Abstract
With the advent of the genome-wide association (GWA) study, a promising new avenue for identifying genetic markers for complex diseases like coronary artery disease (CAD) has been opened. This avenue, however, is not without challenges and limitations, including the need for carefully designed and executed studies and the risk of false positive associations. Nonetheless, new markers have been identified through such studies that could potentially revolutionize the ways that individuals with CAD are identified and managed.
Keywords: atherosclerosis, coronary artery disease, genetics, genome wide association, genomics, myocardial infarction
Background
Coronary artery disease (CAD) and its associated complication, myocardial infarction (MI), is a leading cause of morbidity and mortality worldwide. CAD is a multifactorial disease that can be influenced by many different environmental and heritable risk factors. The progression of atherosclerotic disease involves a complex series of events, each event (e.g. foam cell formation, smooth muscle cell recruitment, etc.) involving multiple biological pathways and genes. Risk factors for CAD include hypertension, smoking status, male gender, age, body mass index, type 2 diabetes mellitus, and heredity. Analyte risk markers associated with CAD include lipid levels, cholesterol particle size and number, C-reactive protein, homocysteine, fibrinogen, and lipoprotein (a). However, many of the current traditional and novel risk markers are unable to fully predict who is at risk for CAD. For example, 35% of CAD has been shown to occur in people with total cholesterol levels <200 mg/dL [1]. Thus, other novel risk markers, including genetic risk markers, may be important for better refining individuals at risk for CAD and CV events.
Both twin and family studies have demonstrated that CAD has a strong heritable component. In one study examining 20,966 twins over a 36-year period, the heritability (h2) for fatal CAD events was 57% for men and 38% for women [2]. Many studies have demonstrated that family history of CAD is an independent predictor of CAD [3–6]. Others have shown that distinct morphological features of CAD display high heritability [7, 8]. In addition, individuals with a family history of CAD generally have earlier onset, are male, have a history of smoking, and are hyperlipidemic [9]. The majority (72%) of early CAD cases (men < 55 years, women < 65 years) and 48% of all CAD cases, regardless of age, have a family history of CAD [10].
While multiple studies have concurred that CAD is highly heritable, the mechanisms underlying the heritable basis of CAD have been elusive. This is likely due to the complex nature of CAD. While a very small proportion of CAD is monogenic, or Mendelian in nature (e.g. familial hypercholesterolemia, familial defective apoB-100), the vast majority of CAD is genetically complex. Multiple genes are thought to contribute to CAD, each gene contributing to only a small percent of the phenotype. In addition, many of the risk factors associated with CAD (e.g. blood pressure, diabetes) are themselves polygenic, further adding to the complexity, and hundreds of genes may be involved in CAD susceptibility [11]. Thus, while some genetic parameters, such as heritability, can be quantified for CAD, determining the number and scope of genes involved in CAD is challenging.
Many different family and population-based studies have identified candidate genes potentially associated with CAD. Unfortunately, the vast majority of results from linkage and association studies have not been reproducible or statistically significant, bringing to question the clinical utility of these markers [12]. However, recent technological and scientific advances have now made possible a new type of study: the genome-wide association (GWA) study. The advent of GWA studies has produced some novel and replicated associations in many disease states, including CAD.
Genome-wide association studies and CAD
GWA studies are essentially unbiased large-scale population-based studies evaluating the association of hundreds of thousands of markers (generally single nucleotide polymorphisms, or SNPs) across the genome with a particular phenotype. Part of the beauty of these types of studies is that they are not hypothesis-driven, allowing for the discovery of novel genetic markers. However, GWA studies are not without challenges and limitations and they must be carefully designed and executed. In the design of a GWA study, it is important to keep phenotypic heterogeneity of cases and misclassification of controls to a minimum [13]. Cases and controls should be well-matched to avoid confounders, such as population stratification. Furthermore, sample sizes should be large, generally in the thousands, and replication sample sizes in an independent population should be even larger. The risk for false positive or spurious associations is high in GWA studies, and strict quality control is essential.
One of the interesting aspects about GWA studies is that usually, loci are identified that have not been previously suspected as candidate genes/loci [14]. Several reasons could be behind this. First, the marker may be a spurious association. Second, the identified marker may be in linkage disequilibrium (LD) with the etiological marker. In this case, the effect of different populations with different LD structures must be taken into account [15]. Third, the identified marker may be an etiological marker, but has not been previously targeted as having a role in the disease. In the latter two cases, novel biological pathways involved in the disease state may be uncovered, leading to a better understanding of disease processes and, potentially, the development of novel therapies.
A search of the National Cancer Institute (NCI)-National Human Genome Research Institute (NHGRI)'s Catalog of Published GWA Studies (http://www.genome.gov/26525834) and the literature uncovered a total of 48 GWA studies examining CAD and/or associated traits or markers with p ≤ 10−5. Eight GWA studies with risk factor associations of p ≤ 10−5 have been published looking specifically at CAD or MI (Table 1). The remainder of the 40 identified GWA studies have been published examining traits or markers associated with CAD including coronary artery calcification, blood pressure, LDL cholesterol, HDL cholesterol, triglycerides, Lp(a), C-reactive protein, type 2 diabetes mellitus, and body mass index.
Table 1.
Genome-Wide Association Studies of CAD and MI (p<5 × 10−5)
| Phenotype | Chromo-somal Region | Reported Gene(s) | SNP | Risk Allele Frequency | P-value | Odds ratio | 95% CI | Reference |
|---|---|---|---|---|---|---|---|---|
| CAD | 1p13.3 | CELSR2-PSRC1-SORT1 | rs5999839 | 0.23 | 4 × 10−19 | 1.29 | 1.18–1.40 | [18] |
| early-onset MI | 1p13 | CELSR2-PSRC1-SORT1 | rs646776 | 0.81 | 1.5 × 10−8 | 1.17 | 1.11–1.24 | [21] |
| early-onset MI | 1p32 | PCSK9 | rs11206510 | 0.81 | 9.6 × 10−9 | 1.15 | 1.10–1.21 | [21] |
| early-onset MI | 1q41 | MIA3 | rs1746048 | 0.72 | 5.9 × 10−7 | 1.13 | 1.08–1.18 | [21] |
| CAD | 1q41 | MIA3 | rs17465637 | 0.29 | 1 × 10−6 | 1.2 | 1.12–1.30 | [18] |
| CAD | 1q43 | NA | rs17672135 | 0.87 | 2 × 10−6 | 1.43 | 1.23–1.64 | [16] |
| early-onset MI | 2q33 | WDR12 | rs6725887 | 0.14 | 1.3 × 10−8 | 1.17 | 1.11–1.23 | [21] |
| CAD | 2q36.3 | pseudogene | rs2943634 | 0.65 | 2 × 10−7 | 1.21 | 1.13–1.30 | [18] |
| CAD | 3q22.3 | MRAS | rs9818870 | NA | 7.4 × 10−13 | 1.15 | 1.11–1.19 | [92] |
| CAD | 5q21 | NA | rs383830 | 0.22 | 1 × 10−5 | 1.60 | 1.16–2.21 | [16] |
| early-onset MI | 6p24 | PHACTR1 | rs12526453 | 0.65 | 1.3 × 10−9 | 1.12 | 1.08–1.27 | [21] |
| CAD | 6q25 | MTHFD1L | rs6922269 | 0.25 | 2 × 10−5 | 1.17 | 1.04–1.32 | [16] |
| CAD | 6q25.1 | MTHFD1L | rs6922269 | 0.25 | 3 × 10−8 | 1.23 | 1.15–1.33 | [18] |
| CAD | 6q27 | SCL22A3-LPAL2-LPA | rs2048327, rs3127599, rs7767084, rs10755578 (CTTG and CCTC haplotype) | NA | 1.2 × 10−9 (CTTG), 4.2 × 10−15 (CCTC) | 1.20 (CTTG) 1.82 (CCTC) | 1.13–1.28 (CTTG) 1.57–2.12 (CCTC) | [86]* |
| early-onset MI | 9p21 | CDKN2A, CDKN2B | rs4977574 | 0.56 | 1.1 × 10−30 | 1.28 | 1.23–1.33 | [21] |
| MI | 9p21.3 | CDKN2A, CDKN2B | rs10757278 | 0.45 | 1 × 10−20 | 1.28 | 1.22–1.35 | [19] |
| CAD | 9p21.3 | intergenic | rs1333049 | 0.47 | 3 × 10−19 | 1.36 | 1.27 – 1.46 | [18] |
| CAD | 9p21.3 | CDKN2A, CDKN2B | rs1333049 | 0.47 | 1 × 10−13 | 1.47 | 1.27–1.70 | [16] |
| CAD | 9p21.3 | NA | rs10757274 and rs2383206 | NA | NA | NA | NA | [17] |
| CAD | 10q11.21 | CXCL12 | rs501120 | 0.13 | 9 × 10−8 | 1.33 | 1.20–1.48 | [18] |
| early-onset MI | 10q11 | CXCL12 | rs1746048 | 0.84 | 3.4 × 10−5 | 1.14 | 1.08–1.21 | [21] |
| MI | 12q24 | SH2B3 | rs3184504 | 0.38 | 8.6 × 10−8 | 1.13 | 1.08–1.18 | [82] |
| CAD | 12q24.41 | HNF1A-C12orf43 | rs2259816 | NA | 5 × 10−7 | 1.08 | 1.05–1.11 | [92] |
| CAD | 15q22.33 | SMAD3 | rs17228212 | 0.30 | 2 × 10−7 | 1.21 | 1.13–1.30 | [18] |
| CAD | 16q23 | NA | rs8055236 | 0.20 | 6 × 10−6 | 1.91 | 1.33–2.74 | [16] |
| early-onset MI | 19p13 | LDLR | rs1122608 | 0.75 | 1.9 × 10−9 | 1.15 | 1.10–1.20 | [21] |
| CAD | 19q12 | NA | rs7250581 | 0.22 | 3 × 10−5 | 1.06 | 0.79–1.43 | [16] |
| early-onset MI | 21q22 | SLC5A3-MRPS6-KCNE2 | rs9982601 | 0.13 | 6.4 × 10−11 | 1.20 | 1.14–1.27 | [21] |
| CAD | 22q12 | NA | rs688034 | 0.31 | 4 × 10−6 | 1.11 | 0.99–1.25 | [16] |
NA = not available
Genome-wide haplotype analysis
Chromosome 9, band p21.3
Multiple GWA studies have shown a highly significant association with various SNPs within a large LD block on chromosome 9 at band p21.3 (9p21.3) and CAD [16–21]. While results did not reach statistical significance to be included in the NHGRI catalog of published GWA studies, a GWA study involving the Framingham Heart Study also found an association with a 13 kb region on 9p21.3 with major CHD (p 2.5–3.5 × 10−4) [22]. Additionally, they observed that 7 SNPs in a 76 kb region around 9p21 had p<10−5 for major CHD and/or major cardiovascular disease (CVD).
Markers at the 9p21 locus have been shown to lead to a 15–20% risk for CAD in the 50% of Caucasian individuals heterozygous for the allele, and a 30–40% increased risk of CAD in the 25% of individuals homozygous for the allele [17]. Numerous follow-up case-control analyses have been performed and confirm the association of 9p21 and CAD [22–28]. A meta-analysis of case-control studies showed that the odds ratio per copy of the 9p21 risk allele was 1.29 (95% CI 1.22–1.37, p=0.0079) [29]. Abdullah et al demonstrated that four SNPs within the 9p21 region were significantly (p=6.61 × 10−7 to 1.87 × 10−8) associated with premature and familial MI and CAD (average age of onset 40.3 +/- 5.1 years) [26]. Another group similarly made the connection between 9p21 and familial CAD in a high-risk population with familial hypercholesterolemia [25]. Although one study found an association between 9p21 and CAD, the association did not hold up with incident events or prevalent MI [27]. Additionally, the Rotterdam Study did not observe an association between 9p21 genotype and coronary heart disease or myocardial infarction in a large cohort of individuals aged 55 years and older [30]. Thus, while the vast majority of studies have identified a strong association with 9p21 and CAD, an association with this locus and MI may be population-dependent and garners further investigation.
The primary GWA studies were done in cohorts of individuals of Caucasian, Northern European, and Canadian descent. Follow-up studies in Korean, Japanese, and Italian populations also established the 9p21-CAD association in those populations [31–34]. Additionally, a multi-ethnic Atherosclerotic Disease, Vascular Function, and Genetic Epidemiology (ADVANCE) study confirmed the association in whites and extended it to U.S. Hispanics and U.S. East Asians, but not African Americans [35]. The lack of association in African Americans was consistent with what was observed by McPherson, et al. [17]. However, in both studies examining the association with African Americans and in some of the other non-Caucasian-based studies, the sample sizes were not large, thus potentially limiting the statistical power of the studies. Nonetheless, the studies imply a consistent association of the 9p21 risk allele with CAD in individuals from a wide range of Asian and Caucasian backgrounds.
While the 9p21 association with CAD has been replicated on multiple occasions, associations between surrogate markers of atherosclerotic cardiovascular disease and 9p21 have remained less conclusive. Two studies did not identify an association between the 9p21 locus and carotid intima media thickness (IMT) [36, 37]. An additional study also did not demonstrate an association with 9p21 and abdominal aortic IMT [38]. Samani et al. postulated that CAD risk influenced by 9p21 occurs by a mechanism independent of carotid intimal thickening or endothelial dysfunction, and may instead affect coronary plaque stability [36]. Anderson et al. found that while 9p21 correlated with prevalence of angiographic coronary disease, it was unable to predict extent of disease [39]. The 9p21 locus was significantly correlated with angiographically characterized CAD in another study [24]. Horne et al., however, found that while 9p21 genotype was associated with the CAD phenotype, it was not associated with the severity or extent of CAD assessed angiographically [27]. They also did not find a strong association between history of MI and the 9p21 locus, as described above. They hypothesized that the 9p21 locus acted in the early stages, but not in the progression, of atherosclerosis. On the other hand, Ye et al. observed that the 9p21 genotype was associated with established carotid atherosclerosis and progression of atherosclerotic plaques as determined by carotid duplex scanning [37]. Thus, further investigations are necessary to help define where 9p21 fits into the picture of initiation or progression of atherosclerosis.
Other phenotypic correlations have been investigated with the 9p21 locus and CAD. In the Heart and Soul Study, 9p21 genotype was not associated with any echocardiographic parameter of cardiovascular structure and function (left ventricular hypertrophy, systolic dysfunction, diastolic disfunction, inducible ischemia, exercise capacity, mitral annular calcification, and aortic plaque) in individuals with known CAD [40]. Additionally, Chen et al. observed no significant association between 9p21 and quantitative indices of coronary atherosclerosis, such as minimal lumen diameter and number of coronary lesions or occlusions [41]. However, the 9p21 locus has been associated with coronary artery calcification, a reasonable marker of subclinical atherosclerosis [42].
In addition to CAD, the 9p21.3 locus has been associated with stroke, abdominal aortic aneurysms, and intracranial aneurysms [23, 28, 29, 43–46]. In part because of the relationship between the risk allele and stroke, and as discussed above, it has been suggested that the 9p21 risk allele may be related to plaque stability [36, 45]. The association of 9p21 with aneurysms also suggests an involvement in processes related to vessel wall integrity [23]. Bjorck et al. observed that the 9p21 locus was associated with abdominal aortic compliance and distensibility coefficients (measurements of arterial stiffness), further supporting a link between 9p21 and arterial wall diseases [38]. However, a recent GWA study of a Sardinian cohort did not report an association between 9p21 and arterial stiffness as determined by carotid-femoral pulse wave velocity [47].
Biology of 9p21
While the 9p21 association with CAD has been replicated on multiple occasions, the biological relevance of 9p21 is unclear at this time. The 9p21 region has been commonly implicated in the tumorigenesis of a variety of malignancies [48–51]. However, neither mouse models nor in vitro analyses have specifically implicated atherosclerotic processes with deletion or mutation of the 9p21 locus [52–55].
The risk-allele SNPs within the 9p21.3 locus, which spans approximately 50–60kb, have been defined as being in a “desert zone of the genome” (Figure 1) [56]. The 3' end of CDKN2B, encoding the cyclin-dependent kinase inhibitor tumor suppressor p15INK4B, is in a LD block with a cluster of tightly linked SNPS at the 9p21 locus. Weaker linkage disequilibrium extends through CDKN2B to CDKN2A, which encodes another tumor suppressor p16INK4B. The cyclin dependent kinases are involved in cell cycle regulation and transforming growth factor-β (TGF-β) cell cycle arrest [57]. In smooth muscle cells, TGF-β has been shown to have impaired signaling and reduced expression in atherosclerotic lesions and overexpression in abdominal aortic aneurysms [58–60]. Other studies have shown increased TGF-β levels in different stages of plaque development [61–63]. In fact, TGF-β1 has been proposed as a marker and potential therapeutic target for cardiovascular disease [60].
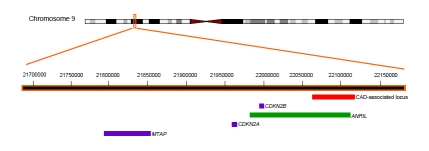
Figure 1.
The chromosome 9, band p21 region. The location of the CAD-associated region is shown in red, annotated genes are shown in purple, and the ANRIL anti-sense non-coding RNA is shown in green.
A gene encoding a large antisense non-coding RNA (ANRIL) spans almost the entire 9p21-CAD association region (Figure 1) [55]. ANRIL was discovered through deletion analysis of a family with hereditary melanoma-neural system tumors [64]. It was found that the expression of ANRIL coclusters with the expression of ARF, which is encoded for by an alternative exon 1 and exons 2 and 3 of CDKN2A. Atheromatous human vessels, abdominal aortic aneurysm walls, vascular endothelial cells, monocyte-derived macrophages, and coronary smooth muscle cells have all been shown to express ANRIL [43]. It has been speculated that transcription of the cyclin-dependent kinase genes may be regulated, at least in part, by ANRIL through RNA interference or some other mechanism [43, 44]. In fact, a p15INK4B antisense construct located in the 5' region of ANRIL, was shown to cause silencing of p15INK4B (encoded for by CDKN2B) in mouse embryonic stem cells through heterochromatin formation and DNA methylation [65]. Thus, a speculated mechanism for the 9p21 risk allele involves antisense regulation of CDKN2B (and/or CDKN2A), which could then affect signaling of TGF-β and/or additional cytokine(s) involved in cell cycle arrest/proliferation.
Another gene, MTAP, encoding methylthio-adenosine phosphorylase, is part of a different LD block that is closer to the chromosome 9 telomere. MTAP encodes for methylthio-adenosine phosphorylase (MTAP), an enzyme involved in polyamine metabolism that is deleted (along with p16INK4B) in many cancers. Whether or not this enzyme is involved in CAD requires further investigation.
Clinical utility of genotyping 9p21.3
While the specific phenotypic associations of the 9p21 genotype are somewhat conflicting and not well defined at present, whether or not 9p21 has proven clinical utility in predicting cardiovascular risk is of significant importance. As described above, an approximate 15–40% increased risk for CAD has been observed (depending on number of 9p21 risk alleles carried by the individual). However the Women's Genome Health Study demonstrated that 9p21 genotype did not add significantly to prediction of cardiovascular risk compared to what can be assessed via established risk markers, high-sensitivity C-reactive protein (CRP), and family history of premature myocardial infarction [66]. Thus, while a 20% increased risk (for heterozygous 9p21 risk allele carriers) for CAD may be relevant in some populations, middle-aged women (who are generally at low risk for CAD) who carry the 9p21 allele will not have a high, or even intermediate, risk for cardiovascular disease, despite the additional risk due to the presence of the 9p21 allele. Additionally, Talmud et al. demonstrated that 9p21 genotype did not add significantly to the CHD predictive utility by conventional risk factors in the Framingham risk score algorithm in a cohort of healthy middle-aged Caucasian men [67]. They did note, however, that 9p21 genotype did improve reclassification of CHD risk. Accordingly, based on studies performed in patients with early-onset angiographic CAD, Anderson et al. concluded that the clinical utility of 9p21 genetic assessment might be in refining CHD risk classification [39]. Thus, overall, it is suggested that 9p21 genotype may not be useful in stratifying risk in some low-risk populations but may provide discrimination in intermediate-risk individuals.
The launch of a clinical genotyping assay for 9p21, in October, 2007, was met with mixed enthusiasm (http://www.theheart.org/article/817629.do). Concerns regarding genetic testing for 9p21 include inherent problems with testing for the 9p21 locus in isolation, lack of biological knowledge of this locus and how it relates to CAD, uncertainty of the value and applicability of information obtained from genotyping, and potential for false negative reassurance in individuals who do not carry the risk allele. Thus, while 9p21 has been repeatedly associated with CAD, demonstrates a risk that is independent of traditional risk factors, and may be a useful marker in some populations (e.g. intermediate risk individuals), 9p21 genotyping as standard of care is debatable.
Chromosome 1p, band 13.3
A risk allele at 1p13 was found to be associated with CAD and early-onset MI, and this correlation was confirmed in a large scale association analysis [18, 20, 21]. This risk allele is in a 97-kb region of LD containing the CELSR2, PSRC1, and SORT1 genes, and has been found to be strongly associated with low-density lipoprotein (LDL) and total cholesterol concentrations [68–74]. Based on expression studies correlated with LDL cholesterol levels, it was concluded that SORT1 and CELSR2 were the most likely candidate susceptibility genes at the 1p13.3 locus [75]. Sortilin is a pro-neurotrophin receptor encoded for by the SORT1 gene, and is involved in adipocyte and muscle glucose metabolism. Sortilin expression is downregulated in obesity and has been implicated in insulin resistance [76]. Sortilin has also been shown to be involved in uptake and degradation of lipoprotein lipase, an important enzyme for lipid hydrolysis [77]. In relation to this, the gene encoding lipoprotein lipase (LPL) at 8p21 has also been implicated in multiple GWA studies as associated with high-density lipoprotein (HDL) cholesterol and triglyceride levels and other metabolic traits [69, 70, 73, 74, 78–80]. CELSR2 encodes for cadherin EGF LAG seven-pass G-type receptor 2, but very little is known about its function. The third gene in the 1p13.3 gene complex is PSRC1, which encodes for proline/serine-rich coiled-coil 1, a microtubule-associated protein. The functional connection between PSRC1 and CAD and/or lipid metabolism is not known. Thus, while there is a potential connection between the biology of SORT1 and cholesterol levels and CAD, the picture involving CELSR2 and PSRC1 is less clear and further studies are needed to elucidate those mechanisms.
Other loci
Other loci that have been identified through multiple, independent GWA studies and confirmed in follow-up CAD association analyses include 1q41 and 10q11 (Table 1) [16, 18, 20, 21]. Interestingly, 10q11 was shown to have a significant relationship to CAD in women but not in men [20]. Another locus, 6q25, was also identified to be associated with CAD in multiple, independent GWA studies, but the correlation was not statistically significant in a large-scale association analysis [20]. Loci within the 12q23-24 region were found to be associated with CAD, MI, and coronary artery calcification in three separate GWA studies [42, 81, 82]. The reported genes in these studies are DRIM (12q23), SH2B3 (12q24), and HNF1A-C12orf43 (12q24.41). The HNF1A gene has also been implicated for its association with LDL cholesterol and C-reactive protein in other GWA studies [79, 83, 84].
Two genes, LDLR and PCSK9, in which mutations can lead to autosomal dominant hypercholesterolemia, were reported in a GWA study of early-onset myocardial infarction [21]. Mutations in LDLR (on 19p13), which encodes for the LDL receptor, leads to familial hypercholesterolemia. Gain of function mutations in PCSK9 (on 1p32), which encodes for proprotein convertase, subtilisin/kesin-type 9 and is involved in recycling of the LDL receptor, leads to autosomal dominant hypercholesterolemia 3. SNPs in these genes have also been identified in other GWA studies as associated with LDL cholesterol [69, 70, 74, 79].
Copy number variant and haplotype GWA analyses
Genome-wide association studies involving the univariate analysis of single nucleotide polymorphisms (SNPs) have been the standard to date. In contrast to polymorphisms at single base positions (SNPs), copy number variants (CNVs) are polymorphic deletions or duplications of large, (1 kilobase to several megabase) regions of the genome, and could likewise be associated with specific disease states. In that regard, The Myocardial Infarction Genetics Consortium explored the possibility of an association between copy number variants (CNVs) and early-onset myocardial infarction [21]. However, they did not detect a significant difference in CNVs in cases vs. controls, in genes vs. the genome, or at any individual locus.
In addition to univariate analysis of SNPs, there is the potential for SNPs in haplotype to be associated with certain phenotypes. Tregouet et al. recently performed a genome-wide haplotype association study in CAD, using a sliding-windows approach [85]. Using this type of analysis, a haplotype of four SNPs in the SLC22A3-LPAL2-LPA gene cluster was found to be associated with CAD. Of particular interest in this gene cluster is LPA, which encodes apolipoprotein (a), the protein component of lipoprotein (a) [Lp(a)]. Elevated levels of Lp(a) have been associated with an increased risk for CAD and MI [86–88].
Conclusion
GWA analyses are a powerful tool for evaluating genetic associations in complex disease. However, as described above, careful design and execution of GWA and follow-up association studies are imperative to providing robust, meaningful data. A major challenge with GWA analyses is separating the true markers from the spurious associations.
Having stringent p-values is important, as is replicating results in independent cohorts. Additionally, comprehensive phenotyping of the cohort is also beneficial for understanding biological pathways. Extensively characterized cohorts will provide the foundation for ascertaining genetic and environmental risk factors in an integrated, interdependent manner.
For the vast majority of published GWA studies, the ethnic background of the primary study population has been European Caucasian. However, there is a need to extend association analyses to populations with other ethnic backgrounds. Linkage disequilibrium blocks and allele frequencies can vary widely between European Caucasians, African Americans, Asians, Hispanics, and other ethnic groups. Thus, associations found in one ethnic group may not translate to the same association in other ethnic groups. A case in point is 9p21.3, in which an association between this locus and CAD was observed in many different populations, but the association was not present in African Americans [17, 35].
In addition to confounding issues in association studies due to ethnicity, age also should be taken into account. In fact, a stronger impact of genetic factors on CAD is observed in younger individuals, possibly due to heterogeneity of effect associated with advancing age [2]. The 9p21 risk allele had a more pronounced effect in individuals with earlier onset MI, as observed by Helgadottir et al. [19]. Additionally, the Rotterdam Study was unable to make a correlation with 9p21 and CHD or MI in a prospective cohort study of nearly 8000 participants aged 55 years and older [30]. The authors cautioned, however, that the results of the study may be too underpowered to generalize them to an elderly population. Proper characterization of cohorts, as well as matching for age, gender, and ethnicity, is important to prevent misrepresentation of the findings.
While GWA studies have identified novel potential markers for CAD and other complex diseases, there are several limitations to the results of these studies [89]. These limitations include the risk of false positive results, lack of detection of rare variants due to the insensitive nature of the assay, biases from incomplete phenotyping of cases and controls, and confounders due to population stratification. Furthermore, very few functional variants have been identified through GWA studies. Thus, further studies are necessary to ascertain the relationship of the identified variants to functional variants and biology of the disease.
Because of the nature of complex diseases, it is likely that multiple genetic risk alleles are needed to accurately assess risk. The risk alleles may be identified through association and linkage studies, but many of them may be identified by downstream studies investigating interactions between multiple risk loci. One study, using computer simulation, estimated that over 200 alleles were required to provide a reasonable assessment of CAD risk [90]. Thus, while GWA studies may be useful in identifying potential novel markers for complex disease, essential follow-up investigations involve determining the biology of the markers, interactions between loci, effect of risk markers in subclinical phenotypes, and association in other populations.
Acknowledgments
The author acknowledges helpful comments by Dr. Allan Jaffe.
References
- 1.Castelli WP. Lipids, risk factors and ischaemic heart disease. Atherosclerosis. 1996;124(Suppl):S1–9. doi: 10.1016/0021-9150(96)05851-0. [DOI] [PubMed] [Google Scholar]
- 2.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Jones DM, Nam BH, D'Agostino RB, Levy D, Sr, Murabito JM, Wang TJ, Wilson PW, O'Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 4.Assmann G, Cullen P, Schulte H. Simple scoring scheme for calculating the risk of acute coronary events based on the 10-year follow-up of the prospective cardiovascular Munster (PROCAM) study. Circulation. 2002;105:310–315. doi: 10.1161/hc0302.102575. [DOI] [PubMed] [Google Scholar]
- 5.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, Lisheng L. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 6.Nasir K, Michos ED, Rumberger JA, Braunstein JB, Post WS, Budoff MJ, Blumenthal RS. Coronary artery calcification and family history of premature coronary heart disease: sibling history is more strongly associated than parental history. Circulation. 2004;110:2150–2156. doi: 10.1161/01.CIR.0000144464.11080.14. [DOI] [PubMed] [Google Scholar]
- 7.Fischer M, Broeckel U, Holmer S, Baessler A, Hengstenberg C, Mayer B, Erdmann J, Klein G, Riegger G, Jacob HJ, Schunkert H. Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation. 2005;111:855–862. doi: 10.1161/01.CIR.0000155611.41961.BB. [DOI] [PubMed] [Google Scholar]
- 8.Fischer M, Mayer B, Baessler A, Riegger G, Erdmann J, Hengstenberg C, Schunkert H. Familial aggregation of left main coronary artery disease and future risk of coronary events in asymptomatic siblings of affected patients. Eur Heart J. 2007;28:2432–2437. doi: 10.1093/eurheartj/ehm377. [DOI] [PubMed] [Google Scholar]
- 9.Harpaz D, Behar S, Rozenman Y, Boyko V, Gottlieb S. Family history of coronary artery disease and prognosis after first acute myocardial infarction in a national survey. Cardiology. 2004;102:140–146. doi: 10.1159/000080481. [DOI] [PubMed] [Google Scholar]
- 10.Williams RR, Hunt SC, Heiss G, Province MA, Bensen JT, Higgins M, Chamberlain RM, Ware J, Hopkins PN. Usefulness of cardiovascular family history data for population-based preventive medicine and medical research (the Health Family Tree Study and the NHLBI Family Heart Study) Am J Cardiol. 2001;87:129–135. doi: 10.1016/s0002-9149(00)01303-5. [DOI] [PubMed] [Google Scholar]
- 11.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 12.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297:1551–1561. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 15.Ioannidis JP. Prediction of cardiovascular disease outcomes and established cardiovascular risk factors by genome-wide association markers. Circulation Cardiovascular Genetics. 2009 doi: 10.1161/CIRCGENETICS.108.833392. published online Jan 23, 2009. [DOI] [PubMed] [Google Scholar]
- 16.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 20.Consortium CAD. Large Scale Association Analysis of Novel Genetic Loci for Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2009 doi: 10.1161/ATVBAHA.108.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, Macrae CA, O'Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown PP, Patterson CC, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O'Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D'Agostino RB, Fox CS, Sr, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, Murabito JM, Newton-Cheh C, O'Donnell CJ, Seshadri S, Vasan RS, Wang TJ, Wolf PA, Levy D. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8(Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 24.Muendlein A, Saely CH, Rhomberh S, Sonderegger G, Loacker S, Rein P, Beer S, Vonbank A, Winder T, Drexel H. Evaluation of the association of genetic variants on the chromosomal loci 9p21.3, 6q25.1, and 2q36.3 with angiographically characterized coronary artery disease. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2008.10.035. doi:10.1016/j.atherosclerosis.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 25.van der Net JB, Oosterveer DM, Versmissen J, Defesche JC, Yazdanpanah M, Aouizerat BE, Steyerberg EW, Malloy MJ, Pullinger CR, Kastelein JJ, Kane JP, Sijbrands EJ. Replication study of 10 genetic polymorphisms associated with coronary heart disease in a specific high-risk population with familial hypercholesterolemia. Eur Heart J. 2008;29:2195–2201. doi: 10.1093/eurheartj/ehn303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdullah KG, Li L, Shen GQ, Hu Y, Yang Y, MacKinlay KG, Topol EJ, Wang QK. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest) Ann Hum Genet. 2008;72:654–657. doi: 10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horne BD, Carlquist JF, Muhlestein JB, Bair TL, Anderson JL. Association of Variation in the Chromosome 9p21 Locus With Myocardial Infarction Versus Chronic Coronary Artery Disease. Circulation Cardiovascular Genetics. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karvanen J, Silander K, Kee F, Tiret L, Salomaa V, Kuulasmaa K, Wiklund PG, Virtamo J, Saarela O, Perret C, Perola M, Peltonen L, Cambien F, Erdmann J, Samani NJ, Schunkert H, Evans A. The impact of newly identified loci on coronary heart disease, stroke and total mortality in the MORGAM prospective cohorts. Genet Epidemiol. 2008 doi: 10.1002/gepi.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, Blankenberg S, Tiret L, Ducimetiere P, Keniry A, Ghori MJ, Schreiber S, El Mokhtari NE, Hall AS, Dixon RJ, Goodall AH, Liptau H, Pollard H, Schwarz DF, Hothorn LA, Wichmann HE, Konig IR, Fischer M, Meisinger C, Ouwehand W, Deloukas P, Thompson JR, Erdmann J, Ziegler A, Samani NJ. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dehghan A, van Hoek M, Sijbrands EJ, Oostra BA, Hofman A, van Duijn CM, Witteman JC. Lack of association of two common polymorphisms on 9p21 with risk of coronary heart disease and myocardial infarction; results from a prospective cohort study. BMC Med. 2008;6:30. doi: 10.1186/1741-7015-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen GQ, Li L, Rao S, Abdullah KG, Ban JM, Lee BS, Park JE, Wang QK. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008;28:360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 32.Shen GQ, Rao S, Martinelli N, Li L, Olivieri O, Corrocher R, Abdullah KG, Hazen SL, Smith J, Barnard J, Plow EF, Girelli D, Wang QK. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008;53:144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 33.Hiura Y, Fukushima Y, Yuno M, Sawamura H, Kokubo Y, Okamura T, Tomoike H, Goto Y, Nonogi H, Takahashi R, Iwai N. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ J. 2008;72:1213–1217. doi: 10.1253/circj.72.1213. [DOI] [PubMed] [Google Scholar]
- 34.Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, Chida K, Sawabe M, Arimura T, Sato A, Lee BS, Ban JM, Yasunami M, Park JE, Izumi T, Kimura A. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008;53:357–359. doi: 10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 35.Assimes TL, Knowles JW, Basu A, Iribarren C, Southwick A, Tang H, Absher D, Li J, Fair JM, Rubin GD, Sidney S, Fortmann SP, Go AS, Hlatky MA, Myers RM, Risch N, Quertermous T. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008;17:2320–2328. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samani NJ, Raitakari OT, Sipila K, Tobin MD, Schunkert H, Juonala M, Braund PS, Erdmann J, Viikari J, Moilanen L, Taittonen L, Jula A, Jokinen E, Laitinen T, Hutri-Kahonen N, Nieminen MS, Kesaniemi YA, Hall AS, Hulkkonen J, Kahonen M, Lehtimaki T. Coronary artery disease-associated locus on chromosome 9p21 and early markers of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:1679–1683. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- 37.Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol. 2008;52:378–384. doi: 10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 38.Bjorck HM, Lanne T, Alehagen U, Persson K, Rundkvist L, Hamsten A, Dahlstrom U, Eriksson P. Association of genetic variation on chromosome 9p21.3 and arterial stiffness. J Intern Med. 2009;265:373–381. doi: 10.1111/j.1365-2796.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 39.Anderson JL, Horne BD, Kolek MJ, Muhlestein JB, Mower CP, Park JJ, May HT, Camp NJ, Carlquist JF. Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J. 2008;156:1155–1162. e1152. doi: 10.1016/j.ahj.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Farzaneh-Far R, Na B, Schiller NB, Whooley MA. Lack of association of chromosome 9p21.3 genotype with cardiovascular structure and function in persons with stable coronary artery disease: The Heart and Soul Study. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2008.12.026. doi:10.1016/j.athersclerosis.2008/12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen SN, Ballantyne CM, Gotto AM, Jr, Marian AJ. The 9p21 susceptibility locus for coronary artery disease and the severity of coronary atherosclerosis. BMC Cardiovasc Disord. 2009;9:3. doi: 10.1186/1471-2261-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Donnell CJ, Cupples LA, D'Agostino RB, Fox CS, Hoffmann U, Hwang SJ, Ingellson E, Liu C, Murabito JM, Polak JF, Wolf PA, Demissie S. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI's Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, Seedorf U, Rust S, Eriksson P, Hamsten A, Farrall M, Watkins H. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 44.Bown MJ, Braund PS, Thompson J, London NJM, Samani NJ, Sayers RD. Association between the coronary artery disease risk locus on chromosome 9p21.3 and abdominal aortic aneurysm. Circ Cardiovasc Genet. 2008;1:39–42. doi: 10.1161/CIRCGENETICS.108.789727. [DOI] [PubMed] [Google Scholar]
- 45.Smith JG, Melander O, Lovkist H, Heblad B, Engstrom G, Nilsson P, Carlson J, Berglund G, Norrving B, Lindgren A. Common genetic variants on chromosome 9p21 confers risk of ischemic stroke: a large-scale genetic association study. Circ Cardiovasc Genet. 2009 doi: 10.1161/CIRCGENETICS.108.835173. 10.1161/CIRCGENETICS.108.835173. [DOI] [PubMed] [Google Scholar]
- 46.Thompson AR, Golledge J, Cooper JA, Hafez H, Norman PE, Humphries SE. Sequence variant on 9p21 is associated with the presence of abdominal aortic aneurysm disease but does not have an impact on aneurysmal expansion. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarasov KV, Sanna S, Scuteri A, Strait JB, Orru M, Parsa A, Lin P-I, Maschio A, Lai S, Piras MG, Masala M, Tanaka T, Post WS, O'Connell JR, Schlessinger D, Cao A, Nagaraja R, Mitchell BD, Abecasis GR, Shuldiner AR, Uda M, Lakatta E, Najjar SS. COL4A1 is associated with arterial stiffness by genome wide association scan. Circ Cardiovasc Genet. 2009 doi: 10.1161/CIRCGENETICS.108.823245. DOI: 10.1161/CIRCGENETICS.108.823245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollock PM, Pearson JV, Hayward NK. Compilation of somatic mutations of the CDKN2 gene in human cancers: non-random distribution of base substitutions. Genes Chromosomes Cancer. 1996;15:77–88. doi: 10.1002/(SICI)1098-2264(199602)15:2<77::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 49.Smith-Sorensen B, Hovig E. CDKN2A (p16INK4A) somatic and germline mutations. Hum Mutat. 1996;7:294–303. doi: 10.1002/(SICI)1098-1004(1996)7:4<294::AID-HUMU2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 50.Foulkes WD, Flanders TY, Pollock PM, Hayward NK. The CDKN2A (p16) gene and human cancer. Mol Med. 1997;3:5–20. [PMC free article] [PubMed] [Google Scholar]
- 51.Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16:1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 52.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 53.Sharpless E, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22:3092–3098. doi: 10.1038/sj.onc.1206461. [DOI] [PubMed] [Google Scholar]
- 54.Chin L, Pomerantz J, DePinho RA. The INK4a/ARF tumor suppressor: one gene–two products–two pathways. Trends Biochem Sci. 1998;23:291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 55.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 56.Topol EJ, Murray SS, Frazer KA. The genomics gold rush. JAMA. 2007;298:218–221. doi: 10.1001/jama.298.2.218. [DOI] [PubMed] [Google Scholar]
- 57.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 58.Kalinina N, Agrotis A, Antropova Y, Ilyinskaya O, Smirnov V, Tararak E, Bobik A. Smad expression in human atherosclerotic lesions: evidence for impaired TGF-beta/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler Thromb Vasc Biol. 2004;24:1391–1396. doi: 10.1161/01.ATV.0000133605.89421.79. [DOI] [PubMed] [Google Scholar]
- 59.Fukui D, Miyagawa S, Soeda J, Tanaka K, Urayama H, Kawasaki S. Overexpression of transforming growth factor beta1 in smooth muscle cells of human abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2003;25:540–545. doi: 10.1053/ejvs.2002.1857. [DOI] [PubMed] [Google Scholar]
- 60.Redondo S, Santos-Gallego CG, Tejerina T. TGF-beta1: a novel target for cardiovascular pharmacology. Cytokine Growth Factor Rev. 2007;18:279–286. doi: 10.1016/j.cytogfr.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 61.Bobik A, Agrotis A, Kanellakis P, Dilley R, Krushinsky A, Smirnov V, Tararak E, Condron M, Kostolias G. Distinct patterns of transforming growth factor-beta isoform and receptor expression in human atherosclerotic lesions. Colocalization implicates TGF-beta in fibrofatty lesion development. Circulation. 1999;99:2883–2891. doi: 10.1161/01.cir.99.22.2883. [DOI] [PubMed] [Google Scholar]
- 62.Piao M, Tokunaga O. Significant expression of endoglin (CD105), TGFbeta-1 and TGFbeta R-2 in the atherosclerotic aorta: an immunohistological study. J Atheroscler Thromb. 2006;13:82–89. doi: 10.5551/jat.13.82. [DOI] [PubMed] [Google Scholar]
- 63.Panutsopulos D, Papalambros E, Sigala F, Zafiropoulos A, Arvanitis DL, Spandidos DA. Protein and mRNA expression levels of VEGF-A and TGF-beta1 in different types of human coronary atherosclerotic lesions. Int J Mol Med. 2005;15:603–610. [PubMed] [Google Scholar]
- 64.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 65.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, Humphries SE. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008;54:467–474. doi: 10.1373/clinchem.2007.095489. [DOI] [PubMed] [Google Scholar]
- 68.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marcano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, Inouye M, Luben R, Sims M, Hadley D, McArdle W, Barter P, Kesaniemi YA, Mahley RW, McPherson R, Grundy SM, Bingham SA, Khaw KT, Loos RJ, Waeber G, Barroso I, Strachan DP, Deloukas P, Vollenweider P, Wareham NJ, Mooser V. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samani NJ, Braund PS, Erdmann J, Gotz A, Tomaszewski M, Linsel-Nitschke P, Hajat C, Mangino M, Hengstenberg C, Stark K, Ziegler A, Caulfield M, Burton PR, Schunkert H, Tobin MD. The novel genetic variant predisposing to coronary artery disease in the region of the PSRC1 and CELSR2 genes on chromosome 1 associates with serum cholesterol. J Mol Med. 2008;86:1233–1241. doi: 10.1007/s00109-008-0387-2. [DOI] [PubMed] [Google Scholar]
- 73.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Doring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, Jones CG, Zaitlen NA, Varilo T, Kaakinen M, Sovio U, Ruokonen A, Laitinen J, Jakkula E, Coin L, Hoggart C, Collins A, Turunen H, Gabriel S, Elliot P, McCarthy MI, Daly MJ, Jarvelin MR, Freimer NB, Peltonen L. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaddai V, Jager J, Gonzalez T, Najem-Lendom R, Bonnafous S, Tran A, Le Marchand-Brustel Y, Gual P, Tanti JF, Cormont M. Involvement of TNF-alpha in abnormal adipocyte and muscle sortilin expression in obese mice and humans. Diabetologia. 2009 doi: 10.1007/s00125-009-1273-3. [DOI] [PubMed] [Google Scholar]
- 77.Nielsen MS, Jacobsen C, Olivecrona G, Gliemann J, Petersen CM. Sortilin/neurotensin receptor-3 binds and mediates degradation of lipoprotein lipase. J Biol Chem. 1999;274:8832–8836. doi: 10.1074/jbc.274.13.8832. [DOI] [PubMed] [Google Scholar]
- 78.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, Balding D, Scott J, Kooner JS. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 79.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gomez Perez FJ, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 81.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, Cambien F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O'Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, Mokhtari NE, Schafer A, Marz W, Renner W, Bugert P, Kluter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, Williams C, Hui J, Beilby J, Warrington NM, James A, Palmer LJ, Koppelman GH, Heinzmann A, Krueger M, Boezen HM, Wheatley A, Altmuller J, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Gislason D, Park CS, Rasmussen LM, Porsbjerg C, Hansen JW, Backer V, Werge T, Janson C, Jonsson UB, Ng MC, Chan J, So WY, Ma R, Shah SH, Granger CB, Quyyumi AA, Levey AI, Vaccarino V, Reilly MP, Rader DJ, Williams MJ, van Rij AM, Jones GT, Trabetti E, Malerba G, Pignatti PF, Boner A, Pescollderungg L, Girelli D, Olivieri O, Martinelli N, Ludviksson BR, Ludviksdottir D, Eyjolfsson GI, Arnar D, Thorgeirsson G, Deichmann K, Thompson PJ, Wjst M, Hall IP, Postma DS, Gislason T, Gulcher J, Kong A, Jonsdottir I, Thorsteinsdottir U, Stefansson K. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 83.Reiner AP, Barber MJ, Guan Y, Ridker PM, Lange LA, Chasman DI, Walston JD, Cooper GM, Jenny NS, Rieder MJ, Durda JP, Smith JD, Novembre J, Tracy RP, Rotter JI, Stephens M, Nickerson DA, Krauss RM. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ridker PM, Pare G, Parker A, Zee RY, Danik JS, Buring JE, Kwiatkowski D, Cook NR, Miletich JP, Chasman DI. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protethe Women's Genome Health Study. Am J Hum Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tregouet DA, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Grosshennig A, Linsel-Nitschke P, Perret C, Desuremain M, Meitinger T, Wright BJ, Preuss M, Balmforth AJ, Ball SG, Meisinger C, Germain C, Evans A, Arveiler D, Luc G, Ruidavets JB, Morrison C, van der Harst P, Schreiber S, Neureuther K, Schafer A, Bugert P, El Mokhtari NE, Schrezenmeir J, Stark K, Rubin D, Wichmann HE, Hengstenberg C, Ouwehand W, Ziegler A, Tiret L, Thompson JR, Cambien F, Schunkert H, Samani NJ. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 86.Rhoads GG, Dahlen G, Berg K, Morton NE, Dannenberg AL. Lp(a) lipoprotein as a risk factor for myocardial infarction. JAMA. 1986;256:2540–2544. [PubMed] [Google Scholar]
- 87.Bostom AG, Cupples LA, Jenner JL, Ordovas JM, Seman LJ, Wilson PW, Schaefer EJ, Castelli WP. Elevated plasma lipoprotein(a) and coronary heart disease in men aged 55 years and younger. A prospective study. JAMA. 1996;276:544–548. doi: 10.1001/jama.1996.03540070040028. [DOI] [PubMed] [Google Scholar]
- 88.Armstrong VW, Cremer P, Eberle E, Manke A, Schulze F, Wieland H, Kreuzer H, Seidel D. The association between serum Lp(a) concentrations and angiographically assessed coronary atherosclerosis. Dependence on serum LDL levels. Atherosclerosis. 1986;62:249–257. doi: 10.1016/0021-9150(86)90099-7. [DOI] [PubMed] [Google Scholar]
- 89.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 90.Ortlepp JR, Lauscher J, Janssens U, Minken-berg R, Hanrath P, Hoffmann R. Analysis of several hundred genetic polymorphisms may improve assessment of the individual genetic burden for coronary artery disease. Eur J Intern Med. 2002;13:485–492. doi: 10.1016/s0953-6205(02)00182-6. [DOI] [PubMed] [Google Scholar]