Abstract
Nitric oxide (NO) enhances human sperm motility and capacitation associated with increased protein phosphorylation. NO activates soluble guanylyl cyclase, but can also modify protein function covalently via S-nitrosylation of cysteine. Remarkably, this mechanism remains unexplored in sperm although they depend on post-translational protein modification to achieve changes in function required for fertilisation. Our objective was to identify targets for S-nitrosylation in human sperm. Spermatozoa were incubated with NO donors and S-nitrosylated proteins were identified using the biotin switch assay and a proteomic approach using tandem mass spectrometry. 240 S-nitrosylated proteins were detected in sperm incubated with S-nitrosoglutathione. Minimal levels were observed in glutathione or untreated samples. Proteins identified consistently based on multiple peptides included established targets for S-nitrosylation in other cells e.g. tubulin,, glutathione-S-transferase and heat shock proteins but also novel targets including A-kinase anchoring protein (AKAP) types 3 and 4, voltage-dependent anion-selective channel protein 3 and semenogelin 1 and 2. In situ localisation revealed S-nitrosylated targets on the post-acrosomal region of the head and throughout the flagellum. Potential targets for S-nitrosylation in human sperm include physiologically significant proteins not previously reported in other cells. Their identification will provide novel insight into the mechanism of action of NO in spermatozoa.
Keywords: Human, Nitric Oxide, Signalling, S-Nitrosylation, Spermatozoa
1. Introduction
The mature mammalian spermatozoon is a ‘minimalist’ cell, specialised for its sole purpose of delivering the haploid nucleus and activating factors to the female gamete. To this end ‘surplus’ cytoplasm, including all endoplasmic reticulum, is jettisoned during spermiogenesis (the final stage of differentiation). The differentiated sperm thus lacks functional apparatus for DNA transcription or protein expression. However, fully-differentiated spermatozoa must undergo profound functional changes, both during maturation in the epididymis and upon interaction with the female reproductive tract [1]. These events, which are vital for fertilisation to occur, depend on post-translational modification of proteins (e.g. tyrosine phosphorylation) [2] in the mature cells.
Human spermatozoa undergo extensive redox regulated signalling [3, 4] and a plethora of studies have documented a significant positive effect on sperm function upon the addition of exogenous nitric oxide (NO) [5-7]. For example, NO is involved in sperm motility [8], capacitation (a maturational process spermatozoa must undergo before they can fertilize oocytes) [9], acrosome reaction [9], and enhancement of sperm-zona pellucida binding ability [10]. Human spermatozoa have both endothelial and neuronal forms of nitric oxide synthase (endothelial, eNOS and neuronal, nNOS) [8, 11-15]. However, the significance of these enzymes in signalling in mature sperm is debatable (see discussion). Perhaps more significantly, spermatozoa are exposed to a significant array of highly effective NO producing cells in the human female tract (concentration from 5nM to 4μM; Marcondes et al. [16] with which they have intimate and prolonged [hours] contact. The exact site(s) of production of NO in the female tract and the temporal sequence of events remains to be investigated but evidence from our laboratory suggests that cumulus cells surrounding the oocyte produce easily detectable amounts of NO [17] (unpublished data), implying that NO from this source greatly exceeds endogenous production by sperm.
The question is, ‘how does NO affect human sperm function?’ Until recently, it was generally accepted that NO effects were mediated principally through cGMP-related mechanisms. Stimulation of soluble guanylyl cyclase is a well-characterised mode of action for NO and the importance of cGMP signalling in the sperm of marine invertebrates is also well established [18]. However, the concentration of cGMP and the activities of cGMP-specific phosphodiesterase (type 5) and protein kinase G are low or undetectable in human spermatozoa [12, 19]. To date, the primary role of cGMP in human sperm (as shown by use of specific kinase inhibitors and addition of dibutyryl cGMP) is induction of the acrosome reaction [18, 20]. Studies on capacitation have shown that NO also modifies activities of other kinases. Application of exogenous NO is associated with an increased cAMP levels (leading to tyrosine phosphorylation) [21] and NO is also involved in activation of a protein extra cellular signal regulated kinases (ERKs) [22, 23]. However, the exact role and site of action of NO remains speculative and the molecular mechanisms incompletely understood [23].
An alternative, cGMP-independent and potentially crucial action of NO is the covalent modification of proteins via S-nitrosylation - the formation of S-NO bond by cysteine thiol nitrosylation to form a nitrosothiol [24, 25]. S-nitrosylation is a selective, temporal and spatially regulated post-translational protein modification which regulates physiological cellular signalling, analogous to phosphorylation and acetylation [25, 26]. Over 120 proteins, from both animal and plant cells have been shown to undergo S-nitrosylation [25, 27-30] and an increasing number of proteins are shown to be functionally regulated by S-nitrosylation e.g. estrogen receptor [31] connexin 43 hemichannels [32], dynamin [29]. Remarkably, this mechanism of action remains unexplored in sperm even though NO is a well know modulator of sperm function and these remarkable cells are exclusively dependent on post-translational protein modification to effect physiological changes in function necessary for fertilisation.
The objective of our study was to perform a systematic assessment of potential targets for protein S nitrosylation in human spermatozoa using the biotin switch assay. We identified 240 S-nitrosylated proteins. This global assessment allows critical insight into the role of NO in sperm physiology, identifies potential novel mechanisms of action for S-nitrosylation and opens up a new signalling regimen in the spermatozoon which will act as a platform for further detailed studies.
2. Materials and Methods
2.1 Materials
Spermine NONOate was purchased from Molecular Probes (distributed by Invitrogen Ltd, Paisley, UK), S-Nitrosoglutathione from Merck Biosciences Ltd. (Beeston, Nottingham, UK), protease inhibitor cocktail tablets from Roche Diagnostics Ltd. (Lewes, East Sussex, UK) and EZ-Link Biotin-N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (EZ-Link Biotin-HPDP) from Perbio Science UK Ltd, (Cramlington, Northumberland, UK). Nitrocellulose membrane was supplied by GE Healthcare UK Ltd. (St.Giles, Bucks, UK), IgG Fraction Monoclonal Mouse Anti-Biotin and FITC-donkey anti-rabbit IgG conjugated to fluorescein isothiocyanate by Jackson Immunoresearch Laboratories (Stratech Scientific, Soham, Cambrigshire, UK) and Lumi-GLO, an enhanced chemiluminescence kit, from Insight Biotechnology Ltd. (Wembley, Middlesex, UK). The Silver Stain Plus was purchased from Bio-Rad Laboratories Ltd. (Hempstead, Hertfordshire, UK). Texas Red®-2-sulfonamidoethyl methanethiosulfonate (TEXAS RED®-MTSEA) was obtained from Toronto Research Chemicals Inc. (North York, Ontario, Canada) and the fluorescence mounting medium from Dako Cytomation Ltd. (Ely, Cambridgeshire, UK). All other chemicals were purchased from Sigma (Poole, Dorset, UK).
2.2 Donors
Semen samples were provided by research donors who attended the Assisted Conception Unit, Birmingham Women's Hospital (Human Fertilisation and Embryology Authority Centre #0119). Donors gave informed consent under COREC ethical approval (South Birmingham REC #2003/239). All procedures were in accordance with HFEA 6th Code of Practice. Donors had normal semen characteristics as assessed by the World Health Organisation guidelines [33].
2.3 Sperm Preparation
Semen samples were obtained after 2–3 days of sexual abstinence. After liquefaction, 1 ml of semen was layered over 1 ml fractions of 45 and 90% Percoll-made isotonic with M medium (1X: 137 mM NaCl, 2.5 mM KCl, 20 mM HEPES, 10 mM glucose). Samples were centrifuged at 2000 g for 20 min. Sperm concentration was determined using an improved Neubauer haemocytometer and at least 400 cells were scored [34]. Percoll-washed spermatozoa were further washed with Phosphate Buffered Saline (PBS) to remove Percoll. Spermatozoa were then diluted and incubated in PBS.
2.4 Biotin-Switch Assay
S-nitrosylated proteins in human spermatozoa were detected using the biotin switch assay as described previously [26] with minor modifications. Briefly intact spermatozoa (50 × 106 cells/ml) were incubated in the presence of nitric oxide donors of different classes, S-nitroso-glutathione (GSNO), L-nitroso-cysteine (CSNO) and N–(2–aminoethyl)–N–(2–hydroxy–nitrosohydrazino)–1, 2–ethylenediamine (spermine NONOate), control compounds (glutathione [GSH] and exhausted CSNO) or no treatment for 1 hr at 37°C. Following incubation, spermatozoa were centrifuged at 2000g for 5 min and supernatant removed, spermatozoa were then resuspended in lysis buffer (250 mM Hepes pH 7.7, 1 mM EDTA, 0.1 mM neocuproine, 1% Triton, 2.5% SDS) and protease inhibitors added to the cells. Samples were incubated for 5 min at RT and then centrifuged at 2000g for 5 min. Protein concentration was assessed and adjusted to less than 0.5mg/ml. Proteins were then precipitated using 4 volumes of ice-cold acetone for 20 min at −20°C, centrifuged at 2000g for 5min at 4°C, washed twice with 70% acetone and dried out. Precipitate was resuspended in HEN medium (250 mM Hepes pH 7.7, 1 mM EDTA, 0.1 mM neocuproine) containing 2.5% SDS. Free thiols were blocked with a rapidly thiol-reactive agent such as methyl methanethiosulfonate (MMTS; 20 mM) for 30 min at 50°C. After blocking, extracts were precipitated with acetone as described above to remove MMTS and resuspended in HEN medium containing 1% SDS. Finally, 1mM biotin-HPDP and 1mM ascorbate were added and incubated for 1hr at 25°C to achieve biotinylation. Samples were further acetone precipitated as described above. Pellets were resuspended in 1X SDS-PAGE loading buffer (final concentration: 2% sodium dodecyl sulphate (SDS), 10% glycerol, 62.5 mM Tris–HCl, pH 6.8) without reducing agents and the samples were resolved by SDS-PAGE (10%) and transferred for immunoblotting. All steps preceding SDS-PAGE were carried out in the dark. This experiment was repeated six times. Each time either a single or a pool of two ejaculates was used. Different donors were used on each occasion.
2.5 Immunodetection of Proteins by Western Blotting
Proteins were separated by electrophoresis on SDS–PAGE (10%) gels and electrotransferred onto nitrocellulose membrane. Nonspecific binding sites on membranes were blocked with 5% (w/v) dry skimmed milk in Tris-buffered saline (0.9% NaCl, 20 mM Tris–HCl, pH 7.8) supplemented with 0.1% Tween-20 (TTBS) for the detection of biotinylated proteins. The membranes were incubated for 1 h at room temperature with the anti-biotin antibody (1/10 000). The membranes were then extensively washed with TTBS, incubated with corresponding secondary antibody conjugated with horseradish peroxidase for 1 h and again extensively washed with TTBS. Positive immunoreactive bands were detected by chemiluminescence using Lumi-GLO, according to the manufacturer's instructions. Silver staining of the proteins transferred on the nitrocellulose membrane was performed after the detection to confirm equal protein loading for all samples [35].
2.6 In Situ S-Nitrosoprotein Detection
Localisation of S-nitrosoproteins in situ was done using a method adapted from Yang and Loscalzo [36]. This method depends on blocking thiols with a thiol-reactive agent such as MMTS, followed by reducing the S-nitrosothiols with ascorbate, after which the thiols generated by ascorbate reduction are labelled with a fluorescent derivative of methanethiosulfonate (MTSEA). Briefly, spermatozoa were incubated with or without GSH or GSNO for 1 hr at 37°C. Cells were then fixed on slides using 4% formaldehyde at room temperature for 6 min and slides were washed 3 times with HEN containing 0.1% Triton X-100 for 5 min to permeabilise the cells. Thiol groups were then blocked with 20mM MMTS in HEN at 50°C for 30 min. The cells were then washed four times with HEN, after which they were incubated with 1 mM ascorbate and 0.4 μM MTSEA-Texas red in HEN at room temperature for 1 h. Excess dye was removed by washing the cells repeatedly with HEN. Stained cells were then treated with Prolonged Antifade Mounting Medium. All images were taken using an imaging system running Openlab (v. 3.1.7, Improvision Ltd, Coventry UK) with a Hammamatsu Orca Camera attached to a Nikon TE2000 microscope. Fluorescence and Hoffman Modulation Contrast images were take with LWD40X/0.55 MC3 HMC and fluorescence with 100X/1.30 oil Plan Fluor DIC H lens.
2.7 Purification of S-Nitrosylated Proteins
Biotin-labelled proteins were purified according to Foster and Stamler [26] with modifications. Briefly, after biotinylation, proteins were submitted to two round of acetone precipitation and pellets were resuspended in 1/10 diluted HEN/10 media (10X dilution of HEN) containing 1% SDS per mg of proteins. Three volumes of neutralisation buffer (20 mM Hepes pH 7.7, 100 mM NaCl, 1mM EDTA and 0.5% Triton X-100) were added. The mixture was incubated overnight at 4°C with 50μl of streptavidin agarose (50% slurry) per mg of initial proteins. Beads were previously washed twice with Neutralisation buffer and centrifuged at 200g for 10 sec. Once the incubation terminated, the beads were washed 10 times with 500μl of Wash buffer (neutralisation buffer containing 600mM NaCl). Proteins were eluted with 1X SDS sample buffer containing 90 mM of DTT. Samples were boiled for 5 min at 100°C and centrifuged at 14 000 g for 5 min. The eluate was collected and proteins were separated by SDS-PAGE (10%) and visualised by silver staining kit as described in the manufacturer's instructions. Major protein bands (between 10 and 31 bands) were excised and proteomic analyses were performed as described below. The data represents 5 independent experiments, for each of these experiments 4-6 ejaculates (from different donors) were pooled.
2.8 Mass Spectrometry/In-Gel Protein Digestion
Bands from stained 1D SDS gels were cut out and treated with trypsin as previously described [37]. Tryptic peptides obtained from individual bands were subjected to tandem mass spectrometry (MS/MS) on a nanoESI Q-Tof mass spectrometer following separation of peptides using capillary liquid chromatography with a 15 cm C18 PepMap column as previously described [37, 38]. Following MS/MS the raw data were processed using MassLynx 3.5 (Micromass). The resulting tryptic peptide de novo sequences data were then compared with the MSDB non-identical protein sequence and SwissProt curated protein databases using MASCOT software (Matrix Science Ltd, London, UK). Following matching of a peptide to a protein, the quality of the raw MS/MS data was validated (this required the presence of good quality y-ion data). Finally, each of the peptides was used to BLAST search to confirm that the protein identified by MASCOT was the only relevant match in the non-redundant protein database for a particular peptide sequence [39].
2.9 Protein distribution and function
The Gene Ontology database (www.geneontology.org/) was searched for each of the proteins identified as being S-nitrosylated in human sperm as listed in Table 1. For each protein the cellular compartment and function was noted. Gene Ontology entries were not available (March 2007) for all the proteins we identified and these have been reported as ‘No data’.
Table 1.
| Protein Name | Swiss-Prot | Mol. mass (kDa) |
No. of Unique Peptides |
Sequence Coverage (%) |
No. of Exp.c |
Previously Identified as S-Nitrosylated Targets |
|
|---|---|---|---|---|---|---|---|
| Entry Name | Accession No. |
||||||
| Signalling/Regulating Proteins | |||||||
| Leucine-rich repeat protein SHOC-2 | SHOC2_HUMAN | Q9UQ13 | 65.3 | 3 | 5.0 | 4 | |
| Phosphoinositide 3-kinase regulatory subunit 5 |
PI3R5_HUMAN | Q8WYR1 | 98.6 | 2 | 1.1 | 1 | |
| Protein kinase A-anchoring protein 3 | AKAP3_HUMAN | O75969 | 95.4 | 16 | 25.8 | 3 | |
| Protein kinase A-anchoring protein 4 | AKAP4_HUMAN | Q5JQC9 | 95.8 | 28 | 39.9 | 3 | |
| Protein kinase A-anchoring protein 9 | AKAP9_HUMAN | Q99996 | 455.7 | 2 | 0.5 | 2 | |
| 14-3-3 protein zeta/delta | 1433Z_HUMAN | P63104 | 27.9 | 3 | 17.1 | 1 | [27, 28, 41, 91] |
| FKBP12-rapamycin complex- associated protein |
FRAP_HUMAN | P42345 | 290.8 | 2 | 0.7 | 1 | |
| Ras GTPase-activating-like protein IQGAP2 |
IQGA2_HUMAN | Q13576 | 180.9 | 3 | 1.8 | 1 | |
| Ras GTPase-activating protein 4 | RASL2_HUMAN | O43374 | 91.4 | 3 | 3.9 | 2 | |
| Ras-related protein Rab-2A | RAB2A_HUMAN | P61019 | 23.7 | 3 | 22.2 | 1 | |
| Ras-related protein Rab-14 | RAB14_HUMAN | P61106 | 23.7 | 2 | 9.3 | 1 | |
| Ribosome-binding protein 1 | RRBP1_HUMAN | Q9P2E9 | 152.8 | 2 | 1.7 | 1 | |
| Signal-regulatory protein gamma precursor |
SIRPG_HUMAN | Q9P1W8 | 42.9 | 3 | 8.8 | 1 | |
| Protein Kinases/Phosphatases | |||||||
| cAMP-dependent protein kinase, alpha-catalytic subunit |
KAPCA_HUMAN | P17612 | 40.5 | 2 | 6.3 | 1 | |
| cAMP-dependent protein kinase type II-alpha regulatory subunit |
KAP2_HUMAN | P13861 | 45.7 | 3 | 8.4 | 1 | |
| Obscurin-myosin light chain kinase | OBSCN_HUMAN | Q5VST9 | 879.6 | 2 | 0.2 | 1 | |
| Protein phosphatase 1 catalytic subunit gamma isoform |
AAX29836.1 | 37.7 | 1 | 3.7 | 1 | ||
| Proto-oncogene tyrosine-protein kinase Yes |
YES_HUMAN | P07947 | 61.1 | 2 | 3.3 | 1 | |
| Serine/threonine-protein kinase ATR |
ATR_HUMAN | Q13535 | 304.8 | 3 | 1.5 | 1 | |
| Serine/threonine-protein kinase 35 |
STK35_HUMAN | Q8TDR2 | 44.9 | 3 | 7.5 | 1 | |
| Serine/threonine-protein kinase SMG1 |
SMG1_HUMAN | Q96Q15 | 414.1 | 2 | 0.6 | 1 | |
| Tyrosine-protein kinase JAK1 | JAK1_HUMAN | P23458 | 133.6 | 2 | 1.8 | 1 | |
| Receptors | |||||||
| Roundabout homolog 4 precursor | ROBO4_HUMAN | Q8WZ75 | 108.4 | 2 | 2.5 | 1 | |
| Ryanodine receptor 2 | RYR2_HUMAN | Q92736 | 569.3 | 3 | 0.7 | 1 | [71, 76, 92] |
| Transport | |||||||
| Alpha-centractin | ACTZ_HUMAN | P61163 | 42.7 | 3 | 9.8 | 1 | |
| Choline transporter-like protein 1 | CTL1_HUMAN | Q8WWI5 | 74.8 | 2 | 4.6 | 1 | |
| Ferritin, mitochondrial precursor | FTMT_HUMAN | Q8N4E7 | 27.8 | 2 | 10.3 | 1 | [92, 93] |
| Nucleoprotein TPR | TPR_HUMAN | P12270 | 265.8 | 3 | 1.3 | 1 | |
| Probable phospholipid-transporting ATPase IA |
AT8A1_HUMAN | Q9Y2Q0 | 132.6 | 2 | 1.7 | 1 | |
| Serum albumin precursor | ALBU_HUMAN | P02768 | 71.3 | 10 | 18.6 | 2 | [92, 94-97] |
| Sodium channel protein type 2 subunit alpha |
SCN2A_HUMAN | Q99250 | 230.0 | 2 | 0.9 | 1 | |
| Sodium channel protein type 4 subunit alpha |
SCN4A_HUMAN | P35499 | 210.1 | 1 | 0.6 | 3 | |
| Sodium channel protein type 5 subunit alpha |
SCN5A_HUMAN | Q14524 | 229.4 | 1 | 0.6 | 3 | |
| Solute carrier family 2, facilitated glucose transporter member 3 |
GTR3_HUMAN | P11169 | 54.3 | 2 | 1.8 | 2 | |
| Solute carrier organic anion transporter family member 1B1 |
SO1B1_HUMAN | Q9Y6L6 | 77.5 | 2 | 3.2 | 2 | |
| Voltage-dependent anion-selective channel protein 2 |
VDAC2_HUMAN | P45880 | 38.6 | 1 | 4.3 | 2 | |
| Voltage-dependent anion-selective channel protein 3 |
VDAC3_HUMAN | Q9Y277 | 31.0 | 1 | 3.5 | 1 | |
| Structural Proteins | |||||||
| Actin, aortic smooth muscle | ACTA_HUMAN | P62736 | 42.4 | 6 | 20.7 | 2 | |
| Actin, cytoplasmic 1 (Beta-actin) | ACTB_HUMAN | P60709 | 42.1 | 4 | 12.5 | 1 | [40, 91, 97, 98] |
| Actin-related protein T2 | ACTT2_HUMAN | Q8TDY3 | 42.1 | 3 | 5.8 | 1 | |
| Axonemal dynein light intermediate polypeptide 1 |
IDLC_HUMAN | O14645 | 29.9 | 1 | 4.7 | 1 | |
| Calicin | CALI_HUMAN | Q13939 | 67.539 | 1 | 2.2 | 1 | |
| Catenin alpha-2 | CTNA2_HUMAN | P26232 | 105.9 | 3 | 3.4 | 1 | |
| Ciliary dynein heavy chain 9 | DYH9_HUMAN | Q9NYC9 | 515.6 | 1 | 0.3 | 1 | |
| Cytoskeleton-associated protein 2 | CKAP2_HUMAN | Q8WWK9 | 77.5 | 2 | 2.8 | 1 | |
| Desmoplakin | DESP_HUMAN | P15924 | 331.8 | 2 | 0.7 | 1 | |
| Dynein intermediate chain 1, axonemal |
DNAI1_HUMAN | Q9UI46 | 79.7 | 1 | 1.7 | 1 | |
| Gelsolin precursor | GELS_HUMAN | P06396 | 86.0 | 1 | 1.7 | 1 | |
| Hornerin | HORN_HUMAN | Q86YZ3 | 283.1 | 3 | 1.6 | 3 | |
| Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 |
MACF1_HUMAN | Q9UPN3 | 623.6 | 5 | 1.0 | 1 | |
| Microtubule-actin cross linking factor 1, isoform 4 |
MACF4_HUMAN | Q96PK2 | 673.7 | 4 | 1.0 | 2 | |
| Myosin heavy chain, cardiac muscle beta isoform |
MYH7_HUMAN | P12883 | 223.8 | 2 | 1.5 | 1 | |
| Periplakin | PEPL_HUMAN | O60437 | 205.1 | 4 | 2.2 | 1 | |
| Plectin-1 | PLEC1_HUMAN | Q15149 | 531.7 | 3 | 0.9 | 1 | |
| Protein piccolo | PCLO_HUMAN | Q9Y6V0 | 568.3 | 5 | 1.2 | 1 | |
| Rootletin | CROCC_HUMAN | Q5TZA2 | 228.8 | 2 | 0.9 | 1 | |
| Smoothelin | SMOO_HUMAN | P53814 | 100.0 | 3 | 3.2 | 3 | |
| Titin (Connectin) | TITIN_HUMAN | Q8WZ42 | 3843.6 | 7 | 0.3 | 1 | |
| Tubulin alpha-2 chain | TBA2_HUMAN | Q13748 | 50.6 | 9 | 26.7 | 3 | [28, 40, 98] |
| Tubulin alpha-3 chain | TBA3_HUMAN | Q71U36 | 50.8 | 7 | 20.8 | 1 | [28, 40, 98] |
| Tubulin alpha-ubiquitous chain | TBAK_HUMAN | P68363 | 50.8 | 5 | 13.5 | 4 | [28, 40, 98] |
| Tubulin beta-1 chain | TBB1_HUMAN | Q9H4B7 | 50.9 | 2 | 6.9 | 1 | [28, 40] |
| Tubulin beta-2C chain | TBB2C_HUMAN | P68371 | 50.3 | 12 | 32.4 | 5 | [28, 40] |
| Tubulin beta-3 chain | TBB3_HUMAN | Q13509 | 50.9 | 9 | 23.1 | 4 | [28, 40] |
| Tubulin beta-4q chain | TBB4Q_HUMAN | Q99867 | 48.9 | 4 | 11.1 | 1 | [28, 40, 99, 100] |
| Tubulin beta-6 chain | TBB6_HUMAN | Q9BUF5 | 50.3 | 6 | 18.2 | 2 | [28, 40] |
| Stress-Related Proteins | |||||||
| Chaperonin containing TCP1, subunit 3 (gamma) variant |
BAD92119.1 | 61.1 | 4 | 7.8 | 2 | ||
| Chaperonin containing TCP1, subunit 8 |
NP_006576.2 | 60.0 | 3 | 6.9 | 2 | ||
| Endoplasmin precursor | ENPL_HUMAN | P14625 | 92.696 | 4 | 4.9 | 2 | [101] |
| 78 kDa glucose-regulated protein precursor |
GRP78_HUMAN | P11021 | 72.4 | 2 | 3.7 | 1 | [101, 102] |
| Glutathione S-transferase A3 | GSTA3_HUMAN | Q16772 | 25.3 | 2 | 10.8 | 1 | |
| Glutathione S-transferase Mu 2 | GSTM2_HUMAN | P28161 | 25.8 | 1 | 5.2 | 1 | |
| Glutathione S-transferase Mu 3 | GSTM3_HUMAN | P21266 | 26.9 | 7 | 37.3 | 4 | |
| Heat shock 70 kDa protein 1 | HSP71_HUMAN | P08107 | 71.082 | 5 | 8.1 | 1 | |
| Heat shock 70 kDa protein 1L | HS70L_HUMAN | P34931 | 70.73 | 5 | 9.5 | 1 | |
| Heat shock 70 kDa protein 6 | HSP76_HUMAN | P17066 | 71.44 | 7 | 10.4 | 4 | |
| Heat shock cognate 71 kDa protein | HSP7C_HUMAN | P11142 | 71.082 | 8 | 11.3 | 4 | [97, 101] |
| Heat shock protein 60 kDa | CH60_HUMAN | P10809 | 61.187 | 3 | 7.2 | 2 | [28, 97, 102, 103] |
| Heat shock protein 75 kDa | TRAP1_HUMAN | Q12931 | 80.345 | 1 | 2.3 | 2 | |
| Heat shock protein HSP 90-alpha | HS90A_HUMAN | P07900 | 84.875 | 23 | 25.4 | 4 | [40, 99, 100, 103-105] |
| Heat shock protein HSP 90-beta | HS90B_HUMAN | P08238 | 83.423 | 12 | 18.5 | 2 | [40, 99, 100, 103-105] |
| Heat shock-related 70 kDa protein 2 | HSP72_HUMAN | P54652 | 70.263 | 22 | 36.2 | 4 | |
| 2-hydroxyacyl-CoA lyase 1 | HACL1_HUMAN | Q9UJ83 | 64.4 | 2 | 3.6 | 1 | |
| KIAA0098 | BAA07894.2 | 60.1 | 4 | 8.3 | 2 | ||
| Peroxisome biogenesis factor 1 | PEX1_HUMAN | O43933 | 143.8 | 2 | 2.0 | 4 | |
| Stress-70 protein, mitochondrial precursor |
GRP75_HUMAN | P38646 | 73.9 | 5 | 9.3 | 1 | [27, 102] |
| T-complex 1 | AAP36354.1 | 60.8 | 1 | 2.2 | 1 | [103] | |
| T-complex protein 1 subunit zeta-2 | TCPW_HUMAN | Q92526 | 58.2 | 1 | 2.1 | 1 | [27] |
| Metabolic Enzymes | |||||||
| Acetyl-CoA acetyltransferase, mitochondrial precursor |
THIL_HUMAN | P24752 | 45.5 | 6 | 15.9 | 1 | [102] |
| Adenylate kinase isoenzyme 1 | KAD1_HUMAN | P00568 | 21.7 | 3 | 16.0 | 2 | |
| Alpha-enolase | ENOA_HUMAN | P06733 | 47.3 | 10 | 31.6 | 3 | [30, 97-101, 103, 105] |
| Aspartate aminotransferase, cytoplasmic |
AATC_HUMAN | P17174 | 46.3 | 2 | 6.5 | 1 | |
| Aspartate aminotransferase, mitochondrial precursor |
AATM_HUMAN | P00505 | 47.8 | 1 | 2.6 | 1 | [26] |
| 3′(2′), 5′-biphosphate nucleotidase 1 | BPNT1_HUMAN | O95861 | 33.7 | 1 | 3.3 | 1 | |
| Carbonic anhydrase 2 | CAH2_HUMAN | P00918 | 29.154 | 1 | 4.2 | 1 | [100, 106, 107] |
| Carnitine O-acetyltransferase | CACP_HUMAN | P43155 | 71.3 | 3 | 4.8 | 1 | |
| 11-cis retinol dehydrogenase | RDH1_HUMAN | Q92781 | 35.3 | 1 | 4.4 | 1 | |
| Citrate synthase, mitochondrial precursor |
CISY_HUMAN | O75390 | 51.9 | 6 | 15.0 | 3 | |
| Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor |
ECH1_HUMAN | Q13011 | 36.1 | 3 | 9.2 | 1 | |
| 2,4-dienoyl-CoA reductase, mitochondrial precursor |
DECR_HUMAN | Q16698 | 36.3 | 5 | 16.1 | 1 | [102] |
| Dihydrolipoyl dehydrogenase, mitochondrial precursor |
DLDH_HUMAN | P09622 | 54.7 | 3 | 8.3 | 1 | [26] |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor |
ODP2_HUMAN | P10515 | 66.2 | 2 | 3.6 | 2 | |
| Dihydroxyacetone kinase | DAK_HUMAN | Q3LXA3 | 59.3 | 2 | 3.5 | 1 | |
| Ethanolamine-phosphate cytiddylyltransferase |
PCY2_HUMAN | Q99447 | 44.3 | 1 | 4.1 | 1 | |
| Fructose-bisphosphate aldolase A | ALDOA_HUMAN | P04075 | 39.7 | 11 | 34.3 | 3 | [99, 100, 103, 105, 108] |
| Fumarate hydratase, mitochondrial precursor |
FUMH_HUMAN | P07954 | 54.8 | 6 | 14.1 | 1 | |
| Gamma-glutamyl hydrolase precursor |
GGH_HUMAN | Q92820 | 36.3 | 2 | 9.8 | 1 | |
| Glucose-6-phosphate isomerase | G6PI_HUMAN | P06744 | 63.2 | 6 | 44.2 | 2 | |
| Glutamine-dependent NAD (+) synthetase |
NADE1_HUMAN | Q6IA69 | 80.5 | 1 | 1.6 | 1 | |
| Glutamine synthetase | GLNA_HUMAN | P15104 | 42.5 | 4 | 11.1 | 2 | [30] |
| Glyceraldehyde-3-phosphate dehydrogenase |
G3P_HUMAN | P04406 | 36.1 | 1 | 3.6 | 2 | [28, 40, 41, 91, 92, 97, 98, 105, 107, 109-114] |
| Hexokinase-1 | HXK1_HUMAN | P19367 | 103.6 | 16 | 18.3 | 2 | [40, 92] |
| Histidine ammonia-lyase | HUTH_HUMAN | P42357 | 73.3 | 1 | 1.6 | 1 | |
| 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor |
3HIDH_HUMAN | P31937 | 35.7 | 1 | 4.5 | 1 | [102] |
| Kinesin family member 21B | KI21B_HUMAN | O75037 | 184.3 | 3 | 2.3 | 1 | |
| Lactotransferrin | TRFL_HUMAN | P02788 | 80.0 | 8 | 13.2 | 2 | |
| L-lactate dehydrogenase A chain | LDHA_HUMAN | P00338 | 36.8 | 7 | 27.1 | 3 | |
| L-Lactate dehydrogenase B chain | LDHB_HUMAN | P07195 | 36.8 | 4 | 14.7 | 3 | [101] |
| L-lactate dehydrogenase A-like 6A | LDH6A_HUMAN | Q6ZMR3 | 36.8 | 2 | 7.2 | 1 | |
| L-lactate dehydrogenase A-like 6B | LDH6B_HUMAN | Q9BYZ2 | 42.4 | 3 | 8.7 | 2 | |
| Long-chain-fatty-acid-CoA ligase 6 |
ACSL6_HUMAN | Q9UKU0 | 78.8 | 1 | 1.9 | 1 | |
| Malate dehydrogenase, cytoplasmic |
MDHC_HUMAN | P40925 | 36.5 | 1 | 3.3 | 1 | [28, 97, 100, 115] |
| Malate dehydrogenase, mitochondrial precursor |
MDHM_HUMAN | P40926 | 36.0 | 13 | 43.8 | 3 | [26, 100, 102] |
| Mitochondrial dicarboxylate carrier |
DIC_HUMAN | Q9UBX3 | 31.7 | 1 | 3.5 | 1 | |
| NAD-dependent malic enzyme, mitochondrial precursor |
MAOM_HUMAN | P23368 | 66.0 | 2 | 1.5 | 1 | |
| 6-phosphofructokinase type C | K6PP_HUMAN | Q01813 | 86.4 | 11 | 13.5 | 2 | [28] |
| Phosphoglycerate kinase 1 | PGK1_HUMAN | P00558 | 44.8 | 2 | 4.6 | 1 | [100, 103] |
| Phosphoglycerate mutase 1 | PGAM1_HUMAN | P18669 | 28.8 | 1 | 4.7 | 1 | |
| Phosphoglycerate mutase 2 | PGAM2_HUMAN | P15259 | 28.8 | 4 | 18.6 | 2 | |
| Probable saccharopine dehydrogenase |
SCPDH_HUMAN | Q8NBX0 | 47.5 | 7 | 20.8 | 2 | |
| Protein disulfide-isomerase A3 precursor |
PDIA3_HUMAN | P30101 | 57.1 | 2 | 5.5 | 1 | [97, 102, 103] |
| Pyruvate dehydrogenase E1 component alpha subunit, testis specific form, mitochondrial precursor |
ODPAT_HUMAN | P29803 | 43.6 | 3 | 8.5 | 1 | |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial precursor |
ODPB_HUMAN | P11177 | 39.5 | 2 | 6.4 | 1 | |
| Pyruvate kinase isozymes M1/M2 | KPYM_HUMAN | P14618 | 58.3 | 14 | 29.2 | 3 | [28, 98] |
| Transaldolase | TALDO_HUMAN | P37837 | 37.7 | 2 | 6.8 | 1 | |
| Triosephosphate isomerase | TPIS_HUMAN | P60174 | 26.8 | 9 | 44.2 | 3 | [98-100, 103, 105, 108] |
| Cell Cycle | |||||||
| Apoptosis-stimulating of p53 protein 2 |
ASPP2_HUMAN | Q13625 | 126.2 | 1 | 0.9 | 3 | |
| Histone H1.3 | H13_HUMAN | P16402 | 22.2 | 2 | 10.0 | 1 | |
| Microtubule-associated protein 1A |
MAP1A_HUMAN | P78559 | 307.6 | 1 | 0.3 | 1 | |
| Nicotinamide phosphoribosyltransferase |
NAMPT_HUMAN | P43490 | 55.8 | 1 | 2.7 | 1 | |
| Peroxiredoxin-1 | PRDX1_HUMAN | Q06830 | 22.3 | 1 | 6.0 | 1 | [91, 103] |
| Translin | TSN_HUMAN | Q15631 | 26.3 | 2 | 9.7 | 2 | |
| Transcription Factors | |||||||
| Centromere protein F | CENPF_HUMAN | P49454 | 357.4 | 2 | 0.8 | 1 | |
| HIV Tat-specific factor 1 | HTSF1_HUMAN | O43719 | 86.4 | 3 | 4.4 | 2 | |
| Protein bassoon | BSN_HUMAN | Q9UPA5 | 418.2 | 6 | 1.6 | 2 | [28] |
| SW1/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 |
SMRC1_HUMAN | Q92922 | 123.2 | 2 | 2.4 | 1 | |
| Zinc finger protein 395 | ZN395_HUMAN | Q9H8N7 | 55.7 | 1 | 1.6 | 1 | |
| Zinc finger protein 512 | ZN512_HUMAN | Q96ME7 | 65.7 | 3 | 5.6 | 1 | |
| Zinc finger protein 696 | ZN696_HUMAN | Q9H7X3 | 41.7 | 2 | 5.9 | 1 | |
| Zinc finger protein 740 | ZN740_HUMAN | Q8NDX6 | 22.5 | 1 | 5.7 | 1 | |
| Protein Binding | |||||||
| Ankyrin-3 | ANK3_HUMAN | Q12955 | 482.4 | 5 | 1.4 | 1 | |
| Ankyrin repeat domain-containing protein 11 |
ANR11_HUMAN | Q6UB99 | 299.8 | 4 | 1.6 | 2 | |
| Calcium-binding tyrosine phosphorylation-regulated protein |
CABYR_HUMAN | O75952 | 52.9 | 10 | 19.7 | 4 | |
| Coiled-coil alpha-helical rod protein 1 |
CCHCR_HUMAN | Q8TD31 | 88.9 | 1 | 1.0 | 1 | |
| Coiled-coil domain-containing protein 71 |
CCD71_HUMAN | Q8IV32 | 49.8 | 3 | 6.4 | 2 | |
| Collagen alpha-2 (VI) chain precursor |
CO6A2_HUMAN | P12110 | 109.7 | 2 | 2.2 | 1 | |
| Collagen alpha-3 (IV) chain precursor |
CO4A3_HUMAN | Q01955 | 163.0 | 2 | 1.7 | 1 | |
| Cullin-3 | CUL3_HUMAN | Q13618 | 89.4 | 3 | 4.4 | 3 | |
| Cytoskeleton-associated protein 5 | CKAP5_HUMAN | Q14008 | 225.5 | 2 | 0.9 | 1 | |
| DNA replication licensing factor MCM6 |
MCM6_HUMAN | Q14566 | 93.8 | 4 | 5.9 | 1 | |
| Elongation factor 1-gamma | EF1G_HUMAN | P26641 | 50.3 | 4 | 10.2 | 1 | |
| Elongation factor 2 | EF2_HUMAN | P13639 | 96.1 | 5 | 6.1 | 1 | [27, 98, 100, 101, 103, 105] |
| Extracellular matrix protein 2 precursor |
ECM2_HUMAN | O94769 | 80.6 | 3 | 5.2 | 1 | |
| Liprin-alpha-1 | LIPA1_HUMAN | Q13136 | 136.3 | 2 | 1.7 | 1 | |
| Neurexin-1-alpha precursor | NRX1A_HUMAN | Q9ULB1 | 164.1 | 3 | 2.2 | 1 | |
| Plasminogen | AAH60513.1 | 93.3 | 1 | 1.5 | 2 | [95, 116] | |
| Ran-binding protein 2-like 3 | RBP23_HUMAN | O14715 | 84.9 | 3 | 4.9 | 1 | |
| RuvB-like 1 | RUVB1_HUMAN | Q9Y265 | 50.5 | 3 | 8.6 | 1 | |
| RuvB-like 2 | RUVB2_HUMAN | Q9Y230 | 51.2 | 4 | 10.2 | 1 | |
| Tankyrase-1 | TNKS1_HUMAN | O95271 | 143.7 | 3 | 1.7 | 1 | |
| Transitional endoplasmic reticulum ATPase |
TERA_HUMAN | AAC07984.1 | 89.8 | 5 | 7.7 | 1 | [100] |
| Vang-like protein 2 | VANG2_HUMAN | Q9ULK5 | 59.9 | 2 | 4.2 | 1 | |
| Protein turnover | |||||||
| Cytosol aminopeptidase | AMPL_HUMAN | P28838 | 53.0 | 2 | 5.5 | 2 | |
| E3 Ubiquitin-protein ligase UBR1 | UBR1_HUMAN | Q8IWV7 | 203.4 | 2 | 1.0 | 1 | |
| Matrix metalloproteinase-24 precursor |
MMP24_HUMAN | Q9Y5R2 | 73.6 | 1 | 1.9 | 1 | |
| Probable aminopeptidase NPEPL1 |
PEPL1_HUMAN | Q8NDH3 | 56.7 | 1 | 2.7 | 1 | |
| Probable serine carboxypeptidase CPVL precursor |
CPVL_HUMAN | Q9H3G5 | 54.4 | 2 | 5.9 | 1 | |
| 26S protease regulatory subunit 4 | PRS4_HUMAN | P62191 | 49.3 | 2 | 5.7 | 1 | |
| Proteasome subunit alpha type 2 | PSA2_HUMAN | P25787 | 25.9 | 1 | 5.6 | 2 | |
| Proteasome subunit alpha type 3 | PSA3_HUMAN | P25788 | 28.5 | 1 | 5.5 | 1 | |
| Proteasome subunit alpha type 5 | PSA5_HUMAN | P28066 | 26.6 | 2 | 9.5 | 1 | |
| Proteasome subunit alpha type 6 | PSA6_HUMAN | P60900 | 27.8 | 1 | 6.1 | 1 | |
| Proteasome subunit alpha type 7-like | PSA7L_HUMAN | Q8TAA3 | 28.7 | 2 | 11.3 | 1 | |
| Proteasome subunit beta type 3 | PSB3_HUMAN | P49720 | 23.2 | 1 | 5.4 | 1 | |
| Proteasome subunit beta type 5 precursor |
PSB5_HUMAN | P28074 | 23.0 | 1 | 7.2 | 1 | |
| Proteasome subunit beta type 6 precursor |
PSB6_HUMAN | P28072 | 25.6 | 3 | 9.6 | 2 | |
| Serine protease inhibitor Kazal-type 5 precursor |
ISK5_HUMAN | Q9NQ38 | 124.4 | 2 | 2.1 | 1 | |
| Energy | |||||||
| ATP synthase subunit alpha, mitochondrial precursor |
ATPA_HUMAN | P25705 | 59.8 | 5 | 10.1 | 2 | [30, 100, 102, 117] |
| ATP synthase B Chain, mitochondrial precursor |
AT5F1_HUMAN | P24539 | 28.9 | 1 | 3.9 | 1 | [100, 102] |
| ATP synthase gamma chain, mitochondrial precursor |
ATPG_HUMAN | P36542 | 33.0 | 2 | 8.4 | 2 | |
| ATP synthase O subunit, mitochondrial precursor |
ATPO_HUMAN | P48047 | 23.4 | 2 | 12.6 | 1 | |
| Electron transfer flavoprotein subunit beta |
ETFB_HUMAN | P38117 | 27.9 | 6 | 25.9 | 2 | [102] |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor |
DHSA_HUMAN | P31040 | 73.7 | 3 | 6.2 | 2 | [102] |
| Trifunctoinal enzyme subunit alpha, mitochondrial precursor |
ECHA_HUMAN | P40939 | 83.7 | 5 | 7.2 | 1 | |
| Trifunctional enzyme subunit beta, mitochondrial precursor |
ECHB_HUMAN | P55084 | 51.5 | 3 | 7.2 | 1 | |
| 4-trimethylaminobutyraldehyde dehydrogenase |
AL9A1_HUMAN | P49189 | 54.7 | 1 | 2.6 | 1 | [92, 118] |
| Testis/Sperm Proteins | |||||||
| Acrosin-binding protein precursor | ACRBP_HUMAN | Q8NEB7 | 62.4 | 5 | 9.8 | 3 | |
| Acrosin precursor | ACRO_HUMAN | P10323 | 25.5 | 4 | 11.9 | 3 | |
| Glyceraldehyde-3-phosphate dehydrogenase, testis specific |
G3PT_HUMAN | O14556 | 44.5 | 11 | 36.3 | 4 | |
| L-lactate dehydrogenase C chain | LDHC_HUMAN | P07864 | 36.5 | 17 | 53.0 | 4 | |
| Nucleoside diphosphate kinase homolog 5 |
NDK5_HUMAN | P56597 | 24.4 | 1 | 4.7 | 1 | |
| Outer dense fiber of sperm tails 2 isoform 3 |
AAP83847.1 | 76.1 | 5 | 8.8 | 1 | ||
| Outer dense fiber protein | ODFP_HUMAN | Q14990 | 30.4 | 3 | 11.2 | 2 | |
| Phosphoglycerate kinase, testis specific |
PGK2_HUMAN | P07205 | 45.0 | 10 | 29.5 | 3 | |
| RANBP2-like and GRIP domain- containing protein 8 |
RGPD8_HUMAN | Q99666 | 200.0 | 3 | 1.8 | 1 | |
| Semenogelin-1 precursor | SEMG1_HUMAN | P04279 | 52.2 | 3 | 9.1 | 2 | |
| Semenogelin-2 precursor | SEMG2_HUMAN | Q02383 | 65.5 | 4 | 8.6 | 2 | |
| Sperm protein associated with the nucleus on the X chromosome A |
SPNXA_HUMAN | Q9NS26 | 11.1 | 1 | 13.4 | 2 | |
| Sperm protein associated with the nucleus on the X chromosome B/F |
SPNXB_HUMAN | Q9NS25 | 11.9 | 3 | 47.6 | 3 | |
| Testis-specific Y-encoded-like protein 3 |
TSYL3_HUMAN | Q9H489 | 39.8 | 2 | 4.5 | 1 | |
| Zona pellucida-binding protein 1 | ZPBP1_HUMAN | Q9BS86 | 41.0 | 5 | 12.5 | 3 | |
| Others | |||||||
| Dermcidin precursor | DCD_HUMAN | P81605 | 11.4 | 2 | 22.7 | 1 | |
| Disks large-associated protein 3 | DLGP3_HUMAN | O95886 | 106.9 | 1 | 1.0 | 1 | |
| DNA (cytosine-5)- methyltransferase 1 |
DNMT1_HUMAN | P26358 | 185.4 | 3 | 2.3 | 1 | [92, 119, 120] |
| DnaJ homolog subfamily B member 6 |
DNJB6_HUMAN | O75190 | 36.122 | 3 | 9.5 | 2 | |
| ES1 protein homolog, mitochondrial precursor |
ES1_HUMAN | P30042 | 28.5 | 3 | 13.1 | 2 | |
| FERM domain-containing protein 4A |
FRM4A_HUMAN | Q9P2Q2 | 114.5 | 3 | 3.2 | 1 | |
| GRAM domain-containing protein 3 | GRAM3_HUMAN | Q96HH9 | 48.3 | 2 | 5.1 | 1 | |
| Hemicentin-1 precursor | HMCN1_HUMAN | Q96RW7 | 62.3 | 4 | 1.0 | 1 | |
| Kelch-like protein 10 | KLH10_HUMAN | Q6JEL2 | 69.9 | 2 | 3.6 | 1 | |
| Leucine-rich repeat-containing protein 37A |
LR37A_HUMAN | O60309 | 181.6 | 1 | 0.6 | 1 | |
| Leucine-rich repeat-containing protein 37B precursor |
LR37B_HUMAN | Q96QE4 | 106.4 | 6 | 8.3 | 3 | |
| Myeloid leukaemia factor 1 | MLF1_HUMAN | P58340 | 30.7 | 3 | 11.9 | 2 | |
| Myeloid/lymphoid or mixed-lineage leukaemia protein 3 homolog |
MLL3_HUMAN | Q8NEZ4 | 548.2 | 5 | 1.0 | 2 | |
| Nesprin-2 | SYNE2_HUMAN | Q8WXH0 | 801.8 | 3 | 0.5 | 1 | |
| Periaxin | PRAX_HUMAN | Q9BXM0 | 155.2 | 2 | 1.4 | 1 | |
| Poly [ADP-ribose] polymerase 14 | PAR14_HUMAN | Q460N5 | 195.6 | 3 | 1.6 | 1 | |
| Prostate-specific antigen precursor |
KLK3_HUMAN | P07288 | 29.3 | 2 | 8.8 | 1 | |
| Protein FAM29A | FA29A_HUMAN | Q7Z4H7 | 109.6 | 2 | 2.5 | 1 | |
| Regulating synaptic membrane exocytosis protein 1 |
RIMS1_HUMAN | Q86UR5 | 189.0 | 4 | 2.5 | 3 | |
| Scaffold attachment factor B | SAFB1_HUMAN | Q15424 | 103.0 | 3 | 3.7 | 1 | |
| SH3 domain and tetratricopeptide repeats-containing protein 1 |
S3TC1_HUMAN | Q8TE82 | 148.5 | 2 | 2.0 | 2 | |
| Spindle assembly abnormal protein 6 homolog |
SAS6_HUMAN | Q6UVJ0 | 74.8 | 2 | 4.1 | 1 | |
| Stathmin-2 | STMN2_HUMAN | Q93045 | 20.9 | 2 | 11.7 | 1 | |
| Targeting protein for Xklp2 | TPX2_HUMAN | Q9ULW0 | 86.2 | 2 | 2.4 | 1 | |
| Thioredoxin domain-containing protein 3 |
TXND3_HUMAN | Q8N427 | 67.7 | 3 | 6.6 | 1 | |
| Thioredoxin domain-containing protein 11 |
TXD11_HUMAN | Q6PKC3 | 111.8 | 2 | 2.4 | 2 | |
| Tripartite motif-containing protein 34 |
TRI34_HUMAN | Q9BYJ4 | 58.1 | 2 | 1.9 | 1 | |
| Tryptophane aspartate-containing coat protein |
AF495470_1 | AAM18516 | 51.5 | 1 | 3.9 | 1 | |
| Uncharacterized protein C6orf54 | CF054_HUMAN | Q9Y6Z4 | 20.1 | 2 | 12.2 | 1 | |
MOWSE scores >33
See supplementary Table S1 for additional information.
Number of experiment refers to how many times a particular protein was identified out of a total of 5 independent experiments.
We interrogated available literature for previous reports of S-nitrosylation for each protein we identified and we believe these to be correct at the time of writing (March 2007).
3. Results and Discussion
3.1 Detection of S-Nitrosylated Proteins in Human Spermatozoa
To detect S-nitrosylated proteins, we used the biotin-switch assay developed by Jaffrey et al. [40] which is a sensitive and selective methods that is based on the labelling of S-nitrosylated cysteine on targeted proteins with a biotin moiety. This assay is based on the ability of ascorbate selectively to reduce S-nitrosothiols without reducing disulfide bonds. The method involves three sequential steps: blockage of the cellular free thiols by incubation with a thiol-specific methylthiolating agent (MMTS), specific reduction of S-nitrosothiols to thiols by ascorbate, and labelling of the nascent thiol with biotin. Detection of biotinylated proteins was performed by immunoblotting with an anti-biotin antibody [40].
After sperm were incubated with GSNO (Figure 1) or with other NO donors such as S-nitrosocysteine [CSNO] or spermine NONOate (data not shown), over 30 protein bands comprising a wide range of molecular weights were detected with the biotin switch assay (Figure 1). By contrast, in untreated samples or samples treated with GSH, only a few weak protein bands could be seen, confirming that the detected bands observed in the GSNO-treated samples can be attributed to NO (Figure 1). The very low levels of staining in control preparations (no treatment, GSH or exhausted CSNO) confirm that the vast majority of thiols were successfully blocked by MMTS and were unable to react with biotin-HPDP. Moreover, analyses of GSNO-treated extracts that underwent the biotin switch method without a blocking step (no MMTS) resulted in high levels of non-specific unspecific biotinylation (Figure 1A). The low levels of S-nitrosylated proteins observed in untreated, GSH and exhausted CSNO-treated cells may represent endogenous S-nitrosylated proteins.
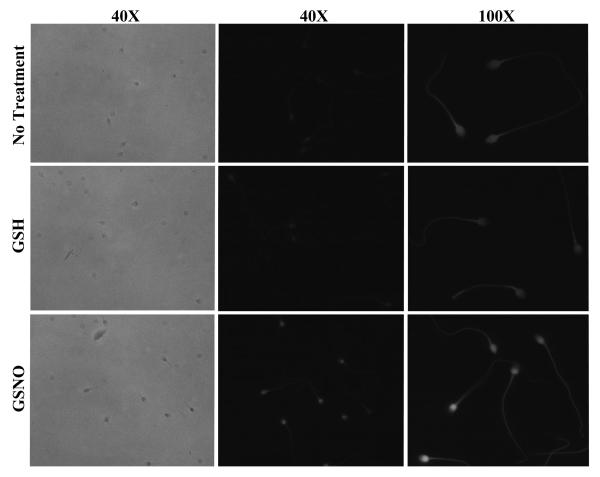
Figure 1. Detection of S-nitrosylated proteins in human spermatozoa with biotin switch assay.
Intact human sperm were treated with GSNO, GSH, CSNO, exhausted CSNO or oxidised GSH (GSSG) or incubated with no treatment (no treatment) for 1hr at 37°C prior biotin switch assay as described in Materials and Methods. A series of controls were performed to assess the specificity of the biotin switch assay. A) Firstly where indicated, the assay was tested in the absence of biotin-HPDP to show if any endogenously biotinylated proteins could be detected. Secondly where indicated, labelling with biotin-HPDP was performed with or without ascorbate, MMTS or in presence of DTT. B) GSSG was used to assess that the assay was specific for S-nitrosylation and not S-glutathionylation. Proteins were separated by SDS-PAGE (10%) and transferred onto nitrocellulose membrane. Detection of biotinylated proteins was achieved using anti-biotin antibody. The relative masses of protein standards are shown on the left. Results of 1 experiment presented are representative of 6 others performed with different sperm donors.
No reactivity was detected in GSNO-treated spermatozoa in which biotin-HPDP was omitted (Figure 1A). Furthermore, no protein bands could be detected in GSNO-treated samples that underwent the biotin switch procedure followed by DTT treatment (Figure 1A). This data demonstrate that the anti-biotin antibody shows no non-specific cross reaction with unlabeled proteins and that no in vivo biotinylated proteins were detected in this study.
When the ascorbate reduction step was omitted, GSNO-treated cells showed very low but detectable levels of biotinylation, similar to those observed in untreated or GSH-treated samples. In this assay, ascorbate is used selectively to reduce nitrosothiols in order that they can be labelled by biotin. However, this may also occur spontaneously during the biotinylation step, an effect observed in another study [26] which may explain the low levels of S-nitrosylation detected in the absence of ascorbate reduction.
Finally, the specificity of the assay for S-nitrosylation was verified by treating spermatozoa with oxidised GSH (GSSG) as it has been suggested that the biotin switch assay may also detect protein S-glutathionylation [41]. Only faint bands were detected with GSSG-dependent labelling (Figure 1B) similar to the ones observed in control samples. Taken together, these results confirm the specificity of the biotin switch method for detection of S-nitrosylated proteins in human spermatozoa.
The above experiments were carried out on spermatozoa exposed to non-capacitating conditions. We performed several experiments where the same procedures (detection and identification of S-nitrosylated proteins, localisation of S-nitrosylated proteins) were carried out on spermatozoa incubated under capacitating conditions but could not detect any consistent differences compared to spermatozoa incubated under non-capacitating conditions (data not shown). Thus, the results presented herein are from spermatozoa incubated under non-capacitating conditions.
3.2 Localisation S-Nitrosoproteins in Human Spermatozoa
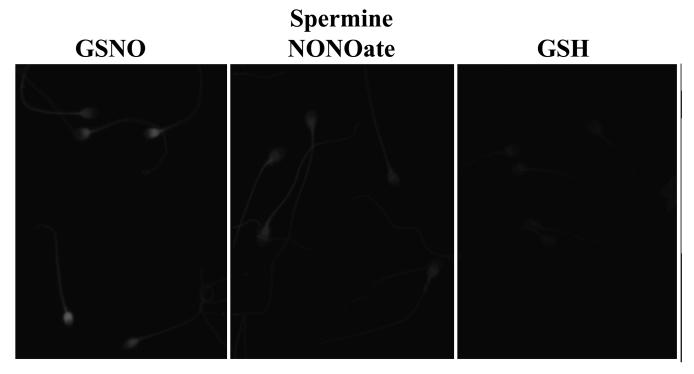
Spermatozoa treated with GSNO or CSNO (≥25 μM) and labelled with MTSEA-Texas red showed bright fluorescence over the head, mid and principal piece of the tail but with the greatest intensities over the post-acosomal region of the sperm head (Figure 2 and 3). The pattern was similar in sperm treated with spermine NONOate (Figure 3) or with lower concentrations of GSNO or CSNO (data not shown) but staining in the post acrosomal region was less intense. By contrast, control slides (untreated or GSH-treated spermatozoa) showed faint fluorescence on the entire length of the sperm (head and tail; Figure 2 and 3). Fluorescence was reduced to control levels by the omission of ascorbate (data not shown) confirming the specificity of this assay. In spermatozoa treated without MMTS to block thiol groups there was strong fluorescence throughout the entire cell (data not shown). To determine if the weak staining observed in control cells was due to endogenous S-nitrosylation we treated samples with the NOS-inhibitor, N(G)-nitro-L-arginine methyl ester hydrochloride (L-NAME) or with DTT. This failed to decrease the level of fluoresence suggesting that the fluorescence observed in controls was due to non-specific labelling (data not shown).
Figure 2. In situ detection of S-nitrosylated targets in human spermatozoa.
Spermatozoa were incubated without (no treatment; top) or with GSH (middle) or GSNO (bottom) for 1 hr prior S-nitrosylated proteins labelling with Texas-red fluorescence-MTSEA as described in Materials and Methods. Left and middle columns represent phase and fluorescence images, respectively, using the 40X objective. On the right column, a more detailed localisation of sperm S-nitrosylation labelling is shown using 100X objective. Results of 1 experiment presented are representative of 6 others performed with different sperm donors.
Figure 3. In situ detection of S-nitrosylated targets using different nitric oxide donors.
Spermatozoa were incubated with GSH (top), spermine NONOate (middle) or GSNO (bottom) for 1 hr prior S-nitrosylated proteins labelling with Texas-red fluorescence-MTSEA as described in Materials and Methods. Fluorescence images were obtained using using 100X objective. Results of 1 experiment presented are representative of 3 others performed with different sperm donors.
The general distribution of labelling is to be expected in view of the large number of S-nitrosylation targets identified (see Table 1; Supplementary Table S1). The more intense labelling produced by GSNO (Figure 2 and 3) and CSNO (data not shown) was probably because these agents can induce protein S-nitrosylation by transnitrosylation as well as by acting as NO donors. The former mechanism is likely to predominate since transnitrosylation reactions are faster than NO release [42]. By contrast, spermine NONOate acts as a pure NO donor and NO probably has to be oxidised to N2O3, a relatively slow reaction except at high NO concentrations [43] before it can react with thiols to produce S-nitrosothiols.
3.3 Proteomic Identification of S-Nitrosylated Protein from GSNO-Treated Human Spermatozoa
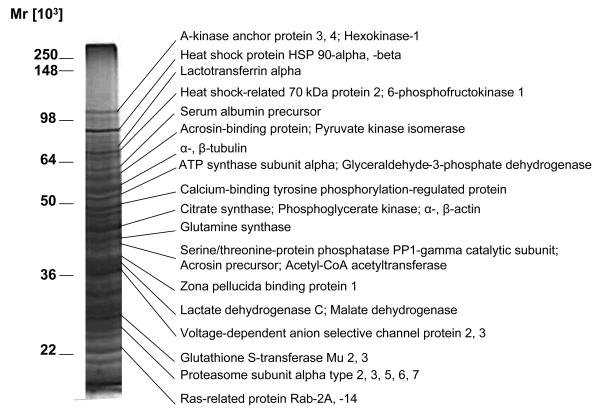
Following treatment of sperm by NO donors and the biotin switch assay, biotinylated proteins were affinity purified using streptavidin-agarose and proteins selectively eluted with DTT, which cleaved the disulphide bonds formed by biotin-HPDP. Proteins were then separated by SDS-PAGE, visualised by silver staining and the protein bands of interest excised from the gel, digested with trypsin, and analysed by MALDI-TOF mass spectrometry (Figure 4; Table 1; Supplementary Table S1).
Figure 4. Isolation and identification of S-nitrosoproteins in human spermatozoa.
Biotinylated sperm proteins obtained from the biotin switch assay were purified by affinity chromatography using streptavidin-agarose and eluted with sample buffer containing DTT as described in Materials and Methods. Proteins were separated by SDS-PAGE (10%) and visualized by silver staining. The relative masses of protein standards are shown on the left. Individual bands were trypsinised and submitted to MS/MS for protein identification. More than 1 protein was found for each bands (see Table 1). Results of 1 experiment are representative of 5 others performed with different pools of sperm donors. Proteins shown on the right side of the gel are a sample of known sperm proteins that are shown in Table 1.
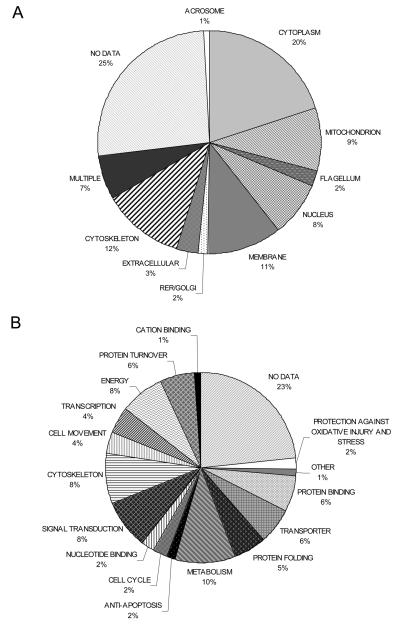
In this study we have identified 240 proteins present in sperm from normozoospermic men which, in the presence of nitric oxide donors, undergo S-nitrosylation at cysteine residues. These proteins are listed in Table 1 and Supplementary Table S1. Comparison with the only publicly available human sperm proteome found only 30 (12%) in common [44]. An earlier study estimated that the human sperm proteome comprises at least 1760 proteins [45] indicating that the capacity of proteins to become S-nitrosylated upon exposure to NO is not ubiquitous; an essential characteristic of a modification if it is to have specific downstream effects. Analysis of both the cellular localisation and the cellular function of the identified proteins were performed. In both cases the largest group identified was that for which there was no information available in the Gene Ontology (GO) database; 23% and 22% of proteins, respectively. This is in keeping with the findings of the Johnson et al., [45] and also with a recent microarray analysis of gene expression in the testis of a set of infertile men where, together “spermatogenesis” and “unknown” were the most significant GO categories enriched in germ cell genes [46].
The cellular distribution of the 240 identified proteins is shown in Figure 5A. Discounting those proteins with no associated gene ontology, the most commonly identified location was the cytoplasm (21%). This was somewhat surprising given that mature spermatozoa shed the majority of their cytoplasm in a residual body during spermiation. This most likely is a reflection of the morphological differences between sperm and somatic cells. Sperm are highly specialised cells which contain a number of unique, specialised organelles and it is likely that the cellular location of many proteins differs between sperm and other cell types. For example, in sperm many of the glycolytic enzymes are found in the flagellum compared to the cytoplasm in somatic cells [47]. It should also be noted that there is only a small amount of sperm-specific data in collections such as the Gene Ontology database. The next most populated compartments were cytoskeleton (13%), membrane (10%), mitochondria (10%) and nucleus (8%). We saw an enrichment of membranous and nuclear proteins and a decrease in the proportion of mitochondrial proteins compared with the study of Martinez-Heredia et al. [44].
Figure 5. Ontology of the proteins identified as being S-nitrosylated in human spermatozoa.
A) Distribution of the proteins in the different compartments of the spermatozoon. B) Biological function of the proteins.
The functional analysis of the proteins identified is shown in Figure 5B. Metabolic proteins and protein associated with energy generation and cell movement were found to be abundant (22% when combined) suggesting a role for S-nitrosylation in sperm motility. Another abundant group of proteins were those involved in signal transduction (9%), fitting with a role for S-nitrosylation in the modulation of sperm function. Surprisingly, given that sperm are generally assumed to be transcriptionally inactive, 4% of the proteins that we identified as being S-nitrosylated were associated with transcription. The presence of proteins involved in transcription and protein synthesis has been previously observed in sperm proteomic studies [44, 48] and is further discussed in Lefièvre et al. [49]. We were interested to note that when we compared the human sperm S-nitrosoproteome with proteins identified during a proteomic analysis of sperm-egg interaction we found only 3 proteins in common suggesting that S-nitrosylation is not a regulatory mechanism employed during fertilisation [50].
We next interrogated the available literature to assess the proportion of the proteins that we identified which had previously been shown to be S-nitrosylated in other cells. Out of the 240 proteins we identified only 52 (22%) had been documented to undergo S-nitrosylation in other cell types. The remaining proteins have not yet been reported to be S-nitrosylated and as such represent a considerable addition to the number of identified target proteins modified by NO.
3.4 Biological significance
In this study we have demonstrated that a large number of human sperm proteins are potential targets for S-nitrosylation. Whilst it is important as a first step to determine the S-nitrosoproteome of a cell, three key questions must be addressed: 1) Are spermatozoa exposed to sufficient levels of NO in vivo? 2) How does S-nitrosylation affect bioactivity of sperm proteins? 3) What is the biological significance of sperm protein S-nitrosylation for fertilisation?
3.4.1 Are spermatozoa exposed to significant levels of NO in vivo?
It is known that sperm can synthesise NO, but the evidence that synthesis is sufficient to be physiologically significant is equivocal [6, 7]. Measured NO production in human sperm suspensions is decreased when in the presence of NOS inhibitors [11, 51] at doses that are reported to reduce sperm motility [8], serine-threonine and tyrosine phosphorylation [9, 52], acrosome reaction [9, 11] and sperm-oocyte fusion [53]. In contrast, it is well known that a number of cell types in the mammalian female reproductive tract, including oviductal cells, generate significant levels of NO [5] and our own studies using the fluorescent sensor DAF-FM diacetate (4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate) have shown that NO is present in human cumulus at far higher concentrations than in sperm suspensions (unpublished data). Immunolocalisation of eNOS and nNOS in cumulus cells suggest that the NO detected are produced by these cells and not from oocytes. Nitrosylation of sperm proteins in vivo is thus more likely to occur in response to NO generated by cells of the female tract than by autocrine effects of sperm NO generation. Recent studies that showed that one-by-one deletion of the three types of NOS (eNOS, nNOS, inducible, iNOS) in KO mice had no discernible effect on male or female fertility, save some positive effects of iNOS deletion [54], do not exclude an important role for NO in fertilisation because of possible compensation by other sources of NO and the high degree of redundancy in the regulatory cascades that control capacitation/fertilisation.
3.4.2 How does S-nitrosylation affect bioactivity of sperm proteins?
To understand fully the significance of our observations it will be necessary to investigate, for each S-nitrosylated protein, whether S-nitrosylation alters protein bioactivity, to what degree, its reversibility and which cysteine (s) are involved. For spermatozoa, this is likely to involve in vitro transfection of somatic cells with specific proteins e.g. AKAPs in order to obtain a detailed understanding. However, a number of proteins merit discussion because of their known significance in sperm biology.
A-Kinase anchoring proteins (AKAPs)
AKAPs have never before been shown to be S-nitrosylated. AKAP3 (O75969, Table 1) and AKAP4 (Q5JQC9, Table 1) are believed to localise signalling complexes in the spermatozoa but, compared to their functional significance in other cell types relatively little is know [55, 56]. AKAP4 is a major protein present in the fibrous sheath of the sperm tail which binds both PRKAR1 (R1α) and PRKAR2A (R11α) regulatory subunit of PKA and is one of the key proteins phosphorylated during the process of capacitation [2, 57]. AKAP3 is also present in the fibrous sheath. In bovine sperm AKAP3 binds to PDE4A isoforms, potentially controlling PKA activity [58], and is tyrosine phosphorylated during capacitation in humans. AKAP complexes in the sperm tail modulate the motility of the cells. For example, specific inhibitors of PDE's (RS23544 –PDE4A5 inhibitor; Fisch et al. [59] noticeably increase the motility of the cell and protein kinase A-anchoring inhibitor peptides arrest sperm motility [60].
Our identification of three sperm AKAP proteins as being capable of undergoing S-nitrosylation is further supported by the presence of a consensus S-nitrosylation motif in their primary protein sequences. The acid-Cys-base motif was reported by Stamler et al. as being present, initially in haemoglobin [61], and subsequently in a number of other proteins known to undergo S-nitrosylation in vivo [62]. The redox environment need not necessarily to be created by acidic and basic residues adjacent to the target cysteine residue in the primary sequence and may instead be placed in close proximity in the tertiary structures [63]. In a number of proteins the presence of a Cys-(Asp,Glu) motif appears to be sufficient [62]. A role for further polar amino acid residues in the vicinity of the modified cysteine has also been proposed [61]. In the context of the well documented effects of NO on sperm motility, S-nitrosylation of AKAP's presents a novel form of modulation which should be the focus of further studies.
Heat shock proteins (HSPs)
In this study we showed that a number of heat shock proteins, belonging to the major 90kDa, 70kDa and 60kDa families, were S-nitrosylated. A number of these have recently been reported to play a very significant role in sperm capacitation and determination of fertilising ability. For example; heat shock protein 1 (chaperonin; heat shock protein 60kDa family) (homologue of human 60 kDa heat shock protein; P10809, Table 1) and endoplasmin (ERP99; heat shock protein 90kDa family) (homologue of human endoplasmin; P10809, Table 1) are tyrosine phosphorylated during capacitation in mouse sperm [64] where it is suggested that these proteins form part of a zona receptor complex during capacitation allowing the successful binding of the sperm to the egg [65]. Heat shock 70 kDa protein 8 (P11142, Table 1) and heat shock protein 90α (P7900, Table 1) are tyrosine phosphorylated during capacitation in human spermatozoa [2] but whether they function in a zona receptor complex is as yet unknown. The bovine homologue of heat shock protein 1 (HSP60) was found to become associated with the sperm surface during transit through the oviduct and is suggested to have a role in sperm capacitation [66]. HspA2 (also known as heat shock-related 70 kDa protein 2) (P54652, Table 1) has been shown to be a marker of sperm maturity [67] with lower levels of HspA2 gene expression in oligoteratoasthenozoospermic men [68]. We have found S-nitrosylation to be a common modification of heat shock proteins in human spermatozoa. This has also been showed in other cell types (see Table 1) suggesting that S-nitrosylation may be a universal mechanism for the modification of heat shock protein function.
Ryanodine receptors (RyRs)
RyRs are intracellular Ca2+ channels that play a central role in [Ca2+]i signalling, particularly signal amplification by Ca2+-induced Ca2+ release. These proteins contain a large number of sulfhydryls and are thus subject to S-nitrosylation [69, 70]. NO or NO-related proteins modulate activity of RyR1 and RyR2 [71-74], critical SH residues in RyR1 having been identified [73, 75] and NO donors rapidly mobilise Ca2+ from skeletal muscle and cardiac microsomes, an effect that can be reversed by thiol reducing agents [76]. In the present study we have identified RyR2 by MS/MS as being S-nitrosylated by NO treatment of human spermatozoa (Q92736, Table 1). The literature on expression of RyRs in the male gamete is inconsistent. Both the presence and (when detected) type of RyRs in mouse germ cells is controversial [77-79]. No RyR expression was detected with antibodies or with a fluorescent analogue of ryanodine (BODIPY-FL-X-ryanodine) in bull sperm [80]. However, BODIPY-FL-X-ryanodine stains live human sperm, primarily in the area of the sperm neck and mid-piece section of the flagellum [81]. Immunolocalisation using polyclonal antibodies specific for RyR1 and RyR2 confirm the presence of both receptors in the neck and mid-piece region of the human spermatozoa (unpublished data). Cyclic mobilisation of stored Ca2+ in this region, which regulates flagellar activity, is sensitive to caffeine and ryanodine [81]. Our findings confirm the presence of RyRs in human sperm but the low incidence of detection (Table 1, Supplementary Table S1) suggests that they may be present in low abundance (which may partly explain the difficulty in their detection and characterisation). This is consistent with their restricted cellular localisation [81] and might be expected for expression of a high conductance ion channel in a cell with a very small cytoplasmic volume. Significantly, the ryanodine-sensitive Ca2+-store in human sperm is mobilised upon exposure to NO [82] (Machado-Oliveira and Publicover, personal communication).
Glycolytic enzymes
It is interesting that a number of glycolytic enzymes are targets for S-nitrosylation (see Table 1). Human sperm require glucose (or some other glycolysable sugar) for optimum motility although unlike mouse sperm they do not require it for capacitation [83]. The sperm-specific isoform of glyceraldehyde 3-phosphate dehydrogenase (GAPDHs; O14556, Table 1) is of particular interest. This enzyme is bound to the fibrous sheath of the flagellum [84] and is required for sperm motility in the mouse [85]. GAPDHs has a very similar primary sequence to the somatic isoenzymes with the addition of a proline rich N-terminal extension [86]. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; P04406, Table 1) is rich in sulphydryl groups and is a known target for S-nitrosylation. S nitrosylated GAPDH has decreased enzymatic activity and as part of the cellular response to NO stress, it associates with Siah (an E3 ubiquitin lyase) and the complex is transported to the nucleus where it promotes apoptosis [87]. Since GAPDHs is anchored to the fibrous sheath it will be unable to act in this way but nitrosylation could affect its interaction with other fibrous sheath proteins. Another interesting enzyme is sperm specific lactate dehydrogenase (LDH-C4; P07864, Table 1). Alone among lactate dehydrogenase isoforms it is expressed in the mitochondrion as well as the cytoplasm offering the sperm a unique lactate-pyruvate redox shuttle [88].
3.4.3 What is the biological significance of sperm protein S-nitrosylation for fertilisation?
We need to understand the physiological importance of S-nitrosylation modification in regulating the competence of sperm to fertilise. Although poorly understood, the movement of sperm through the female reproductive tract is known to be a finely controlled, complex and dynamic process [1, 89]. During most of this journey, the female reproductive tract ‘holds back’ the sperm from being ready to fertilise, yet, as the sperm approach the egg they have to become fully competent to fertilise, which includes increasing the ‘power’ of the cell so that it can penetrate the cumulus and the zona pellucida [90]. The cumulus is a logical source of external signals to activate sperm prior to fertilisation in vivo and is a likely source of NO which would have a maximal effect on the sperm. It is our hypothesis that sperm will experience a modest tonic NO stimulus from endogenous generation (and possible sources in the female tract) but encounter a steeply increasing concentration of NO on entering the cumulus [17, 82]. Further experiments are warranted to test this concept.
4. Concluding Remarks
Despite the well known effects of NO on human spermatozoa, the identification of S-nitrosylated proteins until now remained unexplored. In this study, using a series of NO donors combined with the biotin switch assay we report the identification of S-nitrosylated proteins in human spermatozoa. This global assessment allows critical insight into the role of NO in sperm physiology identifies potential novel mechanisms of action of S-nitrosylation and opens up a new signalling regimen in the spermatozoon which will act as a platform for further detailed studies.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the donors who took part in our research and the staff in the Assisted Conception Unit for assistance in collecting semen samples. We would also like to thank Prof F.A. Lai (Cardiff University) for the polyclonal anti-RyR1 (2142) and anti-RyR2 (1093) antibodies. This study was supported Wellcome Trust Grant 078905 [L.L, C.L.R.B.], Fonds de la recherche en santé du Québec [L.L.] and the Functional Genomics Laboratory.
Abbreviations
- AKAP
A-kinase anchor proteins
- Biotin-HPDP
N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide
- CSNO
L-nitroso-cysteine
- DAF-FM diacetate
4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate
- ERKs
Protein extracellular signal regulated kinases
- eNOS
endothelial nitric oxide synthase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GAPDHs
sperm isoform specific glyceraldehyde 3-phosphate dehydrogenase
- GSH
Glutathione
- GSNO
S-nitroso-glutathione
- GSSG
oxidised glutathione
- HSPs
Heat shock proteins
- iNOS
inducible nitric oxide synthase
- LDH-C4
sperm specific lactate dehydrogenase
- L-NAME
N(G)-Nitro-L-arginine methyl ester
- MMTS
Methanethiosulfonate
- nNOS
neuronal nitric oxide synthase
- NO
Nitric oxide
- NOS
nitric oxide synthase
- TEXAS RED®-MTSEA
Texas Red®-2-sulfonamidoethyl methanethiosulfonate
- TTBS
Tris-buffered saline supplemented with 0.1% Tween-20
- PKA
protein kinase A
- PDE
phosphodiesterase
- RyR
ryanodine receptor
- Spermine NONOate
N-(2-aminoethyl)-N-(2-hydroxy-nitrosohydrazino)-1,2-ethylenediamine
5. References
- 1.Suarez SS, Pacey AA. Hum. Reprod. Update. 2006;12:23–37. doi: 10.1093/humupd/dmi047. [DOI] [PubMed] [Google Scholar]
- 2.Ficarro S, Chertihin O, Westbrook VA, White F, et al. J. Biol. Chem. 2003;278:11579–11589. doi: 10.1074/jbc.M202325200. [DOI] [PubMed] [Google Scholar]
- 3.Ford WC. Hum. Reprod. Update. 2004;10:387–399. doi: 10.1093/humupd/dmh034. [DOI] [PubMed] [Google Scholar]
- 4.Baker MA, Aitken RJ. Mol. Cell. Endocrinol. 2004;216:47–54. doi: 10.1016/j.mce.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 5.Rosselli M, Keller PJ, Dubey RK. Hum. Reprod. Update. 1998;4:3–24. doi: 10.1093/humupd/4.1.3. [DOI] [PubMed] [Google Scholar]
- 6.Herrero MB, Gagnon C. J. Androl. 2001;22:349–356. [PubMed] [Google Scholar]
- 7.Herrero MB, de Lamirande E, Gagnon C. Curr. Pharm. Des. 2003;9:419–425. doi: 10.2174/1381612033391720. [DOI] [PubMed] [Google Scholar]
- 8.Lewis SEM, Donnelly ET, Sterling ESL, Kennedy MS, et al. Mol. Hum. Reprod. 1996;2:873–878. doi: 10.1093/molehr/2.11.873. [DOI] [PubMed] [Google Scholar]
- 9.Herrero MB, de Lamirande E, Gagnon C. Biol. Reprod. 1999;61:575–581. doi: 10.1095/biolreprod61.3.575. [DOI] [PubMed] [Google Scholar]
- 10.Sengoku K, Tamate K, Yoshida T, Takaoka Y, et al. Fértil. Steril. 1998;69:522–527. doi: 10.1016/s0015-0282(97)00537-2. [DOI] [PubMed] [Google Scholar]
- 11.Revelli A, Soldati G, Costamagna C, Pellerey O, et al. J. Cell. Physiol. 1999;178:85–92. doi: 10.1002/(SICI)1097-4652(199901)178:1<85::AID-JCP11>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 12.Willipinski-Stapelfeldt B, Lubberstedt J, Stelter S, Vogt K, et al. Mol. Hum. Reprod. 2004;10:543–552. doi: 10.1093/molehr/gah065. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Zhou QM, Li XD, Xie Y, et al. Arch. Pharm. Res. 2006;29:145–151. doi: 10.1007/BF02974276. [DOI] [PubMed] [Google Scholar]
- 14.Herrero MB, Perez Martinez S, Viggiano JM, Polar JM, de Gimeno MF. Reprod. Fertil. Dev. 1996;8:931–934. doi: 10.1071/rd9960931. [DOI] [PubMed] [Google Scholar]
- 15.O'Bryan MK, Zini A, Cheng CY, Schlegel PN. Fertil. Steril. 1998;70:1143–1147. doi: 10.1016/s0015-0282(98)00382-3. [DOI] [PubMed] [Google Scholar]
- 16.Marcondes S, Cardoso MH, Morganti RP, Thomazzi SM, et al. Proc. Natl. Acad. Sci. USA. 2006;103:3434–3439. doi: 10.1073/pnas.0509397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Publicover SJ, Harper CV, Barratt CLR. Nat. Cell. Biol. 2007;9:235–242. doi: 10.1038/ncb0307-235. [DOI] [PubMed] [Google Scholar]
- 18.Revelli A, Ghigo D, Moffa F, Massobrio M, Tur-Kaspa I. Endocr. Rev. 2002;23:484–494. doi: 10.1210/er.2001-0020. [DOI] [PubMed] [Google Scholar]
- 19.Lefièvre L, De Lamirande E, Gagnon C. J. Androl. 2000;21:929–937. [PubMed] [Google Scholar]
- 20.Bielfeld P, Faridi A, Zaneveld LJ, De Jonge CJ. Fertil. Steril. 1994;61:536–541. [PubMed] [Google Scholar]
- 21.Herrero MB, Chatterjee S, Lefièvre L, de Lamirande E, Gagnon C. Free Radic. Biol. Med. 2000;29:522–536. doi: 10.1016/s0891-5849(00)00339-7. [DOI] [PubMed] [Google Scholar]
- 22.Thundathil J, de Lamirande E, Gagnon C. Biol. Reprod. 2003;68:1291–1298. doi: 10.1095/biolreprod.102.008276. [DOI] [PubMed] [Google Scholar]
- 23.O'Flaherty C, de Lamirande E, Gagnon C. Free Radic. Biol. Med. 2006;40:1045–1055. doi: 10.1016/j.freeradbiomed.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 24.Stamler JS, Simon DI, Osborne JA, Mullins ME, et al. Proc. Natl. Acad. Sci. USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 26.Foster MW, Stamler JS. J. Biol. Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 27.Greco TM, Hodara R, Parastatidis I, Heijnen HF, et al. Proc. Natl. Acad. Sci. USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. Proc. Natl. Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Proc. Natl. Acad. Sci. USA. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. Proc. Natl. Acad. Sci. USA. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garban HJ, Marquez-Garban DC, Piedras RJ, Ignarro LJ. Proc. Natl. Acad. Sci. USA. 2005;102:2632–2636. doi: 10.1073/pnas.0409854102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Retamal MA, Cortes CJ, Reuss L, Bennett MV, Saez JC. Proc. Natl. Acad. Sci. USA. 2006;103:4475–4480. doi: 10.1073/pnas.0511118103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization Laboratory Manual for the Examination of Human Semen and Semen–Cervical Mucus Interaction. 4th edn Cambridge University Press; Cambridge: 1999. [Google Scholar]
- 34.Kvist U, Björndahl L. Manual on Basic Semen Analysis. ESHRE Monographs #2. Oxford University Press; Oxford: 2002. [Google Scholar]
- 35.Jacobson G, Karsnas P. Electrophoresis. 1990;11:46–52. doi: 10.1002/elps.1150110111. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Loscalzo J. Proc. Natl. Acad. Sci. USA. 2005;102:117–122. doi: 10.1073/pnas.0405989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pixton KL, Deeks ED, Flesch FM, Moseley FL, et al. Hum. Reprod. 2004;19:1438–1447. doi: 10.1093/humrep/deh224. [DOI] [PubMed] [Google Scholar]
- 38.Lefièvre L, Conner SJ, Salpekar A, Olufowobi O, et al. Hum. Reprod. 2004;19:1580–1586. doi: 10.1093/humrep/deh301. [DOI] [PubMed] [Google Scholar]
- 39.Kinter M, Sherman NE. Protein Sequencing and Identification Using Tandem Mass Spectrometry. John Wiley; New York: 2000. [Google Scholar]
- 40.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 41.Kuncewicz T, Sheta EA, Goldknopf IL, Kone BC. Mol. Cell. Proteomics. 2003;2:156–163. doi: 10.1074/mcp.M300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Arnelle DR, Stamler JS. Arch. Biochem. Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 43.Gow AJ, Ischiropoulos H. J. Cell. Physiol. 2001;187:277–282. doi: 10.1002/jcp.1085. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Heredia J, Estanyol JM, Ballesca JL, Oliva R. Proteomics. 2006;6:4356–4369. doi: 10.1002/pmic.200600094. [DOI] [PubMed] [Google Scholar]
- 45.Johnston DS, Wooters J, Kopf GS, Qiu Y, Roberts KP. Ann. N. Y. Acad. Sci. 2005;1061:190–202. doi: 10.1196/annals.1336.021. [DOI] [PubMed] [Google Scholar]
- 46.Ellis PJI, Furlong RA, Conner SJ, Kirkman-Brown JC, Afnan M, Barratt CLR, Griffin DK, Affara NA. Hum. Mol. Genet. Submitted [Google Scholar]
- 47.Eddy EM, Toshimori K, O'Brien DA. Microsc. Res. Tech. 2003;61:103–115. doi: 10.1002/jemt.10320. [DOI] [PubMed] [Google Scholar]
- 48.Lalancette C, Faure RL, Leclerc P. Proteomics. 2006;6:4523–4540. doi: 10.1002/pmic.200500578. [DOI] [PubMed] [Google Scholar]
- 49.Lefièvre L, Bedu-Addo K, Conner SJ, Machado-Oliveira GSM, et al. Reproduction. In Press. [Google Scholar]
- 50.Stein KK, Go JC, Lane WS, Primakoff P, Myles DG. Proteomics. 2006;6:3533–3543. doi: 10.1002/pmic.200500845. [DOI] [PubMed] [Google Scholar]
- 51.Donnelly ET, Lewis SEM, Thompson W, Chaakravarthy U. Mol. Hum. Reprod. 1997;3:755–762. doi: 10.1093/molehr/3.9.755. [DOI] [PubMed] [Google Scholar]
- 52.O'Flaherty C, de Lamirande E, Gagnon C. Mol. Hum. Reprod. 2004;10:355–363. doi: 10.1093/molehr/gah046. [DOI] [PubMed] [Google Scholar]
- 53.Francavilla F, Santucci R, Macerola B, Ruvolo G, Romano R. Biol. Reprod. 2000;63:425–429. doi: 10.1095/biolreprod63.2.425. [DOI] [PubMed] [Google Scholar]
- 54.Yang JZ, Ajonuma LC, Rowlands DK, Tsang LL, Ho LS, et al. Cell Biol. Int. 2005;29:785–791. doi: 10.1016/j.cellbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Smith FD, Langeberg LK, Scott JD. Trends Biochem. Sci. 2006;31:316–323. doi: 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 56.McConnachie G, Langeberg LK, Scott JD. Trends Mol. Med. 2006;12:317–323. doi: 10.1016/j.molmed.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 57.Visconti PE, Johnson LR, Oyaski M, Fornes M, et al. Dev. Biol. 1997;192:351–363. doi: 10.1006/dbio.1997.8768. [DOI] [PubMed] [Google Scholar]
- 58.Bajpai M, Fiedler SE, Huang Z, Vijayaraghavan S, et al. Biol. Reprod. 2006;74:109–118. doi: 10.1095/biolreprod.105.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisch JD, Behr B, Conti M. Hum. Reprod. 1998;13:1248–1254. doi: 10.1093/humrep/13.5.1248. [DOI] [PubMed] [Google Scholar]
- 60.Vijayaraghavan S, Goueli SA, Davey MP, Carr DW. J. Biol. Chem. 1997;272:4747–4752. doi: 10.1074/jbc.272.8.4747. [DOI] [PubMed] [Google Scholar]
- 61.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 62.Stamler JS. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 63.Perez-Mato I, Castro C, Ruiz FA, Corrales FJ, Mato JM. J. Biol. Chem. 1999;274:17075–17079. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 64.Asquith KL, Baleato RM, McLaughlin EA, Nixon B, Aitken RJ. J. Cell Sci. 2004;117:3645–3657. doi: 10.1242/jcs.01214. [DOI] [PubMed] [Google Scholar]
- 65.Nixon B, Asquith KL, Aitken RJ. Mol. Cell. Endocrinol. 2005;240:1–10. doi: 10.1016/j.mce.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 66.Boilard M, Reyes-Moreno C, Lachance C, Massicotte L, et al. Biol. Reprod. 2004;71:1879–1889. doi: 10.1095/biolreprod.103.026849. [DOI] [PubMed] [Google Scholar]
- 67.Ergur AR, Dokras A, Giraldo JL, Habana A, et al. Fertil. Steril. 2002;77:910–918. doi: 10.1016/s0015-0282(02)03073-x. [DOI] [PubMed] [Google Scholar]
- 68.Cedenho AP, Lima SB, Cenedeze MA, Spaine DM, et al. Hum. Reprod. 2006;21:1791–1794. doi: 10.1093/humrep/del055. [DOI] [PubMed] [Google Scholar]
- 69.Takeshima H, Nishimura S, Matsumoto T, Ishida H, et al. Nature. 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- 70.Otsu K, Willard HF, Khanna VK, Zorzato F, et al. J. Biol. Chem. 1990;265:13472–13483. [PubMed] [Google Scholar]
- 71.Xu L, Eu JP, Meissner G, Stamler JS. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 72.Zahradnikova A, Minarovic I, Venema RC, Meszaros LG. Cell Calcium. 1997;22:447–454. doi: 10.1016/s0143-4160(97)90072-5. [DOI] [PubMed] [Google Scholar]
- 73.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Proc. Natl. Acad. Sci. USA. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun J, Xu L, Eu JP, Stamler JS, Meissner G. J. Biol. Chem. 2003;278:8184–8189. doi: 10.1074/jbc.M211940200. [DOI] [PubMed] [Google Scholar]
- 75.Aracena-Parks P, Goonasekera SA, Gilman C, Dirksen RT, et al. J. Biol. Chem. 2006;281:40354–40368. doi: 10.1074/jbc.M600876200. [DOI] [PubMed] [Google Scholar]
- 76.Stoyanovsky D, Murphy T, Anno PR, Kim YM, Salama G. Cell Calcium. 1997;21:19–29. doi: 10.1016/s0143-4160(97)90093-2. [DOI] [PubMed] [Google Scholar]
- 77.Giannini G, Conti A, Mammarella S, Scrobogna M, Sorrentino V. J. Cell Biol. 1995;128:893–904. doi: 10.1083/jcb.128.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trevino CL, Santi CM, Beltran C, Hernandez-Cruz A, et al. Zygote. 1998;6:159–172. doi: 10.1017/s0967199498000094. [DOI] [PubMed] [Google Scholar]
- 79.Chiarella P, Puglisi R, Sorrentino V, Boitani C, Stefanini M. J. Cell Sci. 2004;117:4127–4134. doi: 10.1242/jcs.01283. [DOI] [PubMed] [Google Scholar]
- 80.Ho HC, Suarez SS. Biol. Reprod. 2001;65:1606–1615. doi: 10.1095/biolreprod65.5.1606. [DOI] [PubMed] [Google Scholar]
- 81.Harper CV, Barratt CL, Publicover SJ. J. Biol. Chem. 2004;279:46315–46325. doi: 10.1074/jbc.M401194200. [DOI] [PubMed] [Google Scholar]
- 82.Conner SJ, Lefièvre L, Kirkman-Brown JC, Michelangeli F, et al. Soc. Reprod. Suppl. 2007;63:237–256. [PubMed] [Google Scholar]
- 83.Williams AC, Ford WC. J. Androl. 2001;22:680–695. [PubMed] [Google Scholar]
- 84.Eddy EM, Toshimori K, O'Brien DA. J. Androl. 2003;22:680–695. [Google Scholar]
- 85.Miki K, Qu W, Goulding EH, Willis WD, et al. Proc. Natl. Acad. Sci. USA. 2004;101:16501–16506. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Welch JE, Schatte EC, O'Brien DA, Eddy EM. Biol. Reprod. 1992;46:869–878. doi: 10.1095/biolreprod46.5.869. [DOI] [PubMed] [Google Scholar]
- 87.Hara MR, Cascio MB, Sawa A. Biochim. Biophys. Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 88.Ford WCL, Rees JM. In: Controls of sperm motility: biological and clinical aspects. Gagnon C, editor. CRC Press; Boca Raton: 1990. pp. 175–202. [Google Scholar]
- 89.De Jonge C. Hum. Reprod. Update. 2005;11:205–214. doi: 10.1093/humupd/dmi010. [DOI] [PubMed] [Google Scholar]
- 90.Quill TA, Sugden SA, Rossi KL, Doolittle LK, et al. Proc. Natl Acad Sci U S A. 2003;100:14869–14874. doi: 10.1073/pnas.2136654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez-Ruiz A, Lamas S. Arch. Biochem. Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 92.Stamler JS, Lamas S, Fang FC. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 93.Lee M, Arosio P, Cozzi A, Chasteen ND. Biochemistry. 1994;33:3679–3687. doi: 10.1021/bi00178a026. [DOI] [PubMed] [Google Scholar]
- 94.Nedospasov A, Rafikov R, Beda N, Nudler E. Proc. Natl. Acad. Sci. USA. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stamler JS, Jaraki O, Osborne J, Simon D, et al. Proc. Natl. Acad. Sci. USA. 1992a;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stamler JS, Simon DI, Osborne JA, Mullins ME, et al. Proc. Natl. Acad. Sci. USA. 1992c;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Belluoccio D, Wilson R, Thornton DJ, Wallis TP, et al. Proteomics. 2006;6:6549–6553. doi: 10.1002/pmic.200600191. [DOI] [PubMed] [Google Scholar]
- 98.Gao C, Guo H, Wei J, Mi Z, et al. Nitric Oxide. 2005;12:121–126. doi: 10.1016/j.niox.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 99.Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, et al. Arch. Biochem. Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 100.Lindermayr C, Saalbach G, Durner J. Plant Physiol. 2005;137:921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dall'Agnol M, Bernstein C, Bernstein H, Garewal H, Payne CM. Proteomics. 2006;6:1654–1662. doi: 10.1002/pmic.200500240. [DOI] [PubMed] [Google Scholar]
- 102.Moon KH, Hood BL, Kim BJ, Hardwick JP, et al. Hepatology. 2006;44:1218–1230. doi: 10.1002/hep.21372. [DOI] [PubMed] [Google Scholar]
- 103.Fratelli M, Demol H, Puype M, Casagrande S, et al. Proc. Natl. Acad. Sci. USA. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fratelli M, Demol H, Puype M, Casagrande S, et al. Proteomics. 2003;3:1154–1161. doi: 10.1002/pmic.200300436. [DOI] [PubMed] [Google Scholar]
- 105.Shenton D, Grant CM. Biochem. J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ji Y, Akerboom TP, Sies H, Thomas JA. Arch. Biochem. Biophys. 1999;362:67–78. doi: 10.1006/abbi.1998.1013. [DOI] [PubMed] [Google Scholar]
- 107.Klatt P, Pineda Molina E, Perez-Sala D, Lamas S. Biochem. J. 2000;349:567–578. doi: 10.1042/0264-6021:3490567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ito H, Iwabuchi M, Ogawa K. Plant Cell Physiol. 2003;44:655–660. doi: 10.1093/pcp/pcg098. [DOI] [PubMed] [Google Scholar]
- 109.Molina y Vedia L, McDonald B, Reep B, Brune B, et al. J. Biol. Chem. 1992;267:24929–24932. [PubMed] [Google Scholar]
- 110.Padgett CM, Whorton AR. Am. J. Physiol. 1995;269:C739–749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- 111.Mohr S, Stamler JS, Brune B. J. Biol. Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 112.Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B. J. Biol. Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 113.Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Proc. Natl. Acad. Sci. USA. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hara MR, Agrawal N, Kim SF, Cascio MB, et al. Nat. Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 115.Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. Plant Cell Physiol. 2004;45:18–27. doi: 10.1093/pcp/pch019. [DOI] [PubMed] [Google Scholar]
- 116.Stamler JS, Simon DI, Jaraki O, Osborne JA, et al. Proc. Natl. Acad. Sci. USA. 1992b;89:8087–8091. doi: 10.1073/pnas.89.17.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eaton P, Jones ME, McGregor E, Dunn MJ, et al. J. Am. Soc. Nephrol. 2003;14:S290–296. doi: 10.1097/01.asn.0000078024.50060.c6. [DOI] [PubMed] [Google Scholar]
- 118.DeMaster EG, Redfern B, Quast BJ, Dahlseid T, Nagasawa HT. Alcohol. 1997;14:181–189. doi: 10.1016/s0741-8329(96)00142-5. [DOI] [PubMed] [Google Scholar]
- 119.Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. J. Exp. Med. 1999;190:1595–1604. doi: 10.1084/jem.190.11.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laval F, Wink DA. Carcinogenesis. 1994;15:443–447. doi: 10.1093/carcin/15.3.443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.