Abstract
Summary
Studies in model organisms have identified regulatory processes that profoundly influence aging, many of which modulate resistance against environmental or metabolic stresses. In C. elegans the transcription regulator SKN-1 is important for oxidative stress resistance and acts in multiple longevity pathways. SKN-1 is the ortholog of mammalian Nrf proteins, which induce Phase 2 detoxification genes in response to stress. Phase 2 enzymes defend against oxygen radicals and conjugate electrophiles that are produced by Phase 1 detoxification enzymes, which metabolize lipophilic compounds. Here we have used expression profiling to identify genes and processes that are regulated by SKN-1 under normal and stress-response conditions. Under non-stressed conditions SKN-1 upregulates numerous genes involved in detoxification, cellular repair, and other functions, and downregulates a set of genes that reduce stress resistance and lifespan. Many of these genes appear to be direct SKN-1 targets, based upon presence of predicted SKN-binding sites in their promoters. The metalloid sodium arsenite induces skn-1-dependent activation of certain detoxification gene groups, including some that were not SKN-1-upregulated under normal conditions. An organic peroxide also triggers induction of a discrete Phase 2 gene set, but additionally stimulates a broad SKN-1-independent response. We conclude that under normal conditions SKN-1 has a wide range of functions in detoxification and other processes, including modulating mechanisms that reduce lifespan. In response to stress, SKN-1 and other regulators tailor transcription programs to meet the challenge at hand. Our findings reveal striking complexity in SKN-1 functions and the regulation of systemic detoxification defenses.
Keywords: Aging, oxidative stress, detoxification, SKN-1, insulin signaling, C. elegans
Introduction
Living organisms are subjected to stress caused by reactive oxygen species (ROS) or electrophiles that are derived from metabolism of various compounds. Cellular damage caused by oxidative stress has been implicated in conditions that include diabetes, atherosclerosis, many neurodegenerative syndromes, and aging (Droge 2002). It is important to understand how organisms defend themselves against this damage at the systemic level. For example, several transcription factors that promote resistance to free radicals have been associated with extended longevity in model organisms, including worms, flies, and mice (Lithgow & Walker 2002; Kenyon 2005; Guarente 2007; Tullet et al. 2008).
Eukaryotes defend themselves from toxic or reactive compounds through a three-phase detoxification system (Xu et al. 2005; Sarkadi et al. 2006). During Phase 1, lipophilic endobiotics or xenobiotics are solubilized through modification by enzymes such as Cytochrome P450s (CYPs) and short-chain dehydrogenases/reductases (SDRs). This process allows these compounds to be excreted, but may also produce damaging reactive compounds. The Phase 2 enzymes defend cells against such compounds, as well as ROS. They encompass a diverse group of enzymes that metabolize free radicals, repair cellular structures, or directly conjugate xenobiotics and peroxidized lipids, including glutathione-S-transferases (GSTs) and UDP-glucuronosyl/glucosyl transferases (UGTs). In Phase 3, conjugated toxins are pumped out of the cell by ATP-binding cassette (ABC) or other transporters (Sarkadi et al. 2006). How these systems contribute to the functions of different tissues, how they are regulated in the context of an organism, and how this regulation might be adapted to different stress scenarios are all important questions.
Current data suggest that Phase 2 genes may be regulated as a coordinated network. In mammals many Phase 2 genes are induced directly by the Nrf1 and Nrf2 (NF-E2-related factor) proteins (Nguyen et al. 2003; Kobayashi & Yamamoto 2006). In the cell types examined thus far Nrf proteins are predominantly cytoplasmic, but in response to stress they accumulate in nuclei and upregulate Phase 2 gene expression. Mice that lack Nrf2 are sensitive to ROS and other toxic insults, but it is problematic to evaluate how a complete lack of Nrf proteins affects the intact mouse because Nrf1−/−; Nrf2−/− mice embryos die by day 10 (Leung et al. 2003).
Since the Phase 2 network is broadly conserved (Jasper 2008), it is possible to employ simpler model organisms to study its regulation and functions. In the nematode C. elegans the Nrf ortholog SKN-1 inducibly regulates expression of candidate Phase 2 genes in the intestine, the digestive system equivalent, and skn-1 mutants are highly sensitive to oxidative stress (An & Blackwell 2003; An et al. 2005; Inoue et al. 2005). SKN-1 accumulates in intestinal nuclei in response to stress and is inhibited from doing so constitutively by mechanisms that include phosphorylation by glycogen synthase kinase-3 (GSK-3) and the conserved insulin/IGF-1-like signaling (IIS) pathway (An et al. 2005; Tullet et al. 2008). In C. elegans, IIS is initiated by binding of insulin-like peptides to the receptor DAF-2, which leads eventually to activation of the downstream IIS kinases AKT-1/2 and SGK-1. These kinases phosphorylate and inhibit SKN-1 in parallel to the FOXO transcription factor DAF-16 (Tullet, et al., 2008), which regulates genes involved in numerous biological processes, including stress resistance (Murphy et al. 2003; Kenyon & Murphy 2006; Oh et al. 2006; Dong et al. 2007; McElwee et al. 2007; Samuelson et al. 2007). It is still unknown whether SKN-1 might simply regulate a suite of Phase 2 genes, or is involved more broadly in control of stress defense or other genes.
Multiple lines of evidence implicate SKN-1 in C. elegans longevity. For example, reductions in IIS delay aging and increase stress resistance in diverse organisms (Kenyon 2005). While it is well established that in C. elegans these benefits of reduced IIS require DAF-16, it has been shown recently that SKN-1 also contributes to these effects (Tullet et al. 2008). In addition, SKN-1 delays aging under normal conditions, at least in part through its action in the intestine (An & Blackwell 2003; Tullet et al. 2008). Finally, skn-1 is required for lifespan extension by calorie restriction (CR), a condition that promotes longevity in all eukaryotes tested thus far (Bishop & Guarente 2007b). This last SKN-1 function is mediated by its expression in the two ASI neurons (Bishop & Guarente 2007b), which sense or regulate food intake (You et al. 2008). These observations indicate that SKN-1 has important functions under non-stressed as well as stress conditions. It remains to be determined whether SKN-1 regulates similar sets of genes under normal and stress-response conditions, and how these genes influence stress resistance and longevity.
Here we have used expression profiling to investigate how SKN-1 influences C. elegans gene expression under normal conditions, and in response to two different sources of oxidative stress, the metalloid sodium arsenite (As) and tert-butyl hydrogen peroxide (t-BOOH). Arsenite is a highly toxic trivalent form of the environmentally pervasive metalloid arsenic. It attacks thiol groups on glutathione and other polypeptides, and stimulates ROS production (Hughes 2002). The stable organoperoxide t-BOOH attacks cellular proteins and lipids, and is also scavenged by glutathione (Mathews et al. 1994). skn-1 mutants are sensitive to each of these stresses (An et al. 2005; Inoue et al. 2005). We find that under normal conditions SKN-1 regulates expression of numerous genes, many of which may be direct targets. These genes are involved in processes that include detoxification and stress resistance, lysosome and proteasome function, metabolism, and cell-surface recognition. Interestingly, SKN-1 also suppresses expression of many genes that decrease stress resistance and lifespan, including the insulin-like peptide ins-7 and the IIS pathway kinase pdk-1. Treatment with As results in activation of a particular group of SKN-1 dependent detoxification genes. By contrast, t-BOOH treatment also mobilizes a broad SKN-1-independent stress response. Some functional clusters of genes are regulated by SKN-1 specifically under normal or particular stress conditions, indicating that unknown signals interact with SKN-1 to restrict its activities. Our findings identify a complex set of processes that are regulated by SKN-1 under normal conditions, and reveal that SKN-1 acts together with other regulators in specialized responses to exogenous stresses.
Results
We used oligonucleotide microarrays to compare expression profiles of worms that had been treated with RNA interference (RNAi) against skn-1 (skn-1(−)), or control (gfp) RNAi (skn-1(+)) from hatching (Fig. 1). We examined synchronized L4 stage larvae, in which stress robustly induces intestinal expression of the SKN-1 target gene gcs-1 (An & Blackwell 2003). To investigate how SKN-1 responds to stress, worms were exposed to arsenite (As) or tert-Butyl hydroperoxide (t-BOOH) or incubated under the corresponding control conditions (NGM agar plates or M9 liquid media, respectively). In applying stress-inducing agents, we titrated the concentrations used and the time of exposure so that gcs-1 was induced comparably (Fig. S1), and at least 95% of the animals consistently survived the treatment (not shown).
Fig. 1. Identification of SKN-1-regulated genes.
mRNA samples were generated under the indicated conditions, with skn-1(−) referring to skn-1 RNAi and skn-1(+) to RNAi control. Pairs of samples designated by arrows were compared on Agilent 44X oligonucleotide microarrays to identify genes that are regulated by SKN-1 under normal conditions (red arrow), and in response to treatment with Arsenite (As) (dark blue arrows), or tert-Butyl hydroperoxide (t-BOOH) (teal arrows). Genes that are regulated by SKN-1 under normal conditions were identified by both SAM and hierarchical clustering (red arrow). skn-1-dependent and –independent genes that respond to As or t-BOOH stress were identified by hierarchical clustering. While the As-induced response seemed to be entirely dependent upon skn-1, t-BOOH induced both skn-1-dependent and independent gene sets (see text). Some gene categories that we identified as being prominent in stress- and SKN-1-upregulated gene sets are listed in bold, with those that were overrepresented in only one or two sets indicated in italics.
SKN-1 regulates stress-related and other genes under normal conditions
We first searched for genes that are regulated by SKN-1 under normal (non-stressed) conditions, by comparing the expression profiles of skn-1(+) and skn-1(−) wild-type (N2) animals that served as controls for our stress-treatment experiments (Fig. 1, red arrow). We compared these seven sets of skn-1(+) and skn-1(−) samples using hierarchical clustering (Eisen et al. 1998) and statistical analysis of microarrays (SAM) (Tusher et al. 2001). In performing SAM we adjusted the delta value to 1.023, resulting in an expected false positives rate of 1.7%. These analyses identified 233 genes for which expression was significantly reduced in skn-1(−) animals (SKN-1-upregulated genes; Fig. 2A; Table 1, S1). The extent of this SKN-1-upregulated profile was unexpected, because under non-stressed conditions SKN-1 is seen at relatively low levels in intestinal nuclei (An & Blackwell 2003). We also identified 63 genes for which expression was increased in skn-1(−) animals, indicating that they are downregulated by SKN-1 (SKN-1-downregulated genes; Fig. 3A; Table 2, S2). As an independent test of these results, we assessed the relative levels of representative SKN-1-regulated mRNAs in skn-1(+) and skn-1(−) animals using quantitative (q)RT-PCR. These results concurred with our microarray experiments for both SKN-1-upregulated (13 of 13) and -downregulated (10 of 10) genes (Table S3, S4).
Fig. 2. Genes that are up-regulated by SKN-1 under normal conditions.
(A) Hierarchical clustering of gfp control (skn-1(+)) and skn-1(−) RNAi samples that were analyzed on microarrays (7 sample sets in total). Incubation conditions under which these samples were obtained are indicated. A representative subset of SKN-1-upregulated genes is shown. (B) Representation of functional group categories among the 233 SKN-1-upregulated genes that were identified by hierarchical clustering and SAM (Table S1). Genes were classified according to their molecular or biological function, based upon GO terms. The CUB/CUB-like group was classified by presence of these motifs (Blanc et al. 2007). The following broad categories were created by combining GO-terms: Detoxification/Stress response, Defense/Immunity, Signaling/Transcriptional regulation, and Protein folding/degradation. (C) Enrichment of SKN-1-binding motifs at SKN-1-upregulated genes. RSAT and Weeder Web were used to identify novel sequence motifs that are overrepresented in the predicted promoters of SKN-1-up-regulated genes, as defined by the 2 Kb or less of intergenic sequence upstream of each ORF. The consensus identified by Weeder Web is represented by WebLogo (Crooks et al. 2004). (D) Enrichment of functional gene categories among SKN-1-upregulated genes, compared to a set of Nrf2-upregulated genes. Highly represented GO terms are graphed for SKN-1-upregulated genes, and for Nrf2-dependent genes that were identified by expression profiling of primary cortical astrocytes from Nrf2 −/− and Nrf2 +/+ mice under non-stressed conditions (Lee et al. 2003a).
Table 1. SKN-1-upregulated genes identified under non-stressed conditions.
The top 35 genes for which expression was differentially decreased in skn-1(RNAi) animals under non-stressed conditions (SKN-1-upregulated genes), as ranked by SAM score. These and the other SKN-1-upregulated genes we identified (233 total) are listed by functional group in Table S1.
| Sequence Name | Gene Name | KOG Title or Protein Domain | Score |
|---|---|---|---|
| K08F4.7 | gst-4 | Glutathione S-transferase | 15.351 |
| C32H11.4 | Cub-like domain/Cub-like region | 8.778 | |
| C32H11.12 | dod-24 | Cub-like domain/Cub-like region | 8.711 |
| ZK1058.6 | nit-1 | Carbon-nitrogen hydrolase | 8.506 |
| Y45G12C.2 | gst-10 | Glutathione S-transferase | 7.765 |
| K10D11.1 | dod-17 | Cub-like domain/Cub-like region | 7.664 |
| C32H11.3 | Cub-like domain/Cub-like region | 7.342 | |
| Y102A11A.3 | RNAi causes Ste and Sck | 7.268 | |
| T26C5.1 | gst-13 | Glutathione S-transferase | 7.087 |
| F56D5.3 | NADH:flavin oxidoreductase/12-oxophytodienoate reductase | 6.667 | |
| F56A4.4 | Glutathione S-transferase | 6.551 | |
| K10C2.3 | Aspartyl protease | 6.349 | |
| F23B2.12 | pcp-2 | Hydrolytic enzymes of the alpha/beta hydrolase fold | 6.166 |
| F55G11.2 | Cub-like domain/Cub-like region | 6.153 | |
| C35B1.5 | Thioredoxin, nucleoredoxin and related proteins | 5.811 | |
| ZK896.4 | Cub-like domain/Cub-like region | 5.625 | |
| T25B6.2 | M13 family peptidase, neprilysin, metallopeptidase | 5.568 | |
| C55A6.7 | Predicted short chain-type dehydrogenase | 5.455 | |
| C09B8.4 | Protein of unknown function DUF829 | 5.444 | |
| ZC443.6 | ugt-16 | UDP-glucuronosyl and UDP-glucosyl transferase | 5.366 |
| H20E11.3 | Cub-like domain/Cub-like region | 5.354 | |
| F14D7.6 | Predicted transporter/transmembrane protein | 5.331 | |
| K11H12.4 | Protein of unknown function DUF274 | 5.087 | |
| Y32F6A.5 | Serine carboxypeptidases | 5.028 | |
| K04A8.10 | UDP-glucuronosyl and UDP-glucosyl transferase | 4.922 | |
| B0041.6 | ptps-1 | 6-pyruvoyl tetrahydrobiopterin synthase | 4.906 |
| K10H10.2 | Cystathionine beta-synthase and related enzymes | 4.905 | |
| F32G8.6 | cat-4 | GTP cyclohydrolase I | 4.814 |
| F01D5.3 | Secreted surface protein | 4.602 | |
| C07D8.6 | Aldo/keto reductase family proteins | 4.545 | |
| C02D5.3 | Glutathione S-transferase | 4.471 | |
| F35E8.8 | gst-38 | Glutathione S-transferase | 4.434 |
| M03F8.4 | Protein of unknown function DUF23 | 4.411 | |
| F 08G5.6 | Cub-like domain/Cub-like region | 4.344 |
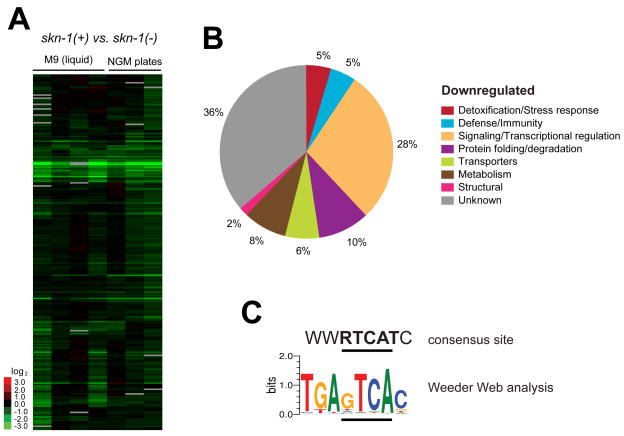
Fig. 3. Genes that are down-regulated by SKN-1 under normal conditions.
(A) Hierarchical clustering (pictured) and SAM analysis of 7 sample sets identified a set of 63 SKN-1-downregulated genes (Table S2). (B) Representation of functional gene groups among the SKN-1-downregulated genes, analyzed as in Fig. 2B. (C) Enrichment of SKN-1 binding motifs in SKN-1-downregulated genes, analyzed as in Fig. 2C.
Table 2. SKN-1-downregulated genes identified under non-stressed conditions.
The top 35 genes for which expression was differentially increased in skn-1(RNAi) animals under non-stressed conditions (SKN-1-downregulated genes), as ranked by SAM score. These and the other SKN-1-downregulated genes we identified (63 total) are listed by functional group in Table S2.
| Sequence Name | Gene Name | KOG Title or Protein Domain | Score |
|---|---|---|---|
| F42G2.4 | fbxa-182 | Protein containing an F-box motif | −9.335 |
| T26F2.2 | Uncharacterized protein | −6.858 | |
| C31B8.4 | Uncharacterized protein | −6.611 | |
| Y39B6A.24 | Aspartyl protease | −6.555 | |
| ZC196.4 | Protein of unknown function DUF713 | −6.389 | |
| C17H1.7 | Uncharacterized protein | −6.218 | |
| Y47H9C.1 | Protein of unknown function DUF274 | −6.137 | |
| C06E1.1 | Protein involved in membrane traffic (YOP1/TB2/DP1/HVA22 family) | −6.065 | |
| Y69A2AR.12 | Uncharacterized protein | −5.562 | |
| F15B9.6 | Uncharacterized protein | −5.402 | |
| Y75B8A.32 | Predicted DNA-binding protein | −5.093 | |
| F15D3.8 | Uncharacterized protein | −5.031 | |
| F47H4.8 | fbxa-188 | Protein containing an F-box motif | −5.009 |
| M01G12.12 | rrf-2 | Putative RNA-directed RNA polymerase QDE-1 required for posttranscriptional gene silencing and RNAi | −4.913 |
| K08H10.1 | lea-1 | Protein that is predicted to be hydrophilic and heat-resistant | −4.785 |
| Y46G5A.20 | Uncharacterized protein | −4.639 | |
| Y6E2A.4 | Protein of unknown function DUF713 | −4.611 | |
| Y41C4A.11 | Vesicle coat complex COPI, beta’ subunit | −4.424 | |
| F45E4.1 | arf-1.1 | GTP-binding ADP-ribosylation factor Arf1 | −4.408 |
| F47H4.10 | skr-5 | SCF ubiquitin ligase, Skp1 component | −4.021 |
| C54G6.5 | spp-17 | Saposin-like Protein family | −4.009 |
| Y43C5A.3 | Uncharacterized protein | −3.989 | |
| B0024.4 | Protein of unknown function DUF274 | −3.929 | |
| W03D2.6 | C-type lectin | −3.826 | |
| F58B3.3 | lys-6 | N-acetylmuraminidase/lysozyme | −3.758 |
| Y51B9A.9 | Jun-N-terminal kinase (JNK) | −3.546 | |
| C06E4.8 | Uncharacterized protein | −3.527 | |
| F23F12.3 | Synaptic vesicle transporter SVOP and related transporters (major facilitator superfamily) | −3.474 | |
| F25B3.5 | Uncharacterized protein | −3.464 | |
| F58F9.7 | Pristanoyl-CoA/acyl-CoA oxidase | −3.452 | |
| C01G6.7 | Acyl-CoA synthetase | −3.428 | |
| ZK1251.2 | ins-7 | Insulin-like peptides | −3.423 |
| C45B11.3 | dhs-18 | Reductases with broad range of substrate specificities | −3.295 |
| F02C12.5 | cyp-13B1 | Cytochrome P450 CYP3/CYP5/CYP6/CYP9 subfamilies | −3.276 |
| C04F12.1 | Translation initiation factor 2C (eIF-2C) and related proteins | −3.263 |
We investigated the functions of SKN-1-upregulated genes by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) to analyze the statistical representation of functional gene categories, as defined by gene ontology (GO) terms or predicted protein domains. The GO categories for glutathione transferase, xenobiotic metabolism, thioredoxin fold, and UDP glucuronosyl transferase (UGT) were highly overrepresented among the SKN-1-upregulated genes (Fig. 2D), 19% of which overall are involved in detoxification or stress responses (Fig. 2B; Table S1). In addition, numerous Phase 2 genes were among the SKN-1-upregulated genes with the highest SAM scores, including the known SKN-1 target gst-4 (Kahn et al. 2008; Tullet et al. 2008), other GSTs (gst-10, -13, -38, F56A4.4, C02D5.3), and UGTs (ugt-16, K04A8.10)(Table 1). The SKN-1-upregulated genes also included some known or predicted Phase 1 detoxification genes (aldo/keto reductase proteins (C07D8.6, T08H10.1), a cytochrome P450 (CYP) (cyp-14A.1), and multiple short-chain dehydrogenases (SDRs) (dhs-8, C55A6.6, C55A6.7, F20G2.1, F20G2.2, R08H2.1, F25D1.5), Table S1)(McElwee, et al., 2007). In addition, short chain dehydrogenase was one of the most highly enriched GO terms in this gene set (Fig. 2D). We conclude that SKN-1 not only controls expression of numerous Phase 2 detoxification genes, but also upregulates some Phase 1 genes.
The SKN-1-upregulated genes also included gene groups that represent other biological processes, some of which are stress-related. We identified numerous genes involved in protein folding or degradation, some of which have lysosomal functions (Fig. 2B; Table S1). Among the latter group were vacuolar H+ATPases that translocate protons into lysosomes and other organelles (vha-2, vha-6, vha-8, vha-16 and vha-17), serine carboxypeptidases, and the ortholog of LYST (T01H10.8), which has been implicated in the lysosomal storage disease Chediak–Higashi syndrome (Kaplan et al. 2008). We also identified many genes that encode transporters for metals, small molecules, ions, or water (Fig. 2B; Table S1). Many SKN-1-upregulated genes are involved in metabolic processes (Fig. 2B, 2D; Table S1). These include cystathionine beta-synthase and cystathionine beta-lyases, which convert homocysteine to cysteine, a precursor of glutathione (Banerjee & Zou 2005). Two prominent groups of SKN-1-upregulated genes are involved in cell surface processes (C-type lectins and CUB-like domain proteins)(Fig. 2B, 2D; Tables 1 and S1). In other species the CUB domain has been implicated in cell-surface functions such as complement activation, tissue repair, axon guidance, inflammation, and receptor-mediated endocytosis (Blanc et al. 2007).
SKN-1 binds preferentially in vitro to the consensus WWTRTCAT (W=A/T, R=G/A), and upregulates gcs-1 by interacting with this motif (Blackwell et al. 1994; An & Blackwell 2003). The underlined RTCAT motif is most critical for binding affinity and specificity, because SKN-1 directly contacts the RTCA base pairs in the major groove (Rupert et al. 1998; Kophengnavong et al. 1999). The WWTRTCAT motif should occur randomly only once every 2048 bp in the genome, but 49% of the SKN-1-upregulated genes (115/233) contained 2–6 copies of this element within their putative promoters, as defined by 5′ intergenic sequence up to 2 kb (Table S1; Fig. S2). An analysis of SKN-1-upregulated gene promoters for novel overrepresented sequences identified a motif that is similar to the canonical SKN-1 consensus (TTDTCATC, (D=A/G/T); Fig. 2C; see Experimental Procedures), and in many instances corresponds to the same element within these putative promoters (data not shown). This novel motif, which is more restrictive than the WWTRTCAT consensus (1/10923 bp randomly), was present in 110 (47%) of the SKN-1-upregulated gene promoters (≤ 2 kb) (Table S1; Fig. S2). Taken together, the data indicate that SKN-1 may directly control the expression of many of the SKN-1-upregulated genes we identified.
The functional parallels between SKN-1 and mammalian Nrf proteins (An & Blackwell 2003) predicts that these proteins should regulate similar categories of genes. Accordingly, many of the GO terms that we identified in SKN-1-upregulated genes were also enriched in a set of Nrf2-dependent genes that were identified in murine primary cortical astrocytes (Lee et al. 2003a), including glutathione transferase, xenobiotic metabolism, and other stress response groups (Fig. 2D). In addition, UGTs were identified in two sets of stress-induced Nrf2-dependent genes (Kwak et al. 2003; Lee et al. 2003a). Interestingly, the SKN-1- and Nrf2-upregulated genes also included some GO term groups that did not overlap (Fig. 2D). Apparent C. elegans homologs exist for more than half of the Nrf2-regulated genes in the GO terms that were unique to Nrf2 (not shown), suggesting that their lack of detectable regulation by SKN-1 might derive from tissue- or organism-specific differences.
In contrast to the SKN-1-upregulated genes, no particular biological function predominated among the 63 genes that were down-regulated by SKN-1 under normal conditions (Fig. 3B; Table S2). Two of these genes function in the IIS pathway. ins-7 encodes an insulin-like peptide and DAF-2 agonist (Murphy et al. 2003), and pdk-1 encodes a kinase that functions downstream of DAF-2 to activate the AKT-1/2 and SGK-1 kinases, which inhibit DAF-16 and SKN-1 through phosphorylation (Paradis & Ruvkun 1998; Hertweck et al. 2004; Tullet et al. 2008). Other SKN-1-downregulated genes encode regulatory proteins that function in signaling, ubiquitin-mediated proteolysis, or gene regulation, including four that may be involved in RNAi (a putative RNA-directed RNA polymerase (RRF-2) and the Argonaute-related proteins PPW-1, SAGO-2, and C04F12.1 (Yigit et al. 2006)). Of 14 genes that had been identified as downregulated by Nrf2 in cortical astrocytes under normal conditions (Lee et al. 2003a), only 4 have apparent C. elegans homologs (not shown). The SKN-1-downregulated genes did not include any of these 4 genes, a group that is too small for a conclusive comparison.
Multiple copies of the canonical in vitro SKN-1 binding site WWTRTCAT were present within predicted promoters at 17 SKN-1-downregulated genes (27%), including ins-7 (Fig. S2; Table S2), suggesting that many of these genes might be repressed directly by SKN-1. In addition, within these 63 promoters we identified the novel motif TGAGTCAC (Fig. 3C), which may be a variant of the canonical motif. Interestingly, only 10 of 233 SKN-1-upregulated genes (4.3%) displayed this new motif, compared to 31% of the SKN-1 downregulated genes (Fig. S2, Table S2), suggesting that it might mediate transcriptional inhibition by SKN-1.
SKN-1 mediates the transcriptional response to Arsenite (As)
We next examined how SKN-1 contributes to stress responses, first by investigating its role in the As response (Fig. 1, dark blue arrows). Hierarchical clustering identified 118 genes that are upregulated in a skn-1-dependent manner upon As exposure (As/SKN-1-dependent genes), but did not detect any genes that were down-regulated by SKN-1 in response to As, or induced by As independently of SKN-1 (Fig. 4A; Table S5). A qRT-PCR analysis confirmed the skn-1-dependence of a set of our As/SKN-1-dependent genes, supporting the microarray data (Table S6). An analysis of the predicted promoters of these As/SKN-1-dependent genes for novel shared motifs identified essentially the same elements we had earlier detected in the SKN-1-upregulated gene promoters (Fig. 2C, 4B; Table S5), suggesting that a high proportion of these genes are likely to be direct SKN-1 targets. The bulk of the transcriptional response to As therefore appears to be mobilized by SKN-1.
Fig. 4. SKN-1 regulation of overlapping gene groups under normal and Arsenite stress conditions.
(A) Hierarchical clustering of genes that are differentially regulated in response to As treatment (4 sample sets, see Experimental Procedures). Genes were identified that are As-upregulated and skn-1-dependent, but none were identified that are As-upregulated and skn-1-independent. A subset of the genes identified by hierarchical clustering is shown. (B) Enrichment of SKN-1-binding motifs in SKN-1-downregulated genes, analyzed as in Fig. 2C. (C) Venn diagram showing overlap among genes that were upregulated by SKN-1 under normal and As stress conditions. (D) Comparison of SKN-1-upregulated genes identified under normal, As-treatment, and t-BOOH-treatment conditions, grouped by GO terms. Note that some GO terms are overrepresented among only one or two of these gene groups.
When we compared the As/SKN-1-dependent genes to the SKN-1-upregulated genes we had identified under normal conditions, we found that the majority of the As transcriptional response (83 genes) was common to both sets (Fig. 4C). These gene sets also shared many prominent GO terms, including glutathione transferase, thioredoxin fold, lyase activity, and short-chain dehydrogenase (Fig. 4D). The SKN-1-upregulated genes that were identified under normal and As-induction conditions also differed in important respects. Within the GO terms that these gene sets had in common, the As/SKN-1-dependent set included potentially important As-specific genes such as hmt-1, an ABC-type transporter that is critical for C. elegans heavy metal tolerance (Vatamaniuk et al. 2005). In addition, the As/SKN-1-dependent genes included some new GO terms, such as Alcohol dehydrogenase, as well as a new set of genes involved in glutathione synthesis (Table S5). Importantly, these As-induced genes also lacked some GO terms that were prominent among the SKN-1-upregulated genes we had identified under normal conditions (i.e. UGTs and C-type lectins)(Fig. 4D). We conclude that the response to As does not consist of simply a broad induction of the genes that are upregulated by SKN-1 in the absence of stress, but instead involves induction of particular sets of those genes, along with additional targets.
We also used hierarchical clustering across the As-treated SKN-1 (+) and (−) samples and their controls (dark blue arrows, Fig. 1) to identify genes that were upregulated by SKN-1 under both normal and As-exposed conditions, but were not upregulated by As treatment (As-independent; Table S7). As would be predicted, these new genes did not include any of our As-/SKN-1-dependent genes, and included few or no UGTs, C-type lectins, or alcohol dehydrogenases, groups that were prominent among SKN-1-upregulated genes under either normal or As-treated conditions, but not both (Fig. 4C; Tables S1, S5, S7). This further supports the idea that As treatment activates distinct subsets of SKN-1 target genes. In addition, by analyzing samples obtained under non-stressed and As-treatment conditions simultaneously, this analysis detected many SKN-1-regulated genes that we had not identified in our study of skn-1(+) and skn-1(−) control samples only, including 15 genes that encode proteasome subunits (Table S7). Proteasome genes have been implicated as Nrf2 targets (Kwak et al. 2003), suggesting that the regulation of these genes by SKN-1/Nrf proteins is conserved.
SKN-1-dependent and -independent responses to tert-Butyl hydroperoxide (t-BOOH)
To test further the notion that SKN-1 upregulates particular subsets of its target genes in response to stress, we investigated how SKN-1 contributes to the transcriptional response to t-BOOH treatment, again using hierarchical clustering (Fig. 1, teal arrows; Fig. 5A). Here, in striking contrast to the effects of As treatment, we observed that t-BOOH not only upregulates a set of SKN-1-dependent genes, but also induces a broad SKN-independent response (Fig. 5B; Tables S8, S9, S10).
Fig. 5. SKN-1-dependent and -independent responses to an organoperoxide.
(A) t-BOOH treatment affects regulation of skn-1-dependent and skn-1-independent gene programs. Hierarchical clustering identified genes that are up- or down-regulated in response to t-BOOH treatment, and unaffected by skn-1 RNAi (skn-1(−)) (SKN-1-independent genes). A subset of the genes identified from 3 sample sets by hierarchical clustering is shown, along with motifs that were identified as being overrepresented in their predicted upstream promoters (determined as in Fig. 2C). NGM corresponds to normal conditions (see Experimental Procedures). (B) Venn diagram of genes that were upregulated by SKN-1 under normal conditions and t-BOOH treatment. (C) Genes that were upregulated by t-BOOH treatment (Tables S8, S9), graphed as in Fig. 4D. GO terms that are overrepresented among t-BOOH-induced SKN-1-upregulated and SKN-1-independent genes are compared.
Only a minority of the t-BOOH response appeared to require skn-1, as skn-1 RNAi impaired induction of only 64 (22%) of the 285 t-BOOH-upregulated genes we detected. Interestingly, we had previously identified only 12 (19%) of the SKN-1-dependent t-BOOH-induced genes as being SKN-1-upregulated under normal conditions (Fig. 5B). The other 52 SKN-1-dependent t-BOOH-induced genes encompassed some gene classes or GO terms that were not detected under either normal or As-induced conditions (i.e. BTB/POZ-like, casein kinase, hydrolase activity)(Table S1, S5, S8; Fig. 5C). Analysis of the t-BOOH-induced SKN-1-dependent gene promoters for novel motifs identified an element that is consistent with the SKN-1 in vitro consensus and was not overrepresented at SKN-1-independent t-BOOH-induced genes (TKTCATCA, Fig. 5A), suggesting that many of these genes might be direct SKN-1 targets.
We identified a much larger number of genes that were up- or down-regulated by t-BOOH under both control and skn-1 RNAi conditions (referred to as SKN-1-independent). 109 genes were downregulated by t-BOOH, including many metabolism genes (Table S10). Importantly, the 221 genes that were upregulated by t-BOOH independently of skn-1 encompassed many functional groups that were not prominent in the As-induced set. They included a greater number of Phase 1 detoxification genes (Cytochrome P450 enzymes and other monooxygenases), nuclear hormone receptors, additional signaling or transcription regulators, and many lipid metabolism genes (Fig. 5C; Table S9).
It is unlikely that the striking differences between the As and t-BOOH responses simply reflect different stress “levels,” because our stress conditions were adjusted to comparable gcs-1 induction and resulted in only a residual frequency of death (Fig. S1; see Experimental Procedures). Furthermore, under conditions where As treatment resulted in substantially greater toxicity than t-BOOH, a representative SKN-1-independent, t-BOOH-upregulated gene (fmo-2) was induced by t-BOOH but not As (Fig. S3). A qRT-PCR analysis showed that many t-BOOH-upregulated genes were induced more robustly in a predicted null skn-1 mutant than in N2, thereby confirming their independence from skn-1 and suggesting that their induction is stronger when SKN-1-mediated stress defenses are impaired (Fig. S4). Surprisingly, we had previously identified some SKN-1-independent t-BOOH-upregulated genes as being SKN-1-dependent under normal or As-induction conditions (including gst-14 and gst-39; Table S1, S5, S9). A qRT-PCR analysis confirmed that gst-14 and gst-39 were induced by t-BOOH in the absence of SKN-1, although their induction was more robust in N2 (Fig. S4).
Taken together, the data demonstrate that the organismal transcriptional response to t-BOOH is more complex than the As response. The t-BOOH response involves induction of particular Phase 2 genes by SKN-1, along with a broad skn-1-independent response that includes upregulation of some Phase 2 genes that were skn-1-dependent under As-induction or normal conditions.
SKN-1 downregulates genes that decrease stress resistance or lifespan
The SKN-1-regulated gene profiles that we identified under normal and stress conditions were surprisingly complex, suggesting that SKN-1 not only responds acutely to stress, but also may regulate many genes under normal conditions that could be important for stress resistance and longevity. To test this idea, we investigated how genes that are regulated by SKN-1 under normal conditions influence the organism’s capacity for stress resistance. We first examined how six SKN-1-upregulated genes affect As resistance, by inhibiting their expression using RNAi (Fig. 6A). These genes were selected from among those that showed the most statistical significance by SAM (Table 1). They each encoded known stress-defense enzymes, with the exception of the CUB-like genes C32H11.3 and C32H11.4. RNAi of each gene that we tested decreased As resistance but did not impair movement or fertility of control animals (Fig. 6A; not shown), suggesting that many SKN-1-upregulated genes contribute to stress resistance. In no case did knockdown of these genes compromise As resistance comparably to skn-1 RNAi (Fig. 6A), consistent with the idea that SKN-1 coordinates many defense mechanisms.
Fig. 6. SKN-1-regulated genes influence oxidative stress resistance and lifespan.
(A) Many SKN-1-upregulated genes promote oxidative stress resistance. SKN-1-upregulated genes (Table S1) were knocked down by RNAi, then survival of young adults (8–9 hr) was assayed at the indicated times after introduction into 4 mM As. A representative experiment is shown in which 5 wells of 10 worms each were examined. Error bars indicate the SEM, and p values (Student’s t-test) indicate comparison to control RNAi. *p ≤ 0.0008; **p ≤ 0.008 (Student’s t-test). (B) Many SKN-1-downregulated genes reduce oxidative stress resistance. Resistance to As was analyzed after RNAi of the indicated genes (Table S2) as in (A). Other experiments and analyses of additional genes are described in (C) and Fig. S5. *p ≤ 0.0008; **p ≤ 0.008 (Student’s t-test). (C) Analysis of As resistance in young adults (2–6 hr). Experimental and control RNAi worms were placed in 5mM As, then the fraction surviving was counted 16, 24, and 40 hrs later. Results are presented as a graph from which we calculated the approximate fraction of animals in each set that were alive when 20% of the control animals were still surviving (black vertical line). A comparison of this fraction to control is plotted in Fig. S5. 6 samples of 10 worms each were examined for every condition. p-value of fraction alive compared to control at 20% control survival is < 0.05 for all genes shown (Student’s t-test performed across samples). Error bars = SEM. (D) Many SKN-1-downregulated genes decrease lifespan. A set of SKN-1-downregulated genes was analyzed for effects on longevity using a feeding RNAi longevity assay in RNAi-sensitive rrf-3(pk1426) worms at 20°C. Genes for which RNAi extended lifespan significantly in 3/3 trials (p<0.01, log-rank) are diagrammed, with data from a single trial shown (Table S11B, Experiment 2). Control is empty RNAi feeding vector L4440. Data and statistical analyses for all experiments and genes tested are provided in Table S11A-D.
We also asked how 22 of the SKN-1-downregulated genes affect stress resistance. One possible model is that these genes might protect against stress under normal conditions, and were upregulated after skn-1 RNAi as a secondary defensive response to stress resulting from SKN-1 loss. Alternatively, if these SKN-1-downregulated genes are actively repressed by SKN-1, through SKN-1 either acting directly at their promoters or triggering a repressive signal, they might be predicted to decrease stress resistance. Consistent with the latter model, the SKN-1-downregulated genes pdk-1 and ins-7 (Table S2) each act in the IIS pathway to decrease lifespan (Paradis & Ruvkun 1998; Murphy et al. 2003). These examples suggested that other SKN-1-downregulated genes might also decrease stress resistance or lifespan. Accordingly, As resistance was increased consistently in young adult animals after RNAi knockdown of 15 of 22 SKN-1-downregulated genes that we tested, including ins-7 (Fig. 6B, 6C, S5). The observation that these SKN-1 downregulated genes act to decrease stress resistance suggests that they might be actively repressed by SKN-1.
We next examined how 26 of the SKN-1-downregulated genes affect lifespan under non-stressed conditions. These genes were selected among those with the most significant SAM scores (Table S2). RNAi knockdown of 11 of these 26 genes significantly extended lifespan in each of three independent experiments, (p<0.01; log-rank) (Fig. 6D; Table S11A-D). These 11 genes included ins-7, along with 10 other genes that are newly identified here as longevity-affecting genes, as none had been shown previously to influence lifespan (WormBase). This number is probably an underestimate, as RNAi of several additional genes extended lifespan in two of three trials (Table S11D). Of the 15 genes for which RNAi increased As stress resistance (Fig. S5), 7 were associated with increased lifespan in all three of our longevity trials, and 5 in two of these trials (Table S2). We conclude that under normal conditions SKN-1 inhibits many genes that reduce stress resistance and/or longevity (Fig. 7).
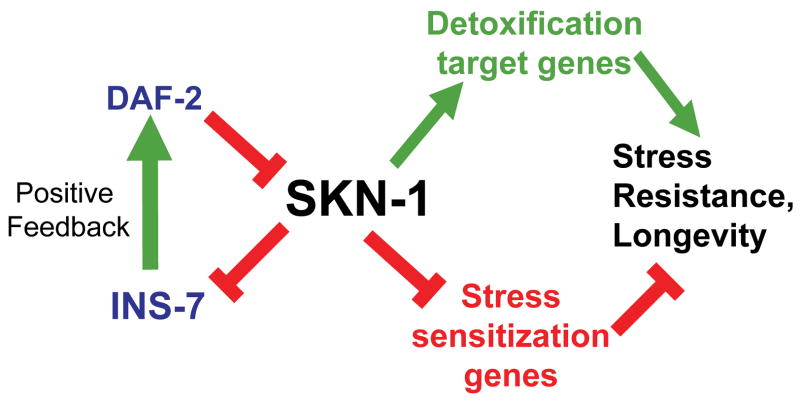
Fig. 7. A model for SKN-1 functions under normal conditions.
A positive feedback interaction with ins-7 and the DAF-2 pathway is featured. SKN-1 upregulates many genes that promote detoxification and stress resistance, and also downregulates genes that decrease stress resistance, lifespan, or both. Among the SKN-1-downregulated genes are both ins-7 and pdk-1 (not shown), each of which promotes DAF-2 pathway signaling (see text). The DAF-2 pathway in turn inhibits SKN-1 (Tullet et al. 2008).
Discussion
SKN-1 is required for oxidative stress resistance and has important functions in the absence of acute stress, as it promotes longevity under normal, reduced IIS, and CR conditions (An & Blackwell 2003; Bishop & Guarente 2007a; Tullet et al. 2008). Here we show that under non-stressed conditions SKN-1 upregulates numerous detoxification genes, along with other genes with functions that may be related to stress defenses. We also found that SKN-1 inhibits genes that reduce stress resistance and longevity. Finally, we observed that SKN-1 induces discrete target gene groups in response to stress, and that some stresses activate SKN-1-independent stress defense mechanisms in parallel. Many of the genes we identified in each SKN-1-dependent gene set are likely to be direct SKN-1 targets, as suggested by the prevalence of SKN-1 binding sites in their promoters. Our results reveal a notable degree of complexity in SKN-1 functions and C. elegans stress responses.
Multiple SKN-1 functions under normal conditions
Many of the 233 genes that we identified as SKN-1-upregulated under normal conditions are involved directly in stress-related processes (Table 1, S1). These included GST, UGT, and other Phase 2 genes that are involved in conjugation of toxic compounds, ROS metabolism, or glutathione production (Fig. 2B, 2D, Table S1). Interestingly, some of these GSTs might be involved in functions besides detoxification. For instance, the most highly upregulated SKN-1 target (gst-4, Table 1) seems to function not only as a GST, but also as a glutathione-dependent prostaglandin D synthase (Kubagawa et al. 2006). Besides Phase 2 genes, we also detected significant representation of Phase 1 (SDR, CYP) and Phase 3 (Transporter) genes, indicating that SKN-1 plays a broad role in systemic detoxification. Interestingly, our SKN-upregulated gene sets did not include superoxide dismutase (SOD) or catalase genes, and we found previously that sod-3 is upregulated independently of skn-1 in the context of reduced IIS (Tullet et al. 2008). Taken together, our data suggest that SKN-1 does not regulate a primary response to endogenously produced superoxide or hydrogen peroxide, and instead promotes detoxification, cellular repair, and activity of the many antioxidant and stress-defense systems that depend upon glutathione.
Numerous SKN-1-upregulated genes are involved in lysosomal or proteasomal functions (Table S1, S5, S8; see Results), suggesting that SKN-1 may promote recycling of damaged cellular components. RNAi knockdown of many proteasome component genes has been shown to result in accumulation of SKN-1 in nuclei, through an unknown mechanism (Kahn, et al., 2008). Together with this observation, our results suggest the existence of a feedback mechanism whereby SKN-1 might limit its own activity by upregulating proteasome gene expression.
Other genes we identified implicate SKN-1 in additional activities. Many SKN-1-upregulated genes encode cell-surface proteins, including CUB-like proteins (Fig. 2B; Table S1). Some CUB-like genes are also regulated by p38 MAPK signaling, which is important for SKN-1 function in the intestine (Inoue et al. 2005; Troemel et al. 2006). RNAi of the CUB-like genes C32H11.3 and C32H11.4 modestly reduced As resistance (Fig. 6A), indicating that some CUB-like proteins affect stress resistance. SKN-1-upregulated genes are involved in additional diverse molecular functions that include transcription, signaling, ubiquitination, and metabolism (Fig. 2B, 2D; Table S1), indicating that SKN-1 is involved in a complex group of processes. It will be interesting to elucidate which of these processes might indirectly affect detoxification or stress resistance.
It seems likely that the SKN-1-regulated genes we identified under normal and other conditions primarily reflect SKN-1 functioning in the intestine, as opposed to the ASI neurons, because SKN-1 expression is more prominent in the intestine (An and Blackwell, 2003). In addition, we reduced SKN-1 expression by RNAi, which works comparatively poorly in neurons (Timmons et al. 2001). We were therefore very surprised to find that SKN-1 controls so many genes under normal conditions, because SKN-1 is present in intestinal nuclei at comparatively low levels in the absence of stress (An & Blackwell 2003). It is even possible that we might have underestimated the breadth of SKN-1 activity, because these analyses of whole worms could have missed some genes that are regulated by SKN-1 in only subsets of tissues. Importantly, most of the individual genes that were controlled by SKN-1 under normal conditions were not upregulated in As or t-BOOH stress responses (Fig, 2, 4, 5), arguing against the idea that the SKN-1-dependent gene activity detected under normal conditions derives simply from animals being mildly “stressed.” We conclude that under normal conditions SKN-1 is important for fine-tuning of genes involved in many stress-related and other functions.
Many of the functional categories that are characteristic of SKN-1-upregulated genes (GST, UGT, SDR, CYT, CUB domain, drug transporters) are also prominent among C. elegans or yeast genes that depend upon the general mRNA transcription factor MDT-15 (MED-15) (Taubert et al. 2008; Thakur et al. 2008). MDT-15 is a subunit of Mediator, a large multiprotein complex that must be brought to promoters for transcription to initiate. MDT-15 is required for function of the transcription regulators SBP-1 (SREBP) and NHR-49 (PPARα), which are critical for lipid homeostasis and metabolic regulation (Taubert et al. 2006; Yang et al. 2006). Those two regulators seem to activate transcription at least in part by binding to MDT-15, and thereby recruiting Mediator to promoters. MDT-15 is also required for xenobiotic defense and has been proposed to coordinate multiple transcriptional responses to food, toxins, and other ingested materials (Taubert et al. 2008). Our results suggest the intriguing model that SKN-1 might interact functionally or physically with MDT-15 to regulate some detoxification genes.
SKN-1-dependent suppression of stress-sensitization and anti-longevity genes
It was striking that SKN-1 downregulates numerous genes under non-stressed conditions, and that in many cases RNAi of these genes increased stress resistance and/or lifespan (Fig. 6; Table S2, S11D). Many of these SKN-1-downregulated genes contain predicted SKN-1 binding sites in their putative promoters, predicting that some might be repressed directly by SKN-1 (Table S2). This seems surprising, because SKN-1 is a powerful activator of transcription (Walker et al. 2000). However, other examples have been identified of transcription regulators that seem to function as both activators and repressors, including DAF-16 (Murphy et al. 2003). An important implication of our findings is that the previously described functions of SKN-1 in promoting stress resistance and longevity (An, et al., 2003; Bishop and Guarente, 2007; Tullet, et al., 2008) might be attributable not only to SKN-1 upregulating stress defense and other genes, but also to its inhibiting genes that have the opposite effect.
The stress-sensitization and anti-longevity genes we identified among the SKN-1-downregulated genes are involved in diverse functions (Table S2). For example, lea-1 decreases As resistance (Fig. 6B) but is predicted to protect against dessication (Browne et al. 2002). Perhaps adaptations to some conditions are not beneficial in the setting of stresses that would activate a SKN-1 response. Other SKN-1-downregulated genes that reduce stress resistance or lifespan encode regulatory proteins, including an F-box protein (FBXA-188) and the predicted SCF ubiquitin ligase component SKR-5 (Fig. 6B, 6C, Table S11D). It is particularly noteworthy that SKN-1 downregulates genes that encode the IIS pathway kinase PDK-1 and the DAF-2 agonist INS-7, each of which had previously been shown to reduce longevity (Paradis & Ruvkun 1998). INS-7 coordinates IIS and DAF-16 activity among tissues (Murphy et al. 2003; Murphy et al. 2007). DAF-16 inhibits ins-7 expression in a positive feedback loop, thereby relieving negative regulation of itself by IIS. Our results indicate that SKN-1 and INS-7 are involved in a similar feedback loop that could magnify the effects of upregulating IIS on the one hand, or either DAF-16 or SKN-1 on the other (See model in Fig. 7).
SKN-1 functions analogously to DAF-16 in three intriguing ways. Firstly, both proteins are inhibited directly by IIS (Tullet et al. 2008). Secondly, our new results reveal that SKN-1, like DAF-16, down-regulates multiple mechanisms that reduce stress-resistance or longevity (Murphy, et al., 2003)(Fig. 7). Why would such mechanisms exist, and why would they respond to SKN-1? Perhaps it is advantageous to hold some stress defense mechanisms in check under normal conditions; for example, enzymes that metabolize endobiotics or free radicals could have profound effects on hormonal and cell signaling pathways. Thirdly, like SKN-1, DAF-16 also upregulates many stress resistance genes, as indicated by transcription profiling and proteomics performed under conditions of reduced IIS (Murphy et al. 2003; Dong et al. 2007; McElwee et al. 2007), comparative genomics and bioinformatics (Lee et al. 2003b), and chromatin immunoprecipitation studies (Oh et al. 2006). DAF-16 upregulates many CYP and other Phase 1 genes (Murphy et al. 2003; McElwee et al. 2007), and one analysis suggests that some GSTs are upregulated by DAF-16 and are associated with IIS regulation in other species (McElwee et al. 2007). In addition, we earlier observed that SKN-1 and DAF-16 together increase activity of particular GST genes in the context of a daf-2 mutant (Tullet et al. 2008). In the future, it will be interesting to elucidate the extent to which SKN-1 and DAF-16 might function cooperatively under particular conditions.
Customized skn-1-dependent and -independent responses to stress
How animals respond to metabolic or environmental stresses and how these responses are regulated are fundamentally important questions. By analyzing the transcriptional responses of C. elegans to As and t-BOOH we have obtained new insights into SKN-1 functions, its role in these stress responses, and how C. elegans adapts to stresses. For example, it was striking that only a subset of SKN-1-responsive genes were upregulated by each of these stresses. Multiple GO terms that were prominent among SKN-1-upregulated genes under normal conditions were not represented among the As or t-BOOH induced genes (i. e. UGT, C-type lectin), and new GO terms appeared among the SKN-1-dependent genes that were upregulated by As (i.e. Alcohol dehydrogenase) and t-BOOH (Hydrolase) (Fig. 4D). The apparent specificity of these responses indicates that SKN-1 does not simply regulate its target genes in tandem in response to stress levels. Instead, SKN-1 must integrate multiple signals, so that in response to a given stimulus some genes are induced and others are left unaffected. Our results suggest that mammalian Nrf proteins may have a similarly complex set of functions that might not be apparent from analyses of single cell types or tissues.
Another interesting observation was that while the entire As response we detected required skn-1, t-BOOH stimulated a skn-1-independent response that included induction of large numbers of Phase 1 detoxification, nuclear receptor, and lipid metabolism genes, along with upregulation of some genes that were skn-1-dependent under other conditions (Fig. 5B, 5C; Table S9). As these findings were obtained with skn-1 RNAi (Fig. 1), it is impossible to establish that all of these genes were induced by t-BOOH independently of skn-1. However, this was true for each of the seven genes that we analyzed in a predicted null skn-1 mutant (Fig. S4), indicating that t-BOOH induces a broad skn-1 independent response. Interestingly, RNAi knockdown of the 2-Cys peroxiredoxin prdx-2 results in skn-1-independent activation of the SKN-1 target gene gcs-1 (Olahova et al. 2008), further supporting the idea that some signals induce Phase 2 genes independently of skn-1. Together, our findings suggest that the transcriptional responses to oxidative stresses may be highly specific, and adapted to the challenge faced by the organism.
Why would the responses to As and t-BOOH be so different? Arsenite is a metalloid that attacks thiols, depletes glutathione, and induces ROS formation, whereas t-BOOH is a stable lipid soluble peroxide that attacks lipids and proteins. Perhaps the As-induced genes represent a “simpler” response to stress arising from excess ROS or a need for glutathione-related defenses. We speculate that the more complex t-BOOH response could additionally involve stress from phospholipid damage, or a global response to lipophilic toxins. Further supporting the notion that C. elegans stress responses are “tailored,” the list of gene groups induced by acrylamide is very similar to our As list (GSTs, UGTs, SDRs, glutathione metabolism), but also includes some distinct categories (collagens, major sperm proteins) (Hasegawa et al. 2008). C. elegans could prove to be valuable for elucidating signals that lie upstream of different stress responses, and the role of individual tissues in mobilizing these signals and defending the organism against stress.
Our results demonstrate that SKN-1 plays a number of roles besides inducing Phase 2 detoxification genes, and that multiple factors influence its transcriptional output under normal and stress conditions. It will now be important to delineate how its regulation of detoxification and regulatory genes contributes to the effects of SKN-1 on longevity under normal and reduced IIS conditions, and to identify how SKN-1 acts in different tissues to influence regulation of these genes at the organismal level.
Experimental Procedures
C. elegans growth and RNAi for microarray experiments
C. elegans were maintained on Nematode Growth Medium (NGM) and E. coli OP50 as described (Brenner 1974). For microarray experiments, a synchronous population of wild type (N2) animals was obtained by hypochlorite treatment of embryos. Synchronized L1 larvae were placed at 20°C on E. coli HT115 that expressed either skn-1 or control dsRNA for 46h, until they reached the L4 stage. For As exposure, worms were incubated for 30 min in 5mM Sodium Arsenite (Sigma-Aldrich) in M9 medium, or in M9 alone. Worms were exposed to t-BOOH (12mM, Sigma-Aldrich) for 1 hour on NGM plates, or incubated on NGM control plates. In each case, worms were then allowed to recover for 1 hour on OP50-seeded NGM plates. These stress treatment conditions were established by titrating As or t-BOOH concentrations and incubation times, and scoring for induction of the SKN-1 target gene reporter gcs-1::GFP (Fig. S1)(An & Blackwell 2003). Under the conditions used for microarray analysis this reporter was induced robustly in the intestine, but for each stress tested worms that appeared sick or or dead were observed at only a low frequency (0–5% across samples).
The skn-1 RNAi plasmid consisted of a full length SKN-1c isoform cDNA subcloned into pPD129.36 (gift of A. Fire). The control plasmid was pPD128.110 (gift of A. Fire) which contains the GFP gene flanked by T7 promoters (Timmons et al. 2001). RNAi was performed by feeding as described (Kamath & Ahringer 2003).
RNA preparation and microarray data collection
For each microarray experiment, total RNA was isolated from 50000 animals using Trizol (Invitrogen). cDNA was synthesized and linearly amplified from 325 ng RNA using the Low RNA Input Linear Amplification Kit (Agilent), and labeled with Cy3- or Cy5-CTP (Perkin Elmer). A dye swap analysis was performed for each set of biological replicate samples. Samples were fragmented according to Agilent protocols and hybridized overnight at 60°C to Agilent-015061: C. elegans oligonucleotide Microarray 4×44 arrays (covering 21,481-genes). Array scanning was performed using a DNA Microarray Scanner (Agilent) at 5μm resolution. The output image was processed by Feature Extractor (Agilent) and normalized for dye bias by linear correction using rank consistent probes. Prior to hierarchical clustering, values from spots on the microarray that represented the same gene were averaged to a single value. Spots flagged by the Feature Extractor software as having Red and Green intensities well above background were omitted. Finally, genes that did not have an observed absolute value of 0.4 for the log(base2) ratio of Red/Green intensities for at least one array were omitted, as were any genes that lacked information for >20% of the arrays. After filtering, the remaining genes were submitted for downstream analysis. Raw microarray data will be available via the Princeton University MicroaAray database: http://puma.princeton.edu/.
Hierarchical clustering and SAM analysis
Average linkage gene clustering was performed with an uncentered correlation similarity metric using Cluster (Eisen et al. 1998; de Hoon et al. 2004). One-class analysis in SAM (Tusher et al. 2001) was performed to identify genes that had statistically significant changes in expression regardless of the magnitude of change.
Promoter analysis
We analyzed up to 1.5 kb of intergenic sequence upstream of SKN-1-regulated genes for the presence of novel regulatory elements. Sequence elements that were statistically overrepresented in these regions were identified using Regulatory Sequence Analysis Tools (RSAT) oligo-analysis (Thomas-Chollier et al. 2008) and Weeder (Pavesi et al. 2004), in each case specifying an oligonucleotide length of 8 bases. We later searched directly for these novel consensus elements and the consensus in vitro SKN-1 binding site (Blackwell et al. 1994) within up to 2 kb of upstream intergenic sequences. WebLogo (Crooks et al. 2004) was used to display consensus motifs.
Gene ontology analysis
WormBase gene names were converted to NCBI Protein Gene Info (GI) numbers using WormMart (Schwarz et al. 2006), then analyzed using DAVID (Dennis et al. 2003). Functional clusters of SKN-1-regulated genes were identified using DAVID’s Functional Annotational Clustering tool, with the exception of the CUB-like domain genes and other annotated open reading frames (ORFs) that lacked a GI entry. The Enrichment Score was used to predict whether representation of a gene group among SKN-1-regulated genes was biologically significant. The Enrichment Score of a cluster of genes or GO terms derives from the geometric mean (in negative log scale) of the p-values for members of that cluster. If the geometric mean of the p-values = 1e−10, then the Enrichment Score is 10. These p-values correspond to the probability that the members of the cluster are present together randomly in the gene list.
Lifespan analysis
In analyses of lifespan under normal conditions the first day of adulthood was defined as t=0, standard Kaplan-Meier survival curves were generated from the data, and the log-rank (Mantel-Cox) method was used to test the null hypothesis (StatView). These analyses were performed using the RNAi-sensitive strain rrf-3(pk1426) (Sijen et al. 2001). In two assays (Experiments 1 and 2), n >100 worms (see Table S11 for n) were transferred at L4 to 100mm HGM plates (1mM IPTG, 100 μg/mL carbenicillin, and 50 μM FUdR, 20°C) that had been inoculated with the indicated RNAi bacteria. Live/dead counts were made approximately every other day. The experiments were terminated upon contamination after approximately 3 weeks, and the surviving animals were censored from the assay on that day. A third assay (Experiment 3) was also performed using rrf-3(pk1426) worms at 20°C, but in this case, ~75 eggs (see Table S11C for n) were transferred onto 6×60mm NG plates (1mM IPTG, 100 μg/mL carbenicillin) that had been inoculated with the indicated RNAi bacteria. Worms were transferred to fresh RNAi plates every four days, and animals that were missing, exploded, or bagged were censored from the data on the day of the event.
Stress resistance assays
For stress assays, N2 or rrf-3(pk1426) worms that had been arrested at L1 were grown for 48–55 hrs at 20°C on either RNAi or control bacteria. RNAi clones were obtained from published libraries (Kamath & Ahringer 2003; Rual et al. 2004). RNAi was performed essentially according to Protocol 2 in (Kamath et al. 2001). Worms were then placed in 4 or 5 mM Sodium Arsenite (in M9) and periodically tested for survival, with 3–6 wells of ≥10 worms each examined in each experimental measurement. Worms were prodded with a platinum wire and scored as dead if they displayed no pharyngeal pumping or movement. Control wells of M9 always displayed 100% survival for all time points examined. The assays represented in Figure S3 were carried out on NGM agar plates containing either As or t-BOOH.
qRT-PCR
Stress- and control-treated worms were collected as for the microarray analysis samples. RNA was isolated and purified using Tri Reagent (Sigma). cDNA was synthesized using Superscript III Reverse Transcriptase (Invitrogen). SYBR GreenR (Invitrogen) Real Time PCR was performed in an ABI 7700 machine in duplicate and the data was analyzed using the comparative Ct method with the exception of data in Fig. S3B, which were analyzed by normalization to a standard curve. Relative mRNA levels were normalized to act-1 mRNA levels, and calculated from at least three biological replicates. Primers were designed to be intron-spanning, with sequences available upon request.
Supplementary Material
Table 3. Summary of Lifespan Effects of SKN-1 Downregulated Genes.
For details of lifespan assay conditions and analysis see Experimental Procedures, Table S11, and Table S12.
| Expt 1 | Expt 2 | Expt 3 | |||||
|---|---|---|---|---|---|---|---|
| Gene | Gene name | % of control | p-value | % of control | p-value | % of control | p-value |
| C54G6.5** | spp-17 | 121.7 | <0.0001 | 125.2 | <0.0001 | 119.3 | <0.0001 |
| C01G6.7** | acs-7 | 112.6 | <0.0001 | 116.1 | <0.0001 | 121.4 | <0.0001 |
| Y43C5A.3** | 121.7 | <0.0001 | 114.0 | <0.0001 | 113.6 | 0.006 | |
| F25B3.5* | 126.6 | <0.0001 | 113.3 | <0.0001 | 110.9 | 0.0209 | |
| F23F12.3** | 116.1 | <0.0001 | 113.3 | 0.0002 | 125.1 | <0.0001 | |
| Y47H9C.1** | 113.3 | <0.0001 | 112.6 | <0.0001 | 119.8 | <0.0001 | |
| ZK1251.2** | ins-7 | 111.2 | 0.0006 | 112.6 | <0.0001 | 122.3 | <0.0001 |
| B0024.4* | 120.3 | <0.0001 | 110.5 | <0.0001 | 103.4 | 0.5417 | |
| F58F9.7* | 118.2 | <0.0001 | 110.5 | <0.0001 | 112.6 | 0.0151 | |
| F58B3.3 | lys-6 | 116.8 | <0.0001 | 110.5 | 0.0128 | ||
| F02C12.5* | cyp-13B | 127.3 | <0.0001 | 109.1 | <0.0001 | 110.4 | 0.039 |
| C31B8.4** | 116.8 | <0.0001 | 109.1 | <0.0001 | 121.6 | <0.0001 | |
| F15B9.6** | 125.2 | <0.0001 | 108.4 | <0.0001 | 122.7 | <0.0001 | |
| ZC196.4* | 101.4 | 0.898 | 108.4 | 0.0003 | 120.6 | 0.0001 | |
| Y46G5A.20** | 130.8 | <0.0001 | 107.7 | 0.0001 | 120.0 | <0.0001 | |
| F45E4.1** | arf-1.1 | 111.2 | 0.0002 | 107.7 | <0.0001 | 117.5 | 0.0008 |
| F47H4.8* | fbxa-188 | 117.5 | <0.0001 | 107.0 | 0.0208 | 120.0 | 0.0002 |
| F15D3.8* | 112.6 | <0.0001 | 107.0 | 0.0087 | 100.0 | ||
| Y41C4A.11** | 121.7 | <0.0001 | 105.6 | 0.0005 | 123.4 | <0.0001 | |
| Y6E2A.4 | 113.3 | <0.0001 | 105.6 | 0.8425 | |||
| C17H1.7* | 124.5 | <0.0001 | 104.9 | 0.0004 | 109.9 | 0.0435 | |
| F47H4.10* | skr-5 | 116.1 | <0.0001 | 104.2 | 0.0079 | ||
| Y51B9A.9 | 104.2 | 0.0858 | 104.2 | 0.0106 | |||
| Y39B6A.24 | 112.6 | <0.0001 | 102.8 | 0.0508 | |||
| K08H10.1* | lea-1 | 120.3 | <0.0001 | 97.2 | 0.867 | 112.9 | 0.007 |
| M01G12.12 | rrf-2 | 114.0 | <0.0001 | 93.7 | 0.6139 | ||
Significant (p<0.01) in all three trials.
Significant (p<0.01) in two trials.
% of control refers to the mean lifespan. Functional information for these genes is available in Table 2.
Acknowledgments
We thank Elizabeth Veal and Blackwell lab members for critically reading this manuscript, and Joe Baker and Aileen Zhen for early contributions to the study. Supported by a Kirchstein fellowship to RO (GM70088), training grant funding to RO and JPA (DK07260), NIH grant GM62891 to TKB, Pew Biomedical Scholar and Sloan Fellow funding to CTM, and an NSF Pre-doctoral fellowship to JL. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Footnotes
Note added in proof
While this manuscript was under review, it was reported online that skn-1 is required for a substantial proportion of the transcriptional response to hyperbaric oxygen (Park, S. K., Tedesco, P. M., and Johnson, T. E., Aging Cell, Accepted Article). Several of these potential SKN-1 target genes overlapped with those identified here. A contemporaneous study identified many of our SKN-1-dependent genes as being induced in an age-dependent manner by the oxygen-generating stressor juglone (Pryzbysz, et al., Mech. Aging Dev. (2009) 130, 357-369).
Author Contributions
Conceived and designed the experiments: RPO, JPA, KD, CTM, TKB. Performed the experiments: RPO, JPA, KD, JL, JA. Analyzed the data: RPO, JPA, KD, JL, CTM, TKB. Wrote the paper: RPO, JPA, CTM, TKB.
Supporting Information
Additional Supporting Information: Raw microarray data will be available via the Princeton University MicroArray database: http://puma.princeton.edu/.
References
- An JH, Blackwell TK. SKN-1 links Celegans mesendodermal specification to a conserved oxidative stress response . Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, Matsumoto K, Blackwell TK. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Zou CG. Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein. Arch Biochem Biophys. 2005;433:144–156. doi: 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007a;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in Celegans. Nature. 2007b;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Blanc G, Font B, Eichenberger D, Moreau C, Ricard-Blum S, Hulmes DJ, Moali C. Insights into how CUB domains can exert specific functions while sharing a common fold: conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J Biol Chem. 2007;282:16924–16933. doi: 10.1074/jbc.M701610200. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne J, Tunnacliffe A, Burnell A. Anhydrobiosis: plant desiccation gene found in a nematode. Nature. 2002;416:38. doi: 10.1038/416038a. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in Celegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Miwa S, Isomura K, Tsutsumiuchi K, Taniguchi H, Miwa J. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol Sci. 2008;101:215–225. doi: 10.1093/toxsci/kfm276. [DOI] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. Celegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Hughes MF. Arsenic toxicity and potential mechanisms of action. Toxicol Lett. 2002;133:1–16. doi: 10.1016/s0378-4274(02)00084-x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. The Celegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response . Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper H. SKNy worms and long life. Cell. 2008;132:915–916. doi: 10.1016/j.cell.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, De Domenico I, Ward DM. Chediak-Higashi syndrome. Curr Opin Hematol. 2008;15:22–29. doi: 10.1097/MOH.0b013e3282f2bcce. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Murphy CT. Enrichment of regulatory motifs upstream of predicted DAF-16 targets. Nat Genet. 2006;38:397–398. doi: 10.1038/ng0406-397. author reply 398. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Kophengnavong T, Carroll AS, Blackwell TK. The SKN-1 amino terminal arm is a DNA specificity segment. Mol Cell Biol. 1999;19:3039–3050. doi: 10.1128/mcb.19.4.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubagawa HM, Watts JL, Corrigan C, Edmonds JW, Sztul E, Browse J, Miller MA. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat Cell Biol. 2006;8:1143–1148. doi: 10.1038/ncb1476. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003a;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 Target Genes That Control Celegans Life-Span and Metabolism. Science. 2003b;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, Walker GA. Stress resistance as a determinate of Celegans lifespan. Mech Ageing Dev. 2002;123:765–771. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- Mathews WR, Guido DM, Fisher MA, Jaeschke H. Lipid peroxidation as molecular mechanism of liver cell injury during reperfusion after ischemia. Free Radic Biol Med. 1994;16:763–770. doi: 10.1016/0891-5849(94)90191-0. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D. Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8:R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Lee SJ, Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Olahova M, Taylor SR, Khazaipoul S, Wang J, Morgan BA, Matsumoto K, Blackwell TK, Veal EA. A redox-sensitive peroxiredoxin that is important for longevity has tissue- and stress-specific roles in stress resistance. Proc Natl Acad Sci U S A. 2008;105:19839–19844. doi: 10.1073/pnas.0805507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert PB, Daughdrill GW, Bowerman B, Matthews BW. The binding domain of Skn-1 in complex with DNA: a new DNA-binding motif. Nature Struct Biol. 1998;5:484–491. doi: 10.1038/nsb0698-484. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of Celegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkadi B, Homolya L, Szakacs G, Varadi A. Human multidrug resistance ABCB and ABCG transporters: participation in a chemoimmunity defense system. Physiol Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Canaran P, Chan J, Chen N, Chen WJ, Davis P, Fiedler TJ, Girard L, Harris TW, Kenny EE, Kishore R, Lawson D, Lee R, Muller HM, Nakamura C, Ozersky P, Petcherski A, Rogers A, Spooner W, Tuli MA, Van Auken K, Wang D, Durbin R, Spieth J, Stein LD, Sternberg PW. WormBase: better software, richer content. Nucleic Acids Res. 2006;34:D475–478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Taubert S, Hansen M, Van Gilst MR, Cooper SB, Yamamoto KR. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 2008;4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in Celegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Naar AM. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in Celegans . PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in Celegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk OK, Bucher EA, Sundaram MV, Rea PA. CeHMT-1, a putative phytochelatin transporter, is required for cadmium tolerance in Caenorhabditis elegans. J Biol Chem. 2005;280:23684–23690. doi: 10.1074/jbc.M503362200. [DOI] [PubMed] [Google Scholar]
- Walker AK, See R, Batchelder C, Kophengnavong T, Gronniger JT, Shi Y, Blackwell TK. A conserved transcription motif suggesting functional parallels between Celegans SKN-1 and Cap ‘n’ Collar-related bZIP proteins. J Biol Chem. 2000;275:22166–22171. doi: 10.1074/jbc.M001746200. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, Debeaumont R, Mako Saito R, Hyberts SG, Yang S, Macol C, Iyer L, Tjian R, van den Heuvel S, Hart AC, Wagner G, Naar AM. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006 doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the Celegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi . Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in Celegans: a model for satiety . Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.