Abstract
DNA-DNA hybridization (DDH) has been used for nearly 50 years as the gold standard for prokaryotic species circumscriptions at the genomic level. It has been the only taxonomic method that offered a numerical and relatively stable species boundary, and its use has had a paramount influence on how the current classification has been constructed. However, now, in the era of genomics, DDH appears to be an outdated method for classification that needs to be substituted. The average nucleotide identity (ANI) between two genomes seems the most promising method since it mirrors DDH closely. Here we examine the work package JSpecies as a user-friendly, biologist-oriented interface to calculate ANI and the correlation of the tetranucleotide signatures between pairwise genomic comparisons. The results agreed with the use of ANI to substitute DDH, with a narrowed boundary that could be set at ≈95–96%. In addition, the JSpecies package implemented the tetranucleotide signature correlation index, an alignment-free parameter that generally correlates with ANI and that can be of help in deciding when a given pair of organisms should be classified in the same species. Moreover, for taxonomic purposes, the analyses can be produced by simply randomly sequencing at least 20% of the genome of the query strains rather than obtaining their full sequence.
Keywords: average nucleotide identity, DNA-DNA hybridization, genome-based taxonomy, tetranucleotide regression
The concept of species was conceived first by Aristotle ≈2,400 years ago, and since then taxonomists of all disciplines have been trying to find those premises that would help to circumscribe the biological units observed in nature. Ever since, the idea behind this term has been a topic of considerable interest that has caused great controversy with difficult reconciliation (1). Prokaryotes are not exempt from this problem, and even the existence of discrete biological units is being questioned (2). However, for pragmatic reasons, microbiologists need to deal with a classification of the organisms that they isolate. The ultimate goal of taxonomy is to construct a classification that is of operative and predictive use for any discipline in microbiology and that is also essentially stable. From among the serious classifications, spanning nearly one century, taxonomists have obtained a sound system by circumscribing prokaryotes based on their phylogenetic, genomic, and phenotypic coherence (3, 4).
The early classification of prokaryotes was based solely on phenotypic similarities, but in the late 1960s some genome-based methods were developed to evaluate genomic interrelationships. Among them, DNA-DNA hybridization (DDH) techniques applied to determine crude genome similarities became popular. DDH tended to reproduce and even improve phenotypically circumscribed organism clusters that were considered to be species (3). Over the years that followed, the construction of the classification system was based on the fact that DDH could reveal coherent genomic groups (genospecies) of strains generally sharing DDH values with greater than 70% similarity (5). The comparative study of the different methods, prone to distinct experimental error, indicated that the value of 70% could not be used as absolute boundary, but still a gap between 60 and 70% similarity seemed to embrace clear-cut clusters of organisms (6). Given the large extent of diversity among prokaryotes, the circumscription of each genospecies would, in addition, be dependent on each group being studied (2, 6). Nevertheless, the use of DDH has mainly driven the construction of the current prokaryotic taxonomy, as it has become the gold standard for genomically circumscribing species. This parameter has had a similar impact in prokaryotic taxonomy as the interbreeding premise that is the basis for the biological species concept for animal and plant taxonomies (1). In the late 1980's (5), taxonomists already believed that the reference standard for determining taxonomy would be full genome sequences.
Despite being a traditional method, DDH has been often criticized as being inappropriate to circumscribe prokaryotic taxa because of the complex and time-consuming nature of the technique (7), although these are facts that themselves should not be scientific constraints. However, the impossibility of building cumulative databases based on DDH results is indeed a major drawback in the bioinformatics era. For this major reason, the scientific community has expressed the need to substitute DDH by other methods that offer a similar resolution and simultaneously allow the construction of databases that permit the retrieval of any information for comparative purposes (4). The major hope was vested in the use of the 16S rRNA gene as a putative marker for species circumscription (8), but the conservative nature of the gene did not show enough resolution on such a taxonomic scale. Moreover, single protein coding genes have also been evaluated as substitutes for DDH, and it does in fact seem that for certain groups the resolution power of a given gene may equal the genospecies drawn by reassociation experiments (9). However, the analysis of the genealogical relationships based on concatenating several housekeeping genes, a technique known as multilocus sequence analysis, has been suggested as the primary approach for substituting DDH (7, 4). Despite this approach being successful for specific groups, such as Burkholderia spp (10), it has major drawbacks that arise from a putative bias in gene selection and amplification primer availability.
In the era of genomics, in which high-quality genetic information can be retrieved from public databases, DDH seems to be an obsolete approach that urgently needs substitution (4). Among the different attempts to find an alternative, the average nucleotide identity (ANI) between a given pair of genomes seems currently to be the best alternative for a gold standard. The first attempt to evaluate the meaning of ANI was based on pairwise genome comparison of all shared orthologous protein coding genes (11). The evaluation showed that the resulting averages reflected the degree of evolutionary distance between the compared genomes, and a value of 94% identity could represent the DDH boundary of 70%. An advance in the comparison that may better reflect the degree of reassociation between the DNA stretches of two genomes, was achieved by comparing artificially sectioned genomes in 1,020 nucleotide fragments with independence, whether or not they responded to real open reading frames (ORFs) (12). This approach produced similar results as the former method based on predicted protein-coding sequences. Other parameters, such as maximal unique matches (MUM), have been evaluated to circumscribe species (13). However, despite the fact that this parameter seemed to help in embracing species, given that it correlates nicely with ANI, this method needs to work with fully sequenced genomes and does not work with only draft incomplete genomes.
The aim of this study was to find a way to reconcile the genomic information with the current knowledge on the taxonomy of prokaryotes to recommend an immediate shift from the traditional DDH to the modern ANI parameters. A software tool (JSpecies) was designed that easily allowed the calculation of ANI based on the BLAST algorithm (14), as well as on the MUMmer ultra-rapid aligning tool (15). Both methods are evaluated here. In addition, a statistical calculation was implemented in the program based on tetranucleotide frequencies, an alignment-free parameter that has been successfully applied to phylogenetically sort metagenome inserts (16). Finally, and because all hitherto ANI measurements have been made on complete genome sequences, the use of the pyrosequencing 454 technique was evaluated to obtain random partial genome coverages for evaluating whether stable values can be achieved through a reduction in sequencing costs, as previously required (17).
Results and Discussion
Taxonomy and the Genome Database.
Species descriptions tend to present the genotypic, phenotypic, and sometimes ecologic properties of what has been regarded as a unit by the taxonomist. One of the most important premises when classifying new taxa is the designation of one of the strains as being the type material that should be used as reference for any further taxonomic work. In this regard, it is required that the designated type strain is deposited in two international strain collections to make it publicly available (18). For any kind of comparative study that implies the use of taxonomic categories (e.g., evolutionary or ecological discussions), it is of the utmost importance to ensure that any observation is made with the type strain, or with material that has been proved by taxonomic studies. However, one of the major drawbacks that taxonomists may find in the current genome database (www.ncbi.nlm.nih.gov) is that it relies on the identification of the strains that have been sequenced. The authors submitting their sequences tag them with a putative specific name together with a strain designation. In most cases, the strain code corresponds to that given in the original isolation, and only about 10% of the entries are tagged with one of the international strain collection numbers.
To track the identity of the deposited strains, all strain designations in the genome database were verified by crosschecking with the Straininfo bioportal (19) and the List of Prokaryotic names with Standing in Nomenclature (LPSN) (20) databases, and those corresponding to species type strains were recognized. Our observations indicated that less than 30% of the sequenced genomes (≈50% of the validly published names listed) belonged to the type strain of the species for which they were identified (Table S1 and Table S2). This fact represents a major problem when trying to implement genomic data into microbial taxonomy. For example, from the ≈797 genomes identified with a validly published name, only ≈255 were from a type strain. However, none of the remaining 256 validly published names (corresponding to 683 strains) listed as sequenced genomes were represented by the corresponding type strain (Table S1 and Table S2). In addition, ≈50 listed names had never been validly published (Table S2). Incorrect identifications will lead to mistaken observations. Perhaps (as will be described below), these sequenced strains are not even members of the species carrying the given name. This major drawback can be overcome by making a sequencing effort to obtain all of the genomes of most of the type material available, an effort that was clearly identified by an ad hoc committee of scientists in 2006 (21). Once this catalog is achieved, the identification of new organisms as members of a given species may be easily based only on database matches.
ANIb and ANIm.
Among the various candidate methods for substituting DDH (4), ANI may be the best choice, as it is the best in silico parameter that could represent DDH, as has been experimentally demonstrated (1, 11, 12). So far the results on genome comparisons for taxonomic purposes have been made by basing the calculations on BLAST (14). The pairwise comparisons were preceded by either first finding the shared orthologous protein coding genes (11) or then by artificially cutting the genomes in pieces of 1,020 nucleotide stretches (12). However, there are new and more efficient algorithms for large DNA sequences, such as the MUMmer software package, for example (15). This uses an efficient data structure named suffix trees to calculate alignments. These suffix trees can rapidly align sequences containing millions of nucleotides with precision. To facilitate the calculation of ANIb and ANIm, we wrapped both algorithms within the software tool JSpecies that was specially designed to calculate and compare species specific signatures. The calculation time for the evaluation of the ANIm algorithm was shown to be much faster, with nearly similar precision (Fig. S1). Moreover, the speed enhancement, which is an important factor when it comes to large comparisons, of the ANIm calculation does not require previous slicing of the genomes into pieces or sieving of shared orthologous genes. Hereafter, “ANIb” will be used to refer to those results calculated with the BLAST algorithm and “ANIm” to those calculated with the MUMmer algorithm.
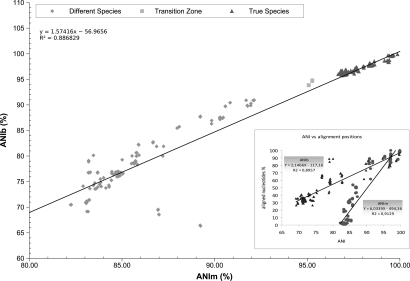
Both parameters were calculated and the results were compared by determining 200 pairwise comparisons on the available full genomes for which we could obtain DDH values in the literature (Table S3 and SI). Our calculations corresponded to an ≈80% increase in data compared with previous calculations (12). As can be seen in Fig. 1, the general picture is that both parameters correlate very precisely, especially in the high ANI value zone, where almost no differences between the ANIm and ANIb calculations could be seen. Differences started to be more evident once the compared genomes appeared to be divergent (sharing <90% ANI). These discrepancies were basically due to the different sensitivity and sensibility of MUMmer over BLAST, as the former is stricter in detecting matches with the default settings (15). The default parameters used for the results were those that seemed to be best suited for the purpose of the ANI value, to determine whether two organisms were of the same species. In the light of these results, we believe that ANIm provides more robust results when the pair of genomes compared share a high degree of similarity (ANI >90%). However, as the divergence increases (Fig. 1, inset), ANIm is more stringent in the selection of nucleotide stretches for calculation than ANIb using default parameters.
Fig. 1.
Plotted results of ANIb versus ANIm. The triangles show those values that correspond to what taxonomists consider as “true” species according to the DDH values traditionally applied and that have previously been classified. Inset shows the regression lines of the pairwise comparisons of ANIb or ANIm values with their corresponding percentage of aligned stretches (percentage of nucleotides included in the study).
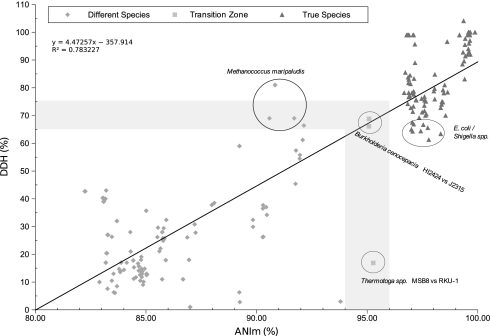
As can be seen in Fig. 2, most of the results obtained from organisms of the same species (i.e., DDH values >60–70% and classified as members of the same taxon) shared an ANIm identity of greater than 96%. In this case, we considered the set of species Escherichia coli, Shigella flexneri, and S. sonnei as a single hybridization group that could be considered as a single species (22). In this regard, it is important to note that, for some cases used for special purposes by taxonomists, they will allow close genospecies to be represented by different taxa, as is the case in maintaining the genus Shigella for medical purposes (23). In the light of our results, the proposed threshold of 94% ANI as the putative boundary for species circumscriptions is reinforced, and seems to work excellently in mirroring the DDH range of ≈60–70%. Actually, the whole database for genomic comparisons of what could be considered as a single species was checked (Table S1) and, in most of the cases, all putative groups could be well circumscribed within the ANIm range of 96–97%. A transition zone could be drawn where still high DDH (>60% similarity) values led to lower ANIm values or vice versa (Fig. 2). The most remarkable cases were (i) the hybridization group of members of the species Methanococcus maripaludis (24), where strains C5, C6, and C7 shared DDH values between 64% and 69% with low ANIm values <91%; or (ii) the pair of species Thermotoga maritima MSB8 and T. petrophila RKU-1 (25) that, despite being divergent in the DDH experiments (<28% similarity), shared high ANIm values (>95%). In both cases, which show important divergence from the expected results, it may perhaps be questioned whether there was experimental error in the DDH determinations or whether the deposition of the sequenced strains was incorrect. Nevertheless, altogether, our calculations based on ≈85 groups of strains putatively representing single species favor the recommendation to use an ANI boundary of ≈95–96% for taxonomically circumscribing prokaryotic species.
Fig. 2.
Plotted values of DDH versus ANIm. Triangles show values that correspond to what taxonomists consider as “true” species according to the DDH values traditionally applied and that have previously been classified. Squares indicate values that appear to be in the transition zone.
Tetranucleotide Signature Frequency Correlation Coefficient.
ANI values are based on pairwise alignment of genome stretches. In contrast, statistical calculations of oligonucleotide frequencies among sequence data are a fast, alignment-free, easy-to-implement, and powerful alternative for a number of different applications (26). Oligonucleotide frequencies carry a species-specific signal, but the evolutionary reasons behind this have not been comprehensive explained so far (27). Longer oligonucleotide signatures carry more signal than shorter ones (17), although the former need higher computational power. In this regard, the use of a tetranucleotide usage pattern has been shown to be a good compromise between computational calculation power and a pronounced phylogenetic signal (28). Here we evaluated tetranucleotide signature frequencies to assess whether an alignment-free genomic feature could be used to circumscribe species. The tetranucleotide calculation was also implemented in JSpecies. The codon usage of each genome type determines a characteristic frequency occurrence for each of the 256 combinations of groups of tetranucleotide sequences. In this regard, it is expected that closely related genomes will show a similar distribution of the usage of these signatures. Pairwise comparisons between genomes can be performed by plotting each corresponding tetranucleotide frequency and then obtaining a regression line. Two very closely related genomes may show very high correlation values where the plotted values follow a clear line (Fig. S2). However, when the genomes show a certain degree of divergence, the plotted values show higher dispersion, and the correlation tends to decrease.
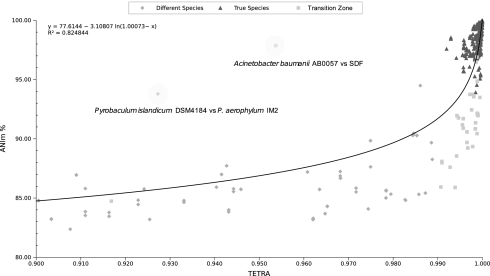
A total of 536 pairwise comparisons were determined among the sequenced genomes in groups of strains putatively belonging to the same species (Table S1). We analyzed the correspondence between the ANIm values and the tetranucleotide frequency correlation coefficients (TETRA) to evaluate the usability of the latter parameter (Fig. 3). As can be seen for most of the intraspecific results, when considered with ANIm values above a 96% identity, they corresponded to very high correlation coefficients >0.99 (triangles). However, there were still cases (6% of the determinations) for which, despite the ANIm values indicating a certain genome divergence, the signature usage was still highly correlated. An explanation could be that evolutionary or environmental forces (29) may impede modifications in the genome signature despite the fact that genetic drift may occur. The rare opposite cases (Fig. 3) in which high gene identities (ANIm >94%) were related to very low TETRA correlations are more difficult to explain. The only case found was Acinetobacter baumannii strain SDF (Table S1) that showed high ANIm values (>97%, similarly to ANIb values, not given) with the rest of the genomes, but TETRA values very divergent (<0.96). However, this strain just aligned about 60% with the remaining genomes, whereas the rest aligned with each other with values above 85%. The difficulties in aligning are perhaps due to intrinsic characteristics of the genome that might be related to lifestyle (30). It is important that the amount of aligned sequence be taken into account. The example of relative TETRA divergence but high ANI between Pyrobaculum spp. strains DSM 4184 and IM2 is explained, as ANIm was calculated by using just >2% of the genome sequence, and thus this value might have been a result of casualty. It is clear from the TETRA value that they cannot be placed in the same taxon. A priori, TETRA values can be an important help in deciding whether a group of strains can be placed in the same species. In this regard, and as a general observation, TETRA values >0.99 may support the species circumscription based on the ANI range >95–96%, but both values should agree. Despite the fact that TETRA may show more fuzziness than ANI, use of the former can be of much help to sieve results on large datasets before the ANI calculation. Further genome sequencing efforts will help in evaluating whether alignment-free parameters are of use in taxonomy and will also clarify the outliers found in the calculations.
Fig. 3.
Plotted values of TETRA versus ANIm. Triangles show those values that correspond to what taxonomists consider as “true” species according to the DDH values traditionally applied and that have previously been classified. Squares indicate values that appear to be in the transition zone. Note that this is an enlarged (zoomed) portion of the graphic, and values <80% ANI and 0.90 TETRA have been skipped.
Checking the Genome Database.
From the available genomes present in the downloadable database at the NCBI (www.ncbi.nlm.nih.gov), we recognized 567 single names representing 511 validly published species (Table S1 and Table S2). Among them, only 255 (45% of the cases) were represented by their type strain. We surveyed the genomic coherence of each single species that was represented by two or more strains in the database and calculated their ANIm and TETRA values (Table S1). According to the results, most of the groups of strains sharing the same specific name appeared to be putatively coherent species by sharing ANIm and TETRA values above the threshold recommended for the circumscription of the taxon. However, we could detect cases in which one or more strains exhibited values to the type strain of the species (or the reference strain if the type strain was absent) below the specific threshold recommended. The strains highlighted in bold in Table S1, such as Bacillus cereus subsp. cytotoxis NVH 391–98 (with TETRA/ANIm of 0.97949/85.33), Rhizobium etli CIAT 652 (with TETRA/ANIm of 0. 99881/90.44), or Xanthomonas campestris pv. vesicatoria str 85–10 (with TETRA/ANIm of 0. 99277/87.6), seem to be wrongly placed in their taxa because of the low similarity to their corresponding type strains. In these cases, the belief that the name truly indicates their affiliation may lead to questioning of the parameters used to circumscribe species (13, 27), rather than to questioning the real identity of the sequenced organism. For example, in the first case, it was tempting to classify strain NVH 391–98 as a new species of Bacillus (31, 32), but this was never formally proposed. However, the other cases may simply correspond to insufficient identification efforts based on phenotypic traits (33). These are just some of the examples that can be detected in the database that may lead to incorrect conclusions. Such observations are of the utmost importance when discussing any comparative genomic study, especially if it deals with important evolutionary, ecological, or taxonomic principles. This is evidence that a name given in a database does not necessarily mean a correct placement and thus the observations should be carried out cautiously.
Pragmatic Approach for Constructing a Stable Taxonomy Based on Genome Data.
Taxonomy, besides its importance in the biological sciences, is still a scientific area of a minority among the scientific community. In general, taxonomic studies are only occurring only as side activities of other major projects for which little financial support exists. Taxonomists deserve the construction of a taxonomy that is fast, database based, stable, and, especially, inexpensive. The primary success will be the achievement of a complete database of all almost closed genomes of all type strains of the classified species (21), for which one would need only to calculate the ANI and TETRA values with the accessible data. However, this will be achieved in the midterm if we consider that all type species still have not had their 16S rRNA gene sequenced (34). For this purpose, we evaluated the approach of calculating ANI and TETRA values based only on a random 454 reads that only partially covered the genomes to be analyzed. Achieving stable ANI and TETRA values by a rough set of ≈250 nucleotide stretches will importantly reduce the sequencing costs when trying to circumscribe species.
To evaluate a relatively inexpensive approach to whole genomics, we have partially sequenced a set of strains for which our group had previously obtained DDH values (35, 36). For this, each genome was tagged with a different multiplex identifier for 454 libraries (Roche Applied Science), and the pooled libraries were randomly sequenced to obtain 123,210 reads covering ≈27.4 Mb (Table S4). The mean of the fragments obtained was ≈221.9 nucleotides, and for each single strain the number of reads ranged from 8,100 to 33,000. A remarkable drawback of the approach was the fact that 31–36% of the reads were multiple copies of identical sequences. The final sieved results rendered between 4,500 and 17,000 single unique reads. This last point should be taken into consideration, as the sequencing effort needs to be almost duplicated until the bias is solved by the company producing the sequencing kits. In addition, we included as an internal control the DNA from E. coli strain K12 that accounted for ≈3,158 reads covering ≈0.7 Mb (≈15% of the genome). The results obtained were clear and satisfactory. For example, just by sequencing 15% of the K12 genome (substr. MG1655), we could obtain a clear and stable ANIm value when comparing the partial sequence with its full genome.
On the other hand, the set of partially sequenced Vibrio spp. or Afifella spp. did not have representatives in the database for comparison. The results obtained (Table S5) were also satisfactory for the set of sequences analyzed and comparable with the DDH values obtained. Within the Vibrio spp. group, ANIm values of ≈97% corresponded to ΔTm measurements of 1.1 °C, and the ANIm of ≈78% with ΔTm of about 7 °C (35). Despite the fact that we did not have reassociation percentages, these negatively correlated with ΔTm and the species threshold could be set at <5 °C (3). In addition, the results based on the set of Afifella spp. were coincident with the ANI thresholds evaluated for species circumscription. The partial sequence comparisons of DDH values of 86–89%, 51–57% and 20% (36) correlated with ANIm values of 97.6%, 92.5% and 83–84%, respectively, and agreed with setting the threshold for circumscribing species at an ANI of 96%. In addition, even though it may seem trivial, the Vibrio spp. set of DNAs was isolated ≈12 years ago and kept frozen at −80 °C. Despite this, we could not detect any problems in the quality of the results, and thus this supports the initiatives of organizing DNA banks as promoted by the DNA Bank Network. Given that the fully sequenced closest relatives to our set of strains (34) harbored a genome with a size ranging between 4 and 6 Mb (www.ncbi.nlm.nih.gov), our partial sequencing rendered at a minimum coverages ranging from 16% to 25% of their genomes. However, despite the low coverage on the Vibrio sp. strains, the interspecies pairwise comparisons rendered nearly identical ANI results (87.16–87.93; Table S5), well in accordance with ΔTm >7 °C. On the other hand, the calculation of the TETRA values showed irregularities when using only draft genomes. For example, the partial random sequence set of E. coli K12 (substr. MG1655) showed a decreased TETRA value of ≈0.96 when using the raw data and ≈0.97 when sieving the 30% sequence multiple identical copies. Similar observations were obtained within the partially sequenced Vibrio spp. group, but not within the alphaproteobacterial group studied (Table S5). The discrepancies are most likey due to the 454 sequencing errors (37). However, an additional source of TETRA biases could arise from using a large number of short fragments that are submitted to calculation. Reducing the number of fragments by increasing the length of the contigs may diminish the bias in TETRA calculations.
As partial sequencing may be the short-term approach to follow while waiting for a reduction in sequencing costs, we wanted to recommend the minimal sequencing effort to achieve reliable results. The number of aligned stretches decreases arithmetically with their ANI values (Fig. 1, inset). Thus, equivalent partial sequence sets of similar genomes will render larger aligned stretches than those that are more divergent. As can be deduced from random comparisons with partial sequences of organisms with different ANI values (Fig. S3), the reliability of the results will depend on the coverage and genome identities. A good compromise now, for taxonomic purposes, will be the recommendation of random sequencing of at least 20% of the genomes of the query strains. For highly similar genomes (i.e., values >94% ANI), with expected aligned stretches close to 4% of their genome sizes, the results may be already reliable, although lower coverages may lead to confusing results. However, reaching at least 50% of genome coverage for both strains, the expected aligned stretches may be close to the 25% that guarantees the values ranging in the species ANI thresholds. In any case, one has to be aware of the artifacts that may be generated by the new sequencing technologies (37) and increase the sequencing efforts.
Circumscribing Uncultured Species.
One of the major constraints of current taxonomic activities is the need to culture the strains that are to be classified. The need arises from the fact that taxonomically workable information can simply be retrieved by manipulating pure cultures in the laboratory. However, for the first time, genomics presents the possibility of obtaining high-quality genomic information by purifying unculturable cells from their original environment (38, 39). To evaluate whether ANI and TETRA would help in advancing the circumscription of uncultured organisms, we compared the available Buchnera aphidicola and Wolbachia spp. endosymbiont genomes (Table S6). The results obtained indicated that, based on ANI and TETRA values, only these endosymbionts of the aphid Acyrthosiphon pisum (strains 5A, Tuc7 and APS) (38) can be considered members of the same species, as they share ANIm values >99%, whereas the endosymbionts originating in other species of aphids (strains Cc, Bp and Sg; 40–42) may each represent single independent species because pairwise ANIm values were always <87%. The same observation applies to the Wolbachia spp. genomes analyzed, as it seems that just those infecting Drosophila spp. may be considered for the same species. The results agree with the fact that these organisms diverged ≈50–70 million years ago, and, despite a highly conserved genomic architecture, their genetic divergence has followed similar rates compared with those of other free-living organisms (42). These observations appear to be fully in accordance with the host-cospeciation theory (43); and the reclassification of the species in at least several independent taxa appears to plausible and, perhaps, in the future, a taxonomically accepted activity.
Shifting the Gold Standard for Prokaryotic Species Circumscriptions Based on Genomic Data.
The extent of diversity of prokaryotes is difficult to evaluate given, their incommensurable cell abundances and ecological niches where they thrive (44). However, the current species catalog of ≈8,000 validly published names (34) is far from the estimated actual number of classifiable taxa, which is expected to be several orders of magnitude higher (44). The species concept for prokaryotes has been constantly criticized because it does not fit with the views that different scientists approach in their understanding of what this category means (1–4, 6, 7, 22, 45). The main problem is not the concept itself (1, 6) but its definition (i.e., the parameters to circumscribe species used). In this regard, DDH has been criticized as being too conservative, embracing multiple species from the point of view of ecologists or evolutionary microbiologists (1, 6, 7, 22, 45). However, there is a conflict between the desire to construct a universal taxonomy and the achievement of an accurate definition that reflects evolutionary and ecological constraints (1, 6). Prokaryote taxonomy must be established by taking into account both genomic and phenotypic information (8). Narrowing the species boundaries could lead to difficulty regarding the main purposes of taxonomy: namely, operationality and predictivity. For pragmatic reasons, finding standards to circumscribe species is required to speed up the process of cataloging prokaryotes. It is foreseen that once a stable framework for classifying prokaryote species is achieved, the views of taxonomists, ecologists, and evolutionary microbiologists may be easier to reconcile.
Genomics has brought an important advance to the species definition. The comparative efforts undertaken to evaluate ANI as a mirror for DDH led us to ascertain that ANI, with the support of TETRA values, is the parameter that can immediately substitute for DDH. We have demonstrated that ANI can be calculated only by partially sequencing the query strains by at least 20% of their genome or by producing an alignment equivalent >4% of their genome sizes. However, reaching 50% of the genome coverage is recommended. The ideal situation will be the achievement of a complete database of all type strains of the validly published species (21), which would simplify the recognition of a strain as a member of a given species and reduce the sequencing effort.
Assuming that ANI is to be successful as an alternative to DDH, it would seem that 95% would be a plausible and narrow enough threshold to help in circumscribing prokaryotic species. In light of our study, this threshold could even be raised to 96%, as most of the clear genospecies evaluated fell within this range. Narrowing the genomic circumscription of species (2) would obviate the need for discussion in areas such as the consideration that some Neisseria species cluster as a single “fuzzy species” (45). Our calculations with the deposited genomes of N. gonhorroeae and N. meningitidis (Table S6) show that both groups of strains are distanced by >5% ANIm. Thus, by strengthening the species boundary to 96%, the classification of both species as different taxa would be justified. On the other hand, special-purpose classifications such as as Shigella spp./Escherichia coli (Fig. 2), or Burkholderia mallei/pseudomallei and Bordetella bronchiseptica/parapertussis/pertussis (Table S6), which share ANI values that would indicate taxonomic synonymy, should be maintained just for medical purposes (6, 23).
We think that it is already time to shift the circumscription gold standards from the traditional DDH to a partial or better full genome sequencing approach. The 95–96% ANI threshold can be readily used as an objective boundary for species circumscription, especially if it is reinforced by high TETRA correlation values. From our observations, it seems that a fuzzy gap exists at this ANI range that can help in circumscribing clusters of organisms that can be assumed to be species (Fig. 2). However, narrowing the boundary to higher identities for a less conservative definition seems not to be pragmatic, as no clear gap is observed. ANI will serve for classifying not only cultured prokaryotes but also for those uncultured strains that exhibit enough additional characters (e.g., ecological, physiological) that allow their identification. The concerns raised by an ad hoc committee several years ago (4), encouraging efforts to substitute DDH, now seem to be nearly addressed. Consequently, it is foreseen that, in the short term, DDH will be an “emeritus” taxonomic tool.
Materials and Methods
DNA Extraction and Sequencing.
Eight organisms were selected to perform a random partial genome pyrosequencing approach. These included three members of the Gammaproteobacteria phylum (35): Vibrio aestuarianus strain LMG 7909T, V. scophthalmi strain A089T and V. scophthalmi strain A107; and four members of the Alphaproteobacteria phylum (36): Afifella marina strain DSM 2698T, A. marina strain C3, “Rhodopseudomonas julia” strain DSM 11549 (pending reclassification to the genus Afifella; 36), and Rhodobium gokarnense strain DSM17935T. Furthermore, Escherichia coli K-12 (substr. MG1655) was used to study the behavior and composition of the pyrosequencing reads. DNAs from V. scophtalmi strains A089T and A107, and V. aestuarianus LMG 7909T were isolated in 1996 with the procedure indicated by Cerdà-Cuéllar et al. (35) and kept for about 12 years at −80 °C. DNAs of the Afifella spp. strains DSM 2697T, C3 and DSM 11549, as well as Rhodobium gokarnense DSM 17935T and E.coli K12 (substr. MG1655), were freshly obtained for this study following the procedure previously reported (36). Sequencing was carried out by “lifesequencing” using the Genome Sequencer FLX System, 454 sequencing instrument (Life Sciences).
Database Information.
All publicly available genome sequences used in this study were obtained from the National Center for Biotechnology Information (NCBI). The genome information was taken directly without removing plasmids or any other DNA stretches.
Software Development.
The JSpecies software tool was primarily designed to analyze and compare innerspecies boundaries between genomes, draft genomes, or partial random genome sequences. JSpecies offers a graphical user interface to assist microbiologists in the exploration of intergenome similarities. JSpecies is written in the platform-independent, object-oriented programming language Java. It can be started using the Java Web Start technology, which automatically downloads and installs the software locally. This ensures the user will always obtain access to the latest version available. Alternatively, it can be downloaded and installed manually. To calculate species relationships, two additional software packages need to be locally installed: BLAST (14) and MUMmer (15). Unfortunately, the MUMmer software is not available for any Microsoft operating systems. JSpecies is freely available from the project web site, where further information and documentation about the tool and how to use it is provided.
BLAST Calculation of ANI (ANIb).
The calculation of ANI values was implemented as described by Goris et al. (12).
MUMmer Calculation of ANI (ANIm).
ANI values were calculated by using the MUMmer software, in particular the NUCmer (NUCleotide MUMmer) tool (15). NUCmer allows DNA sequence alignments to be processed for multiple reference and query sequences. MUMmer was used with standard parameters if not otherwise stated. The tool produces two output files, and the “.delta” file was used for further processing. The .delta file is an encoded representation of the all-vs.-all alignment and was generated by using the default options if not otherwise stated. The .delta file lists the distance between insertions and deletions that produces maximum scoring alignments between sequences. It provides seven values, the start and end in the reference, and the start and end in the query, respectively, the number of errors (nonidentities + indels) and similarity errors (nonpositive match scores). To calculate the ANIm, the number of similarity errors was subtracted from the alignment length to receive the percentage nucleotide identity within all alignments. The value was summed for each entry in a multiFasta.
Calculation of Tetranucleotide Frequencies and Correlation Coefficients.
This was implemented into JSpecies based on a previously described algorithm (16).
Supplementary Material
Acknowledgments.
We are very thankful to K.H. Schleifer, E. Moore, P. Kämpfer, R. Amann, J. Waldmann, and P. Yarza for critical review and helpful discussions. The study was supported by the Spanish Ministry projects Consolider Ingenio 2010 project CE-CSD2007–0005 and Biosal (CLG2006–12714-C02–02) and by the FBBVA project BIOCON05/094.
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906412106/DCSupplemental.
References
- 1.Rosselló-Móra R. Updating prokaryotic taxonomy. J Bacteriol. 2005;187:6255–6257. doi: 10.1128/JB.187.18.6255-6257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Phil Trans Soc B. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosselló-Móra R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 4.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PAD, Kämpfer P, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 5.Wayne L, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 6.Rosselló-Móra R. DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation. In: Stackebrandt E, editor. Molecular Identification, Systematics, and Population Structure of Prokaryotes. Berlin: Springer-Verlag; 2006. pp. 23–50. [Google Scholar]
- 7.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 8.Stackebrandt E, Goebel BM. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 9.Adékambi T, Shinnick TM, Raoult D, Drancourt M. The rpoB gene as a tool for clinical microbiologists. Int J Syst Evol Microbiol. 2008;58:1807–1814. doi: 10.1099/ijs.0.65440-0. [DOI] [PubMed] [Google Scholar]
- 10.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, et al. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp nov and Burkholderia lata sp nov. Int J Syst Evol Microbiol. 2009;59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 11.Konstantinidis K, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2592. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 13.Deloger M, El Karoui M, Petit M-A. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol. 2009;191:91–99. doi: 10.1128/JB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teeling H, Meyerdierks A, Bauer M, Amann R, Glöckner FO. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol. 2004;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- 17.Coenye T, Gevers D, Van de Peer Y, Vandamme P, Swings J. Towards a prokaryotic genomic taxonomy. FEMS Microbiol Rev. 2005;29:147–167. doi: 10.1016/j.femsre.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Tindall BJ, Kämpfer P, Euzéby J, Oren A. Valid publication of names of prokaryotes according to the rules of nomenclature: Past history and current practice. Int J Syst Evol Microbiol. 2006;56:2715–2720. doi: 10.1099/ijs.0.64780-0. [DOI] [PubMed] [Google Scholar]
- 19.Dawyndt P, Vancanneyt M, De Meyer H, Swings J. Knowledge accumulation and resolution of data inconsistencies during the integration of microbial information sources. IEEE Trans Knowl Data Eng. 2005;17:1111–1126. [Google Scholar]
- 20.Euzéby JP. List of bacterial names with standing in nomenclature: A folder available on the internet. Int J Syst Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 21.Buckley M, Roberts R. Reconciling microbial systematics and genomics. The American Academy of Microbiology Reports 2006. 2007 [Google Scholar]
- 22.Lan R, Reeves PR. Escherichia coli in disguise: Molecular origins of Shigella. Microbes Infect. 2002;4:1125–1132. doi: 10.1016/s1286-4579(02)01637-4. [DOI] [PubMed] [Google Scholar]
- 23.Strockbine NA, Maurelli AT. Genus XXXV. In: Brenner DJ, Krieg NR, Staley JT, editors. Shigella. Bergey's Manual of Systematic Bacteriology. 2nd Ed. Part B. Vol 2. New York: Springer; 2005. pp. 811–823. [Google Scholar]
- 24.Keswani J, Orkand S, Premachandran U, Mandelco L, Franklin MJ, Whitman WB. Phylogeny and taxonomy of mesophilic Methanococcus spp. and comparison of rRNA, DNA hybridization, and phenotypic methods. Int J Syst Bacteriol. 1998;46:727–735. doi: 10.1099/00207713-46-3-727. [DOI] [PubMed] [Google Scholar]
- 25.Takahata Y, Nishijima M, Hoaki T, Maruyama T. Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int J Syst Evol Microbiol. 2001;51:1901–1909. doi: 10.1099/00207713-51-5-1901. [DOI] [PubMed] [Google Scholar]
- 26.Bohlin J, Skjerve E, Ussery DW. Reliability and applications of statistical methods based on oligonulcleotide frequencies in bacterial and archaeal genomes. BMC Genomics. 2008;9:104. doi: 10.1186/1471-2164-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Passel MWJ, Kuramae EE, Luyf ACM, Bart A, Boekhout T. The reach of the genome signature in prokaryotes. BCM Evol Biol. 2006;6:84. doi: 10.1186/1471-2148-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003;13:145–158. doi: 10.1101/gr.335003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foerstner KU, Von Mering C, Hooper SD, Bork P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005;12:1208–1213. doi: 10.1038/sj.embor.7400538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fournier P-E, Vallente D, Barbe V, Audic S, Ogata H, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLOS Genet. 2006;2:62–72. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapidus A, Goltsman E, Auger S, Galleron N, Ségurens B, et al. Extending the Bacillus cereus group genomics to putative food-borne pathogens of different toxicity. Chem Biol Interact. 2008;171:236–249. doi: 10.1016/j.cbi.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Auger S, Galleron N, Bidnenko E, Ehrlich SD, Lapidus A, Sorokin A. The genetically remote pathogenic strain NVH391–98 of the Bacillus cereus group is representative of a cluster of thermophilic strains. Appl Environ. 2008;74:1276–1280. doi: 10.1128/AEM.02242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thieme F, Koebnik R, Bekel T, Berger C, Boch J, et al. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris p. vesicatoria revealed by the complete genome sequence. J Bacteriol. 2005;187:7254–66. doi: 10.1128/JB.187.21.7254-7266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarza P, Richter M, Peplies J, Euzéby J, Amann R, et al. The all-species living tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. System Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Cerdà-Cuéllar M, Rosselló-Mora R, Lalucat J, Jofre J, Blanch A. Vibrio scophthalmi sp. nov., a new species from turbot (Scophthalmus maximus) Int J System Bacteriol. 1997;47:58–61. doi: 10.1099/00207713-47-1-58. [DOI] [PubMed] [Google Scholar]
- 36.Urdiain M, López-López A, Gonzalo C, Busse H-J, Langer S, et al. Reclassification of Rhodobium marinum and Rhodobium pfennigii as Afifella marina gen. nov. comb nov. and Afifella pfennigii comb. nov., a new genus of photoheterotrophic Alphaproteobacteria and emended descriptions of Rhodobium, Rhodobium orientis and Rhodobium gokarnense System Appl Microbiol. 2008;31:339–351. doi: 10.1016/j.syapm.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Gómez-Álvarez V, Teal TK, Schmitdt T. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009 doi: 10.1038/ismej.2009.72. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 38.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS Nature. 2002;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 39.Mussmann M, Hu FZ, Richter M, De Beer D, Preisler A, et al. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5:2913–2937. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pérez-Brocal V, Gil R, Ramos S, Lamellas A, Postigo M, et al. A small microbiol genome: The end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 41.Van Ham RCHJ, Kamerbeek J, Palacios C, Rausell C, Abascal F, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamas I, Klasson L, Canbäck B, Näslund AK, Eriksson A-S, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 43.Martínez-Torres D, Buades C, Latorre A, Moya A. Molecular systematics of aphids and their primary endosymbionts. Mol Phyl Evol. 2001;20:437–449. doi: 10.1006/mpev.2001.0983. [DOI] [PubMed] [Google Scholar]
- 44.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staley J. The bacterial dilemma and the genomic—phylogenetic species concept. Phil Trans Soc B. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.