Summary
To enhance our understanding of the function(s) of γ-tubulin-complex proteins (GCPs), we identified and analyzed the functions of the Aspergillus nidulans homologs of GCP2-GCP6 (here designated GCPB-GCBF). The γ-tubulin small complex (γ-TuSC) components, γ-tubulin, GCPB and GCPC, are essential for viability and mitotic spindle formation, whereas GCPD-GCPF are not essential for viability, spindle formation or sexual reproduction. GCPD-GCPF function in reducing the frequency of chromosome mis-segregation and in the assembly of large γ-tubulin complexes. Deletion of any of the γ-TuSC components eliminates the localization of all GCPs to the spindle pole body (SPB), whereas deletion of GCPD-GCPF does not affect localization of γ-TuSC components. Thus, GCPD-GCPF do not tether the γ-TuSC to the SPB, but, rather, the γ-TuSC tethers them to the SPB. GCPD-GCPF exhibit a hierarchy of localization to the SPB. Deletion of GCPF eliminates GCPD-GCPE localization to the SPB, and deletion of GCPD eliminates GCPE (but not GCPF) localization. All GCPs localize normally in a GCPE deletion. We propose a model for the structure of the γ-tubulin complex and its attachment to polar microtubule organizing centers.
Keywords: γ-tubulin complex, Spindle pole body, Centrosome, Microtubule, Microtubule organizing center
Introduction
In many eukaryotic cells, microtubules are organized by microtubule-organizing centers (MTOCs) such as the spindle pole body (SPB) in fungi and the centrosome in metazoa. γ-tubulin is a highly conserved component of MTOCs, playing an essential role in microtubule nucleation and organization (Oakley and Oakley, 1989; Oakley et al., 1990; Zheng et al., 1991; Horio et al., 1991; Moritz et al., 1998; Schnackenberg et al., 1998) [and additional data reviewed by Job and colleagues (Job et al., 2003)]. γ-tubulin forms complexes (γ-tubulin complexes, γ-TuCs) with several proteins (reviewed by Wiese and Zheng, 2006). The majority of γ-TuCs purified from Xenopus laevis, Drosophila melanogaster and humans are large (>25S) complexes that appear to be ring structures with a diameter of 25 nm (γ-tubulin ring complexes, γ-TuRCs) (Stearns and Kirschner, 1994; Zheng et al., 1995; Moritz et al., 1998; Oegema et al., 1999; Meads and Schroer, 1995; Murphy et al., 2001). γ-TuRCs consist of γ-tubulin and at least five GCPs (γ-tubulin complex proteins). GCPs are known as Xgrips (Xenopus γ-tubulin ring proteins) in X. laevis, Dgrips in D. melanogaster and hGCPs in humans (reviewed by Wiese and Zheng, 2006; Raynaud-Messina and Merdes, 2007). In addition to the five GCPs, an additional protein containing WD repeats (GCP-WD, originally called NEDD1 in humans) has been found to associate with the γ-TuRC in D. melanogaster and humans but not in X. laevis (Gunawardane et al., 2003; Haren et al., 2006; Luders et al., 2006; Liu and Wiese, 2008). We will reserve the term γ-TuRC for cases in which the complex has been shown by electron microscopy to be a ring complex, and the more general term `large γ-TuC' for cases in which a large γ-tubulin complex has been shown to exist but the structure has not been studied by electron microscopy. In most cases the γ-TuRC and the large γ-TuC are probably the same.
A smaller (∼9.8S) complex, the γ-tubulin small complex (γ-TuSC), which contains only γ-tubulin, GCP2 (Dgrip84) and GCP3 (Dgrip91), was discovered in D. melanogaster (Moritz et al., 1998; Oegema et al., 1999). It is a component of the γ-TuRC, and γ-TuRCs disassemble in 500 mM KCl, releasing γ-TuSCs (Moritz et al., 1998). A great deal of data indicates that γ-TuRCs are assembled from γ-TuSCs and other GCPs in the cytoplasm and are recruited to the centrosome, where they serve as the major microtubule nucleators in animal cells (Moritz et al., 1995; Moritz et al., 1998; Schnackenberg et al., 1998; Wiese and Zheng, 1999; Wiese and Zheng, 2006). On the basis of these data and electron tomographic data, a structural model has been proposed for the γ-TuRC in which six to seven γ-TuSCs are arranged in a ring and capped by GCPs (Wiese and Zheng, 1999; Moritz et al., 2000; Keating and Borisy, 2000; Wiese and Zheng, 2000).
Each of the γ-TuSC components is essential for viability and for normal mitotic spindle assembly and function (Oakley et al., 1990; Sunkel et al., 1995; Geissler et al., 1996; Spang et al., 1996; Martin et al., 1997; Knop et al., 1997; Knop and Schiebel, 1997; Barbosa et al., 2000; Paluh et al., 2000; Vardy and Toda, 2000; Hannak et al., 2002; Colombie et al., 2006). The functions of GCP4-GCP6 are less clear, however. Although a number of functional analyses have been carried out, the data are contradictory in some cases and show apparent differences among and within species that have prevented the emergence of a coherent model for the function of GCP4-GCP6 (Zhang et al., 2000; Schnorrer et al., 2002; Fujita et al., 2002; Venkatram et al., 2004; Vogt et al., 2006; Verollet et al., 2006; Anders et al., 2006).
A number of important questions about γ-tubulin complexes remain unanswered. For example, the arrangement of the subunits within the larger γ-tubulin complex is not clear. It is not clear which components of the γ-tubulin complex are involved in the attachment of the complex to MTOCs and, as mentioned, the functions of GCP4-GCP6 are still a matter of controversy.
Many of the previous studies have been carried out in different organisms and usually only one or two GCPs have been studied at a time. To cast light on some of the extant questions, and, if possible, create a coherent model for GCP function, we felt that it would be useful to carry out a comprehensive analysis of GCP function in a single organism. We have chosen to carry out our studies in the model filamentous fungus Aspergillus nidulans. This organism has a number of advantages for studying these proteins. First, the sequencing of the genome facilitates identification of genes that encode γ-tubulin-complex proteins. Second, a highly efficient gene targeting system has been developed for A. nidulans (Nayak et al., 2006; Szewczyk et al., 2006) that greatly facilitates tagging or deleting γ-tubulin-complex genes. Third, the heterokaryon rescue technique that can be used with this organism allows one to examine the phenotypes of recessive lethal mutations or gene deletions in vivo (Osmani et al., 1988; Martin et al., 1997; Oakley et al., 1990; Osmani et al., 2006a), and this should allow us to examine the phenotypes of deletions of GCP2 and GCP3 homologs, which are predicted to be lethal. Finally, there is very little data on γ-tubulin complexes in filamentous fungi, and the genus Aspergillus is very important medically and commercially.
Results
GCP2-GCP6 homologs exist in A. nidulans
A blast search of the A. nidulans genome database (http://www.broad.mit.edu/annotation/genome/aspergillus_group) with human GCP protein sequences revealed that strong GCP2-GCP6 homologs exist in A. nidulans. They contain the conserved grip1 and grip2 motifs found in GCPs in other organisms (supplementary material Figs S1, S2) (Gunawardane et al., 2000; Gunawardane et al., 2003; Murphy et al., 2001; Fujita et al., 2002) and we designate the genes gcpB-gcpF and their protein products GCPB-GCPF (Table 1).
Table 1.
Identification of GCP2-6 homologs in A. nidulans by a blast search of the A. nidulans genome database
|
A. nidulans |
|||||||
|---|---|---|---|---|---|---|---|
| γ-TUC protein | Homo sapiens | Cadre/AspGD | Broad | E value | A. nidulans gene symbol | Protein product | Predicted mass of protein (kDa) |
| GCP2 | Q9BSJ2 | AN5873.4 | ANID_05873.1 | 0 | gcpB | GCPB | 98.9 |
| GCP3 | Q96CW5 | AN4867.4 | ANID_04867.1 | 0 | gcpC | GCPC | 108.9 |
| GCP4 | Q9UGJ1 | AN2176.4 | ANID_02176.1 | 1.58E-12 | gcpD | GCPD | 85.6 |
| GCP5 | AAK77662 | AN8120.4 | ANID_08120.1 | 4.09E-18 | gcpE | GCPE | 102.8 |
| GCP6 | AAK82968 | AN1005.4 | ANID_01005.1 | 5.26E-11 | gcpF | GCPF | 107.5 |
The designations of human GCP-encoding genes are from NCBI. The Broad Institute designations for A. nidulans genes have changed recently. We have, therefore, listed the new Broad Institute designations as well as the older designations that are used by the other A. nidulans genome databases, the Central Aspergillus Data Repository (CADRE, http://www.cadre-genomes.org.uk/) and the Aspergillus Genome Database (AspGD, http://www.aspgd.org/)
To confirm that these proteins are GCP homologs, we tagged the C-terminus of each protein with the green fluorescent protein (GFP) and examined their localizations. To avoid artifactual results due to improper expression levels, we used the fusion PCR procedure of Nayak and collegues to produce a single chromosomal copy of the gene of interest, under control of its native promoter, fused in-frame to the GFP coding sequence at its 3′ end (Nayak et al., 2006). Unless otherwise specified, all of the tagging in this paper was performed in this way. All strains carrying GCP fusions grew indistinguishably from wild-type strains at all temperatures tested (20°C, 25°C, 30°C, 37°C and 42°C) (supplementary material Fig. S3). Time-lapse observations revealed that, like γ-tubulin, GCPB-GCPF localize to spindle pole bodies throughout the cell cycle, appearing as one dot at the nuclear periphery during interphase (Fig. 1) and two dots during mitosis. Localization of the GCPs to the SPB was confirmed by colocalization with a tdTomato-tagged version of the A. nidulans homolog of the SPB protein Nud1 (supplementary material Fig. S4). All GCPs localized to septa, but the fluorescence of GCPD-GCPF was barely above background levels. These data are consistent with previous findings that the SPB is the major MTOC throughout the cell cycle (Horio and Oakley, 2005; Sampson and Heath, 2005) and with the finding that microtubules might sometimes grow from septa (Konzack et al., 2005). Konzack and colleagues have suggested that an MTOC might be present at the hyphal apex, on the basis of observations of the movement of a GFP-kinesin-like protein under control of an inducible promoter (Konzack et al., 2005); however, other, more direct, studies (Horio and Oakley, 2005; Sampson and Heath, 2005) have not revealed an apical MTOC and we find no localization of GCPs at the apex.
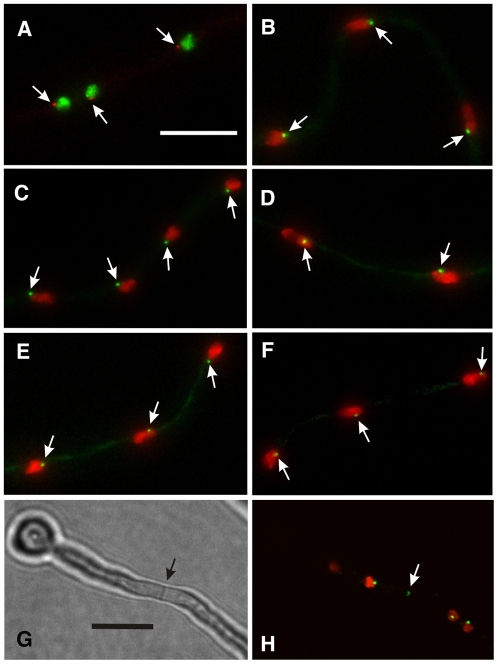
Fig. 1.
Localizations of γ-tubulin and putative GCPs to the SPB and septum. All panels except G are projections of Z-series stacks. A-F were taken with widefield microscopes. (A) γ-tubulin-mCherry and histone H1-GFP. (B) GCPB-GFP, histone H1-mCherry. (C) GCPC-GFP, histone H1-mCherry. (D) GCPD-GFP, histone H1-mCherry. (E) GCPE-GFP, histone H1-mCherry. (F) GCPF-GFP, histone H1-mCherry. GCPF fluorescence is very weak and barely brighter than the hyphal background fluorescence. Arrows in each panel indicate the SPB location of γ-tubulin or the relevant GCP. The variation in nuclear size is due to the cell-cycle stage at which the images were captured. The nuclei in C, for example, are in G1 and the nuclei in D are in late G2. (G,H) Images were taken with an Ultraview Vox spinning disk confocal microscope and show the same field. (G) Bright field image of a portion of a long germling that has formed a septum (arrow). (H) GCPC-GFP and histone H1-mCherry. In addition to the spindle pole bodies, GCPC localizes in relatively faint dots at the septum (arrow). Scale bar in A: 10 μm (A-F). Scale bar in G: 10 μm (G,H).
To further verify that GCPB-GCPF are γ-tubulin complex proteins, we asked whether the GFP fusions of GCPB-GCPF co-immunoprecipitated with γ-tubulin and found that this was, indeed, the case (Fig. 2A). Based on sequence homology, co-localization and co-immunoprecipitations, we can conclude with confidence that GCPB-GCPF are γ-tubulin complex proteins.
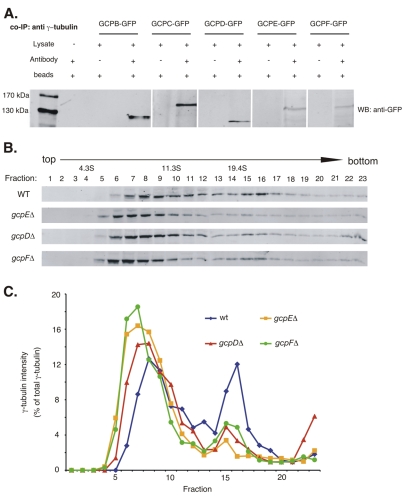
Fig. 2.
Co-immunoprecipitation of putative γ-tubulin complex proteins with γ-tubulin (A) and size distribution of γ-tubulin complexes as determined by sucrose-density centrifugation (B,C). (A) Leftmost lane shows protein size standards. The remaining lanes are from western blots using anti-GFP. For each GCP-GFP fusion strain the left lane shows a mock immunoprecipitation in which the anti-γ-tubulin antibody was omitted. The right lane shows an immunoprecipitation with the anti-γ-tubulin antibody. The anti-GFP recognizes a band of the expected size in each case, indicating that GCPB-GCPF co-immunoprecipitate with γ-tubulin. (B) One set of representative results of western blots, using an anti-γ-tubulin antibody as a probe, of sucrose-density gradients of cytoplasmic extracts of strains wild-type for the GCP-encoding genes (WT) or carrying gcpD, gcpE and gcpF deletions. (C) Plot of the intensity of the γ-tubulin signal for each fraction from the gradients. For each strain, the signal intensity of each fraction is expressed as the percentage of the total γ-tubulin in the sample. In the wild-type strain γ-tubulin is present in a broad peak between approximately 7S and 14S and in a sharper peak at approximately 21S. In the deletion strains, the higher mass peak is greatly diminished and the higher and lower mass peaks are shifted slightly to the left (lower mass).
γ-TuSC components are essential but gcpD-gcpF are not
To determine whether gcpB-gcpF are essential, we deleted each gene by replacing it with the Aspergillus fumigatus riboB gene. Transformants carrying a correct replacement with no additional integration of transforming DNA were confirmed by Southern hybridizations (data not shown). Deletions of mipA, the gene that encodes γ-tubulin, were lethal as shown previously (Oakley et al., 1990; Martin et al., 1997; Jung et al., 2001), as were deletions of gcpB and gcpC. The genes that encode components of the γ-TuSC are, thus, all essential. Single, double or triple deletions of gcpD-gcpF, however, showed a striking absence of phenotype. They exhibited no obvious change in colony growth rates in comparison with the parental control (supplementary material Fig. S5) and the colonies were morphologically normal. Conidiation is often a more sensitive indicator of mitotic defects than colonial growth rate, but deletant colonies produced abundant viable conidia.
Roles of GCPD-GCPF in the assembly of γ-tubulin complexes
The roles of GCPD-GCPF in the assembly of γ-tubulin complexes are somewhat controversial due to the differences in results obtained in various studies (Zhang et al., 2000; Fujita et al., 2002; Venkatram et al., 2004; Verollet et al., 2006; Vogt et al., 2006; Anders et al., 2006). We used sucrose-density centrifugation to determine the size of γ-tubulin complexes in wild-type hyphae and to determine the effects, if any, of GCPD-GCPF deletions on the sizes of the complexes. In cytoplasmic extracts of wild-type hyphae, γ-tubulin sedimented in a broad peak between about 7S and 14S and a sharper peak at about 21S (Fig. 2). A. nidulans, thus, has both large and small γ-tubulin complexes, but the large complexes are somewhat smaller than the γ-TuRCs seen in animal cells (Zheng et al., 1995; Wiese and Zheng, 1999; Job et al., 2003). Deletion of any of the non-essential GCPs greatly reduced the large γ-tubulin complex peak and apparently caused a slight reduction of the sedimentation coefficients of both large and small complexes. These data indicate that GCPD-GCPF play an important role in the assembly or stability of large γ-tubulin complexes in A. nidulans.
GCPs exhibit a hierarchy of localization to the SPB
We were interested in the effects of deletion of GCPs on the localizations of the other GCPs in the hope that this would shed light on which GCPs attach the γ-tubulin complex to the SPB and, perhaps, on the structure of the γ-tubulin complex. A. nidulans has a particular advantage in this regard because it is possible to use the heterokaryon rescue technique to determine the effects of deletion of essential GCPs on the localization of other γ-tubulin complex components. This technique allows deletions or disruptions of essential genes to be maintained in heterokaryons, and the phenotypes of the disruptions or deletions determined in germinating conidia (uninucleate spores) produced by the heterokaryons (Osmani et al., 1988; Oakley et al., 1990; Martin et al., 1997; Osmani et al., 2006b).
We created a comprehensive set of deletions in strains carrying tagged GCPs (Table 2). Deletions were made by replacing the target genes with the A. fumigatus riboB gene. In the case of the essential genes (mipA, gcpB, gcpC), deletant nuclei were maintained in hetereokaryons. For microscopic observations, conidia from the heterokaryons were incubated in selective media (lacking riboflavin). In the absence of riboflavin, conidia with untransformed nuclei did not germinate and conidia with deletant nuclei germinated but had the phenotype caused by the deletion.
Table 2.
Localization of γ-tubulin and GCPs in gcp deletant strains
| Fusion protein | mipAΔ | gcpBΔ | gcpCΔ | gcpDΔ | gcpEΔ | gcpFΔ |
|---|---|---|---|---|---|---|
| γ-tubulin–mCherry | N/A | – | – | + | + | + |
| GCPB-GFP | – | N/A | – | + | + | + |
| GCPC-GFP | – | – | N/A | + | + | + |
| GCPD-GFP | – | – | – | N/A | + | – |
| GCPE-GFP | – | – | – | – | N/A | – |
| GCPF-GFP | – | – | – | + | + | N/A |
The column at the left shows the γ-tubulin and GCP fusion proteins that we tested for localization. Each of the other columns shows the localization results in strains or heterokaryons carrying particular deletions, which are given at the top of each column. + indicates that the fluorescent protein fusion localized to the SPB. – indicates that no localization was visible. Thus, for example, none of the GCP fusion proteins localized to the SPB in germlings carrying the deletion of the mipA (γ-tubulin) gene, whereas, γ-tubulin–mCherry, GCPB-GFP and GCPC-GFP localized normally in the gcpF deletion strain, and GCPD-GFP and GCPE-GFP did not localize to the SPB
N/A, not applicable
Deletion of any of the genes encoding γ-TuSC components (mipA, gcpB, gcpC) resulted in the loss of SPB localization of all γ-tubulin complex components, both the γ-TuSC components and the non-essential GCPs (Fig. 3 and Table 2). The γ-TuSC components are, thus, mutually dependent for SPB localization and the non-essential GCPs are dependent on the γ-TuSC for SPB localization. Deletion of the non-essential GCPs (GCPD-GCPF), however, did not affect the SPB localization of the γ-TuSC components (Fig. 4 and Table 2). The γ-TuSC, thus, does not depend on the non-essential GCPs for the SPB localization. We also examined GCPC localization at the septum in a gcpF deletion and found that it localized normally (our unpublished data).
Fig. 3.
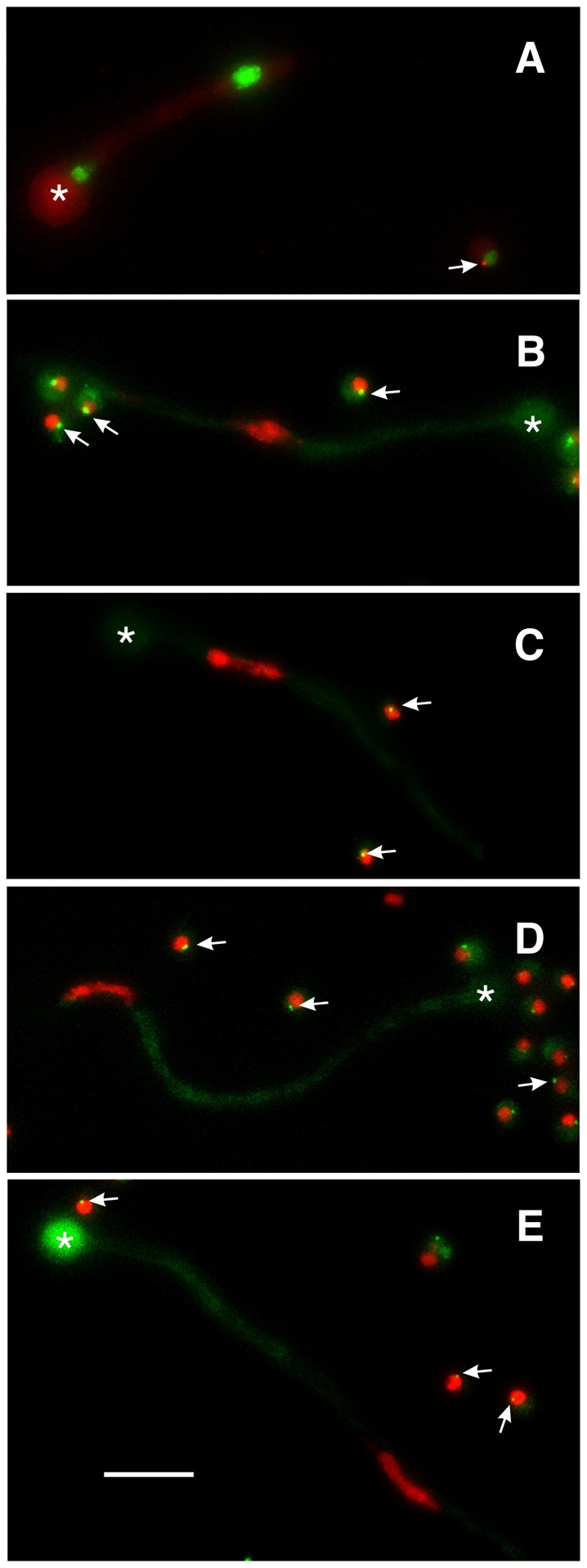
Abolition of γ-tubulin and GCP localization by deletion of gcpC. In each panel a germling carrying a gcpC deletion (gcpCΔ) is shown along with one or more ungerminated conidia that are wild-type for gcpC but carry riboB2 that prevents growth in unsupplemented media. The swollen conidia that gave rise to each germling are marked with asterisks. (A) gcpCΔ in a strain carrying histone H1-GFP and γ-tubulin-mCherry. Two nuclei of unequal sizes are present. γ-tubulin does not show SPB localization in the gcpCΔ germling, but does localize to the SPB of the nucleus in the ungerminated gcpC+ spore (arrow). (B-E) gcpCΔ in strains carrying histone H1-mCherry and GCPB-GFP (B), GCPD-GFP (C), GCPE-GFP (D) and GCPF-GFP (E). Each gcpCΔ germling has one nucleus, reflecting the fact that nuclear division is inhibited in the gcpCΔ strain. Germlings as long as these would normally have many nuclei. In each case, the GCP fails to localize to the SPB in the gcpCΔ germlings (i.e. there is no GCP spot associated with the nucleus), but localizes normally in the ungerminated gcpC+ conidia (arrows). All images are maximum intensity projections of Z-series stacks that include the entire germling so the absence of GCP fluorescence is not due to the SPB being out of the focal plane. Scale bar: 10 μm.
Fig. 4.
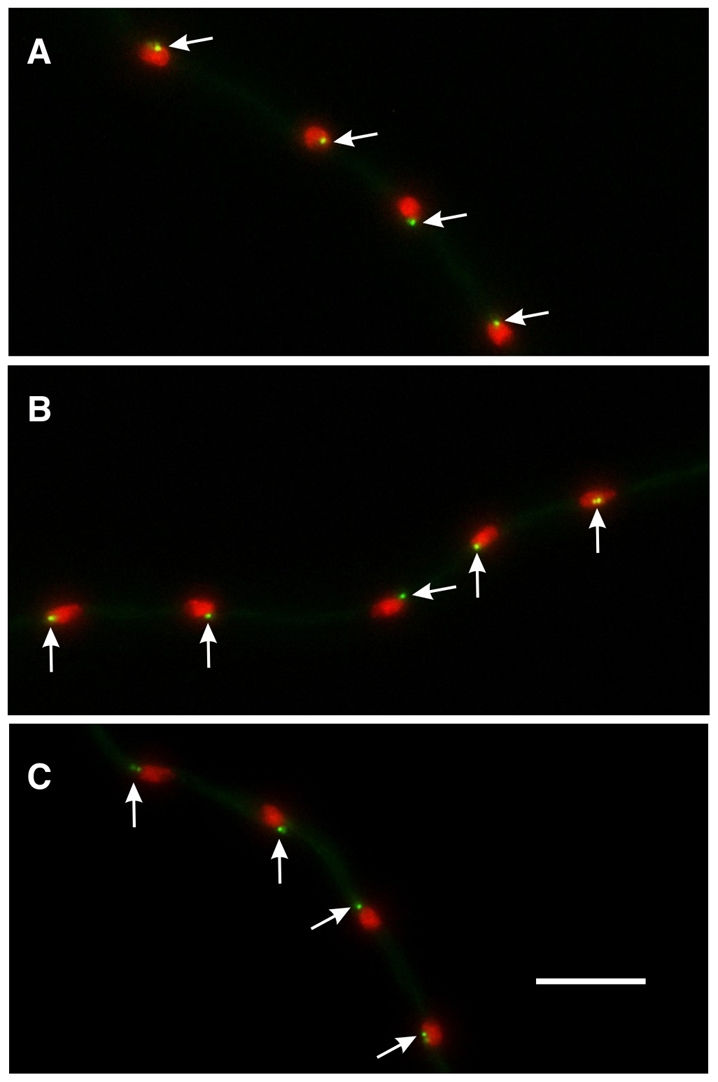
The γ-TuSC does not require GCPD-GCPF for localization to the SPB. (A) GCPC-GFP localization in a gcpD deletion strain. GCPC-GFP localizes normally to SPBs (arrows). Identical results were obtained for gcpEΔ (B) and gcpFΔ (C). GCPB and γ-tubulin also localized normally in gcpD-gcpF deletion strains. Scale bar: 10 μm.
Among the non-essential GCPs, we discovered a hierarchy of SPB-localization dependence. Deletion of gcpF eliminated localization of GCPD and GCPE to the SPB, deletion of gcpD eliminated the localization of GCPE but not GCPF, and deletion of gcpE did not alter the SPB localization of GCPD or GCPF (Fig. 5 and Table 2).
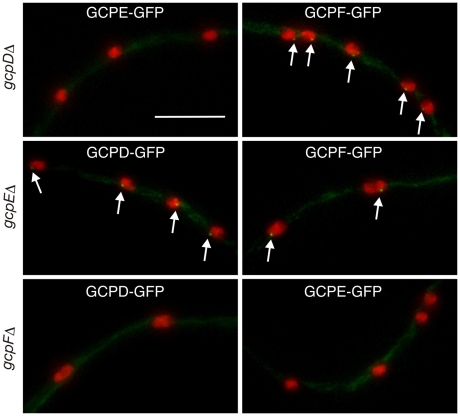
Fig. 5.
Dependency relationships for localization of non-essential GCPs. The gcpD, gcpE and gcpF genes were deleted in strains carrying fusions of GCPD-GCPF with GFP. The GCP-GFP fusion is shown at the top of each panel and the deletion at the left of each pair of panels. SPB localization of the GCP-GFP fusion is indicated with arrows. GCPF localizes to the SPB in gcpD and gcpE deletants. GCPD localizes to the SPB in the gcpE deletion strain but not in the gcpF deletion strain. Neither GCPD nor GCPE localize to the SPB in the gcpF deletion strain. Scale bar: 10 μm.
Effects of gcpB-gcpF deletions on mitotic spindles and cytoplasmic microtubules
To investigate the roles of GCPs in γ-tubulin-mediated microtubule nucleation, we deleted mipA and each of the GCP-encoding genes individually in a strain (LO1915) carrying a GFP-α-tubulin fusion and a histone H1-mCherry fusion. Consistent with our finding that γ-TuSC components are mutually dependent for SPB localization, we found that the gcpB and gcpC deletions phenocopied the mipA deletion (Fig. 6). Functional mitotic spindles were absent and cytoplasmic microtubules were extensively reduced relative to controls, consistent with previously published immunofluorescence data for mipA deletion germlings (Oakley et al., 1990; Martin et al., 1997). Occasionally, we observed abnormal curved microtubule bundles in these deletants, similar to those observed in a previous study of mipA deletants (Martin et al., 1997). Like the disruption of mipA, deletions of either gcpB or gcpC resulted in blockage of nuclear division. Most deletant germlings had one big nucleus after 20 hours of incubation at 25°C (Figs 3, 6). Occasionally, more than one nucleus was observed in germlings after longer incubation, with one extremely large nucleus and some tiny chromosomal masses scattered along the germlings.
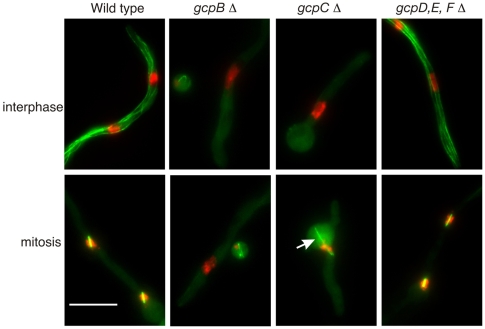
Fig. 6.
Effects of gcp deletions on microtubules. All panels are maximum intensity projections of Z-series stacks. Microtubules are shown with GFP–α-tubulin and chromatin with histone H1-mCherry. Deletion of gcpB or gcpC caused a dramatic reduction in cytoplasmic microtubules and a near absence of mitotic spindles. Some nuclei in gcpB and gcpC deletants had partially condensed nuclei (e.g. gcpBΔ, mitotic), perhaps reflecting partial chromatin decondensation during a slow mitotic exit. Thin, apparently non-functional spindles were seen occasionally in gcpB and gcpC deletants. An example is shown in the gcpCΔ mitotic panel (arrow). The interphase cytoplasmic microtubule array and mitotic spindles were apparently normal in a gcpD, gcpE, gcpF triple-deletant strain. They were also apparently normal in gcpD, gcpE and gcpF single deletions (our unpublished data). Scale bar: 10 μm.
We examined the effects of the deletions on mitosis and microtubule organization by time-lapse microscopy. When germlings carrying any of these three deletions entered mitosis, GFP-α-tubulin entered the nucleoplasm, but functional spindles did not form. Surprisingly, the nuclei underwent stretching and moved vigorously at the late stages of the defective mitosis. We sometimes observed formation of thin, nonfunctional spindles, which, judging from fluorescence intensity, contained only one or a few microtubules (Fig. 6). Thin, non-functional mitotic spindles were observed occasionally in previous studies of mipA deletants (Oakley et al., 1990; Martin et al., 1997) and nucleation of the assembly of the thin spindles could be due to the presence of small amounts of γ-TuSC components inherited from parental hyphae during conidiation (Martin et al., 1997).
GcpD-gcpF deletants in LO1915 grew indistinguishably from the parental strain at all temperatures tested (20°C, 25°C, 30°C, 37°C and 42°C) (supplementary material Fig. S6). We also tested their sensitivities to the microtubule-depolymerizing drug benomyl. In the presence of benomyl concentrations of 0.2-1.2 μg/ml, none of these deletants showed sensitivities different from the parental strains (supplementary material Fig. S6 and additional unpublished data from the authors).
We examined the effects of the gcpD-gcpF deletions on microtubules in vivo and found no obvious defects. Even in the triple-deletion strain (gcpDΔ, gcpEΔ, gcpFΔ), cytoplasmic microtubules and mitotic spindles were not visibly different from the wild type (Fig. 6). Maximum spindle length at anaphase onset was also similar in the control and triple-deletant (wild type, 2.67±0.67 μm; triple-deletant, 2.65±0.41 μm, n=50 for each, P=0.91). Astral microtubules nucleated from SPBs in the triple-deletant were indistinguishable visibly from controls. To determine whether there might be a minor delay in the completion of mitosis that might not lead to an obvious change in growth rate, we captured time-lapse images of mitosis at 20-second intervals and compared the time of completion of mitosis between parental strains and triple-deletion strains. Data were collected at 25°C±1°C. We set the time when the mitotic spindle was first detectable as time zero and when it disappeared completely as the end of mitosis. The difference in the time for mitotic completion between the control LO2064 (361.4±31.9 seconds, n=45) and the triple-deletion strain LO2018 (367.6±49.7 seconds, n=76) was not statistically significant (P=0.7039). However, 10% of triple-deletant nuclei spent longer in mitosis than the longest mitosis (520 seconds) observed in the control strain. This could reflect a meaningful mitotic delay in a small subset of nuclei.
GCPD-GCPF play a nonessential role in chromosome segregation
Although time-lapse microscopy did not reveal a high frequency of chromosomal segregation defects in the triple-deletant, we wondered whether deletion of the genes encoding the GCPs might cause subtle defects that might be revealed by a more sensitive assay. We consequently tested the effects of the deletions on chromosome mis-segregation using a chromosome loss assay. In A. nidulans, vegetative diploids can be created from haploids carrying spore color mutations. Chromosome mis-segregation in diploid cells will result in aneuploid nuclei that lose extra chromosomes rapidly, thus becoming haploid (Käfer, 1977; Clutterbuck, 1992; Rischitor et al., 2004). To test whether deletion of non-essential GCP-encoding genes increases the frequency of chromosome loss, we created diploid strains that were homozygous for deletions of gcpD, gcpE or gcpF and that carried the spore color mutations wA3 and fwA1. Chromosome loss resulted in haploid sectors that were fawn, white and dark green (a haploid sector that does not carry a mutation in a spore color marker gene). The control diploid strain contained the same nutritional and color markers but was wild-type for gcpD-gcpF. The difference in the frequency of chromosome loss became apparent only after the diploid strains were grown for 6 days at 37°C. Compared to the control (33±4 sectors per 20 colonies), the homozygous gcpF deletion diploid produced significantly more haploid sectors (93±10 sectors per 20 colonies; P=0.0048 Student's t-test). Although the homozygous gcpE deletion diploids also showed a significantly higher frequency of haploid sectors on the 6th day of incubation (48±5 sectors per 20 colonies; P=0.0229), the difference between the homozygous gcpD deletion diploid and the control was not significant (29±5 sectors per 20 colonies; P=0.4726).
The spindle assembly checkpoint (SAC) monitors spindle defects and inhibits anaphase until the defects are corrected. If GCPD-GCPF play a role in mitosis, as our chromosome loss data suggest, deletions of gcpD-gcpF might create defects that are detected by the SAC and subsequently corrected. It follows that if the spindle checkpoint were inactivated, enough chromosomal segregation defects might accumulate in gcpD-gcpF deletions to inhibit growth. We consequently created double-mutants, each carrying a deletion of a non-essential GCP-encoding gene as well as a deletion of md2A, the gene that encodes the A. nidulans homolog of Mad2, which is essential for the functioning of the SAC (Prigozhina et al., 2004). Strains carrying deletions of non-essential GCP-encoding genes were indistinguishable in growth rate from a control strain at all temperatures tested (20°C, 25°C, 30°C, 37°C and 42°C), as was a md2A deletion strain. However, gcpFΔ md2AΔ double-deletants were synthetically sick compared to the parental and wild-type strains at 37°C and 42°C (Fig. 7). The gcpDΔ md2AΔ double-deletants and gcpEΔ md2AΔ double-deletants also showed synthetic sickness, although somewhat less severe than gcpFΔ md2AΔ (supplementary material Fig. S7). The stronger synthetic sickness of gcpFΔ and the greater effect of gcpFΔ on chromosome mis-segregation are probably due to the fact that GCPF is required for localization of GCPD and GCPE to the SPB. Together, these data indicate that GCPD-GCPF, although not essential for mitosis, do play a role in chromosome segregation in mitosis.
Fig. 7.
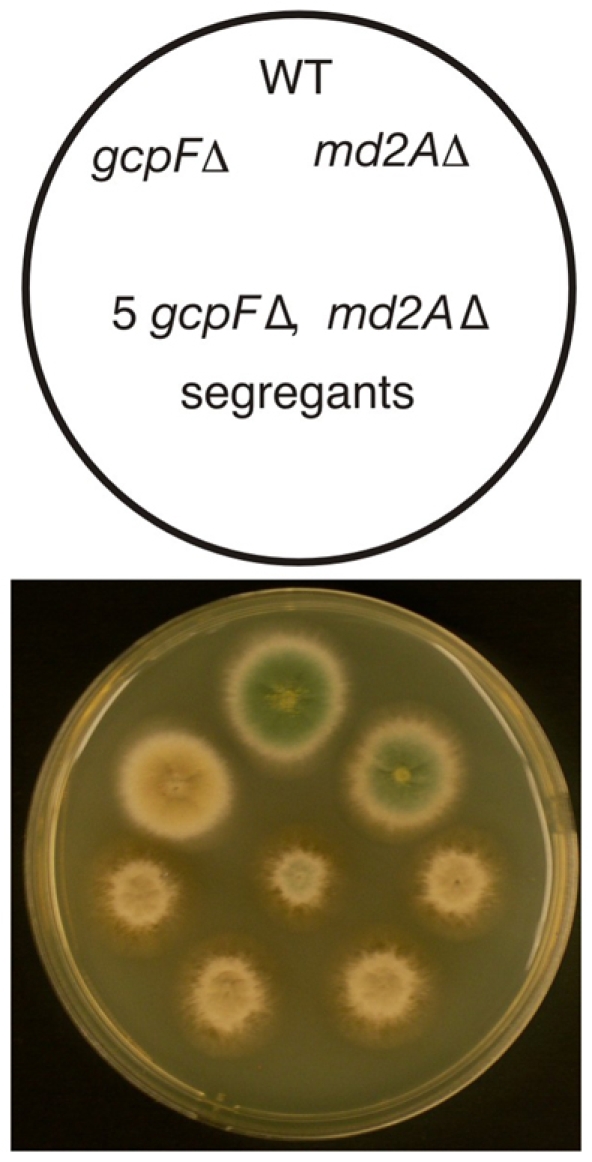
Synthetic interactions of md2A and gcpF deletions. A schematic diagram is shown at the top. Growth was at 37°C. The different colony colors are due to different spore color markers used in the cross to construct the double mutants. The gcpF and md2A deletant strains grow as well as the wild-type control, but the five double-mutant colonies are smaller and have rougher edges, revealing that the gcpF and md2A deletions are synthetically sick.
The nonessential GCPs are not required for meiosis
Because studies in D. melanogaster have shown that Grip75 and Grip128 mutant flies are sterile, suggesting that meiosis is more sensitive to the absence of these proteins than mitosis (Vogt et al., 2006), we tested the ability of triple-deletion strains to undergo meiosis. We created gcpD-gcpF triple deletions in two different strains carrying conidial color mutations, LO2018 carrying wA3, and LO1930 carrying fwA1. Successful mating and gene segregation between the two strains will produce meiotic progeny that are not only white and fawn in color but also the wild-type dark green color. We found that the triple-deletion strains generated cleistothecia with viable ascospores, and that they crossed successfully with each other to produce recombinant progeny (supplementary material Fig. S8). We conclude, therefore, that GCPD-GCPF are not essential for meiosis.
Discussion
GCP4-GCP6 homologs and large γ-TuCs exist in A. nidulans
Our data reveal that A. nidulans contains homologs of GCP2-GCP6. They localize to SPBs and co-immunoprecipitate with γ-tubulin and, thus, are genuine γ-tubulin complex proteins. Although one can identify putative GCPs on the basis of sequence identity in the genomes of a number of filamentous fungi, this represents the first experimental verification that GCPs are present in filamentous fungi. We have also found that A. nidulans possesses distinct large, soluble ∼21S γ-tubulin complexes as well as smaller complexes (Fig. 2). This is of interest because there is controversy as to whether large complexes exist in Saccharomyces cerevisiae and Schizosaccharomyces pombe (Fujita et al., 2002; Vinh et al., 2002; Venkatram et al., 2004; Anders et al., 2006). Large γ-TuCs clearly exist in A. nidulans, although their sedimentation coefficients are somewhat lower than the γ-TuRCs found in animal cells (Stearns and Kirschner, 1994; Zheng et al., 1995; Moritz et al., 1998; Oegema et al., 1999; Meads and Schroer, 1995; Murphy et al., 2001). We have found that A. nidulans also contains smaller soluble γ-tubulin complexes that distribute over a fairly broad peak (Fig. 2). The broad distribution raises the possibility that the small complexes are heterogeneous. Three things are worth noting in this regard. First, this broad distribution has been noted in a previous study and, in high salt, the broad distribution resolved into a much sharper peak that corresponded in size to the 9.8S γ-TuSCs (Akashi et al., 1997). Second, deletion of any of the non-essential GCPs caused a slight apparent reduction in the sedimentation coefficient of the small complexes (Fig. 2), and the greatest reduction occurred with deletion of GCPF. Third, GCPF localizes to the SPB in the absence of GCPD or GCPE, and GCPD localizes to the SPB in the absence of GCPE, revealing that an intact large γ-tubulin complex is probably not required for binding of GCPF and GCPD to the γ-TuSC. These data raise the possibility that the small complexes are not just γ-TuSCs, but perhaps γ-TuSCs bound to one of the non-essential GCPs in a salt-sensitive fashion (such binding could be hierarchical as discussed below).
Roles of the GCPs in binding of γ-TuCs to the SPB
Our deletion experiments reveal that the components of the γ-TuSC (γ-tubulin, GCPB and GCPC) are each essential for the localization of the other γ-TuSC components to the SPB. The structure of the S. cerevisiae γ-TuSC has been determined (Kollman et al., 2008) and, on the basis of this structure and previous experimental data (Nguyen et al., 1998; Takahashi et al., 2002; Zimmerman et al., 2004), it is not surprising that elimination of GCPB or GCPC eliminates the localization of all γ-TuSC components to the SPB. It is more surprising, however, that γ-tubulin is required for localization of GCPB and GCPC to the SPB because the structure of the γ-TuSC (at least when the γ-TuSC is free in solution) does not predict that γ-tubulin would be involved in binding GCPB and GCPC together or in binding the γ-TuSC to the SPB. [Note, however, that, although it is not emphasized in the manuscripts, previous data from D. melanogaster suggest that depletion of individual γ-TuSC components reduces the localization of other γ-TuSC components to the centrosome (Colombie et al., 2006; Raynaud-Messina et al., 2004).] Although there are a number of possible explanations for the failure of GCPB and GCPC to localize to the SPB in a γ-tubulin-deletant, one speculative, but intriguing, possibility is suggested by the work of Kollman and colleagues (Kollman et al., 2008). The structure of the γ-TuSC reveals that, in order for γ-TuSCs to participate in microtubule nucleation, a rotation must occur in GCPB and/or GCPC that moves γ-tubulin into a position to nucleate microtubule assembly. Kollman and colleagues raise the possibility that binding of the γ-TuSC to the SPB proteins is associated with the rotation in GCPB and/or GCPC that brings γ-tubulin molecules into proximity. We speculate that lateral interactions of γ-tubulin molecules, in turn, might help to hold the γ-TuSC in a configuration that binds stably to the SPB and nucleates microtubule assembly. The removal of γ-tubulin would not only eliminate microtubule nucleation, but would prevent the locking of GCPB and GCPC in a configuration in which they bind well to the SPB. In this view, the active microtubule nucleation structure would be formed and stabilized by the cooperative interactions of γ-tubulin, GCPB and GCPC, and one or more SPB proteins.
Our data demonstrate for the first time that the γ-TuSC is required for localization of GCPD-GCPF to the SPB. This, coupled with our finding and the findings of others that the γ-TuSC proteins localize to polar MTOCs in the absence of GCP4-GCP6 homologs (Venkatram et al., 2004; Verollet et al., 2006; Anders et al., 2006), demonstrates that GCPD-GCPF are attached to the SPB via the γ-TuSC rather than vice versa. This finding is apparently at odds with the finding in S. pombe that Gfh1p (GCP4) and Alp16p (GCP6) localize to the SPB in alp4 (GCP2) and alp6 (GCP3) mutants (Venkatram et al., 2004). However, the studies in S. pombe were carried out with temperature-sensitive rather than null alleles, and it is possible that these alleles retain some ability to bind GCP4-GCP6 homologs at the temperatures at which the experiments were carried out.
We have discovered a hierarchy of localization of GCPD-GCPF to the SPB. GCPF requires only the γ-TuSC proteins for localization to the SPB and this indicates that GCPF binds directly to the γ-TuSC. GCPD requires the γ-TuSC and GCPF for localization but not GCPE. This suggests that GCPD binds to GCPF and does not bind (or binds very loosely) to the γ-TuSC. GCPE requires all the other γ-TuC components for localization, which is consistent with observations on Mod21p (GCP5) in S. pombe (Anders et al., 2006). Our results suggest that the γ-TuSC binds to the SPB; GCPF binds to the γ-TuSC; GCPD binds to GCPF; and GCPE binds to GCPD. Although our data are generally consistent with the findings of Verollet and colleagues in D. melanogaster (Verollet et al., 2006), they differ in this instance in that Verrollet and colleagues found that RNAi depletion of Dgrip75 (GCP4) substantially reduced both the total levels of Dgrip128 (GCP5) and Dgrip165 (GCP6) in the cells and at the spindle poles, whereas we found that deletion of GCPD did not affect GCPF localization at the SPB.
Functions of the GCPs in vivo
As expected, we found that the γ-TuSC components are important for mitotic spindle formation and correct organization of cytoplasmic microtubules. In addition, we found that the γ-TuSC is required for localization of all γ-TuC components to the SPB.
GCPD-GCPF, however, are clearly less important. They are not essential and colony growth is not slowed measurably in their absence. GCPD-GCPF homologs are also inessential in D. melanogaster (Verollet et al., 2006; Vogt et al., 2006) and S. pombe (Fujita et al., 2002; Venkatram et al., 2004; Anders et al., 2006). They are quite conserved evolutionarily, however, and this implies that they must have functions important enough to confer a selective advantage.
One function is clearly in the assembly or stability of large γ-TuCs. Large γ-TuC peaks were greatly reduced in sucrose gradients of GCPD-GCPF deletants relative to controls. The simplest interpretation of these data is that GCPD-GCPF collectively stabilize the interactions of γ-TuSCs in solution, helping to form large γ-TuCs, and that GCPD-GCPF are all required for the stabilization. This is consistent with some (Verollet et al., 2006) but not all data from D. melanogaster (Vogt et al., 2006). Consistent with previous structural data (Moritz et al., 2000; Keating and Borisy, 2000; Kollman et al., 2008), the simplest interpretation of our data is that GCPD-GCPF form a cap that helps to stabilize the interactions of γ-TuSCs in solution, forming the large γ-TuC. The elimination of any of GCPD-GCPF weakens the cap and allows the equilibrium to shift from large γ-TuCs to individual γ-TuSCs. The γ-TuSC has an intrinsic tendency to associate laterally with other γ-TuSCs (Kollman et al., 2008) and formation of the large complex might involve interactions between γ-TuSCs, as well as interactions of γ-TuSCs with GCP4-GCP6 homologs. Finally, because the deletion of GCPD-GCPF has little effect on growth and does not cause massive mitotic defects or gross alteration of cytoplasmic microtubules or interfere with meiosis, it follows that large γ-TuCs are not particularly important in A. nidulans.
Although GCPD-GCPF are not essential, we have found that they do have a role in insuring the fidelity of chromosomal segregation. When they are deleted, the frequency of chromosome mis-segregation increases. In addition, GCPD-GCPF deletions are synthetically sick with a deletion of the SAC gene md2A, the mad2 homolog. Although we cannot rule out the possibility that the synthetic sickness has to do with a role for the GCPs in mitotic regulation, the simplest explanation is that deletion of GCPD-GCPF, singly or in combination, results in an increase in mitotic spindle defects. If the SAC mechanism is functional, the defects are repaired and the number of abnormal mitoses is low enough that growth is normal. When the SAC mechanism is rendered dysfunctional by deletion of md2A, however, the defects are not repaired and the resulting mitotic defects cause a significant reduction in growth. It is worth noting that loss of function of GCPD-GCPF homologs also cause mitotic defects in D. melanogaster (Verollet et al., 2006).
We suggest the following model for the function of γ-TuC components in A. nidulans and, perhaps, with only small modifications, in other organisms as well. As previously postulated (Moritz et al., 2000; Keating and Borisy, 2000), GCP4-GCP6 homologs form the `cap' of the large γ-TuC. They promote the assembly of the large complex and in many organisms might be required for its assembly. The γ-TuSC has an intrinsic tendency to associate laterally with other γ-TuSCs (Kollman et al., 2008) and the formation of the large complex might involve interactions between γ-TuSCs as well as interactions of γ-TuSCs with GCP4-GCP6 homologs. The large γ-TuC binds to the SPB, with the γ-TuSC being the site of binding to the SPB. Like the centrosome, the inner face of the A. nidulans SPB is fibrous (supplementary material Fig. S9) and this fibrous material could interact laterally with γ-TuSCs. The large γ-TuC can nucleate microtubule assembly efficiently. In the absence of GCP4-GCP6 homologs, the large complex does not form in the cytoplasm, or forms inefficiently, but the γ-TuSCs can still bind to the SPB and nucleate microtubule assembly. γ-TuSCs nucleate microtubule assembly in vitro very inefficiently (Oegema et al., 1999) and it is probable that they need to interact with SPB or centrosomal proteins and with each other to form complexes that nucleate microtubule assembly efficiently (Kollman et al., 2008). The complexes do not necessarily need to contain as many γ-TuSCs as the normal, large γ-TuC to nucleate microtubule assembly [see Job et al. (Job et al., 2003) for a discussion of the size of microtubule nucleation complexes]. The spindles that assemble in the absence of GCP4-GCP6 homologs are functionally deficient (perhaps there are fewer microtubules than normal or the microtubules have abnormal numbers of protofilaments) but the problems with the spindles are normally corrected if the SAC mechanism is in place.
Materials and Methods
Strains
A. nidulans strains used in this study are listed in supplementary material Table S1.
Gene targeting and transformation
Gene targeting was achieved by transforming with linear DNA molecules that consisted of a selection cassette flanked by two fragments amplified from A. nidulans genomic DNA. The molecules were generated by fusion PCR as previously described (Nayak et al., 2006; Szewczyk et al., 2006). To tag GCPs with a fluorescent protein at the C-terminus, the flanking DNAs were from the coding sequence of the genes encoding GCPs about 500-1000 bp upstream of the stop codon and a similar sized fragment from the 3′ untranslated region. In each case, the central region was a cassette containing a glycine alanine linker (Yang et al., 2004), the fluorescent protein coding sequence and the gene encoding the selectable marker. The GFP–α-tubulin fusion consisted of a selectable marker (A. fumigatus pyroA) followed by the normal tubA (α-tubulin) promoter, followed by the GFP coding sequence fused in frame to a glycine alanine linker that was, in turn, fused to the tubA coding sequence. This construct was integrated at the wA locus. To delete the GCP-encoding genes, the flanking DNAs were from the 5′ untranslated region and 3′ untranslated region. The central cassette was simply the selectable marker gene, A. fumigatus pyrG (AfpyrG) (Weidner et al., 1998), A. fumigatus riboB (AfriboB) (Nayak et al., 2006) or A. fumigatus pyroA (AfpyroA) (Nayak et al., 2006). Transformation was carried out as described previously (Szewczyk et al., 2006).
Diagnostic PCR and Southern hybridizations
A. nidulans genomic DNA of the transformants was prepared as described (Lee and Taylor, 1990). Positive transformants were first confirmed through diagnostic PCR by using outside primers. The presence of correct integrations and the absence of extra integrations of transforming DNAs were also verified by Southern hybridizations in dried agarose gels as described previously (Oakley et al., 1987). The full-length transforming DNA fragments were radioactively labeled and used as probes.
Microscopy and imaging
Cells were grown in selective medium at 25°C and observed using four- or eight-chamber Lab-Tek chambered coverglasses (Nalge Nunc International, Naperville, IL). For imaging we used two Olympus IX71 inverted microscopes with mercury light sources, and a Ultraview Vox (Perkin Elmer) spinning disk confocal system mounted on an Olympus IX71 inverted microscope. The two IX71 microscopes were equipped with Prior shutters and filter wheels. One was equipped with a Hamamatsu ORCA ER camera, and the other with a Hamamatsu ORCA ERAG. We used Semrock GFP/DsRed2X2M-B dual band `Sedat' filter sets [459-481 nm bandpass excitation filter for GFP and a 546-566 nm excitation filter for mCherry and tdTomato, dual reflection band dichroic (457-480 nm and 542-565 nm reflection bands, 500-529 and 584-679 transmission bands) and two separate emission filters (499-529 nm for GFP and 580-654 nm for mCherry and tdTomato)]. Images were acquired with an Olympus 60× 1.42 N.A. planopochromatic objective using Slidebook software (Intelligent Imaging Innovations, Denver, CO) or Volocity software (Perkin Elmer) installed on PowerMac computers. Some Z-series stacks were deconvolved using Slidebook software. For time-lapse two-channel imaging of live cells, Z-series stacks were collected at each time point and maximum intensity projections from all time points were combined to generate movies with Slidebook software. With the Ultraview Vox system an Olympus 60× 1.42 planapochromatic objective was used for image acquisition.
Immunoprecipitation, sucrose-gradient sedimentation and western blotting
Cytoplasm extracts of A. nidulans were prepared as described (Akashi et al., 1997) with minimal modification. Conidia were inoculated at a concentration of 2×106 per ml in YG medium and incubated for 20 hours at 37°C, shaking at 140 rpm. The hyphae were then harvested, frozen immediately in liquid nitrogen, and disrupted by cryoimpaction, producing a frozen powder (Smucker and Pfister, 1975). The powder was suspended in extraction buffer [10 mM HEPES pH 7.4, 1.0 mM MgCl2, 0.1 mM CaCl2, 0.1 M KCl, 50 mM sucrose, 0.5 mM PMSF, supplemented with protease inhibitors (Complete Mini EDTA-free, Roche)]. Extracts were clarified by centrifugation at 10,000 g for 15 minutes at 4°C. The supernatants were collected and the protein concentration determined.
For immunoprecipitation, 4 μl of affinity-purified rabbit polyclonal γ-tubulin antibody (Oakley et al., 1990) was added to 1-3 ml of cytoplasm extract and the mixture incubated with rotation at room temperature for 45 minutes. Forty μl of protein-G agarose beads (Pierce) were then added and the mixture incubated with rotation for an additional hour at room temperature. Beads were washed four times with PBS (58 mM Na2HPO4, 17 mM NaH2PO4, 68 mM NaCl) and boiled in Laemmli sample buffer for western blotting.
For sucrose-gradient sedimentation, 300 μl of cytoplasm extract was loaded onto an 11-ml sucrose-density gradient [10-40% w/v sucrose in gradient buffer: 10 mM HEPES pH 7.4, 1.0 mM MgCl2, 0.1 mM CaCl2, 0.1 M KCl) and centrifuged at 288,000 g for 12 hours at 4°C in a Beckman SW41Ti rotor. Fractions of 500 μl were taken from the top of the gradient by keeping a cut pipette tip on the surface and pipetting carefully. All 23 fractions were analyzed by western blotting. Band intensities were quantified using Image J software (NIH, Bethesda, MD).
Electrophoresis was carried out with a BioRad Mini Protean II apparatus and protein transfer for western blotting was carried out with a BioRad Mini Trans-Blot apparatus following the instructions of the manufacturer. The remainder of the western blotting procedure was carried out following the standard protocol provided by the Antibody Protocol Guide from Clontech. The anti-GFP primary antibody was from Clontech (Living Colors A. v. monoclonal antibody JL-8) and the secondary antibody was AlexaFluor 680 goat anti-mouse IgG (H+L) from Invitrogen. The western blots were scanned using an Odessey Infrared Imaging System from LI-COR Biosciences. The band intensity was analyzed using ImageJ.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/22/4218/DC1
We would like to thank Stephen Osmani and Jian Qiu Wu for the use of their spinning disk microscope systems; Tania Nayak, Elizabeth Oakley and Heather Edgerton for help with strains and transformation and Roger Tsien for the gift of plasmids carrying mCherry and tdTomato. Supported by NIH grant GM031837 and by the University of Kansas Endowment. Deposited in PMC for release after 12 months.
References
- Akashi, T., Yoon, Y. and Oakley, B. R. (1997). Characterization of γ-tubulin complexes in Aspergillus nidulans and detection of putative γ-tubulin interacting proteins. Cell Motil. Cytoskel. 37, 149-158. [DOI] [PubMed] [Google Scholar]
- Anders, A., Lourenco, P. C. and Sawin, K. E. (2006). Noncore components of the fission yeast γ-tubulin complex. Mol. Biol. Cell 17, 5075-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa, V., Yamamoto, R. R., Henderson, D. S. and Glover, D. M. (2000). Mutation of a Drosophila γ tubulin ring complex subunit encoded by discs degenerate-4 differentially disrupts centrosomal protein localization. Genes Dev. 14, 3126-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck, A. J. (1992). Sexual and parasexual genetics of Aspergillus species. Biotechnology 23, 3-18. [PubMed] [Google Scholar]
- Colombie, N., Verollet, C., Sampaio, P., Moisand, A., Sunkel, C., Bourbon, H. M., Wright, M. and Raynaud-Messina, B. (2006). The Drosophila γ-tubulin small complex subunit Dgrip84 is required for structural and functional integrity of the spindle apparatus. Mol. Biol. Cell 17, 272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, A., Vardy, L., Garcia, M. A. and Toda, T. (2002). A fourth component of the fission yeast γ-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when γ-tubulin function is compromised. Mol. Biol. Cell 13, 2360-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler, S., Pereira, G., Spang, A., Knop, M., Soues, S., Kilmartin, J. and Schiebel, E. (1996). The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 15, 3899-3911. [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R. N., Martin, O. C., Cao, K., Zhang, L., Dej, K., Iwamatsu, A. and Zheng, Y. (2000). Characterization and reconstitution of Drosophila γ-tubulin ring complex subunits. J. Cell Biol. 151, 1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R. N., Martin, O. C. and Zheng, Y. (2003). Characterization of a new γTuRC subunit with WD repeats. Mol. Biol. Cell 14, 1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak, E., Oegema, K., Kirkham, M., Gonczy, P., Habermann, B. and Hyman, A. A. (2002). The kinetically dominant assembly pathway for centrosomal asters. J. Cell Biol. 157, 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Remy, M. H., Bazin, I., Callebaut, I., Wright, M. and Merdes, A. (2006). NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172, 505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, T. and Oakley, B. R. (2005). The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol. Biol. Cell 16, 918-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio, T., Uzawa, S., Jung, M. K., Oakley, B. R., Tanaka, K. and Yanagida, M. (1991). The fission yeast γ-tubulin is essential for mitosis and is localized at two different microtubule organizing centers. J. Cell Sci. 99, 693-700. [DOI] [PubMed] [Google Scholar]
- Job, D., Valiron, O. and Oakley, B. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15, 111-117. [DOI] [PubMed] [Google Scholar]
- Jung, M. K., Prigozhina, N., Oakley, C. E., Nogales, E. and Oakley, B. R. (2001). Alanine-scanning mutagenesis of Aspergillus γ-tubulin yields diverse and novel phenotypes. Mol. Biol. Cell 12, 2119-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer, E. (1977). Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19, 33-131. [DOI] [PubMed] [Google Scholar]
- Keating, T. J. and Borisy, G. G. (2000). Immunostructural evidence for the template mechanism of microtubule nucleation. Nat. Cell Biol. 2, 352-357. [DOI] [PubMed] [Google Scholar]
- Knop, M. and Schiebel, E. (1997). Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 16, 6985-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Pereira, G., Geissler, S., Grein, K. and Schiebel, E. (1997). The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16, 1550-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, J. M., Zelter, A., Muller, E. G., Fox, B., Rice, L. M., Davis, T. N. and Agard, D. A. (2008). The structure of the γ-tubulin small complex: Implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell 19, 207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzack, S., Rischitor, P. E., Enke, C. and Fischer, R. (2005). The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell 16, 497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. B. and Taylor, J. W. (1990). Isolation of DNA from fungal mycelia and single spores. In PCR Protocols: A Guide to Methods and Applications (eds M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White), pp. 282-287. San Diego: Academic Press, Inc.
- Liu, L. and Wiese, C. (2008). Xenopus NEDD1 is required for microtubule organization in Xenopus egg extracts. J. Cell Sci. 121, 578-589. [DOI] [PubMed] [Google Scholar]
- Luders, J., Patel, U. K. and Stearns, T. (2006). GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137-147. [DOI] [PubMed] [Google Scholar]
- Martin, M. A., Osmani, S. A. and Oakley, B. R. (1997). The role of γ-tubulin in mitotic spindle formation and cell cycle progression in Aspergillus nidulans. J. Cell Sci. 110, 623-633. [DOI] [PubMed] [Google Scholar]
- Meads, T. and Schroer, T. A. (1995). Polarity and nucleation of microtubules in polarized epithelial cells. Cell Motil. Cytoskeleton 32, 273-288. [DOI] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M. B., Sedat, J. W., Alberts, B. and Agard, D. A. (1995). Microtubule nucleation by γ-tubulin containing rings in the centrosome. Nature 378, 638-640. [DOI] [PubMed] [Google Scholar]
- Moritz, M., Zheng, Y., Alberts, B. M. and Oegema, K. (1998). Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 142, 775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M. B., Guenebaut, V., Heuser, J. and Agard, D. A. (2000). Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365-370. [DOI] [PubMed] [Google Scholar]
- Murphy, S. M., Preble, A. M., Patel, U. K., O'Connell, K. L., Dias, D. P., Moritz, M., Agard, D., Stults, J. T. and Stearns, T. (2001). GCP5 and GCP6: Two new members of the human γ-tubulin complex. Mol. Biol. Cell 12, 3340-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak, T., Szewczyk, E., Oakley, C. E., Osmani, A., Ukil, L., Murray, S. L., Hynes, M. J., Osmani, S. A. and Oakley, B. R. (2006). A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172, 1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T., Vinh, D. B. N., Crawford, D. K. and Davis, T. N. (1998). A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast γ-tubulin complex. Mol. Biol. Cell 9, 2201-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. R. and Morris, N. R. (1983). A mutation Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 96, 1155-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. R., Oakley, C. E., Yoon, Y. and Jung, M. K. (1990). γ tubulin is a component of the spindle-pole-body that is essential for microtubule function in Aspergillus nidulans. Cell 61, 1289-1301. [DOI] [PubMed] [Google Scholar]
- Oakley, C. E. and Oakley, B. R. (1989). Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338, 662-664. [DOI] [PubMed] [Google Scholar]
- Oakley, C. E., Weil, C. F., Kretz, P. L. and Oakley, B. R. (1987). Cloning of the riboB locus of Aspergillus nidulans. Gene 53, 293-298. [DOI] [PubMed] [Google Scholar]
- Oegema, K., Wiese, C., Martin, O. C., Milligan, R. A., Iwamatsu, A., Mitchison, T. J. and Zheng, Y. (1999). Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, A. H., Davies, J., Liu, H. L., Nile, A. and Osmani, S. A. (2006a). Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17, 4946-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, A. H., Oakley, B. R. and Osmani, S. A. (2006b). Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 1, 2517-2526. [DOI] [PubMed] [Google Scholar]
- Osmani, S. A., Engle, D. B., Doonan, J. H. and Morris, N. R. (1988). Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell 52, 241-251. [DOI] [PubMed] [Google Scholar]
- Paluh, J. L., Nogales, E., Oakley, B. R., McDonald, K., Pidoux, A. L. and Cande, W. Z. (2000). A mutation in γ-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11, 1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina, N. L., Oakley, C. E., Lewis, A., Nayak, T., Osmani, S. A. and Oakley, B. R. (2004). γ-Tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15, 1374-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina, B. and Merdes, A. (2007). γ-Tubulin complexes and microtubule organization. Curr. Opin. Cell Biol. 19, 24-30. [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina, B., Mazzolini, L., Moisand, A., Cirinesi, A. M. and Wright, M. (2004). Elongation of centriolar microtubule triplets contributes to the formation of the mitotic spindle in γ-tubulin-depleted cells. J. Cell Sci. 117, 5497-5507. [DOI] [PubMed] [Google Scholar]
- Rischitor, P. E., Konzack, S. and Fischer, R. (2004). The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot. Cell 3, 632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, K. and Heath, I. B. (2005). The dynamic behaviour of microtubules and their contributions to hyphal tip growth in Aspergillus nidulans. Microbiology 151, 1543-1555. [DOI] [PubMed] [Google Scholar]
- Schnackenberg, B. J., Khodjakov, A., Rieder, C. L. and Palazzo, R. E. (1998). The disassembly and reassembly of functional centrosomes in vitro. Proc. Natl. Acad. Sci. USA 95, 9295-9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer, F., Luschnig, S., Koch, I. and Nusslein-Volhard, C. (2002). γ-Tubulin37C and γ-tubulin ring complex protein 75 are essential for bicoid RNA localization during drosophila oogenesis. Dev. Cell 3, 685-696. [DOI] [PubMed] [Google Scholar]
- Smucker, R. A. and Pfister, R. M. (1975). Liquid nitrogen cryo-impacting: a new concept for cell disruption. Appl. Microbiol. 30, 445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., Geissler, S., Grein, K. and Schiebel, E. (1996). γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol. 134, 429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, T. and Kirschner, M. (1994). In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell 76, 623-637. [DOI] [PubMed] [Google Scholar]
- Sunkel, C. E., Gomes, R., Sampaio, P., Perdigao, J. and Gonzalez, C. (1995). γ-Tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO J. 14, 28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., Osmani, S. A. and Oakley, B. R. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111-3120. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Yamagiwa, A., Nishimura, T., Mukai, H. and Ono, Y. (2002). Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-tubulin ring complex. Mol. Biol. Cell 13, 3235-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L. and Toda, T. (2000). The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., Tasto, J. J., Feoktistova, A., Jennings, J. L., Link, A. J. and Gould, K. L. (2004). Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Mol. Biol. Cell 15, 2287-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verollet, C., Colombie, N., Daubon, T., Bourbon, H. M., Wright, M. and Raynaud-Messina, B. (2006). Drosophila melanogaster γ-TuRC is dispensable for targeting γ-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172, 517-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh, D. B., Kern, J. W., Hancock, W. O., Howard, J. and Davis, T. N. (2002). Reconstitution and characterization of budding yeast γ-tubulin complex. Mol. Biol. Cell 13, 1144-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, N., Koch, I., Schwarz, H., Schnorrer, F. and Nusslein-Volhard, C. (2006). The γTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963-3972. [DOI] [PubMed] [Google Scholar]
- Weidner, G., d'Enfert, C., Koch, A., Mol, P. C. and Brakhage, A. A. (1998). Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33, 378-385. [DOI] [PubMed] [Google Scholar]
- Wiese, C. and Zheng, Y. (1999). γ-tubulin complexes and their interaction with microtubule-organizing centers. Curr. Opin. Struct. Biol. 9, 250-259. [DOI] [PubMed] [Google Scholar]
- Wiese, C. and Zheng, Y. (2000). A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell. Biol. 2, 358-364. [DOI] [PubMed] [Google Scholar]
- Wiese, C. and Zheng, Y. (2006). Microtubule nucleation: γ-tubulin and beyond. J. Cell Sci. 119, 4143-4153. [DOI] [PubMed] [Google Scholar]
- Yang, L., Ukil, L., Osmani, A., Nahm, F., Davies, J., De Souza, C. P., Dou, X., Perez-Balaguer, A. and Osmani, S. A. (2004). Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3, 1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Keating, T. J., Wilde, A., Borisy, G. G. and Zheng, Y. (2000). The role of Xgrip210 in γ-tubulin ring complex assembly and centrosome recruitment. J. Cell Biol. 151, 1525-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Jung, M. K. and Oakley, B. R. (1991). γ-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell 65, 817-823. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Wong, M. L., Alberts, B. and Mitchison, T. (1995). Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature 378, 578-583. [DOI] [PubMed] [Google Scholar]
- Zimmerman, W. C., Sillibourne, J., Rosa, J. and Doxsey, S. J. (2004). Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.