Abstract
The Chk1 protein kinase preserves genome integrity in normal proliferating cells and in cells experiencing replicative- and genotoxic-stress. Chk1 is currently being targeted in anti-cancer regimens. Here we identify damaged DNA-binding protein 1 (DDB1) as a novel Chk1 interacting protein. DDB1 is part of an E3 ligase complex that includes the cullin proteins, Cul4A and 4B. We report that Cul4A/DDB1 negatively regulates Chk1 stability in vivo. Chk1 associates with Cul4A/DDB1 during an unperturbed cell division cycle and both Chk1 phosphorylation and replication-stress enhanced these interactions. Cul4A/DDB1 regulates Chk1 ubiquitination in vivo and Chk1 is directly ubiquitinated in vitro in a Cul4A/DDB1-dependent manner. Furthermore, Chk1 is stabilized in cells deficient for Cul4A/DDB1. This study demonstrates that Chk1 abundance is regulated by the Cul4A/DDB1 ubiquitin ligase during an unperturbed cell division cycle, in response to replicative stress and upon HSP90 inhibition and that deregulation of the Chk1/Cul4A/DDB1 pathway perturbs the IR-induced G2 checkpoint.
Keywords: DNA damage, checkpoint, cell cycle, ubiquitination, HSP90
INTRODUCTION
Chk1 is a serine/threonine protein kinase that functions to maintain genome integrity in normal cycling cells and in cells exposed to replicative- and genotoxic-stress (1, 2). Chk1 regulates cell cycle progression, in part, by phosphorylating the Cdc25A protein phosphatase (1–4). Phosphorylation of Cdc25A by Chk1 targets it for ubiquitin-mediated proteolysis and prevents it from binding to and activating Cdk1/cyclin B1 inappropriately during the S- and G2-phases of the cell division cycle (1, 2, 5–7). Chk1 enforces cell cycle delays in cells experiencing replicative- or genotoxic-stress by inhibiting elongation of replication forks; by preventing firing of new origins of replication and by blocking mitotic entry (8, 9). Loss of the Chk1/Cdc25A regulatory pathway disrupts both the DNA replication and DNA damage checkpoints (2). In addition to regulating DNA replication and mitotic entry, Chk1 has also been shown to stabilize stalled replication forks when DNA replication is impeded, to regulate transcription through phosphorylation of histone H3 and to regulate DNA repair by phosphorylating the Fanconi anemia protein FANCE (8, 10–12). Chk1 carries out its functions both in the nucleus and at the centrosome (13). Drugs that block Chk1 kinase activity or enhance its proteolysis by interfering with binding to HSP90 are currently being tested as anti-cancer agents (14–17).
Chk1 is regulated by reversible phosphorylation and by ubiquitin-mediated proteolysis. Under periods of replicative stress, the ATRIP/ATR module binds to single- stranded DNA and together with Rad17 and the 9-1-1 complex activates Chk1 in a Claspin-dependent manner (18–22). ATR directly phosphorylates Chk1 on two C-terminal residues, serines 317 and 345 (23, 24). The dephosphorylation of Chk1 is mediated by at least two phosphatases. These include PP2A, which maintains Chk1 in a hypophosphorylated state in normal cycling cells (25) and PPM1D (Wip1), which dephosphorylates Chk1 during checkpoint recovery (26). In addition, Cul1- and Cul4A-containing E3 ubiquitin ligases have been shown to ubiquitinate Chk1 during periods of prolonged replication stress (27). The Cullins assemble a large number of distinct ubiquitin ligases by binding to ROC1, a RING protein, and to several distinct targeting subunits that recruit substrates for ubiquitination (28). The targeting subunits that recruit Chk1 to Cul1 and Cul4A have not been identified. Here we demonstrate that DDB1, a triple beta propeller adapter protein, targets the Cul4 E3 ubiquitin ligase complex to Chk1.
MATERIALS AND METHODS
Cell lines and lysis buffer
HeLa and HEK 293 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% bovine growth serum. Cells were lysed in MCLB (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, and 2 mM DTT) containing 1 mM sodium orthovanadate, 10 mM β-glycerophosphate, 1 μM microcystin, 2 mM PMSF, 10 μg/ml aprotinin and 5 μg/ml leupeptin (25).
Chemicals and drugs
Cycloheximide (CHX), MG132, hydroxyurea (HU), wortmannin (Wort) and caffeine (Caff) were purchased from Sigma Chemical Co. Gö6976 and Geldanamycin were purchased from Calbiochem. SN38 was purchased from Avachem.
Antibodies and Western blotting
Primary antibodies used in this study included: mouse monoclonal anti-Chk1 (G-4, Santa Cruz Biotechnology); affinity purified rabbit anti-Chk1 (24); mouse anti-β-catenin (BD Biosciences); goat anti-DDB1 (Abcam); rabbit anti-V5 (Abcam); rabbit anti-Cul4A (Rockland); rabbit anti-HA (Santa Cruz Biotechnology); mouse monoclonal anti-Flag (Sigma); mouse monoclonal anti-actin (Santa Cruz Biotechnology); mouse monoclonal anti-PCNA (Santa Cruz Biotechnology) and rabbit anti-pS345 and anti-pS317 Chk1 (Cell Signaling). Anti-Flag M2-Agarose were from Sigma. Secondary antibodies included: anti-rabbit HRP (Zymed), anti-goat HRP (Zymed) and anti-mouse HRP (Jackson ImmunoResearch). Immunoblots were visualized by chemiluminescence using an ECL kit (GE Healthcare) according to the manufacturer’s instructions and quantified by densitometry using Image J (29).
Plasmids
Chk1(WT) was amplified by PCR as a Xho I fragment from pcDNA3-myc-Chk1 (24) and was cloned into the Xho I site of pFlag(subscript)3-CMV (Sigma) to generate pFlag3Chk1 (WT). pFlag3Chk1 (SA/SA), which encodes a mutant form of Chk1 containing alanines in place of serines 317 and 345, was generated by site-directed mutagensis of pFlag3Chk1(WT) using the QuickChange Mutagenesis Kit (Stratagene) as described (24). V5-tagged DDB1 and Cul4A expression plasmids were generated using the Gateway System (Invitrogen) and purchased cDNAs (Invitrogen, ultimate ORF clones) to generate pcDNAnV5-DDB1 and pcDNAnV5-Cul4A. Plasmids encoding HA-ubiquitin have been described (30).
Isolation and identification of Chk1-associated proteins
HEK 293 cells (1.2 × 107) were transfected for 24 h with pFlag3-CMV control plasmid or plasmid encoding Flag3Chk1 (WT) using Superfect (Qiagen) and incubated in the presence of 10 mM HU for 4 h. Lysates were prepared and incubated with anti-Flag M2 affinity beads to generate control-beads and Flag3Chk1-beads. HU treated lysates from HEK 293 cells (4 × 108) were prepared and incubated with control- or Flag3Chk1-beads overnight. Immunocomplexes were washed 6 times with MCLB, boiled and resolved by SDS-PAGE. Gels were stained with Colloidal Commassie Blue (Invitrogen) and selected bands were excised from the gel and analyzed by mass spectrometry as described (31). Briefly, after in-gel trypsinization, peptides were analyzed by microcappillary reverse-phase HPLC nano-electrospray tandem mass spectrometry on a Finnigan LCQ DECA XP Plus quadrupole ion trap mass spectrometer at the Harvard Microchemistry Facility.
RNAi experiments
siRNAs used in this study were purchased from Dharmacon and included a control siRNA (D00121002); or siRNAs specific for Luciferase (D00140001); Cul4A (M01261000); Cul4B (M01796500); and DDB1 (M01289001). Approximately 2 × 105 HeLa cells were seeded in each well of a 6-well tissue culture dish. The following day, cells were transfected for 48 to 72 h with 100 nM siRNAs using DharmaFECT 2 or Dharmafect Duo reagents according to the manufacturer’s instructions. For monitoring the IR-induced G2 checkpoint, asynchronously growing HeLa cells were transfected with siRNAs for 24 h and then incubated with DMSO or 200 nM geldanamycin for another 24 h. Cells were then exposed to 10 Gy IR. In some cases, cells were also incubated with DMSO (vehicle) or Gö6976 (300 nM) and harvested 9 h after IR.
Chk1 immunoprecipitations
Endogenous Chk1 was immunoprecipitated as described (24). For ectopic Flag3Chk1 immunoprecipitations: 1 × 106 HeLa cells were seeded into 60 mm tissue culture dishes and the following day transfected with 2 μg pFlag3Chk1, 4 μg pcDNAnV5-DDB1 and 1.5 μg pcDNAnV5-Cul4A using Lipofectamine 2000 (Invitrogen). Lysates were prepared 24 h later and incubated with Protein A agarose (Pierce) for 1 h. Precleared lysates were then incubated with Flag M2 affinity beads for 4 h at 4°C and precipitates were washed 4 times with MCLB. Flag3Chk1 and associated proteins were eluted with 0.7 μg/μl of purified Flag3 peptide (Sigma) in MCLB. Eluates were subjected to SDS-PAGE followed by Western blotting.
Chk1 ubiquitination in vitro
To generate Chk1 substrate, 1 × 106 HeLa cells were transfected with 5 μg of pFlag3Chk1(WT) using Lipofectamine 2000 (Invitrogen). The following day, transfected cells were treated with 20 mM HU for 1 h to induce phosphorylation of Chk1. Cells were lysed in MCLB and lysates were pre-cleared with Protein A agarose. Flag3Chk1 was immunoprecipitated using 50 μl Anti-Flag M2-Agarose at 4°C for 2 h. Immunoprecipitated Flag3Chk1 was washed 6 times with MCLB, 5 times with LiCl buffer (0.5 M LiCl, 50 mM Tris, pH 8.0) and 2 times with ubiquitination buffer (25 mM Tris, pH 7.5, 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2mM ATP, 0.6 mM DTT). Flag3Chk1 was eluted in ubiquitination buffer containing 0.7μg/μl of Flag3 peptide. To immunoprecipitate Cul4A ligases, cells were treated with 20 mM HU for 1 h, lysed with MCLB and lysates were precleared with Protein A agarose. Cul4A was immunoprecipitated from 1 mg of total cell protein using 2 μl of anti-Cul4A antibody and 50 μl of Protein A agarose at 4°C for 4 h. Cul4A immunocomplexes were washed 5 times with MCLB and 2 times with ubiquitination buffer. Ubiquitin ligase reactions consisted of: Cul4A immunocomplexes, Flag3Chk1, and Sigma reagents: 12 μg of bovine ubiquitin, 1.2 μg of Flag-ubiquitin, 60 ng of E1, 300 ng of E2 (hUbc5c). Reactions were incubated at 37°C for 1 h and terminated by boiling the supernatant in loading buffer. Reactions were resolved by SDS-PAGE and analyzed by Western Blotting.
Chk1 ubiquitination in vivo
3 × 105 HeLa cells were co-transfected for 48 h with plasmids encoding Flag3Chk1 (0.5 μg), HA-Ubiquitin (1.5 μg) and 60 nM of the indicated RNAi using DharmaFECT Duo (Dharmacon). Transfected cells were then treated with 20 mM HU for 4 h. Cell lysates were prepared and incubated with Protein A agarose for 1 h. Precleared lysates were transferred to a fresh tube and incubated with Anti-Flag M2-Agarose. Precipitates were washed, subjected to SDS-PAGE and analyzed by Western blotting.
Cell Synchrony
To synchronize HeLa cells at the G1/S- border, cells were treated with 2 mM thymidine for 16 h. Cells were released from the block by washing twice with PBS, followed by release into complete growth medium containing 24 μM of both thymidine and 2′-deoxycytidine (Sigma). After 8 h, thymidine was added to the medium to a final concentration of 2 mM, and cells were cultured for an additional 16 h. Cells were then rinsed twice with PBS and cultured in complete growth medium. A fraction of the cells was subjected to flow cytometry to determine cell cycle position. Cells were harvested by trypsinization and collected by centrifugation. Cells were washed once with PBS and fixed in 5 ml of 70 % ethanol at 4°C. Cells were washed once with PBS/1% BSA and then incubated with 1 ml of PBS/1% BSA containing 30 μg/ml propidium iodide (PI) and 0.25 mg/ml RNase A for 1 h at room temperature. Cells were analyzed for DNA content by flow cytometry using a FACS Calibur (BD Biosciences). The data were analyzed using CellQuest Analysis software (BD Biosciences).
RESULTS
DDB1 identified as a novel Chk1-interacting protein
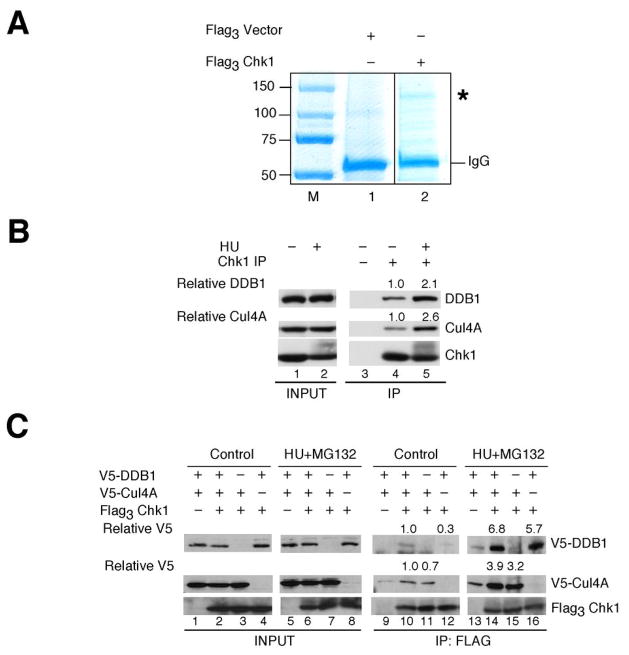
A proteomic approach was employed to identify novel upstream regulators and/or downstream targets of human Chk1. Flag-tagged Chk1 was affinity-purified from HU-treated HEK293 cells and then incubated in lysates prepared from HU-treated HEK293 cells. As a control, cells transfected with the pFlag3-vector were taken through the same procedure. As seen in Fig. 1A, a protein of ~130 kDa selectively co-purified with flag-tagged Chk1 (lane 2) but not control beads (lane 1). Mass spectrometry of this protein identified 26 distinct peptides from the human DDB1 protein (Supp. Fig. 1). These peptides corresponded to sequences spanning the entire DBB1 protein sequence.
Figure 1.
Proteomic screen identifies DDB1 as a novel Chk1 interacting protein. A, lysates prepared from HU-treated HEK293 cells transfected with control plasmid or plasmid encoding Flag3Chk1 were incubated with Anti-Flag M2-Agarose. Washed precipitates were then incubated in lysates prepared from HU-treated HEK293 cells. Bound proteins were resolved by SDS-PAGE and stained with Colloidal Coomassie Blue. The band denoted by an asterisk (*) was present in Flag3 Chk1 but not control precipitates. This protein was excised from the gel and identified as DDB1 by mass spectrometry. B, endogenous Chk1 was immunoprecipitated from control cells or cells treated with 20 mM HU for 6 h. Cells were also treated with 10 μM MG132 for 2 h prior to harvest. Precipitates were resolved by SDS-PAGE and analyzed by Western blotting. Densitometry was performed to determine relative amounts of DDB1 and Cul4A in Chk1 immunoprecipitates (n = 3). C, HeLa cells expressing the indicated tagged proteins were cultured in the presence of 20 mM HU and 50 μM MG132 for 4 h. Flag3Chk1 was immunoprecipitated and precipitates were analyzed by Western blotting. Densitometry was performed to quantitate relative amounts of DDB1 and Cul4A in Chk1 immunoprecipitates (n = 2). Relative levels of DDB1 and Cul4A are indicated.
DDB1 is part of an E3 ligase complex that includes the cullin proteins, Cul4A and Cul4B (28, 32). Interestingly, Cul4A has been reported to target Chk1 for ubiquitin-mediated proteolysis during a DNA damage checkpoint response (27). However, the subunits that target Cul4A to Chk1 has not been identified. Therefore, we tested whether interactions between Chk1 and Cul4A/DDB1 complexes could be detected in vivo. Endogenous Chk1 was immunoprecipitated from vehicle- or HU-treated HeLa cells and precipitates were analyzed for the presence of DDB1 and Cul4A by Western blotting. As seen in Fig. 1B, both DDB1 and Cul4A co-precipitated with Chk1 (lane 4) and associations were enhanced in HU-treated cells (lane 5). Similar observations were made when Chk1 was co-produced with Cul4A and DDB1 in vivo (Fig. 1C). Interestingly, enhanced associations were detected when Chk1 was co-produced with both Cul4A and DDB1 (lane 10) as compared with either subunit alone (lanes 11, 12) and HU-treatment significantly enhanced interactions between Chk1 and Cul4A/DDB1 (lanes 14–16). Upon HU-removal, a decrease in the interactions between Chk1 and Cul4A/DDB1 was observed coincident with a decrease in Chk1 phosphorylation on serine 317 (Supp. Fig. 2A).
Chk1 interactions with Cul4-DDB1 regulated by phosphorylation
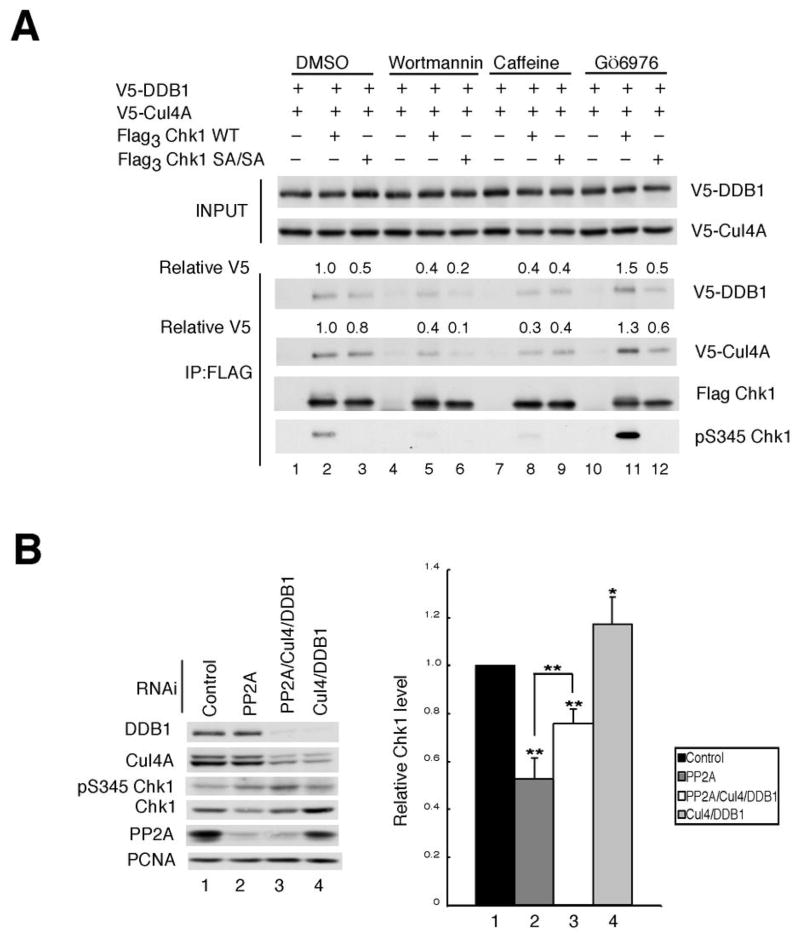
In cells experiencing replicative stress, the ATR protein kinase phosphorylates Chk1 on serines 317 and 345 (S317 and S345) (23, 24, 33–35). This pathway can be blocked by expressing a mutant of Chk1 containing alanine in place of both of these serine residues (SA/SA) or by treating cells with wortmannin or caffeine, inhibitors of the PI3K-related family, which include ATR and ATM (24, 36–38). As seen in Fig. 2A, interactions between Chk1 and Cul4A/DDB1 were impaired but not completely eliminated by mutating both of S317 and S345 (compare lanes 2 and 3). Wortmannin reduced the binding of Cul4/DDB1 to both WT Chk1 and the SA/SA mutant (compare lanes 2 to 5 and 3 to 6), as did caffeine (compare lanes 2 to 8 and 3 to 9). Total levels of Chk1 remained constant and phosphorylation of Chk1 on serine 317 was reduced under these experimental conditions (Supp. Fig 2B). Treatment of cells with Gö6976, a Chk1 inhibitor that enhances Chk1 phosphorylation on S317 and S345 in vivo (25), increased binding of the Cul4A/DDB1 ligase to Chk1 WT but not to Chk1 SA/SA (Fig. 2A, compare lanes 2 to 11 and 3 to 12). Enhanced binding between endogenous Chk1 and endogenous Cul4A/DDB1 was also observed in response to Gö6976-treatment (Supp. Fig. 2C). These results demonstrate that phosphorylation of Chk1 on S317 and S345 contributes to, but is not essential for, its interactions with Cul4A/DDB1.
Figure 2.
Chk1 phosphorylation regulates interactions with Cul4A and DDB1. A, asynchronously growing HeLa cells expressing the indicated tagged proteins were cultured in the presence of vehicle (DMSO), 50 μM wortmannin, 10 mM caffeine or 1 μM Gö6976 for 2 h each. Cells were then exposed to 20 mM HU and 10 μM MG132 for 4 h prior to harvest. Flag3 Chk1 was immunoprecipitated and precipitates were analyzed by Western blotting. Relative levels of DDB1 and Cul4 are indicated (n = 3). B, asynchronously growing HeLa cells were treated with control RNAi or RNAi specific for PP2A, Cul4A/DDB1 or PP2A/Cul4A/DDB1 for 48 h. Lysates were prepared and immunoblotted with antibodies specific for the indicated antibodies (left panel). The data from 4 independent experiments is presented graphically in the right panel. The data is presented as mean +/− standard deviation. P-values from Student’s t-test are shown when significantly different from control. Asterisks indicate significant p-values (* < 0.05; **<0.01)
Next, experiments were performed to monitor the association between Chk1 and the Cul4A/DDB1 complex as a function of the cell division cycle. Cells were arrested at the G1/S-border by a double thymidine block and release protocol. The association between Chk1 and Cul4A/DDB1 was monitored by immunoprecipitating endogenous Chk1 and testing for the co-precipitation of DDB1 and Cul4A by immunoblotting. As seen in Supp. Fig. 3A, enhanced interactions were detected during the S- and G2-phases of the cell division cycle.
We recently reported that Chk1 is continually phosphorylated on S317 and S345 by ATR during the S- and G2-phases of the cell division cycle (25). However, Chk1 phosphorylation on these residues is not readily observed because PP2A continuously dephosphorylates Chk1 on S317 and S345. Furthermore, in cells knocked-down for PP2A, Chk1 phosphorylation increases but Chk1 protein levels decrease (25). We asked if inhibition of the proteosome could restore Chk1 levels in PP2A-deficient cells. MG132-treatment was observed to partially rescue Chk1 levels in PP2A-deficient cells (Supp. Fig. 3B, C). We next asked whether Cul4A/DDB1 was responsible for targeting Chk1 to the proteosome in PP2A-deficient cells (Fig. 2B). As expected, loss of PP2A resulted in enhanced S345-phosphorylation and a concomitant 50% decrease in Chk1 levels (compare lanes 1 and 2). In contrast, Chk1 levels rose in Cul4A/DDB1-deficient cells (compare lanes 1 and 4). Importantly, Chk1 levels were partially restored when PP2A-deficient cells were knocked down for the Cul4A/DDB1 E3 ligase (compare lanes 2 and 3). These results support the conclusion that Chk1 phosphorylation promotes its interactions with Cul4A/DDB1 and that Cul4A/DDB1 targets Chk1 for ubiquitination and proteosomal degradation.
Cul4A and DDB1 required for Chk1 ubiquitination in vitro and in vivo
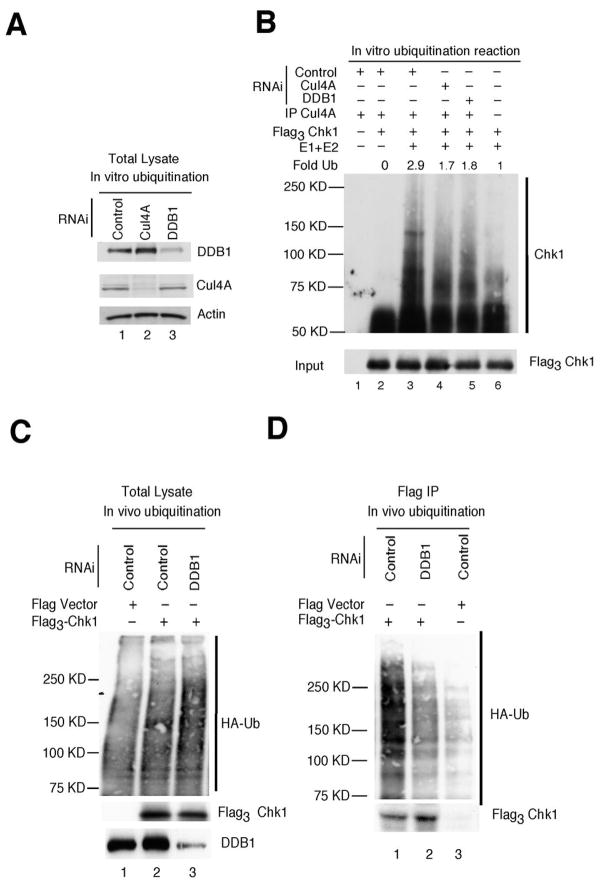
Cul4A is a member of the cullin family of ubiquitin-protein ligases and DDB1 is one of its targeting subunits. We assessed whether Chk1 could be directly ubiquitinated by the Cul4A/DDB1 ligase by performing ubiquitination reactions in vitro (Fig. 3A, B). Purified Flag3Chk1 was used as substrate and RNAi-treated cell lysates were used as a source of Cul4A complexes. Ubiquitination of Chk1 was not observed when Chk1 precipitates were incubated in the presence of Cul4A precipitates but in the absence of purified E1 and E2 (Fig. 3B, lane 2). A low level of Chk1 ubiquitination was observed when Chk1 precipitates were incubated with purified E1 and E2, indicating that Chk1 precipitates also contain E3 ligase activity (lane 6). Chk1 ubiquitination was enhanced in complete reactions containing Cul4A, E1 and E2 (lane 3) and was diminished when Cul4A was precipitated from cells knocked down for either Cul4A or DDB1 (lanes 4, 5). Next, we tested the requirement for Cul4A and DBB1 for Chk1 ubiquitination in vivo. Cells expressing Flag3Chk1 and HA-ubiquitin (HA-Ub) were transfected with siRNAs and Chk1 precipitates were analyzed for the presence of HA-Ub. Chk1 ubiquitation was diminished in cells knocked down for either DDB1 (Fig. 3D) or both DDB1 and Cul4A (Supp Fig. 4A). Taken together, these results demonstrate that Cul4A/DDB1 ubiquitinates Chk1 in vitro and that efficient Chk1 ubiquitination in vivo requires Cul4A and DDB1.
Figure 3.
Chk1 ubiquitination by Cul4A/DDB1. A, asynchronously growing HeLa cells transfected with RNAi targeting luciferase (control, lane 1), Cul4A (lane 2) or DDB1 (lane 3) were cultured in the presence of HU for 1h. Lysates were prepared and resolved directly by SDS-PAGE (A) or were incubated with a Cul4A-specific antibody to precipitate the Cul4A-containing E3 ligase for ubiquitination assays (B). B, Cul4A immunocomplexes were incubated alone (lane 1), with Flag3Chk1 precipitates (lane 2) or with Flag3Chk1 precipitates and purified E1 and E2 (lanes 3, 4, 5). Flag3Chk1 precipitates were also incubated with purified E1 and E2, in the absence of Cul4A immunocomplexes (lane 6). Ubiquitination assays were performed in vitro and reaction products were resolved by SDS-PAGE followed by Western blotting with antibodies specific for Chk1 (top panel) or Flag (bottom panel) (n = 4). C, D, Asynchronously growing HeLa cells were co-transfected with plasmids encoding Flag vector or Flag3Chk1, HA-ubiquitin and RNAi for 48 h and then treated with HU (20 mM) and MG132 (10 μM) for 4 h. Cell lysates were prepared and analyzed directly by Western blotting (panel C) or flag-tagged Chk1 precipitates were isolated prior to SDS-PAGE and subjected to Western blotting with antibodies specific for HA (top panel) and Flag (bottom panel) (n = 3).
Knockdown of Cul4 or DDB1 stabilizes Chk1 in vivo
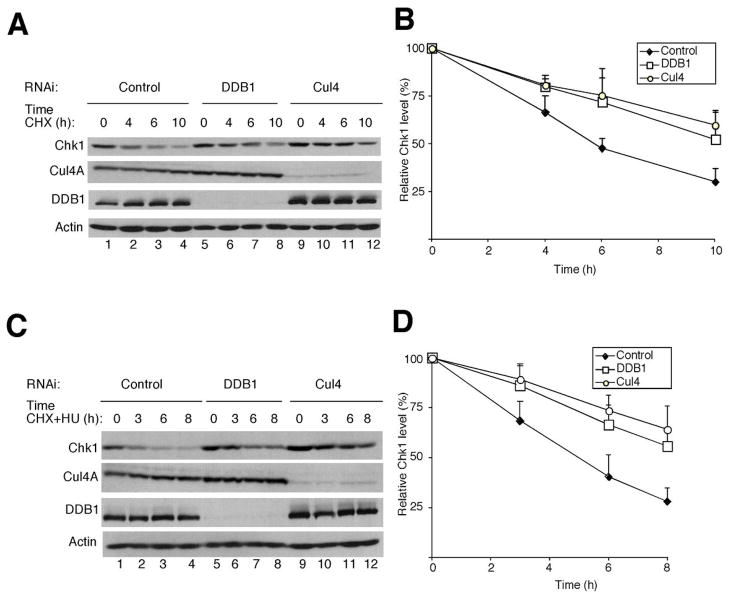
Next, experiments were performed to test whether the half-life of Chk1 is regulated by the Cul4A/DDB1 E3 ligase in vivo. To this end, HeLa cells were treated with control siRNAs or siRNAs specific for either Cul4 or DDB1 and the half-life of Chk1 was measured in the presence of cycloheximide (Fig. 4). These experiments were performed in both normal cycling cells (panels A, B) and in HU-treated cells (panels C, D). The half-life of Chk1 was determined to be ~6 h in normal cycling HeLa cells (Fig. 4B) and ~5 h in HU-treated cells (Fig. 4D). Loss of either Cul4 or DDB1 extended the half-life of Chk1 to greater than 10 h in normal cycling cells (Fig. 4B) and to greater than 8 h in HU-treated cells (Fig. 4D). Additionally, the half-life of Chk1 was extended when DDB1 and/or Cul4A were knocked down in HU-treated A549, HEK293 and HCT116 cells (data not shown).
Figure 4.
Chk1 is stabilized in cells deficient for Cul4 and DDB1. A, B, asynchronously growing HeLa cells were cultured in the presence of the indicated siRNAs for 48 h followed by the addition of 100 μg/ml of CHX. Lysates were prepared at the indicated times after CHX addition. Proteins were resolved by SDS-PAGE and analyzed by Western blotting. A representative Western blot is shown in panel A and quantitation from 3 independent experiments is shown graphically in panel B. Standard deviation is shown as error bars along the y axis. C, D, same as in panels A, B except that cells were cultured in the presence of both CHX and 20 mM HU 48 h after siRNA transfection (n = 3).
Chk1 turnover induced by replication-stress or HSP90 inhibition is dependent on Cul4A/DDB1 E3 ligase
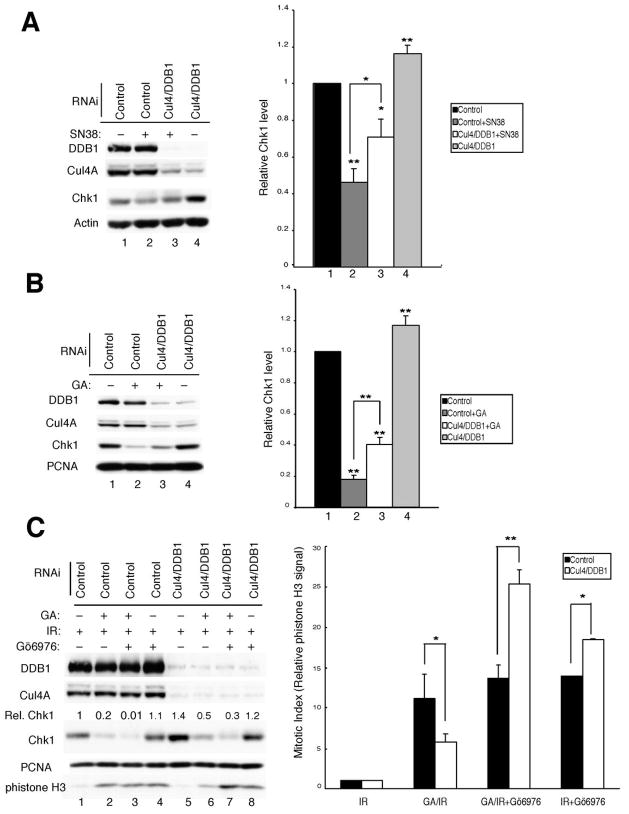
Camptothecin (CPT), a topoisomerase I inhibitor, induces Chk1 degradation in vivo (27). We monitored the effects of SN38, the active metabolite of CPT, on Chk1 levels in control cells and in cells deficient for the Cul4A/DDB1 ligase (Fig. 5A). Importantly, SN38-treatment was less effective at inducing Chk1 loss in cells knocked down for the Cul4 and DDB1. The half-life of Chk1 was ~4 h in control cells treated with SN38 and cycloheximide (Supp. Fig. 4B). In contrast, the half-life of Chk1 was extended to greater than 8 h in DDB1-deficient cells treated with SN38 and cycloheximide (Supp. Fig. 4B).
Figure 5.
Knockdown of Cul4A/DDB1 impairs Chk1 degradation and alters Chk1 function. A, asynchronously growing HeLa cells were transfected with the indicated siRNAs for 48 h and then treated with SN38 (1 μM) for 6 h. Cells lysates were prepared and subjected to Western blotting with antibodies specific for the indicated proteins (left panel). Compiled data from 3 independent experiments is presented graphically in the right panel. B, asynchronously growing HeLa cells were transfected with the indicated siRNAs for 24 h and then incubated with DMSO or 200 nM geldanamycin (GA) for another 24 h. Cells lysates were prepared and subjected to Western blotting with antibodies specific for the indicated proteins (left panel). Compiled data from 5 independent experiments is presented graphically in the right panel. C, Asynchronously growing HeLa cells transfected with the indicated siRNAs for 24 h were treated as described in B and then exposed to 10 Gy IR. In some cases, cells were also incubated with DMSO (vehicle) or Gö6976 (300 nM) and harvested after 9 h. Cells lysates were prepared and subjected to Western blotting with antibodies specific for the indicated proteins (left panel). Compiled data is presented graphically in the right panel. Relative levels of phistone-H3 were determined from 3 independent experiments. All of the graphs are presented as mean +/− standard deviation. P-values from Student’s t-test are shown when significantly different from control (* < 0.05; **<0.01).
Treatment of cells with geldanamycin (GA), an HSP90 inhibitor, also causes Chk1 turnover. HSP90 binds Chk1 (39) and inhibition of Chk1/HSP90 interactions leads to Chk1 proteolysis via the proteosome (Supp. Fig. 4C and reference 40). We tested whether knockdown of Cul4A/DDB1 impaired Chk1 proteolysis in GA-treated cells (Fig. 5B). As expected, knockdown of Cul4A/DDB1 resulted in Chk1 stabilization (compare lanes 1 and 4) and GA-treatment reduced Chk1 levels to ~17% that observed in control RNAi treated cells (compare lanes 1 and 2). Importantly, the ability of GA to induce Chk1 proteolysis was impaired in Cul4A/DDB1-deficient cells (compare lanes 2 and 3).
The Chk1/Cul4A/DDB1 pathway regulates the IR-induced G2 checkpoint
Lastly, we investigated the contribution made by the Chk1/Cul4A/DDB1 regulatory pathway to the IR-induced G2 checkpoint. Knockdown of Cul4A/DDB1 was employed to maintain Chk1 levels in the presence of the HSP90 inhibitor GA. Effects on the IR-induced G2 checkpoint were then evaluated in control cells (low Chk1 levels) and in cells deficient in Cul4A/DDB1 (higher Chk1 levels). The dependency of the checkpoint response on Chk1 was determined by carrying out the experiments in the absence and presence of the Chk1 inhibitor Gö6976. Levels of phosphohistone H3 (phistone H3), a mitosis-specific marker, were monitored to assess the integrity of the checkpoint in these experiments (Fig. 5C).
Cells treated with siRNAs specific for Cul4 and DDB1 (to interfere with Chk1 turnover) or with control-siRNAs were incubated with the HSP90 inhibitor, to induce Chk1 turnover. As expected, Chk1 levels were lower in GA-treated control cells (lane 2) compared with GA-treated Cul4A/DDB1-deficient cells (lane 6), supporting a role for Cul4A/DDB1 in regulating Chk1 turnover. These cells were then exposed to ionizing radiation to activate the IR-induced G2 checkpoint. Since Chk1 is an essential component of this checkpoint, we predicted that GA-treated control cells containing less Chk1 would be impaired in their ability to arrest in G2 following exposure to IR. Indeed, phistone H3 levels were higher in control cells, (lane 2) relative to Cul4A/DDB1-deficient cells (lane 6). To determine if the difference was due to Chk1, the experiment was repeated in the presence and absence of the Chk1 inhibitor Gö6976. Inhibition of Chk1 in control cells did not significantly affect levels of phistone H3, presumably due to the already low levels of Chk1 in these cells (compare Chk1 levels in lanes 2 and 3). In contrast, inhibition of Chk1 in Cul4A/DDB1-deficient cells induced checkpoint bypass as indicated by the higher levels of phistone H3 (compare lanes 6 and 7). These experiments confirm that Chk1 levels can be modulated in vivo by perturbing Cul4A/DDB1 and demonstrate that perturbation of the Chk1/Cul4A/DDB1 regulatory pathway impacts the ability of cells to respond appropriately to DNA double strand breaks.
DISCUSSION
The preservation of genome integrity within individual cells is essential for organismal homeostasis. Cells activate signaling pathways known as checkpoints in order to arrest cell cycle progression when their genome integrity is compromised. This can occur during a normal division cycle when, for example, a replication fork temporarily stalls, or in response to a direct assault on the genome resulting in either single- or double-strand breaks. At some point these signaling pathways must be turned off in order for DNA replication and cell division to resume. As part of the recovery process, Claspin, a Chk1 regulator, is ubiquitinated and degraded (41); Chk1 is inactivated by dephosphoryation of S317 and S345 (26); or alternatively, activated Chk1 is degraded in a proteosome-dependent manner (27).
Here we demonstrate that the Cul4A/DDB1 complex ubiquitinates Chk1 and regulates Chk1 abundance in normal cycling cells and when cells are exposed to exogenous assaults that induce replication stress. Affinity purification of flag-tagged Chk1 coupled with mass spectrometry identified DDB1 as a Chk1-associated protein. Interactions between endogenous Chk1 and endogenous Cul4A/DDB1 were shown to occur in normal cycling cells and to be enhanced by replication stress. In addition, the half-life of Chk1 was extended in cells knocked down for either DDB1 or Cul4A. The Cul4A/DDB1 complex is proposed to target substrates by direct interaction with DDB1 (42) or through a family of adapter proteins called DCAFs for DDB1-and Cul4-Associated Factors (32, 43, 44). Although we demonstrated that Chk1 phosphorylation is important for its interaction with Cul4A/DDB1, we do not yet know whether the interactions between Chk1 and DDB1 are direct or are mediated by a DCAF. To date, our attempts to reconstitute interactions between Chk1 and DDB1 with purified proteins in vitro have failed. Interestingly, two novel WD40-containing proteins were identified in our Chk1 proteomic screen. WD40 domains are a common feature of DCAFs (45) and future experiments will be directed at determining whether either of these proteins mediate interactions between Chk1 and DDB1.
The identification of Chk1 as a Cul4A/DDB1 target adds to a growing list of cellular substrates targeted to Cul4 by DDB1, including the nucleotide excision repair proteins DDB2 and XP-group C (XPC) (46, 47), the DNA replication licensing factor Cdt1 (42, 48, 49), the cell cycle regulator p27 (50), the tumor suppressor proteins TSC2 (51) and Merlin (52), the transcription factors c-Jun and STAT proteins (53, 54) as well as a subset of histones (55, 56). Thus, the Cul4-DDB1-Roc1 complex regulates several critical cellular processes including nucleotide excision repair, transcription, cell cycle progression, checkpoints, and DNA replication. The importance of the Cul4A/DDB1 E3 ligase to the cell division cycle is underscored by the fact that a number of pathogenic viruses subvert the Cul4A/DDB1 complex to benefit their own replicative life cycles and DDB1 expression has been reported to be significantly decreased in adenocortical carcinomas (57).
Chk1 enforces S-phase delays in cells experiencing replicative- or genotoxic-stress by inhibiting elongation of replication forks and by preventing firing of new origins. Thus, Cul4A/DDB1 is predicted to contribute to the checkpoint recovery process by facilitating the ubiquitin-mediated proteolysis of Chk1, thereby enabling faithful resumption of DNA replication and cell cycle progression following recovery from replicative stress and DNA damage.
Supplementary Material
Acknowledgments
V. L.-P. dedicates this work to the memory of Serenity Joy Snyder Leung. The authors thank Janis L. Watkins and Christine E. Ryan for preparing plasmid stocks and manuscript editing. This work was supported by a grant from the National Institutes of Health. H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Sorensen CS, Syluasen RG, Falck J, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–58. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 2.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc NatlAcad Sci USA. 2002;99:14795–800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SH, Chuan L, Maller JL. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis. Dev Biol. 1999;212:381–91. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- 4.Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002;21(14):3694–703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M-S, Ryan CE, Piwnica-Worms H. Chk1 Kinase Negatively Regulates Mitotic Function of Cdc25A Phosphatase through 14-3-3 Binding. Mol Cell Biol. 2003;23:7488–97. doi: 10.1128/MCB.23.21.7488-7497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goloudina A, Yamaguchi H, Chervyakova DB, Appella E, Fornace AJ, Jr, Bulavin DV. Regulation of human Cdc25A stability by serine 75 phosphorylation is not sufficient to activate a S-phase checkpoint. Cell Cycle. 2003;2:473–8. [PubMed] [Google Scholar]
- 7.Hassepass I, Voit R, Hoffmann I. Phosphorylation at serine-75 is required for UV-mediated degradation of human Cdc25A phosphatase at the S-phase checkpoint. J Biol Chem. 2003;278:29824–9. doi: 10.1074/jbc.M302704200. [DOI] [PubMed] [Google Scholar]
- 8.Feijoo C, Hall-Jackson C, Wu R, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154(5):913–23. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol. 2007;27(16):5806–18. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada M, Niida H, Zineldeen DH, et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132(2):221–32. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27(8):3098–108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enders GH. Expanded roles for Chk1 in genome maintenance. J Biol Chem. 2008 doi: 10.1074/jbc.R800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer A, Mailand N, Lukas C, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6(9):884–91. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 14.Kohn EA, Yoo CJ, Eastman A. The protein kinase C inhibitor Go6976 is a potent inhibitor of DNA damage-induced S and G2 cell cycle checkpoints. Cancer Res. 2003;63(1):31–5. [PubMed] [Google Scholar]
- 15.Wang O, Fan S, Eastman A, Worland PJ, Sausville EA, O’Conner PM. UCN-01: a Potent Abrogator of G2 Checkpoint Function in Cancer Cells With Disrupted p53. Journal of the National Cancer Institute. 1996;88:956–65. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 16.Moran DM, Gawlak G, Jayaprakash MS, Mayar S, Maki CG. Geldanamycin promotes premature mitotic entry and micronucleation in irradiated p53/p21 deficient colon carcinoma cells. Oncogene. 2008 doi: 10.1038/onc.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugimoto K, Sasaki M, Isobe Y, et al. Hsp90-inhibitor geldanamycin abrogates G2 arrest in p53-negative leukemia cell lines through the depletion of Chk1. Oncogene. 2008;27(22):3091–101. doi: 10.1038/sj.onc.1210978. [DOI] [PubMed] [Google Scholar]
- 18.Kumagai A, Kim SM, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J Biol Chem. 2004;279(48):49599–608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- 19.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 20.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294(5547):1713–6. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 21.Yang XH, Zou L. Recruitment of ATR-ATRIP, Rad17, and 9-1-1 complexes to DNA damage. Methods Enzymol. 2006;409:118–31. doi: 10.1016/S0076-6879(05)09007-5. [DOI] [PubMed] [Google Scholar]
- 22.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3(7):941–5. [PubMed] [Google Scholar]
- 23.Liu Q, Guntuku S, Cui XS, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–59. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–39. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR Is Antagonized by a Chk1-Regulated Protein Phosphatase 2A Circuit. Mol Cell Biol. 2006;26(20):7529–38. doi: 10.1128/MCB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19(10):1162–74. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YW, Otterness DM, Chiang GG, et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19(5):607–18. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 28.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):920. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 29.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image J. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 30.Kang T, Wei Y, Honaker Y, et al. GSK-3beta Targets Cdc25A for Ubiquitin-Mediated Proteolysis, and GSK-3beta Inactivation Correlates with Cdc25A Overproduction in Human Cancers. Cancer Cell. 2008;13(1):36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meek SE, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279(31):32046–54. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 32.He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20(21):2949–54. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatei M, Sloper K, Sorensen C, et al. ATM and NBS1 dependent phosphorylation of CHK1 on S317 in response to IR. J Biol Chem. 2003;278:14806–11. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 34.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for ATR in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in xenopus egg extracts. Genes Dev. 2000;14(21):2745–56. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8(1):37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 36.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58(19):4375–82. [PubMed] [Google Scholar]
- 37.Blasina A, Price BD, Turenne GA, McGowan CH. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9(19):1135–8. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 38.Sarkaria JN, Busby EC, Tibbetts RS, et al. Inhibition of ATM and ATR kinase activity by the radiosensitizing agent caffeine. Cancer Res. 1999;59:4375–82. [PubMed] [Google Scholar]
- 39.Arlander SJ, Eapen AK, Vroman BT, McDonald RJ, Toft DO, Karnitz LM. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278(52):52572–7. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- 40.Nomura M, Nomura N, Yamashita J. Geldanamycin-induced degradation of Chk1 is mediated by proteasome. Biochem Biophys Res Commun. 2005;335(3):900–5. doi: 10.1016/j.bbrc.2005.07.160. [DOI] [PubMed] [Google Scholar]
- 41.Gewurz BE, Harper JW. DNA-damage control: Claspin destruction turns off the checkpoint. Curr Biol. 2006;16(21):R932–4. doi: 10.1016/j.cub.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 42.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6(10):1003–9. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 43.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8(11):1277–83. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 44.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A Family of Diverse Cul4-Ddb1-Interacting Proteins Includes Cdt2, which Is Required for S Phase Destruction of the Replication Factor Cdt1. Mol Cell. 2006;23(5):709–21. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Lee J, Zhou P. DCAFs, the Missing Link of the CUL4-DDB1 Ubiquitin Ligase. Mol Cell. 2007;26(6):775–80. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Sugasawa K, Okuda Y, Saijo M, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121(3):387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 47.Groisman R, Kuraoka I, Chevallier O, et al. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20(11):1429–34. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5(15):1675–80. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- 49.Higa LA, Mihaylov IS, Banks DP, Zheng J, Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat Cell Biol. 2003;5(11):1008–15. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- 50.Bondar T, Kalinina A, Khair L, et al. Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome. Mol Cell Biol. 2006;26(7):2531–9. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Zacharek S, He YJ, et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22(7):866–71. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Chen J. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene. 2008 doi: 10.1038/onc.2008.44. [DOI] [PubMed] [Google Scholar]
- 53.Wertz IE, O’Rourke KM, Zhang Z, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303(5662):1371–4. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 54.Precious B, Childs K, Fitzpatrick-Swallow V, Goodbourn S, Randall RE. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J Virol. 2005;79(21):13434–41. doi: 10.1128/JVI.79.21.13434-13441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A. 2006;103(8):2588–93. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Zhai L, Xu J, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22(3):383–94. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 57.Fernandez-Ranvier GG, Weng J, Yeh RF, et al. Candidate Diagnostic Markers and Tumor Suppressor Genes for Adrenocortical Carcinoma by Expression Profile of Genes on Chromosome 11q13. World J Surg. 2008 doi: 10.1007/s00268-008-9521-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.