Abstract
The ADP/ATP carrier (AAC) is a very effective membrane protein that mediates the exchange of ADP and ATP across the mitochondrial membrane. In vivo transport measurements on the AAC overexpressed in Escherichia coli demonstrate that this process can be severely inhibited by high-chloride concentrations. Molecular-dynamics simulations reveal a strong modification of the topology of the local electric field related to the number of chloride ions inside the cavity. Halide ions are shown to shield the positive charges lining the internal cavity of the carrier by accurate targeting of key basic residues. These specific amino acids are highly conserved as highlighted by the analysis of multiple AAC sequences. These results strongly suggest that the chloride concentration acts as an electrostatic lock for the mitochondrial AAC family, thereby preventing adenine nucleotides from reaching their dedicated binding sites.
ATP, the energy fuel of the cell, is synthesized from ADP in the mitochondria, using the proton gradient created by the respiratory chain. Alteration of the ADP and ATP exchange across the inner mitochondrial membrane bears various pathological implications (1). The ADP/ATP transport is mediated by a member of the mitochondrial carrier family, e.g., the ADP/ATP carrier (AAC). In the absence of a membrane potential, exchange can occur in both directions (5). The structure of the bovine-heart AAC has been solved in the presence of the inhibitor carboxyatractyloside (6,7). It consists of six α-helices forming a compact transmembrane domain on the matrix side, with a cavity opened toward the intermembrane space. Two patches of basic residues line this cavity. The upper patch, located at the mouth of the AAC, consists of residues K91, K95, R104, K106, R187, and K198. Residues K22, K32, R79, R137, R234, R235, and R279 form the lower basic patch that delineates the bottom of the cavity. Noteworthily, several of these residues are involved in carboxyatractyloside and/or ADP3− binding (6). The latter two basic patches contribute to the unique electrostatic topology of the AAC, shaped in a funnel conducive to drive the substrate downwards to the bottom of the internal cavity (6,8,9). Biochemical experiments have demonstrated that mutation of specific residues abolishes the transport activity of AAC (for a review, see (10)). The function of the protein can also be affected by an appropriate modification of the ionic concentration. Experiments emphasize that high concentrations of either magnesium or chloride ions inhibit the function of the bovine-heart AAC (11–13). Mg2+ is assumed to bind to the substrate, which can no longer be transported in a complexed form. In sharp contrast, chloride is believed to affect directly the properties of the membrane protein (13).
A synergistic experiment-theory study has been carried out here to explain the effects of chloride concentrations on the function of the overall AAC family. Transport activities were measured for Arabidopsis thaliana AAC isoform 1 (AtAAC1) expressed in Escherichia coli to probe how sensitive the inhibition of the AAC family is in response to high-chloride concentrations. Molecular-dynamics (MD) simulations of bovine-heart AAC isoform 1 (bAAC1) were carried out concomitantly to decipher how transport across the AAC is thwarted by chloride concentrations. In the light of the above concerted investigations, supplemented by an evolutionary analysis, congruent conclusions are drawn on chloride inhibition in the AAC family.
Gropp et al. (13) observed the inhibitory effect of anions on purified bAAC1 reconstituted in liposomes. To extend this observation, functional expression of recombinant AACs in E. coli was performed here, thereby providing a unique framework for investigating in vivo the biochemical properties of AACs. Transport of [α-32P]ATP assayed on whole E. coli cells expressing AtAAC1 demonstrates that the carrier is produced in a functional form in the inner membrane of E. coli (Fig. 1, inset). The same experiments performed at low NaCl concentration (0.15 M) do not reveal any significant effect on transport activity, whereas addition of 0.5 M or 0.6 M NaCl markedly reduces transport (Fig. 1). These results are in line with measurements (13) based on bAAC1 sharing 74% of sequence homology with AtAAC1. The direct in vivo measurements of ATP uptake inhibition reported here consequently suggest that the inhibition potency of high-chloride concentrations ought to hold for the entire mitochondrial AAC family.
Figure 1.
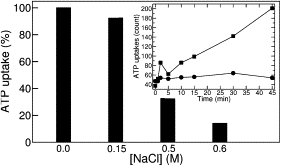
Inhibitory effect of NaCl on the [α-32P]ATP uptake by AtAAC1. (Inset) Time dependence of [α-32P]ATP uptake into intact IPTG-induced E. coli cells harboring plasmid encoding mature AtAAC1 (squares) or the control plasmid (circles). The data shown is the mean of four independent experiments (SE < 10% of the mean values; see Supporting Material).
At the molecular level, chloride ions are envisioned to shield the positive charges lining the internal cavity of the carrier (13). On the theoretical front, the effects of chloride concentrations on the transport properties of the AAC were examined using three different salt conditions as models of low (e.g., 0.1 M and 0.15 M Cl−) and high chloride concentrations (e.g., 0.6 M Cl−). For each concentration, MD trajectories of the bAAC1 embedded in a fully hydrated palmitoyloleylphosphatidylcholine bilayer were generated, with and without ADP3−. All MD simulations were performed employing the NAMD program (14) with the CHARMM27 force field (15,16) and a revision of the latter for lipids (17).
The number of chloride ions in the cavity is a function of the chloride concentration in the solvent (see Supporting Material). At low NaCl concentration, the halide ions primarily interact with residues K22, R79, and R279 of the lower basic patch. Furthermore, a chloride ion transiently binds to R235 (0.1 M NaCl assay) and to R137 (0.15 M NaCl assay). In both assays, no chloride ion is permanently complexed with residues of the upper basic patch. In contrast, in the 0.6 M NaCl assay, several chloride ions interact frequently with residues of the upper patch (i.e., K91, K95, R187, and K198). They also interact with residues K22, R79, and R279 of the lower patch, reminiscent of the simulations at low concentrations. In addition, a chloride ion associates with R234, occupying the same position for as long as 29.5 ns. The significant persistence of this chloride ion, together with the saturation of the two basic patches, results at a high-chloride concentration in a less accessible binding site for ADP3−.
As was highlighted previously (8,9), the protein exhibits a funnellike electrostatic pathway. A cross-sectional view of the three-dimensional map of the electric field (Fig. 2) indicates that the distribution of chloride ions affects the electrostatic signature of the protein (see Supporting Material). At low-salt concentration, the electrostatic funnel ends at the bottom of the cavity, where the electric field points downwards. Under such circumstances, ADP3− has been shown to reach spontaneously its dedicated binding site (8,9). At 0.6 M NaCl, the topology of the electric field differs markedly above the region formed by the lower basic patch, where its direction now points downwards. In this scenario, a negatively charged substrate is prevented from reaching the bottom of the cavity. Thus, even though an electrostatic funnel prevails at a high-chloride concentration, it does not end as deep in the cavity as it would at low-salt concentration.
Figure 2.
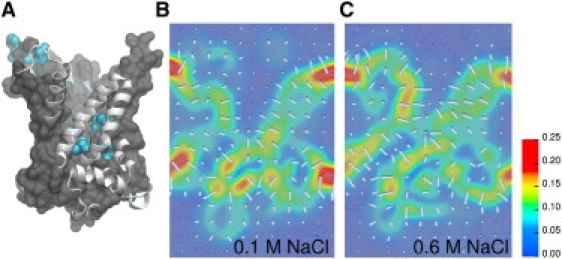
The electric properties of apo-bAAC1 differ as a function of the salt concentration. (A) Orientation of the bAAC1. A molecular-surface rendering and a ribbon representation are used for regions of the protein located, respectively, behind and in front of the plane in which the electric field is shown. The basic patches are highlighted in cyan. The cross-sectional view of the electric field after 30 ns differs between (B) 0.1 M and (C) 0.6 M assay. Maps of the electric field were obtained using the PMEPot (18) module of VMD (19) and OpenDX (http://www.opendx.org).
Binding numerical experiments, where ADP3− is initially located at the mouth of AAC, have revealed that this substrate binds spontaneously to the protein, independently of its starting position (9,8). Simulations featuring three different ligand-association assays carried out at 0.15 M NaCl led to the association of ADP3− (see Supporting Material), consistent with the experimental data (Fig. 1), as well as the previous findings of Dehez et al. (9). Considering nine different starting positions for ADP3−, association was, however, never observed at 0.6 M NaCl (Fig. 3). Nine similar experiments were subsequently repeated in the absence of an ionic concentration, while keeping the same initial position of the substrate. All these simulations invariably led to the association of ADP3− (Fig. 3). Put together, these ligand-association and transport assays remarkably illustrate the crucial role played by chloride concentration on AAC activity.
Figure 3.
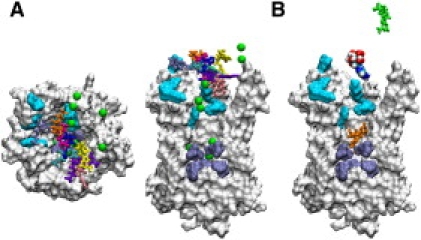
The series of ligand-association assays at 0.6 M NaCl results in the binding of ADP3− only after removal of the ions. (A) The different initial positions of ADP3− are depicted as colored tubes. A molecular-surface rendering is used for the protein (white), the upper (light-blue), and the lower patches (ice-blue). Chloride ions are shown as green van der Waals spheres. (B) Association experiment C. The starting position of ADP3− and its final positions at 0.6 M NaCl and without salt are shown, respectively, as van der Waals spheres, and green and orange tubes. Image rendering using VMD (19).
Inhibition does not appear to be specific to bAAC1. These experiments suggest, on the contrary, that abolition of transport by chloride ions is characteristic of the AAC family. Multiple-sequence alignment was, hence, carried out, based on 74 AAC sequences. Analysis of the alignment reveals a conservation of those residues directly involved in the binding of halide ions (see Supporting Material). It should, therefore, be expected that shielding of dedicated basic residues by chloride ions is a common phenomenon likely to occur in all mitochondrial AACs. The small size of these ions allows each single amino acid essential to the transport to be targeted with optimal accuracy, regardless of the local structure of the carrier.
In this work, the effect of high-chloride concentration on the activity of the mitochondrial AAC family has been unraveled by combining synergistically experimental and theoretical investigations. Uptake experiments of radioactively labeled ATP4− have demonstrated that transport in the plant AtAAC1 is also inhibited at high-chloride concentrations. Molecular detail of the inhibition is unveiled by MD simulations performed on the bAAC1 structure. As a function of the concentration, a large number of ions are prone to gush into the internal cavity of the protein. In turn, ionic saturation of those residues pertaining to the two basic patches of the AAC strongly modulates the topology of the internal electric field, thus, locking the carrier in a state unfavorable to substrate binding. A conservation analysis performed over a large set of AAC sequences shows that the key residues shielded by Cl− are conserved. This observation strongly suggests that inhibition at high-chloride concentrations can occur independently of the AAC sequence. Chloride concentration may, therefore, be viewed as an electrostatic lock for the mitochondrial AAC family, preventing the adenine nucleotide from reaching its target-binding site. Our results emphasize that in the context of functional assays, particularly for experimental nucleotide binding assays, chloride concentration constitutes an important parameter that ought to be optimized carefully.
Supporting Material
Eleven figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01430-1.
Supporting Material
Acknowledgments
The authors are indebted to the Grand Equipement National de Calcul Intensif and the Centre Informatic National de l'Enseignement Supérieur for provision of computer time.
This work is funded by the L'Agence Nationale de la Recherche, the Commissariat à l'Énergie Atomique, the Centre National de la Recherche Scientifique, and the European Drug Initiative on Channels and Transporters program.
Footnotes
Christophe Chipot's present address is Theoretical and Computational Biophysics Group, Beckman Institute, University of Illinois at Urbana-Champaign, 405 North Mathews, Urbana, Illinois 61801.
Contributor Information
François Dehez, Email: francois.dehez@edam.uhp-nancy.fr.
Christophe Chipot, Email: chipot@ks.uiuc.edu.
References and Footnotes
- 1.Dahout-Gonzalez C., Nury H., Trézéguet V., Lauquin G.J.-M., Pebay-Peyroula E. Molecular, functional, and pathological aspects of the mitochondrial ADP/ATP carrier. Physiology (Bethesda) 2006;21:242–249. doi: 10.1152/physiol.00005.2006. [DOI] [PubMed] [Google Scholar]
- 2.Reference deleted in proof.
- 3.Reference deleted in proof.
- 4.Reference deleted in proof.
- 5.Brustovetsky N., Becker A., Klingenberg M., Bamberg E. Electrical currents associated with nucleotide transport by the reconstituted mitochondrial ADP/ATP carrier. Proc. Natl. Acad. Sci. USA. 1996;93:664–668. doi: 10.1073/pnas.93.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G.J.-M. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 7.Nury H., Dahout-Gonzalez C., Trézéguet V., Lauquin G.J.-M., Brandolin G. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579:6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc. Natl. Acad. Sci. USA. 2008;105:9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehez F., Pebay-Peyroula E., Chipot C. Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J. Am. Chem. Soc. 2008;24:12725–12733. doi: 10.1021/ja8033087. [DOI] [PubMed] [Google Scholar]
- 10.Nury H., Dahout-Gonzalez C., Trézéguet V., Lauquin G.J.-M., Brandolin G. Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu. Rev. Biochem. 2006;75:711–741. doi: 10.1146/annurev.biochem.75.103004.142747. [DOI] [PubMed] [Google Scholar]
- 11.Krämer R. Influence of divalent cations on the reconstituted ADP, ATP exchange. Biochim. Biophys. Acta. 1980;592:615–620. doi: 10.1016/0005-2728(80)90104-8. [DOI] [PubMed] [Google Scholar]
- 12.Krämer R., Kürzinger G. The reconstituted ADP/ATP carrier from mitochondria is both inhibited and activated by anions. Biochim. Biophys. Acta. 1984;765:353–362. doi: 10.1016/0005-2728(84)90176-2. [DOI] [PubMed] [Google Scholar]
- 13.Gropp T., Brustovetsky N., Klingenberg M., Müller V., Fendler K. Kinetics of electrogenic transport by the ADP/ATP carrier. Biophys. J. 1999;77:714–726. doi: 10.1016/S0006-3495(99)76926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philipps J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1782. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 16.Feller S.E., MacKerell A.D. An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104:7510–7515. [Google Scholar]
- 17.Hénin J., Shinoda W., Klein M.L. United-atom acyl chains for CHARMM phospholipids. Phys. Chem. B. 2008;112:7008–7015. doi: 10.1021/jp800687p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aksimentiev A., Schulten K. Imaging α-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


