Abstract
Little is known about the molecular development and heterogeneity of callosal projection neurons (CPN), cortical commissural neurons that connect homotopic regions of the two cerebral hemispheres via the corpus callosum and that are critical for bilateral integration of cortical information. Here we report on the identification of a series of genes that individually and in combination define CPN and novel CPN subpopulations during embryonic and postnatal development. We used in situ hybridization analysis, immunocytochemistry, and retrograde labeling to define the layer-specific and neuron-type-specific distribution of these newly identified CPN genes across different stages of maturation. We demonstrate that a subset of these genes (e.g., Hspb3 and Lpl) appear specific to all CPN (in layers II/III and V–VI), whereas others (e.g., Nectin-3, Plexin-D1, and Dkk3) discriminate between CPN of the deep layers and those of the upper layers. Furthermore, the data show that several genes finely subdivide CPN within individual layers and appear to label CPN subpopulations that have not been described previously using anatomical or morphological criteria. The genes identified here likely reflect the existence of distinct programs of gene expression governing the development, maturation, and function of the newly identified subpopulations of CPN. Together, these data define the first set of genes that identify and molecularly subcategorize distinct populations of callosal projection neurons, often located in distinct subdivisions of the canonical cortical laminae.
Introduction
The neocortex is the region of the brain responsible for cognitive function, sensory perception, and consciousness, and, as such, it has undergone pronounced expansion during evolution. Among the different types of neurons of the cortex, a rich variety of glutamatergic projection neurons exist that can be classified into numerous subtypes by the target of their axonal projections to distinct intracortical, subcortical, and subcerebral targets, as well as by their location in specific cortical layers and areas (Molyneaux et al., 2007). The molecular identity of distinct projection neuron types, and the signals that control lineage-restricted neurogenesis, are only beginning to be defined (Weimann et al., 1999; Hevner et al., 2001; Arlotta et al., 2005; B. Chen et al., 2005; J. G. Chen et al., 2005; Molyneaux et al., 2005; Alcamo et al., 2008; Britanova et al., 2008; Joshi et al., 2008; Lai et al., 2008).
In this report, we focus on the molecular development of callosal projection neurons (CPN), a broad population of cortical commissural neurons that connect homotopic regions of the two cerebral hemispheres via the corpus callosum (Richards et al., 2004). CPN show substantial heterogeneity in their axonal projections, with some CPN possessing single projections to the contralateral cortex and others maintaining either dual projections to the contralateral cortex and ipsilateral/contralateral striatum, or dual projections to the contralateral cortex and ipsilateral frontal cortex (Wilson, 1987; Koralek et al., 1990; Reiner et al., 2003; Gao and Zheng, 2004; Mitchell and Macklis, 2005). Adding further to this diversity, CPN are found across multiple cortical layers, with the vast majority located in layers II/III (∼80%) and V (∼20%) and a smaller population present in layer VI (Conti and Manzoni, 1994; Ramos et al., 2008b). This suggests that CPN are highly heterogeneous and are likely composed of multiple molecularly distinct subpopulations not currently resolved at the anatomical level.
To discover subtype-specific molecular controls over the development of CPN and corticofugal populations, we previously purified CPN and two subtypes of subcerebral projection neurons—corticospinal motor neurons (CSMN) and corticotectal projection neurons (CTPN)—from the murine neocortex (Arlotta et al., 2005). Using microarrays, we identified genes that mark CSMN and control development of this neuron type in vivo (Arlotta et al., 2005; Molyneaux et al., 2005; Lai et al., 2008).
Here, we report on the identification of a set of genes that are progressively restricted to CPN and that are novel markers of this projection neuron population. Some of these genes appear specific to all CPN, whereas others discriminate between CPN of the deep layers and the upper layers. Furthermore, we show that a subset of genes finely subdivides CPN within individual layers and appear to label CPN subpopulations that have not been described previously using anatomical and morphologic criteria. Together, these data define the first set of genes that identify and likely play central roles in the subtype-specific development of CPN, providing molecular evidence for the presence of multiple CPN subpopulations often located in distinct subdivisions of the canonical cortical laminae.
Materials and Methods
Retrograde labeling.
All procedures to retrogradely label and purify CPN and the comparative populations CSMN and CTPN have been described previously by Arlotta et al. (2005). In brief, for CPN, neurons were retrogradely labeled at embryonic day 18 (E18), postnatal day 3 (P3), P6, and P14 by injection of green fluorescent microspheres (Lumafluor) into contralateral cortex (E17, P1, P4, and P12) of C57BL/6 mice (Charles River Laboratories), as described previously (Catapano et al., 2001). Cholera toxin subunit B conjugated to Alexa 555 (CTB555; Invitrogen) was used for retrograde labeling of CPN and CSMN for in situ hybridization and immunocytochemistry experiments. Embryonic injections were performed using a Vevo 660 ultrasound system (VisualSonics). All animal studies were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee and were performed in accordance with institutional and federal guidelines.
CPN dissociation and fluorescence-activated cell sorting purification.
Labeled areas of somatosensory cortex were dissociated essentially as described previously (Catapano et al., 2001). In brief, dissociated cortex was enzymatically digested with 0.16 gm/L l-cysteine HCl and 11.7 U/ml papain at 37°C for 30 min. Neurons were mechanically dissociated to create a single-cell suspension by gentle trituration in iced OptiMem (Invitrogen) containing 20 mm glucose and both 0.4 mm kynurenic acid and 0.025 mm APV to protect against glutamate-induced neurotoxicity. Microsphere-labeled CPN were purified from the cortical cell suspension by fluorescence-activated cell sorting (FACS), and neurons were collected directly in RNAlater (Ambion). Additional details and methods to purify CSMN and CTPN have been described previously by Arlotta et al. (2005).
Microarrays.
Microarray probe synthesis and microarray analysis were all performed previously and have been described by Arlotta et al. (2005). To optimize identification of CPN-specific genes, the same hybridization solutions were now applied to the newly available Affymetrix 430 2.0 microarrays and hybridized and processed according to Affymetrix protocols. Probe sets found on both the 430A arrays and the 430 2.0 arrays were found to have a very high degree of correlation. Microarray data were normalized using two independent methods: the RMA function within Bioconductor (Irizarry et al., 2003) and the “error model” method within Rosetta Resolver (version 5.0; Rosetta Biosoftware). Statistical significance of gene expression differences between neuronal subtypes was determined by pairwise comparisons at each age using statistical analysis of microarrays (SAM) (Tusher et al., 2001). Using a SAM D score cutoff of more than 2 or less than −2, we selected significantly differentially expressed genes and further analyzed their temporal dynamics of expression to identify a refined set of genes for additional analysis (Arlotta et al., 2005). For example, genes with similar dynamics of expression that are simply shifted a few days later in CPN compared with CSMN demonstrate statistical difference at each independent time point but likely represent the same biological process in each population, shifted simply as a result of population birthdate and developmental stage. All microarray data have been deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information and are available via accession numbers GSE2039 (Affymetrix 430A microarray data) and GSE17783 (Affymetrix 430 2.0 microarray data).
In situ hybridization and immunocytochemistry.
All clones for in situ hybridization were generated by reverse transcription-PCR. Sequences of all primers used are listed in supplemental Table 1 (available at www.jneurosci.org as supplemental material). Nonradioactive in situ hybridization was performed using reported methods (Berger and Hediger, 2001). Sense probes were used as negative controls in all experiments. For Nectin-3 immunocytochemistry, brains were fixed and stained using standard methods (Fricker-Gates et al., 2002). Briefly, brains were fixed by transcardial perfusion with PBS–heparin (10 U/ml), followed by 4% paraformaldehyde, and postfixed overnight at 4°C in 4% paraformaldehyde. Brains were sectioned coronally at 50 μm on a vibrating microtome (Leica). Sections were blocked in 0.3% BSA (Sigma), 8% goat or donkey serum, and 0.3% Triton X-100 (Sigma) for 1 h at room temperature, before incubation in Nectin-3 primary antibody at 1:100 (Abcam). Secondary antibodies were from the Invitrogen Alexa series. Images were acquired using a Nikon E1000 microscope equipped with a cooled CCD digital camera (Retiga; QImaging) and Openlab acquisition software (Improvision) or via laser confocal analysis using a Bio-Rad Radiance 2100 confocal system attached to a Nikon E800 microscope.
Results
Purification and microarray analysis of callosal projection neurons, corticospinal motor neurons, and corticotectal projection neurons
To identify genes that control the cell-type specification and differentiation of callosal projection neurons, we compared the gene expression profiles of CPN with two other pure populations of cortical projection neurons: corticospinal motor neurons and corticotectal projection neurons. The general approach used and the analysis of data to identify genes involved in the development of the corticospinal motor neuron and related corticofugal populations were previously described (Arlotta et al., 2005). Here, we report on the identification and further characterization of genes that define the broad population and distinct subtypes of interhemispheric callosal projection neurons.
CPN were retrogradely labeled via injections of green fluorescent microspheres into their axonal projection fields in contralateral sensorimotor cortex at four different stages of development, followed by dissociation and FACS of the labeled neurons to typically >99% purity (Catapano et al., 2001, 2004; Arlotta et al., 2005; Ozdinler and Macklis, 2006). Specifically, callosal projection neurons that were purified at multiple stages of development, including E18, P3, P6, and P14, were labeled as schematically depicted in Figure 1. For precise targeting and purification of E18 CPN, fluorescent microspheres were microinjected in the contralateral hemisphere of E17 embryos, in utero, under high-resolution ultrasound backscatter microscopic guidance (Arlotta et al., 2005). Similar methods were used to label corticospinal motor neurons and corticotectal projection neurons (Fig. 1) (Arlotta et al., 2005).
Figure 1.
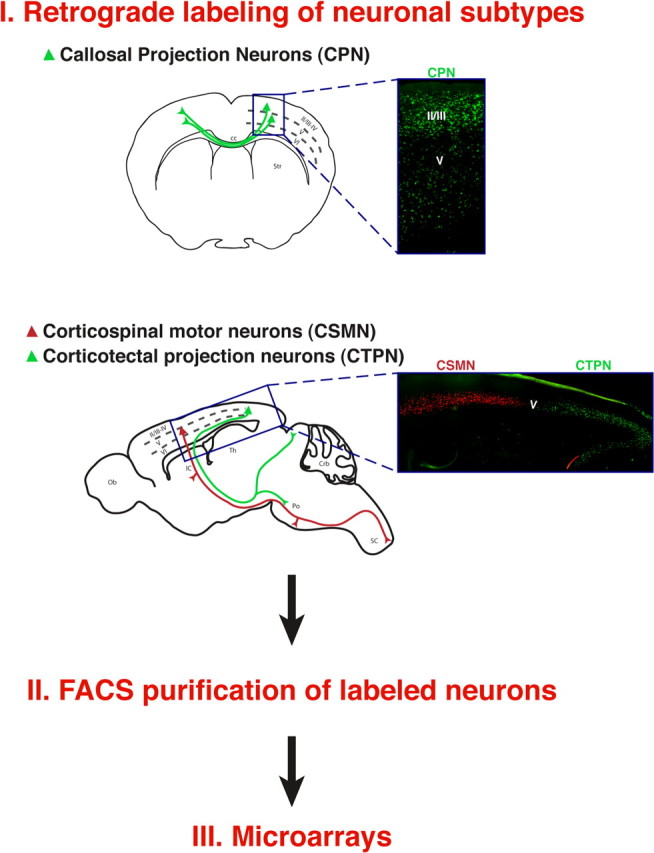
Schematic representation of the experimental approach used to identify CPN-specific genes. CPN, CSMN, and CTPN were retrogradely labeled at distinct stages of development from the contralateral hemisphere, the spinal cord, and the superior colliculus, respectively. Labeled neurons were dissociated, FACS purified, and submitted to comparative microarray analysis.
In the study by Arlotta et al. (2005), we previously used these pure neuronal preparations to compare the molecular development of corticospinal motor neurons and callosal projection neurons using Affymetrix 430A microarrays (data available at NCBI Gene Expression Ominbus accession number GSE2039). To optimize identification of CPN-specific genes and to maximize the number of genes examined by microarray to provide the most inclusive investigation of gene expression through development, we now hybridized samples to the newly available and more inclusive Affymetrix 430 2.0 arrays, which contain a substantially expanded probe set. Using this approach, we identified a large number of genes that are expressed at higher levels in callosal projection neurons compared with CSMN and corticotectal projection neurons. The 40 genes with the most biologically significant and distinctive expression profiles after temporal analysis (see Materials and Methods) are listed in supplemental Table 2 (available at www.jneurosci.org as supplemental material). These genes can be classified in several ontology groups and include, among others, transcription factors (e.g., Cux1, Cux2, Lhx2, Pdzrn3, and Cited 2), cell signaling molecules and receptors (e.g., Gfra2, Gpr6, Gpr88, Ptprk, and TCRβ), and axon guidance molecules (e.g., Chimerin 2, Dcc, EphA3, Plxdc2, and PlxnD1). Although many of these genes are relatively uncharacterized in the cortex and none of them has been previously demonstrated to specifically label the broad CPN population or CPN subtypes, we find that selected genes previously demonstrated to be restricted to the upper layers (in which most CPN are located) are specifically and highly expressed in CPN within those layers [e.g., Cux1 and Cux2 (Nieto et al., 2004; Zimmer et al., 2004)], confirming the validity of our approach in identifying candidates with restricted expression to CPN and distinct CPN subtypes.
Many CPN-specific genes identify anatomic diversity and distinct CPN populations
Previous anatomical and birthdating analyses have shown that the broad CPN population is heterogeneous, including neurons born at different developmental times (e.g., E12.5–E13.5 for layer VI and V CPN, E15.5 for layer II/III CPN), located in different cortical layers (e.g., II/III, V, and VI), with some extending collateral projections to distinct targets (e.g., striatum and ipsilateral frontal cortex) (Wise and Jones, 1976; Mitchell and Macklis, 2005). This is in striking contrast to the much more homogeneous population of corticospinal motor neurons, which are born at ∼E13.5, populate one layer (layer V) and a limited area (sensorimotor) of cortex and extend projections to the spinal cord (although with a range of spinal segmental specificity) (O'Leary and Koester, 1993). Based on such previous anatomical data, we reasoned that CPN-specific genes might fall into at least two general categories: (1) broad CPN “identity genes” that might label most or all CPN; and (2) genes that subparcellate CPN into multiple subtypes, potentially reflecting anatomic, connectivity, and functional heterogeneity. Genes in the second set would only be expressed in distinct subtypes of CPN.
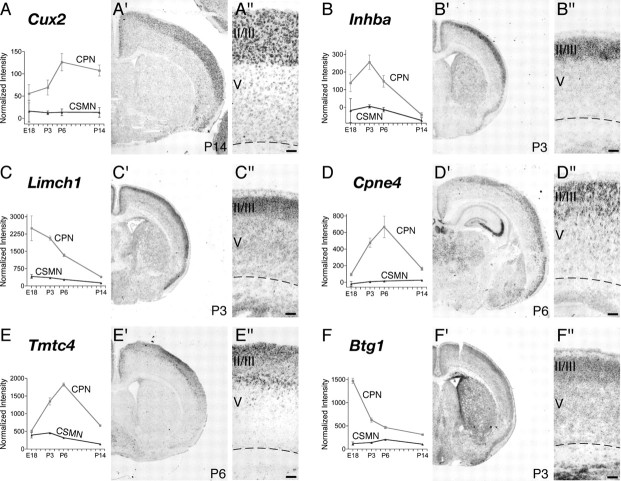
To investigate this possibility and determine whether the anatomical complexity of the broad CPN population is mirrored at the molecular level, we investigated the fine spatial expression of a subset of these newly identified CPN genes via in situ hybridization. We find that a small number of these genes are expressed in a laminar distribution suggestive of their presence in all CPN (Fig. 2), showing high levels of expression in layers II/III, the CPN proportion of V, and in a smaller number of layer VI neurons consistent with CPN. These include the following genes: Lpl (lipoprotein lipase) (Fig. 2A), an enzyme involved in lipoprotein metabolism with unknown function in the brain (Vilaró et al., 1990); Hspb3 (Fig. 2B), a small heat shock protein not described previously in the brain (Sugiyama et al., 2000); and Cited2 (Fig. 2C), thought to function as a transcriptional coactivator and to interact with Lhx2 (Glenn and Maurer, 1999). The spatial expression of these genes matches the distribution of CPN obtained via retrograde tracing of all CPN from the contralateral hemisphere (Fig. 2D). To further investigate the neuronal subtype-specific expression of these genes within CPN, we combined fluorescence in situ hybridization analysis of Lpl and Hspb3 with retrograde labeling of CPN and CSMN from the contralateral cortex and the spinal cord, respectively. We find that both Lpl and Hspb3 are expressed within CPN across layers II/III, V, and VI (Fig. 2E–J). In contrast, Lpl and Hspb3 are not expressed within CSMN (Fig. 2K–N).
Figure 2.
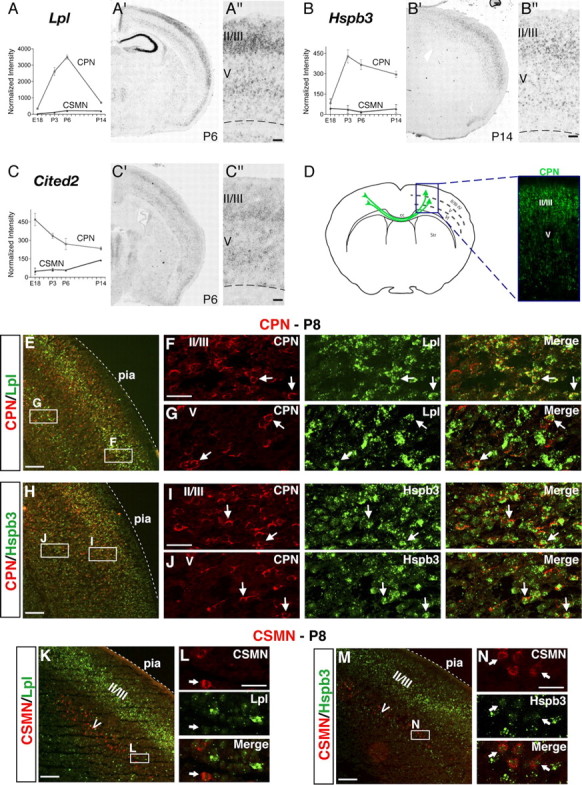
CPN genes that label most callosal projection neurons across layers II/III and V. A–C, Temporal profiles of gene expression from microarray analysis in CPN (gray) versus CSMN (black) during embryonic (E18) and early postnatal (P3, P6, and P14) stages of development. Error bars indicate SEM. A′–C′, In situ hybridization in coronal sections of cortex showing that the expression of selected genes closely resembles the typical distribution of the retrogradely labeled, broad CPN population (D). A″–C″, Magnification of selected areas from A′–C′. Ages are as indicated in A′–C′. E–N, Lpl and Hspb3 are expressed in CPN across layers II/III and V and not in CSMN. E, Fluorescent in situ hybridization for Lpl (green) in a coronal section of P8 cortex demonstrates that it is expressed in CPN identified by retrograde labeling via injection of CTB555 (red) in the contralateral cortex at P6. F, G, Magnification of selected areas in layers II/III and V from E; arrows indicate Lpl-expressing CPN. H, Fluorescent in situ hybridization for Hspb3 (green) in a coronal section of P8 cortex demonstrates that it is expressed in retrogradely labeled CPN (red). I, J, Magnification of selected areas in layers II/III and V from H; arrows indicate Hspb3-expressing CPN. K, Fluorescent in situ hybridization for Lpl (green) in a coronal section of P8 cortex demonstrates that it is not expressed in CSMN identified by retrograde labeling via injection of CTB555 (red) in the spinal cord at P4 (high magnification shown in L). M, Fluorescent in situ hybridization for Hspb3 (green) in a coronal section of P8 cortex demonstrates that it is also not expressed in retrogradely labeled CSMN (red) (high magnification shown in N). Scale bars: A″–C″, 100 μm; E, H, K, M, 200 μm; F, G, I, J, L, N, 50 μm.
In addition to genes that label all CPN and distinguish them from other types of cortical projection neurons within the same layers, in situ hybridization for 20 additional CPN genes reveals that they are distributed in distinct, individual laminar and sublaminar patterns within the cortex (see Figs. 2–6). These data confirm and extend at the molecular level previous anatomical data on the heterogeneity of connections and functional diversity of CPN and provide the first demonstration that distinct CPN populations exist in the cortex that can be identified by the combinatorial expression of newly identified CPN genes.
Figure 3.
CPN genes that selectively label callosal neurons of the superficial layers II/III and IV. A–F, Temporal profiles of gene expression from microarray analysis in CPN (gray) versus CSMN (black) during embryonic (E18) and early postnatal (P3, P6, and P14) stages of development. Error bars indicate SEM. A′–F′, In situ hybridization in coronal sections of cortex showing preferential CPN gene expression in the superficial layers. A″–F″, Magnification of selected areas from A′–F′. Ages are as indicated in A′–F′. Scale bars: A″–F″, 100 μm.
Figure 4.
CPN genes that preferentially label the most superficial portion of layer II/III and layer I. A–C, Temporal profiles of gene expression from microarray analysis in CPN (gray) versus CSMN (black) during embryonic (E18) and early postnatal (P3, P6, and P14) stages of development. Error bars indicate SEM. A′–C′, In situ hybridization in coronal sections of cortex showing preferential CPN gene expression in the most superficial portions of layer II/III. A″–C″, Magnification of selected areas from A′–C′. Ages are as indicated in A′–C′. Scale bars: A″–C″, 100 μm.
Figure 5.
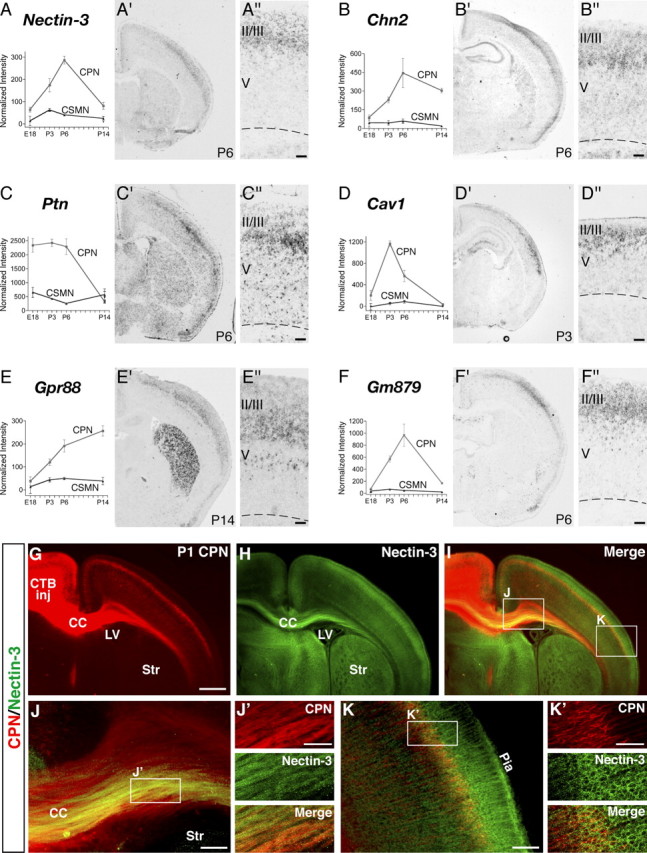
CPN genes that preferentially label the deepest portion of layer II/III and layer IV. A–F, Temporal profiles of gene expression from microarray analysis in CPN (gray) versus CSMN (black) during embryonic (E18) and early postnatal (P3, P6, and P14) stages of development. Error bars indicate SEM. A′–F′, In situ hybridization in coronal sections of cortex showing preferential CPN gene expression in the deepest portions of layer II/III and IV. A″–F″, Magnification of selected areas from A′–F′. G–K, Nectin-3 is expressed in CPN in layer II/III and in CPN axons in the corpus callosum at P1. G, CPN retrogradely labeled with CTB555 (red) via injections into contralateral cortex at P0. H, I, Nectin-3 immunocytochemistry (green) detects Nectin-3 expression in CPN in layer II/III, as well as in CPN axons in the corpus callosum. J, Magnification of selected area in I. J′, Magnification of selected area in J, showing expression of Nectin-3 in a subset of CPN axons in the corpus callosum. K, Magnification of selected area in I. K′, Magnification of selected area in K, showing expression of Nectin-3 in CPN in layer II/III. CTB inj, Site of CTB555 injection; CC, corpus callosum; LV, lateral ventricle; Str, striatum. Ages are as indicated in A′–E′ and G. Scale bars: A″–E″, 100 μm; G–I, 500 μm; J, K, 100 μm; J′, K′, 50 μm.
Figure 6.
Genes that preferentially label CPN of the deep layers V and VI. A–D, Temporal profiles of gene expression from microarray analysis in CPN (gray) versus CSMN (black) during embryonic (E18) and early postnatal (P3, P6, and P14) stages of development. Error bars indicate SEM. A′–D′, In situ hybridization in coronal sections of cortex showing preferential CPN gene expression in the deep layers V and VI. A″–D″, Magnification of the areas boxed in A′–D′. Ages are as indicated in A′–D′. Scale bars: A″–D″, 100 μm.
CPN-specific genes mark distinct temporal stages of CPN development
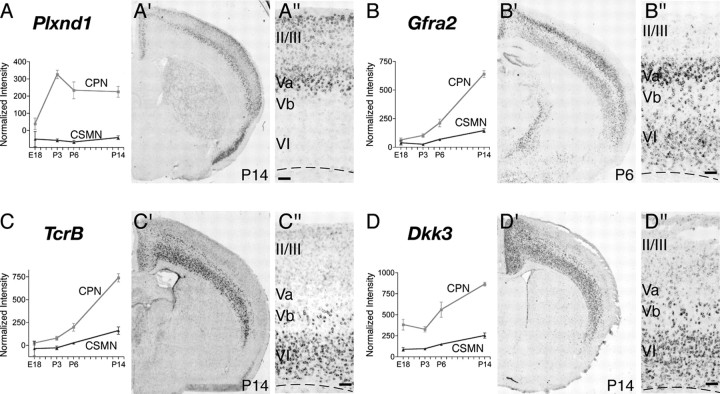
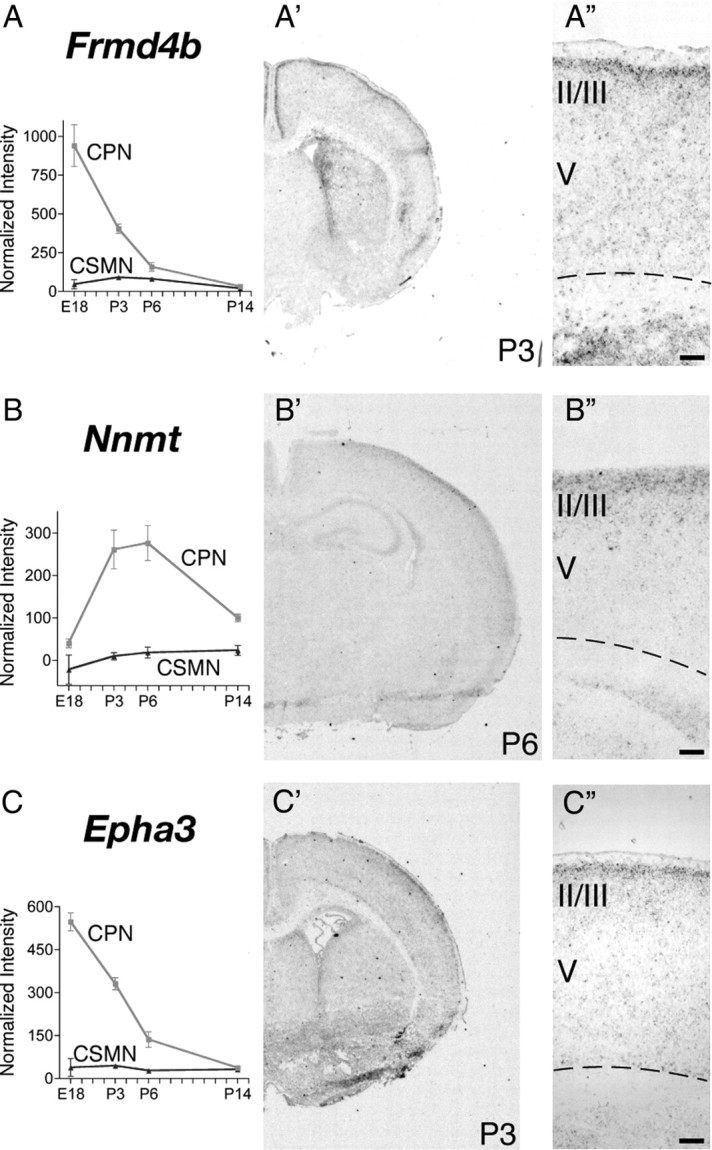
To identify genes expressed by CPN at distinct stages of development and thus likely functioning to control distinct aspects of cell-fate specification and differentiation of this neuronal lineage in vivo, we further investigated by in situ hybridization the fine spatial expression of genes that by microarray appear to be preferentially expressed at early (E18), mid (P3 and P6), or late (P14) stages of CPN development. We identified several genes that, aside from subdividing the CPN population spatially in layers and sublayers, are also expressed selectively at distinct times of development (see Figs. 3–6) (supplemental Table 2, available at www.jneurosci.org as supplemental material). Genes expressed early during CPN development, and thus potentially functioning in lineage specification, neuronal migration, and initial axonal extension, include the following: Cux2, Inhba, and Btg1 (see Fig. 3A,B,F); Frmd4b and EphA3 (see Fig. 4A,C); and Ptn (see Fig. 5C). Other genes are virtually absent at E18 but are highly expressed at P3, by which time CPN have already reached their final location in the neocortex, have sent axonal projections through the corpus callosum and are connecting to targets in the contralateral hemisphere. These genes expressed at mid-stage of CPN development include the following: Cpne4 and Tmtc4 (see Fig. 3D,E); Nnmt and Cav1 (see Figs. 4B, 5D); Nectin-3, Chn2, and Gm879 (see Fig. 5A,B,F). Finally, genes highly expressed at later stages of CPN development and thus that might be important in later CPN maturation, maintenance, and/or control of late events of CPN connectivity include Plexin-D1, Gfra2, TcrB, and Dkk3 (see Fig. 6A–D).
Genes that distinguish upper layer from deep layer CPN
CPN are located in both superficial (II/III) and deep (V and VI) layers of neocortex, but those of layers II/III are very different in their time and place of origin from those of V and VI. CPN of the deep layers are born during early corticogenesis (E11.5–E13.5 in the mouse) from progenitors located in the ventricular zone (VZ) underlying the neocortex (Angevine and Sidman, 1961; Caviness and Takahashi, 1995). In contrast, superficial layer CPN are born later, with a peak of birth at approximately E15.5, and they derive primarily from a second pool of progenitors (basal progenitors; also known as intermediate progenitors) located in the subventricular zone (SVZ), a second germinal layer that develops above the VZ during late corticogenesis (Kriegstein et al., 2006). The SVZ and its intermediate progenitors are thought to be central to the marked expansion of the superficial cortical layers of late evolution (Kriegstein et al., 2006).
To determine whether CPN of the superficial and deep layers are distinct at the molecular level and to identify potential molecular–genetic controls over the subtype-specific differentiation of these distinct CPN populations, we investigated whether any of the newly identified CPN-specific genes are restricted to CPN in either superficial or deep laminae. Indeed, we identified several genes that label only superficial layer CPN (Fig. 3). These include early expressed genes such as Cux2 (Fig. 3A) and Inhba (Fig. 3B), as well as Btg1 (Fig. 3F), a transcriptional coactivator that regulates myoblast differentiation and might play a similar role in CPN development. In addition, later genes such as Cpne4 and Tmtc4 specifically label upper layer CPN (Fig. 3D,E), suggesting potential functional roles in the maturation of these CPN. Although Cux2 was reported previously to label the upper layers of the neocortex (Nieto et al., 2004; Zimmer et al., 2004), acting as a confirmatory positive control for the current analysis, here we refine the previous results by demonstrating that Cux2 is expressed by CPN within the superficial layers. Other genes identified here are both novel markers of CPN and, more broadly, of superficial cortical layers. Together, expression of these genes as superficial layer CPN markers enables the demarcation and molecular classification of CPN of the superficial layers and indicates that upper layer CPN are molecularly distinct even at early stages of differentiation from CPN located in deep layers of neocortex.
A subset of CPN-specific genes defines novel sublamina within the normally unresolved murine upper layer II/III
Although many of the newly identified upper layer CPN genes are broadly expressed across the thickness of layer II/III, we find, interestingly, that several CPN-specific genes label only narrow sublamina, subparcellating what has been traditionally termed layer II/III in rodents. For example, Frmd4b, Nnmt, and EphA3 (Fig. 4A–C) label only a very thin strip of CPN in the most superficial portion of layer II/III. In contrast, other genes label CPN populations in the middle (Nectin-3 and Chn2) (Fig. 5A,B) or deeper (Ptn, Cav1, and Gm879) (Fig. 5C,D,F) portions of nominal layer II/III. Genes that distinguish the most superficial and deepest aspects of nominal layer II/III from the broader middle portion of the layer are of particular interest, because retrograde labeling of CPN from contralateral cortex indicates that the largest number and concentration of CPN are located within the middle portion of layer II/III, suggesting special functional and connectivity roles for the most superficial and deepest layer II/III CPN compared with the predominant layer II/III population.
To further investigate the specificity of expression of these genes within upper layer CPN, we chose Nectin-3 as a prototypical gene and evaluated its cell-type-specific expression. We retrogradely labeled CPN using a cholera toxin subunit B Alexa 555 conjugate, which clearly labels upper layer CPN and their axons coursing through the corpus callosum (Fig. 5G). Immunocytochemistry for Nectin-3 demonstrates colocalization with CPN axons in the corpus callosum, as well as within the somas and apical dendrites of superficial cortical CPN (Fig. 5H–K).
Together, these genes define a previously unrecognized molecular parcellation of the superficial cortical layers, refining previous histological and anatomical definitions and enabling new functional and evolutionary hypotheses regarding distinct CPN populations.
Other CPN-specific genes are expressed only in deep layer CPN and identify distinct populations in layers Va, Vb, and VI
In a complementary manner to genes expressed selectively by CPN of upper layers, many of the newly identified CPN genes are specifically expressed in deep layers V and VI and are excluded from layers II/III, thus defining distinct sets of early-born CPN of the deep layers. These genes include Plexin-D1, Gfra2, Tcrβ, and Dkk3 (Fig. 6A–D). Quite interestingly, layer V and VI CPN can be further defined by the combinatorial expression of selected deep layer CPN genes and thus classified into subpopulations occupying distinct subportions of these canonical layers. For example, in situ hybridization reveals that CPN in layer Va express the unique combination of Plexin-D1 and Gfra2 (Fig. 6A,B); they likely represent callosal neurons that maintain collateral projections to ipsilateral and/or contralateral striatum (Gao and Zheng, 2004). CPN located in deeper positions within layer V (e.g., layer Vb) can be defined by the expression of Gfra2, Tcrβ, and Dkk3 (Fig. 6C,D) and by the absence of Plexin-D1 expression (Fig. 6A). The relatively small population of CPN located in layer VI appears to express higher levels of Gfra2, Tcrβ, and Dkk3 (Fig. 6B–D) and lacks Plexin-D1 (Fig. 6A).
We further investigated cell-type-specific expression of the representative genes Plexin-D1 and Dkk3. We find that Plexin-D1 RNA is expressed at high levels within retrogradely labeled CPN in layer Va and at lower levels within CPN of more superficial layers (Fig. 7A–D). In contrast, retrograde labeling of CSMN demonstrates that they are located in layer Vb, below the Plexin-D1-positive layer Va. Of note, the border between these two layers is indistinct, with CSMN intermingled with PlexinD1-expressing CPN. Even at this interface, CSMN do not express Plexin-D1, clearly demonstrating the true cell-type specificity of this CPN marker. In addition, we examined the cell-type-specific expression of Dkk3 and find that it labels a large portion of layer V and VI retrogradely labeled CPN (Fig. 7I–L), whereas layer V CSMN do not express Dkk3 (Fig. 7M–P).
Figure 7.
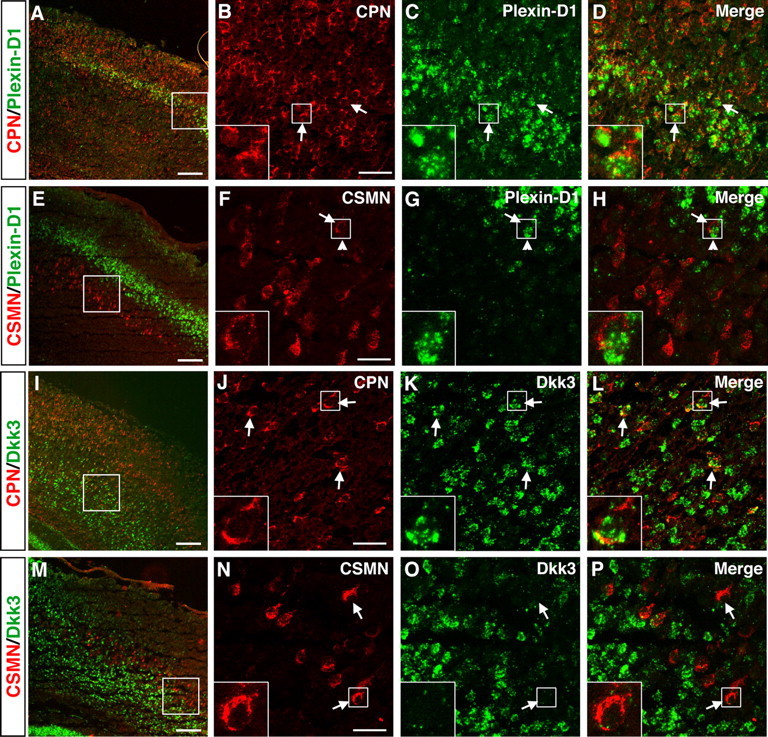
Specific expression of Plexin-D1 and Dkk3 in retrogradely labeled CPN of the deep layers V and VI. A–H, Plexin-D1 is expressed by CPN of the deep layer V and not by CSMN. A, Fluorescent in situ hybridization for Plexin-D1 (green) in a coronal section of P8 cortex demonstrates that it is expressed within CPN (red) identified via injection of CTB555 into contralateral cortex at P6. B–D, Magnification of selected area from A reveals that essentially all Plexin-D1-expressing cells are CPN (arrows). E–H, Fluorescent in situ hybridization for Plexin-D1 (green) in a coronal section of P8 cortex demonstrates that it is not expressed in CSMN (red) identified retrogradely labeled via spinal cord injection of CTB555. F–H, Magnification of selected area from E, with the arrow indicating a CSMN that is adjacent to a PlexinD1-expressing cell (arrowhead). I–P, Dkk3 is expressed by CPN of the deep layers V and VI and not by CSMN. I, Fluorescent in situ hybridization for Dkk3 (green) in a coronal section of P8 cortex demonstrates that it is expressed in CPN (red) identified via injection of CTB555 into contralateral cortex at P6. J–L, Magnification of selected area from I, with arrows indicating Dkk3-positive CPN; notably, a subset of Dkk3-expressing cells are not retrogradely labeled. M, Fluorescent in situ hybridization for Dkk3 (green) in a coronal section of P8 cortex demonstrates that Dkk3 is not expressed in CSMN (red) retrogradely labeled via spinal cord injection of CTB555. N–P, Magnification of selected area from M, with arrows indicating CSMN that do not express Dkk3. Scale bars: A, E, I, M, 200 μm; B–D, F–H, J–L, N–P, 50 μm.
It is intriguing to note that each of the genes expressed highly and somewhat selectively in layer VI appears to label more cells than the number of CPN located in this layer. For example, there is a significant number of Dkk3-expressing cells in layer VI that are not CPN (Fig. 7L). This suggests (1) that each gene also labels other neuronal or glial types within layer VI, including perhaps the predominant population of corticothalamic projection neurons, and (2) that this layer VI subset (and perhaps other subsets) of CPN might be molecularly related to, or even evolutionarily derived from, corticofugal projection neurons. Expression of genes such as Dkk3 with other CPN genes (e.g., Hspb3) may be used to further delineate subtypes of CPN within deep cortical layers.
Combinatorial codes of genes define distinct subpopulations of CPN and identify novel subdivisions of canonical cortical layers
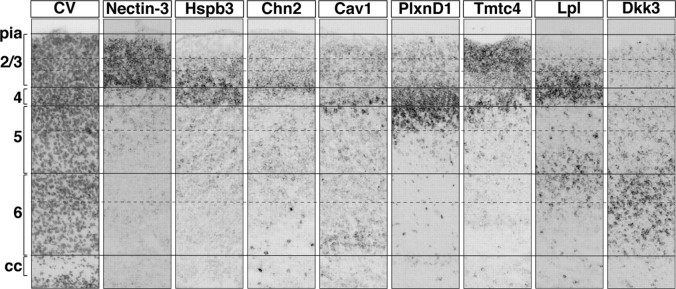
To directly compare the laminar and sublaminar distribution of individual CPN genes and to provide a first spatial map of gene expression by these distinct CPN subpopulations, we investigated the cellular level of expression of a selected set of CPN genes within the same brain. We chose genes that have distinct laminar distributions and similar temporal expression, enabling detection and comparison at the same stage of development. We chose P6 as a midpoint in CPN development at which a large number of CPN-specific genes are expressed, and we performed in situ hybridization for all selected genes on 10-μm-thick serial sections from the same brain (Fig. 8). Cresyl violet staining of the first section of the series was used to distinguish canonical cortical layers at the histological level. For this analysis, we chose eight representative genes: Nectin-3, Cav1, and Chn2 (all expressed in distinct sublamina of layer II/III); Hspb3 and Lpl (both expressed in layers II/III and V); Plexin-D1 (expressed in layer Va); Tmtc4 (expressed in layers II/III and Va); and Dkk3 (expressed in layer VI). This series of in situ hybridizations allowed us to delineate the relative laminar location of the cells expressing each of the genes. For example, we found that, at P6, Cav1-expressing cells are overlapping with Plexin-D1 labeling within layer Va. This analysis reveals that distinct subpopulations of CPN can be identified at the molecular level that express unique combinations of these genes; these distinct CPN populations occupy different layers, and, in some cases, different sublaminae of the same layer (Fig. 8). Together, these results demonstrate that CPN are a molecularly diverse set of populations and suggest that functionally and connectionally distinct subgroups of CPN are defined during their differentiation by distinct combinatorial codes of gene expression that govern their development. These data provide a new “molecular anatomy” of CPN via the expression of genes that classify this highly diverse set of neuronal populations at the molecular level with a precision not previously possible using standard anatomical and histological criteria.
Figure 8.
Combinations of genes reveal subdivisions within canonical cortical laminae. In situ hybridization at P6 for representative genes in sequential coronal sections of cortex from the same mouse, showing molecularly distinct populations of CPN identify subcompartments within the canonical cortical layers. Solid lines identify layers, and dotted lines identify subdivisions of the layers demarcated by in situ hybridization for a subset of the genes identified. Scale bars, 50 μm.
Discussion
Despite knowledge of the developmental and anatomical properties of some broad classes of projection neuron types of the cortex, the genes that distinguish individual neuronal lineages and instruct their lineage-specific development are only beginning to be identified. We previously reported on the identification of genes that mark and control the development of corticospinal motor neurons, a major corticofugal projection neuron population of layer V (Arlotta et al., 2005). Here we report on the identification of genes that identify at the molecular level the lineage of interhemispheric callosal projection neurons of the cortex.
Previous anatomical, histological, and birthdating analyses have highlighted the anatomical complexity and the cellular heterogeneity of this broad population of callosal projection neurons (Koralek et al., 1990; Conti and Manzoni, 1994; Reiner et al., 2003; Mitchell and Macklis, 2005; Ramos et al., 2008a). However, detailed functional and molecular studies of different CPN subpopulations have been complicated by the heterogeneity of neuron types within individual cortical layers and by the scarcity of CPN-specific antigenic markers to identify them as they develop in vivo. Few genes have been reported that label neurons of the upper cortical layers (Molyneaux et al., 2007). These include Satb2, a nuclear matrix protein that induces the formation of callosal connections by CPN during corticogenesis (Britanova et al., 2005; Szemes et al., 2006), and Lmo4, a transcription factor expressed in CPN of all layers (Arlotta et al., 2005). Other genes, such as Cux1 and Cux2 (Nieto et al., 2004; Zimmer et al., 2004) and Unc5d (Tarabykin et al., 2001; Zhong et al., 2004; Sasaki et al., 2008), are expressed in the SVZ during the generation of upper layer neurons and, postnatally, in neurons of the upper layers. Finally, genes with broader patterns of expression have been reported to label neurons in the upper layers of cortex, including Lhx2 (Bulchand et al., 2001, 2003), Gpr6 (Chenn et al., 2001), Brn1 and Brn2 (McEvilly et al., 2002; Sugitani et al., 2002), Kitl, and Dtx4 (Zhong et al., 2004). However, their patterns of expression are typically too broad to identify individual neuronal populations, and it is not known whether they label CPN among other upper layer neuron types. Furthermore, there is a paucity of genes that identify CPN of the deep layers.
Here we provide the first demonstration that distinct classes of callosal neurons can be defined in vivo based on the expression of different molecular markers, suggesting distinct molecular controls over their subtype-specific development, connectivity, and function. Some of the CPN genes are expressed by most callosal neurons, spanning layers II/III, V, and VI (e.g. Lpl and Hspb3), suggesting that they may play broader roles during the development of all callosal neurons. In contrast, other genes are more restricted to only callosal neurons of selected layers: for example, Cpne4, Btg1, Inhba, Gm879, and Tmtc4 are expressed in layer II/III, whereas PlexinD1, Gfra2, TcrB, and Dkk3 mark CPN of the deep layers V and VI. Retrograde labeling experiments reveal that the genes we identified have varying levels of restricted expression to CPN. For example, Plexin-D1 clearly distinguishes the layer V CPN population from CSMN, even at the interface of layer Va and Vb, in which these two neuronal types are intermingled. Additionally, the vast majority of Plexin-D1-expressing cells in the cortex are CPN. In contrast, while still distinguishing CPN from adjacent CSMN in deep layer V, Dkk3 is also expressed in other neuronal populations of layer VI, likely corticothalamic projection neurons. Together, these data reveal the presence of callosal neuron subpopulations that were not previously recognized at the anatomical and histological levels, by demonstrating that different subtypes of callosal projection neurons can be identified by combinations of molecular markers within subdivisions of the same canonical lamina. The identification of genes that uniquely define distinct callosal subpopulations will now make it possible to investigate the function and connectivity of individual subpopulations via genetic manipulation and targeting of distinct callosal projection neuron types.
Interestingly, we find that callosal projection neurons of the upper layers (II/III) exhibit a higher degree of molecular heterogeneity than those located in the deep layers (V and VI), as indicated by the fact that CPN located in different portions of layer II/III express different genes. For example, genes including Grsp1, Nnmt, and EphA3 all selectively label CPN located in the most superficial portion of layer II/III, whereas genes including Nectin-3, Chn2, and Ptn are preferentially restricted to the deeper part of the same layer. Of note, a number of layer-specific genes have been identified via large-scale in situ efforts, including the Allen Brain Atlas (http://www.brain-map.org). Interestingly, examination in the Allen Brain Atlas of the expression profile of the CPN genes identified here reveals that some of these genes maintain layer-specific expression in the adult. For these genes, our data extend these layer-specific expression data to provide an indication of the neuron types that express the individual genes. Many other genes are not identified in the Allen Brain Atlas as layer-specific in the adult, further supporting functional roles during embryonic and early postnatal stages of CPN development. Finally, a small number of the CPN genes reported here (e.g., Lhx2 and COUPTF1) have been shown previously to be expressed only in restricted cortical areas (Nakagawa et al., 1999; Liu et al., 2000), suggesting that additional CPN populations might be distinguished at different rostrocaudal and mediolateral locations. In the future, it will be useful to further define the boundaries of arealization of the CPN genes reported here.
It is interesting to speculate that the presence of a molecularly more diversified population of callosal neurons in the superficial layers of the rodent cortex might reflect the early stages of the expansion and diversification of these layers that occurred during evolution of the primate cortex. Since the divergence of reptiles, birds, and mammals, the cortex has undergone substantial radial expansion, with major addition of new neurons within the superficial cortical layers II–IV of mammalian species (Reiner, 1991; Marin-Padilla, 1992; Aboitiz et al., 2003). For example, the reptilian cortex has only three layers, thought to be homologous to layers I, V, and VI of the six-layer mammalian cortex, and they lack neurons with properties of mammalian upper layer projection neurons (Reiner, 1991). Among mammals, the primate cortex exhibits additional expansion; the six canonical layers seen in rodents and lower mammals have enlarged to include new subdivisions that can be easily distinguished at the histological level (Rakic and Kornack, 2001). Of particular interest is the subdivision of the expanded layer II/III of the rodent into the multiple histologically distinct layers and sublayers seen in primates.
Although these layers cannot be distinguished at the histological level in the rodent, it is intriguing to speculate that our identification of genes that mark neurons located in distinct radial positions within layer II/III might support the hypothesis that the specialization of distinct populations of CPN within upper layers is already occurring in the rodent cortex both molecularly and almost certainly with regard to connectivity and function. Future work that investigates the expression of the genes identified here within the cortex of primates will likely provide important insight into the evolution of the neocortex.
The expansion of the superficial layers during cortical evolution has been accompanied by the expansion of a new germinal zone, the SVZ, and by the appearance of intermediate progenitors within the SVZ that are fated to produce neurons of the upper layers (Smart and McSherry, 1982; Martinez-Cerdeno et al., 2006; Molyneaux et al., 2007). This is in contrast to reptiles and birds, in which cortical neurogenesis occurs only in the VZ (Cheung et al., 2007). In mammals, it is now becoming clear that VZ progenitors give rise to neurons of the deep cortical layers, whereas SVZ progenitors primarily generate the upper cortical layers (Tarabykin et al., 2001; Wu et al., 2005). In agreement with existing data, we find that callosal neurons of the deep cortical layers (V and VI) express a unique set of genes (e.g., TcrB and Dkk3) that are not expressed by callosal neurons of the upper layers, likely reflecting the different evolutionary origin of these two populations.
The data presented here provide the first molecular classification of the callosal projection neuron population in the cortex. Distribution analysis of the newly identified CPN genes reveals the presence of molecularly distinct subpopulations that were not previously described at the histological, morphological, or anatomical levels. This likely reflects the distinct origin of different callosal neuron subpopulations, and it might be predictive of their evolutionary diversification in higher mammalian species. It is likely that distinct combinations of molecular developmental controls define key aspects of CPN diversity—subtype-specific differentiation, axon collateralization, synaptic connectivity, and physiologic function—underlying their central roles in interhemispheric association and connectivity. Together, these data provide the foundation for future studies in which molecular and genetic approaches can be combined with anatomical and cellular data to dissect the mechanisms of development of the diverse populations of callosal projection neurons.
Footnotes
This work was partially supported by National Institutes of Health Grants NS41590 and NS45523 and the Harvard Stem Cell Institute (J.D.M.), with additional infrastructure support by National Institutes of Health Grant NS49553. P.A. was supported by a Claflin Distinguished Scholar Award, the Harvard Stem Cell Institute, and the ALS Association. B.J.M. was supported by the Harvard Medical Science Training Program and the United Sydney Association. We thank A. Meleski and K. Quinn for technical assistance, and J. Menezes and members of the Macklis laboratory for suggestions and critical reading of this manuscript.
References
- Aboitiz F, Morales D, Montiel J. The evolutionary origin of the mammalian isocortex: towards an integrated developmental and functional approach. Behav Brain Sci. 2003;26:535–552. doi: 10.1017/s0140525x03000128. discussion 552–585. [DOI] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Berger UV, Hediger MA. Differential distribution of the glutamate transporters GLT-1 and GLAST in tanycytes of the third ventricle. J Comp Neurol. 2001;433:101–114. doi: 10.1002/cne.1128. [DOI] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Sestan N, Molnár Z, Tarabykin V. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- Catapano LA, Arnold MW, Perez FA, Macklis JD. Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J Neurosci. 2001;21:8863–8872. doi: 10.1523/JNEUROSCI.21-22-08863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catapano LA, Arlotta P, Cage TA, Macklis JD. Stage-specific and opposing roles of BDNF, NT-3 and bFGF in differentiation of purified callosal projection neurons toward cellular repair of complex circuitry. Eur J Neurosci. 2004;19:2421–2434. doi: 10.1111/j.0953-816X.2004.03303.x. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T. Proliferative events in the cerebral ventricular zone. Brain Dev. 1995;17:159–163. doi: 10.1016/0387-7604(95)00029-b. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005a;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005b;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Levin ME, McConnell SK. Temporally and spatially regulated expression of a candidate G-protein-coupled receptor during cerebral cortical development. J Neurobiol. 2001;46:167–177. doi: 10.1002/1097-4695(20010215)46:3<167::aid-neu1000>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Cheung AF, Pollen AA, Tavare A, DeProto J, Molnár Z. Comparative aspects of cortical neurogenesis in vertebrates. J Anat. 2007;211:164–176. doi: 10.1111/j.1469-7580.2007.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti F, Manzoni T. The neurotransmitters and postsynaptic actions of callosally projecting neurons. Behav Brain Res. 1994;64:37–53. doi: 10.1016/0166-4328(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Fricker-Gates RA, Shin JJ, Tai CC, Catapano LA, Macklis JD. Late-stage immature neocortical neurons reconstruct interhemispheric connections and form synaptic contacts with increased efficiency in adult mouse cortex undergoing targeted neurodegeneration. J Neurosci. 2002;22:4045–4056. doi: 10.1523/JNEUROSCI.22-10-04045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao WJ, Zheng ZH. Target-specific differences in somatodendritic morphology of layer V pyramidal neurons in rat motor cortex. J Comp Neurol. 2004;476:174–185. doi: 10.1002/cne.20224. [DOI] [PubMed] [Google Scholar]
- Glenn DJ, Maurer RA. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha-subunit gene expression. J Biol Chem. 1999;274:36159–36167. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II–V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralek KA, Olavarria J, Killackey HP. Areal and laminar organization of corticocortical projections in the rat somatosensory cortex. J Comp Neurol. 1990;299:133–150. doi: 10.1002/cne.902990202. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dwyer ND, O'Leary DD. Differential expression of COUP-TFI, CHL1, and two novel genes in developing neocortex identified by differential display PCR. J Neurosci. 2000;20:7682–7690. doi: 10.1523/JNEUROSCI.20-20-07682.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Padilla M. Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. J Comp Neurol. 1992;321:223–240. doi: 10.1002/cne.903210205. [DOI] [PubMed] [Google Scholar]
- Martinez-Cerdeno V, Noctor SC, Kriegstein AR. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb Cortex. 2006;16(Suppl 1):i152–i161. doi: 10.1093/cercor/bhk017. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Macklis JD. Large-scale maintenance of dual projections by callosal and frontal cortical projection neurons in adult mice. J Comp Neurol. 2005;482:17–32. doi: 10.1002/cne.20428. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Johnson JE, O'Leary DD. Graded and areal expression patterns of regulatory genes and cadherins in embryonic neocortex independent of thalamocortical input. J Neurosci. 1999;19:10877–10885. doi: 10.1523/JNEUROSCI.19-24-10877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Koester SE. Development of projection neuron types, axon pathways, and patterned connections of the mammalian cortex. Neuron. 1993;10:991–1006. doi: 10.1016/0896-6273(93)90049-w. [DOI] [PubMed] [Google Scholar]
- Ozdinler PH, Macklis JD. IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat Neurosci. 2006;9:1371–1381. doi: 10.1038/nn1789. [DOI] [PubMed] [Google Scholar]
- Rakic P, Kornack DR. Neocortical expansion and elaboration during primate evolution: a view from neuroembryology. In: Falk D, Gibson KR, editors. Evolutionary anatomy of the primate cerebral cortex. Cambridge, UK: Cambridge UP; 2001. pp. 30–56. [Google Scholar]
- Ramos RL, Tam DM, Brumberg JC. Physiology and morphology of callosal projection neurons in mouse. Neuroscience. 2008a;153:654–663. doi: 10.1016/j.neuroscience.2008.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos RL, Smith PT, DeCola C, Tam D, Corzo O, Brumberg JC. Cytoarchitecture and transcriptional profiles of neocortical malformations in inbred mice. Cereb Cortex. 2008b;18:2614–2628. doi: 10.1093/cercor/bhn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A. A comparison of neurotransmitter-specific and neuropeptide-specific neuronal cell types present in the dorsal cortex in turtles with those present in the isocortex in mammals: implications for the evolution of isocortex. Brain Behav Evol. 1991;38:53–91. doi: 10.1159/000114379. [DOI] [PubMed] [Google Scholar]
- Reiner A, Jiao Y, Del Mar N, Laverghetta AV, Lei WL. Differential morphology of pyramidal tract-type and intratelencephalically projecting-type corticostriatal neurons and their intrastriatal terminals in rats. J Comp Neurol. 2003;457:420–440. doi: 10.1002/cne.10541. [DOI] [PubMed] [Google Scholar]
- Richards LJ, Plachez C, Ren T. Mechanisms regulating the development of the corpus callosum and its agenesis in mouse and human. Clin Genet. 2004;66:276–289. doi: 10.1111/j.1399-0004.2004.00354.x. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Tabata H, Tachikawa K, Nakajima K. The cortical subventricular zone-specific molecule Svet1 is part of the nuclear RNA coded by the putative Netrin receptor gene Unc5d and is expressed in multipolar migrating cells. Mol Cell Neurosci. 2008;38:474–483. doi: 10.1016/j.mcn.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Smart IH, McSherry GM. Growth patterns in the lateral wall of the mouse telencephalon. II. Histological changes during and subsequent to the period of isocortical neuron production. J Anat. 1982;134:415–442. [PMC free article] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Suzuki A, Kishikawa M, Akutsu R, Hirose T, Waye MM, Tsui SK, Yoshida S, Ohno S. Muscle develops a specific form of small heat shock protein complex composed of MKBP/HSPB2 and HSPB3 during myogenic differentiation. J Biol Chem. 2000;275:1095–1104. doi: 10.1074/jbc.275.2.1095. [DOI] [PubMed] [Google Scholar]
- Szemes M, Gyorgy A, Paweletz C, Dobi A, Agoston DV. Isolation and characterization of SATB2, a novel AT-rich DNA binding protein expressed in development- and cell-specific manner in the rat brain. Neurochem Res. 2006;31:237–246. doi: 10.1007/s11064-005-9012-8. [DOI] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaró S, Camps L, Reina M, Perez-Clausell J, Llobera M, Olivecrona T. Localization of lipoprotein lipase to discrete areas of the guinea pig brain. Brain Res. 1990;506:249–253. doi: 10.1016/0006-8993(90)91258-i. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Zhang YA, Levin ME, Devine WP, Brûlet P, McConnell SK. Cortical neurons require Otx1 for the refinement of exuberant axonal projections to subcortical targets. Neuron. 1999;24:819–831. doi: 10.1016/s0896-6273(00)81030-2. [DOI] [PubMed] [Google Scholar]
- Wilson CJ. Morphology and synaptic connections of crossed corticostriatal neurons in the rat. J Comp Neurol. 1987;263:567–580. doi: 10.1002/cne.902630408. [DOI] [PubMed] [Google Scholar]
- Wise SP, Jones EG. The organization and postnatal development of the commissural projection of the rat somatic sensory cortex. J Comp Neurol. 1976;168:313–343. doi: 10.1002/cne.901680302. [DOI] [PubMed] [Google Scholar]
- Wu SX, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave KA, Tamamaki N. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci U S A. 2005;102:17172–17177. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Takemoto M, Fukuda T, Hattori Y, Murakami F, Nakajima D, Nakayama M, Yamamoto N. Identification of the genes that are expressed in the upper layers of the neocortex. Cereb Cortex. 2004;14:1144–1152. doi: 10.1093/cercor/bhh074. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]