) for ~1 min at room temperature with equal volumes of RNA extraction buffer (50 mM Na-acetate, pH 5.2, 10 mM EDTA, 1% SDS) and water-saturated phenol and then incubated for 4 min at 65°C, followed by ethanol precipitation. The obtained RNA samples were separated on a 1.5% agarose gel and transferred to Hybond-XL membrane (Amersham) before hybridization with DI-72 RNA-specific probe. For detection of plus-strand repRNA, we prepared 32P-labeled RIII/IV(minus) probe generated via T7 transcription from PCR-made template obtained with primers #1165 (AGCGAGTAAGACAGACTCTTCA) and #22 (GTAATACGACTCACTATAGGGCTGCATTTCTGCAATGTTCC) on DI-72 templates (White and Morris, 1994).
Protein analysis
For protein analysis, we used an aliquot of samples used for RNA analysis. A total of 50 ml yeast culture was harvested, the pelleted cells were re-suspended in 150 μl cold extraction buffer (200 mM sorbitol, 50 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma), and 250 μl of glass beads were added to each sample. The cells were broken with a genogrinder for 2 min at 1,500 rpm. Each sample was further mixed with 600 μl pre-chilled extraction buffer, and unbroken cells were removed by centrifugation at 100 × g for 5 min. The supernatant was mixed with 1/2 volume of 3× SDS-PAGE sample buffer followed by SDS-PAGE and Western blot analysis as described previously (Panaviene et al., 2004).
Tombusvirus replicase assays
The “membrane-enriched” replicase preparations, which are suitable to test the replicase activity on the endogenous templates present within the replicase preparation, were obtained as previously described (Panaviene, Panavas, and Nagy, 2005; Panaviene et al., 2004). Briefly, frozen yeast cells were homogenized with Genogrinder for 2 min at 1,500 rpm in 150 μl cold extraction buffer (200 mM sorbitol, 50 mM Tris-HCl, pH 7.5, 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma) plus 250 μl of glass beads. Each sample was further mixed with 600 μl prechilled extraction buffer, and unbroken cells were removed by centrifugation at 100 × g for 5 min at 4°C. The supernatant was centrifuged for 10 min at 21,000 × g at 4°C, and then the pellet was re-suspended and used in a standard tombusvirus replicase assay. Because no template was added to the in vitro reaction, the replicase preparation could only use the endogenous template present within the enriched membrane fraction. The replicase products were phenol-chloroform extracted, precipitated with isopropanol-ammonium acetate, and analyzed under denaturing conditions (5% PAGE containing 8 M urea).
Replication assay using the membrane fraction of the yeast cell-free extract
The replication assay containing 0.5 μg purified recombinant TBSV MBP-p33 and/or TBSV MBP-p92 was done according to (Pogany et al., 2008). The membrane fraction of the cell-free extract was obtained by centrifugation at 4 °C for 10 min to separate the “soluble” (supernatant) and “membrane” (pellet) fraction (21,000 × g). To remove possible contaminating soluble proteins, the pellet was washed with buffer A (30 mM HEPES-KOH pH 7.4, 100 mM potassium acetate, and 2 mM magnesium acetate) followed by centrifugation at 4 °C for 10 min and re-suspension of the pellet in buffer A.
Purification of recombinant tombusvirus replicase proteins from E. coli
TBSV replicase proteins were expressed and purified according to (Rajendran and Nagy, 2003), except using HEPES-KOH buffer (50 mM, pH 7.4) during purification and the elution buffer contained 1 mM DTT instead of the 10 mM β-mercaptoethanol.
Purification of Ssa3p as well as wt and mutant Ssa1p from yeast
The copper-inducible CUP1 promoter was used to express the wt FLAG-tagged Ssa3p, wt FLAG-Ssa1p and the temperature sensitive (ts) FLAG-Ssa1p (Becker et al., 1996) (kindly provided by Elizabeth Craig [University of Wisconsin]) from plasmid pEsc-His/Cup-FLAG/ssa1 wt and pEsc-His/Cup-FLAG/ssa1ts. Ssa1p was affinity purified as described previously (Pogany et al., 2008).
Confocal laser microscopy
To view yeast cells expressing different fluorescent fusion proteins, yeast strains were transformed with pESC-YFP-p33-DI-RNA, in combination with pYES-p92 or GFP-SKL. The confocal microscopy was performed on an Olympus FV1000 (Olympus America Inc., Melville, New York) as described (Jonczyk et al., 2007; Wang and Nagy, 2008; Wang, Stork, and Nagy, 2009).
ACKNOWLEDGEMENTS
We thank Drs. Daniel Barajas and Zhenghe Li for critical reading of the manuscript and for very helpful suggestions. This work was supported by NIH-NIAID and by the Kentucky Tobacco Research and Development Center at the University of Kentucky, awarded to PDN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77(15):8181–6. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsen, Pfanner Molecular chaperones: towards a characterization of the heat-shock protein 70 family. Trends Cell Biol. 1997;7(3):129–33. doi: 10.1016/S0962-8924(96)10056-8. [DOI] [PubMed] [Google Scholar]
- Alzhanova DV, Napuli AJ, Creamer R, Dolja VV. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. Embo J. 2001;20(24):6997–7007. doi: 10.1093/emboj/20.24.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio F, Thomas CL, Lederer C, Niu Y, Wang D, Maule AJ. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 2005;138(1):529–36. doi: 10.1104/pp.104.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda MA, Escaler M, Wang D, Maule AJ. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc Natl Acad Sci U S A. 1996;93(26):15289–93. doi: 10.1073/pnas.93.26.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16(8):4378–86. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Craig EA. Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol Cell Biol. 1990;10(6):3262–7. doi: 10.1128/mcb.10.6.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6(11):1215–25. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- Brown G, Rixon HW, Steel J, McDonald TP, Pitt AR, Graham S, Sugrue RJ. Evidence for an association between heat shock protein 70 and the respiratory syncytial virus polymerase complex within lipid-raft membranes during virus infection. Virology. 2005;338(1):69–80. doi: 10.1016/j.virol.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19(4):445–52. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberte JF, Fortin MG. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology. 2008;374(1):217–27. doi: 10.1016/j.virol.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol. 2004;78(23):13122–31. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275(5298):387–9. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol. 2006;80(15):7394–404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk M, Pathak KB, Sharma M, Nagy PD. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362(2):320–30. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kampmueller KM, Miller DJ. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J Virol. 2005;79(11):6827–37. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F, Langeder A, Raupachova J, Binder M, Hartig A. Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. J Cell Biol. 1993;120(3):665–73. doi: 10.1083/jcb.120.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455(7210):242–5. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100(26):15764–9. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. Cdc34p Ubiquitin-Conjugating Enzyme Is a Component of the Tombusvirus Replicase Complex and Ubiquitinates p33 Replication Protein. J Virol. 2008;82(14):6911–26. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009 doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6(3):201–8. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. Localization of the Tomato Bushy Stunt Virus Replication Protein p33 Reveals a Peroxisome-to-Endoplasmic Reticulum Sorting Pathway. Plant Cell. 2005;17(12):3513–31. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002;277(47):45306–14. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- Monkewich S, Lin HX, Fabian MR, Xu W, Na H, Ray D, Chernysheva OA, Nagy PD, White KA. The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication. J Virol. 2005;79(8):4848–58. doi: 10.1128/JVI.79.8.4848-4858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PD. Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol. 2008;46:217–42. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology. 2000;276(2):279–88. doi: 10.1006/viro.2000.0577. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344(1):211–20. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Nishikiori M, Dohi K, Mori M, Meshi T, Naito S, Ishikawa M. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J Virol. 2006;80(17):8459–68. doi: 10.1128/JVI.00545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005a;338(1):81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T, Nagy PD. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology. 2003;314(1):315–25. doi: 10.1016/s0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Brasher J, Nagy PD. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A. 2005b;102(20):7326–31. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Nagy PD. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J Virol. 2005;79(16):10608–18. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Serva S, Nagy PD. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol. 2004;78(15):8254–63. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379(2):294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Peremyslov VV, Hagiwara Y, Dolja VV. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc Natl Acad Sci U S A. 1999;96(26):14771–6. doi: 10.1073/pnas.96.26.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Nagy PD. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J Virol. 2008;82(12):5967–80. doi: 10.1128/JVI.02737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Stork J, Li Z, Nagy PD. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A. 2008;105(50):19956–61. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol. 2005;79(8):4859–69. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanungo KR, Shaji D, Mathur M, Banerjee AK. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc Natl Acad Sci U S A. 2004;101(16):5952–7. doi: 10.1073/pnas.0401449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran KS, Nagy PD. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J Virol. 2003;77(17):9244–58. doi: 10.1128/JVI.77.17.9244-9258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104(31):12884–9. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–73. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serva S, Nagy PD. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol. 2006;80(5):2162–9. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A. 2005;102(30):10545–50. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ST, Lai MM. Viral and cellular proteins involved in coronavirus replication. Curr Top Microbiol Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Retzlaff M, Nassal M, Beck J. Chaperone activation of the hepadnaviral reverse transcriptase for template RNA binding is established by the Hsp70 and stimulated by the Hsp90 system. Nucleic Acids Res. 2007;35(18):6124–36. doi: 10.1093/nar/gkm628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavis JE, Massey B, Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J Virol. 1998;72(7):5789–96. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Mizuno T, Diez J, Naito S, Ahlquist P, Ishikawa M. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J Virol. 2003;77(5):2990–7. doi: 10.1128/JVI.77.5.2990-2997.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Nagy PD. Tomato bushy stunt virus Co-Opts the RNA-Binding Function of a Host Metabolic Enzyme for Viral Genomic RNA Synthesis. Cell Host Microbe. 2008;3(3):178–87. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Wang RY, Stork J, Nagy PD. A key role for heat shock protein 70 in localization and insertion of the tombusvirus replication proteins to intracellular membranes. J Virol. 2009 doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SA, Miller DJ. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J Virol. 2008;82(4):2004–12. doi: 10.1128/JVI.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Morris TJ. Enhanced competitiveness of tomato bushy stunt virus defective interfering RNAs by segment duplication or nucleotide insertion. J Virol. 1994;68(9):6092–6. doi: 10.1128/jvi.68.9.6092-6096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Nagy PD. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acid Res Mol Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 2003;33(2):271–83. doi: 10.1046/j.1365-313x.2003.01625.x. [DOI] [PubMed] [Google Scholar]
- Whitham SA, Yang C, Goodin MM. Global impact: elucidating plant responses to viral infection. Mol Plant Microbe Interact. 2006;19(11):1207–15. doi: 10.1094/MPMI-19-1207. [DOI] [PubMed] [Google Scholar]