Abstract
Human mitochondrial transcription factor A (TFAM) is a multi-functional protein, involved in different aspects of maintaining mitochondrial genome integrity. In this report, we characterized TFAM and its interaction with tumor suppressor p53 using various biophysical methods. DNA-free TFAM is a thermally unstable protein that is in equilibrium between monomers and dimers. Self-association of TFAM is modulated by its basic C-terminal tail. The DNA-binding ability of TFAM is mainly contributed by its first HMG-box, while the second HMG-box has low-DNA-binding capability. We also obtained backbone resonance assignments from the NMR spectra of both HMG-boxes of TFAM. TFAM binds primarily to the N-terminal transactivation domain of p53, with a Kd of 1.95 ± 0.19 μM. The C-terminal regulatory domain of p53 provides a secondary binding site for TFAM. The TFAM–p53-binding interface involves both TAD1 and TAD2 sub-domains of p53. Helices α1 and α2 of the HMG-box constitute the main p53-binding region. Since both TFAM and p53 binds preferentially to distorted DNA, the TFAM–p53 interaction is implicated in DNA damage and repair. In addition, the DNA-binding mechanism of TFAM and biological relevance of the TFAM–p53 interaction are discussed.
INTRODUCTION
Mitochondria, which possibly have evolved from an endosymbiotically incorporated organism, have their own genome. Human mitochondrial genome (mtDNA) is a circular double-stranded DNA (dsDNA) of 16 569 bp in length (www.mitomap.org). It encodes 13 proteins that are considered essential sub-units of mitochondrial respiratory chain, two rRNAs and 22 tRNAs. The rRNAs and tRNAs are recruited to construct mitochondrial translational machineries, required for synthesis of proteins encoded by mtDNA. Best known for their role in ATP production via the process of oxidative phosphorylation (OXPHOS), mitochondria are vital for an array of fundamental cellular functions including fatty acid oxidation, biosynthesis of amino acids and heme, apoptosis and signal transduction. Maintenance of the mitochondrial genome is naturally crucial for survival. Mitochondrial dysfunctions are increasingly being associated with human diseases (1) such as cancer, diabetes, Parkinson’s disease as well as cellular senescence (2).
Human mitochondrial transcription factor A (TFAM), originally cloned as a transcription factor for mtDNA, is a multi-functional protein (3). Transcription of mitochondrial genomes initiates from three promoters: the light strand promoter (LSP) and two heavy strand promoters (HSP1 and HSP2) that are found in the 1.1 kb displacement loop (D-loop) control region of mtDNA. Transcriptional activation capability of TFAM is mediated through its high affinity to these sequence-specific DNAs and its ability to recruit mitochondrial RNA polymerase (POLRMT) as well as mitochondrial transcription factor B (TFBM) (4). In addition, TFAM serves a role in packaging mtDNA into nucleoid-like structure and its abundance in mitochondria is correlated with mtDNA copy number. According to classic strand-asymmetric model (also termed strand-displacement model) of mtDNA replication (5–7), transcription is coupled to replication. TFAM is, therefore, implicated in mtDNA replication. Accumulating evidences suggest that TFAM is a DNA damage sensor since it binds more strongly to oxidatively damaged DNA and cisplatin adducts. Its interaction with tumor suppressor p53 is believed to play a part in DNA damage and repair (8). mtDNA is physiologically more vulnerable compared with nuclear DNA, in view of its close proximity to OXPHOS sites. It is considered that ∼1–5% of mitochondrially consumed oxygen is converted to reactive oxygen species (ROS) due to electron leaks from respiratory chain. In addition, mtDNA is more prone to chemical damage since lipophilic cations tend to accumulate in mitochondrial matrix, facilitated by negative membrane potential at matrix-side. TFAM has been reported to protect mtDNA from these oxidative and chemical attacks. Moreover, TFAM displayed higher affinity to branched DNA structure (e.g. 4-way junction) and might affect DNA recombination events.
TFAM belongs to a large and diverse superfamily of high-mobility group (HMG) protein (9). Within this superfamily, three structurally distinct classes of HMG proteins have been defined: HMG–nucleosome-binding family (HMGN), HMG-AT-hook family (HMGA) and HMG-box family (HMGB). Mammalian HMG-box containing proteins can further be sub-divided into two major groups. The first group consists of HMGB-type non-sequence-specific DNA-binding proteins with two HMG-box domains and a long highly acidic C-tail (e.g. HMGB1). The second group is highly diverse and consists of mostly sequence-specific proteins having a single HMG-box and no acidic C-tails (e.g. SRY, SOX). Nevertheless, there exist proteins that have up to six copies of the HMG-box domain (e.g. UBF1). As a HMGB protein, TFAM is unique in that it binds specific sequence preferentially, but contains two HMG-box domains. The gene of TFAM spans about 10 kb and consists of seven exons and six introns (10). Exon 5 can splice alternatively, resulting in two TFAM isoforms (11).
In this article, we report biophysical characterization of TFAM and its interaction with tumor suppressor p53. In addition to examinations of TFAM’s oligomerization state and thermal stability, we performed solution-based DNA binding of TFAM using fluorescence spectroscopy and circular dichroism (CD) spectropolarimetry. For the first time, TFAM was studied using nuclear magnetic resonance (NMR). We obtained backbone resonance assignments from the NMR spectra of both HMG-box domains of TFAM. We were, therefore, able to map the p53- and DNA-binding sites on HMG-box domain of TFAM. Similarly, we located binding region of HMG-box on p53. The DNA-binding mechanism of TFAM and biological relevance of TFAM–p53 interaction are also discussed.
MATERIALS AND METHODS
Nucleic acids
The sequences of single-stranded DNAs (ssDNA) and dsDNA used in this study were summarized in Supplementary Table S1. All DNAs were HPLC-purified and were purchased from either Sigma Genosys or Eurogentec.
Cloning and expression of HMG1, HMG2, NEx-HMG2, TFAM and TFAM-ΔC
Gene fragments, corresponding to residues 50–118 (HMG1), 155–219 (HMG2), 144–219 (NEx-HMG2), 50–246 (TFAM) and 50–219 (TFAM-ΔC) of TFAM (NCBI NP_003192), were PCR-amplified and cloned into pET24a-HLTV vector using restriction sites BamHI and EcoRI. N-terminal protein region (residues 1–49) was predicted to be mitochondrial targeting sequence (MTS) using MITOPROT [http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html; (12)], and was, therefore, removed from our protein constructs. For protein expression, plasmid was freshly transformed into Escherichia coli C41 (DE3). Cells were grown in 2xTY media supplemented with 50 µg/ml kanamycin, at 37°C. When OD600 reached 0.5–0.6, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce protein expression and temperature was lowered to 25°C. After 24 h, cells were harvested and pellets were frozen in liquid nitrogen.
Purification of HMG1, HMG2, NEx-HMG2, TFAM and TFAM-ΔC
HMG1, HMG2 and NEx-HMG2
Cells were thawed at 4°C in buffer A [50 mM Tris–HCl pH 7.4, 300 mM NaCl, 5 mM imidazole, 1% (v/v) 2-mercaptoethanol] supplemented with DNase, RNase and protease inhibitor (Roche), and were lysed by high-pressure homogenization (EmulsiFlex-C5, Avestin). After centrifugation, we loaded supernatant onto HisTrap HP column (GE Healthcare) pre-equilibrated with buffer A. After washing with 5 CV of buffer A, protein was eluted with linear gradient of imidazole (0.005–1 M) in buffer A. His-tag + lipoyl domain was cleaved with TEV protease overnight at 4°C. The sample was diluted with buffer B [25 mM Tris–HCl pH 7.4, 5 mM DTT, 10% (v/v) glycerol] to adjust NaCl concentration to ∼100 mM and loaded onto HiTrap Heparin HP column (GE Healthcare) pre-equilibrated with buffer B containing 100 mM NaCl. Upon washing with 5 CV of buffer B containing 100 mM NaCl, protein was eluted with linear gradient of NaCl (0–1 M) in buffer B. Fractions containing protein of interest were pooled and loaded onto HiLoad 26/60 Superdex 75 column (GE Healthcare) pre-equilibrated with buffer C [25 mM Tris–HCl pH 7.4, 150 mM NaCl, 5 mM DTT, 10% (v/v) glycerol]. All chromatographic steps were performed at 4°C. Protein concentration was estimated using extinction coefficient of 16 960/M/cm. 13C,15N-labeled sample for NMR was produced in M9 media using 13C6-D-glucose and 15NH4Cl as carbon source and nitrogen source, respectively.
TFAM and TFAM-ΔC
Both proteins were purified as above, with the following modifications: upon TEV cleavage, TFAM-containing sample was loaded directly onto HiTrap Heparin HP column (GE Healthcare) pre-equilibrated with buffer B containing 300 mM NaCl. After washing with 5 CV of buffer B containing 300 mM NaCl, TFAM was eluted with buffer B containing 1 M NaCl. In the case of TFAM-ΔC, protein sample was diluted with buffer B to adjust NaCl concentration to ∼150 mM after TEV cleavage. TFAM-ΔC-containing sample was loaded onto HiTrap Heparin HP column (GE Healthcare) pre-equilibrated with buffer B containing 150 mM NaCl, followed by washing with 5 CV of identical buffer. TFAM-ΔC was eluted with linear gradient of NaCl (0–1 M) in buffer B. Both TFAM and TFAM-ΔC were subsequently subjected to size exclusion using HiLoad 26/60 Superdex 75 column (GE Healthcare) pre-equilibrated with buffer D [25 mM Tris–HCl pH 7.4, 300 mM NaCl, 5 mM DTT, 10% (v/v) glycerol]. Protein concentrations were estimated using extinction coefficients of 35 410/M/cm (TFAM) and 33 920/M/cm (TFAM-ΔC).
Expression and purification of N-terminal domain of p53
N-terminal domain of p53 (p53N, residues 1–93) was expressed and purified as previously reported (13).
Analytical ultracentrifugation
Equilibrium sedimentation experiments were performed on a Beckman XL-I ultracentrifuge by using Ti-60 rotor and 6-sector cells at the following speeds: [35 000, 40 000 and 45 000 r.p.m.] (HMG1 and HMG2), [25 000, 30 000, 35 000 r.p.m.] (TFAM) and [28 000, 30 000, 34 000 r.p.m] (TFAM-ΔC). Data were collected at 5°C, following absorbance at 230 nm, 280 nm and interference. The sample volume was 110 µl and concentrations tested were 5–400 µM. Buffer conditions were 25 mM Tris–HCl pH 7.4 and 150 mM NaCl. For samples containing TFAM, 1 mM DTT was supplemented. Samples were considered to be at equilibrium as judged by a comparison of several scans at each speed. Data were processed and analyzed by using UltraSpin software, which is available from our web site (www.mrc-cpe.cam.ac.uk).
CD spectropolarimetry and thermal melting
Thermal stabilities of various protein constructs (HMG1, HMG2, NEx-HMG2, TFAM and TFAM-ΔC) were studied through CD spectropolarimetry. Thermostated JASCO J-815 CD spectropolarimeter was used with constant nitrogen gas purged. Protein concentration used was 10–20 µM, in buffer containing 25 mM HEPES pH 7.4 and 150 mM NaCl. For sample containing TFAM, 1 mM 1,4-dithioerythritol was supplemented. Thermal denaturation curves for all proteins were obtained by recording CD spectra at 222 nm, from 2°C to 98°C at a temperature ramp rate of 60°C/h. To study conformational change of TFAM upon DNA binding, CD spectra of TFAM were recorded in the absence or presence of 28-bp dsDNA (Supplementary Table S1). Wavelength scans were performed at 20°C, in 0.2 nm steps from 194 to 260 nm, in 25 mM sodium phosphate pH 7.4, 100 mM KCl and 1 mM 1,4-dithioerythritol. The protein concentration was 10 µM and DNA concentration was 4 µM. The spectra recorded were corrected for the spectra of the buffer and of the DNA. All CD measurements were made in quartz cuvette with 1 mm path length.
DNA- or p53N-binding of HMG1, HMG2, NEx-HMG2, TFAM and TFAM-ΔC
The binding of various protein constructs (HMG1, HMG2, NEx-HMG2, TFAM and TFAM-ΔC) to either ssDNA, dsDNA or p53N was performed by using fluorescence anisotropy. Measurements were recorded on FluoroMax-3 spectrofluorimeter (Jobin–Yvon–Horiba) equipped with a Hamilton Microlab Titrator, at 37°C (unless otherwise stated) in buffer containing 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT. Protein was titrated into a cuvette containing either 1.2-ml fluorescein-labeled DNA [initial concentration 100 nM (HMG1, HMG2 and NEx-HMG2) or 25 nM (TFAM and TFAM-ΔC)] or AlexaFluor 546-labeled p53N (initial concentration 50 nM), as described earlier (13). Excitation/emission wavelengths for fluorescein and AlexaFluor 546 were 480/530 and 540/569 nm, respectively. Fluorescence anisotropy was calculated from the fluorescence intensities and binding analysis was performed using either a simple 1-state binding isotherm or Hill equation (13,14).
Intrinsic fluorescence measurements
Intrinsic fluorescence measurements were performed with a Cary Eclipse fluorescence spectrophotometer (Varian). Scans were recorded using excitation wavelength of 280 nm and emission wavelength ranged from 300 to 400 nm. Slit widths for both excitation and emission were kept at 5 nm, with a photomultiplier voltage of 900 V. Concentrations of TFAM and/or DNA were 500 nM, in buffer containing 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT.
NMR spectroscopy
NMR spectra were acquired with a Bruker Avance-II + 700-MHz spectrometer equipped with a 5 mm cryogenic inverse probe and single-axis gradients.
NMR protein backbone assignment
Backbone 1H, 13C and 15N resonance assignments of HMG1 and HMG2 were recorded at sample temperatures of 15°C and 5°C, respectively. Protein concentration used was 200 µM in buffer containing 25 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM DTT and 5% (v/v) D2O. Sequence-specific backbone assignments were achieved using 2D 1H,15N-heteronuclear single quantum coherence (HSQC), 3D HNCO, 3D HN(CA)CO, 3D CBCA(CO)NH, 3D HNCACB, 3D HN(CO)CA and 3D HNCA. All spectra were processed and analyzed with TopSpin version 2 (Bruker) and Sparky package (http://www.cgl.ucsf.edu/home/sparky/), respectively. The automated assignment program MARS was used to initially assign the backbone resonances (15). Near-complete backbone resonance assignments were achieved for HMG1 (∼97%) and HMG2 (∼88%).
1D 1H-NMR experiment
Effective molecular weights in solution were estimated by measuring 1H T2 relaxation time constants for amide protons, using the 1D-spinecho method (16). Spectra were recorded for ∼75 µM TFAM and TFAM-ΔC in 25 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM DTT and 5% (v/v) D2O, at 5°C. T2 estimates were obtained by comparing peak intensities in spectra recorded with spin-echo delays of 400 µs or 2 ms.
HSQC
To map the HMG1-binding site on p53N, 1H,15N-HSQC spectra were recorded with protein in a buffer of 50 mM MES pH 6.8, 100 mM NaCl, 5 mM DTT and 5% (v/v) D2O at 5°C. Concentrations of 15N-labeled p53N + non-labeled HMG1 were 100 µM. For identifying p53N-binding site or DNA-binding site on HMG1, 1H,15N-HSQC spectra were recorded with protein in a buffer of 25 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM DTT and 5% (v/v) D2O at 15°C. In both control (13C,15N-labeled HMG1) and sample [(13C,15N-labeled HMG1 + non-labeled p53N) or (13C,15N-labeled HMG1 + non-labeled 28-bp dsDNA)] runs, concentrations of HMG1, p53N and DNA were 100 µM. All spectra were externally referenced based on the frequency of the water resonance.
Cell lines and confocal immunofluorescence microscopy
Human colon epithelial cells, HCT116 p53+/+, were grown and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM + GlutaMax-1, Invitrogen), supplemented with 10% FCS. Cells were sub-cultured at a ratio of 1:8 in standard 6-well plate and cells were grown on glass cover slips overnight. Media were replaced with either fresh DMEM media or DMEM supplemented with cisplatin (20/50 µM, freshly prepared in DMSO). After 24 h, cells were subjected to immunofluorescence staining as described earlier (13). Images were collected on a BioRad Radiance confocal microscope.
Chemical shift-based homology modeling
Chemical shift database search was performed using a chemical shift comparison algorithm SimShiftDB (17,18). SimShiftDB analyses each possible pairing of the target protein with each template in the database and retrieves a list of matching proteins, scored by a measure of statistical significance. In addition, SimShiftDB provides a pair-wise chemical shift alignment that maps a set of residues from the target protein to a set of residues from the template structure. We used the chemical shifts data for TFAM HMG1 to scan the PDB template database with SimShiftDB. The search retrieved the structures of UBF1-HMG4 (PDB 1wgf) and TOX-HMG (PDB 2co9) with highest scores of 15e–15 and 14e–14, respectively. A composite alignment derived from the alignments of the top-scoring templates was used as an input for Modeller (19,20). Twenty-five models were generated. The stereo-chemical qualities of all generated models were assessed using PROCHECK (21). The Ramachandran plot shows 98.2% of the residues in the most-favored region, 1.7% in the additional allowed region and 0.2% in the disallowed region. Pair-wise structure superimpositions were performed by using TopMatch (22). PyMOL (http://www.pymol.org/) was used for visualization of the structures.
RESULTS
Domain architecture of TFAM
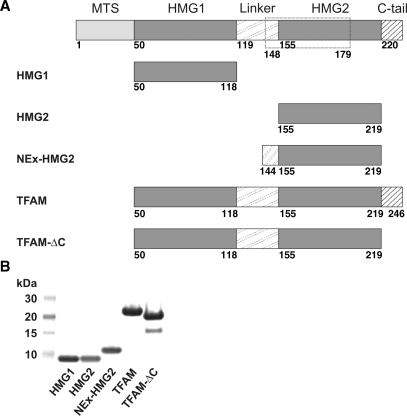
TFAM contains two HMG-box domains (Figure 1A). A basic 36-residue linker connects these tandem HMG-box domains, followed by a 27-residue carboxyl-terminal tail that is also rich in basic amino acid residues. In order to examine the DNA-binding propensity and function of individual domains, we have created a series of TFAM deletion constructs (Figure 1A): ‘HMG1’ (residues 50–118, first HMG-box domain), ‘HMG2’ (155–219, second HMG-box domain), ‘NEx-HMG2’ (144–219, second HMG-box domain with a partial linker at its N-terminus), ‘TFAM’ (50–246, full-length mitochondrial form of TFAM) and ‘TFAM-ΔC’ (50–219, mitochondrial form of TFAM without C-terminal tail). N-terminal protein region (residues 1–49) was predicted to be MTS using MITOPROT [http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html; (12)] and was, therefore, removed from our protein constructs. In other words, proteolytic cleavage site upon mitochondrial transportation of TFAM was assumed to be C49|P50 in the present study. Nevertheless, some research groups regard 42F|43S as the proteolytic cleavage site (23,24). Further, experimental evidences exist indicating that mitochondrial targeting determinants of TFAM may extend beyond the cleavable MTS (24). Thus, the precise presequence of TFAM remains to be clarified. All proteins used in this study were extensively purified to near homogeneity, as judged from SDS–PAGE (Figure 1B). Cell lysate was subjected to DNaseI treatment prior to three chromatographic steps (HisTrap HP column, HiTrap Heparin HP column, HiLoad 26/60 Superdex 75 column) to ensure DNA-free proteins for biophysical studies.
Figure 1.
(A) A schematic diagram of various TFAM deletion constructs used in this study. Dashed box indicates region deleted in the shorter TFAM isoform. (B) SDS–PAGE of various TFAM deletion constructs on a NuPAGE® Novex 4–12% Bis–Tris gel.
DNA-free TFAM exists as monomer–dimer in equilibrium
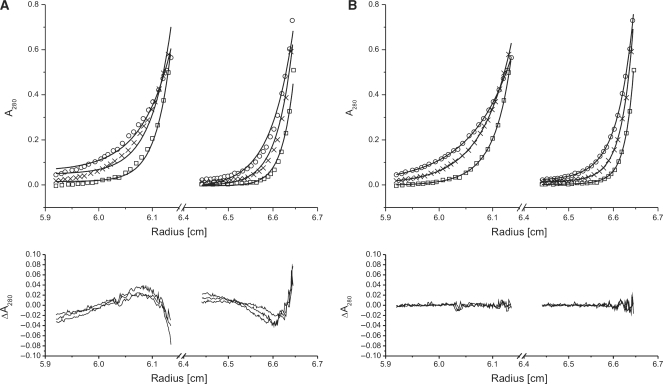
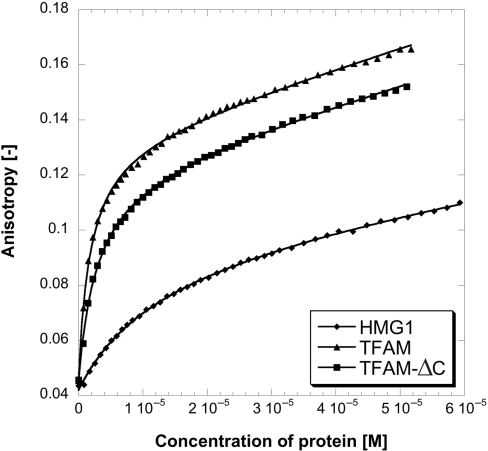
During the purification of HMG1, TFAM and TFAM-ΔC (Figure 1A), we have observed anomalous elution profiles in size exclusion chromatography (Supplementary Figure S1). HMG1 was found to have an elution volume that indicated a size of a tetramer, whereas TFAM and TFAM-ΔC displayed elution volumes consistent with sizes of dimers. In both HMG2 and NEx-HMG2 (Figure 1A), the elution volumes showed approximately sizes of monomers. We have also noticed that elution volume of TFAM was slightly affected by the ionic strength of buffer used (Supplementary Table S2). Increased buffer’s ionic strength resulted in reduced protein mobility, and therefore, a delayed elution from gel filtration column. HMG proteins were named on the basis of their irregular electrophoretic mobility in polyacrylamide gels. As a member of HMG proteins, the observed behavior of TFAM and its deletion constructs (particularly HMG1 and TFAM-ΔC) in Superdex column is probably not surprising. Nevertheless, knowing the stoichiometric state of TFAM is of paramount importance to understand how TFAM functions in promoter or mitochondrial DNA (mtDNA) binding. It has been suggested that TFAM exists as monomer from sedimentation velocity analysis (23) and glycerol gradient sedimentation analysis (25). Kaufman et al. concluded that TFAM was a dimer from size exclusion chromatography (albeit an inappropriate methodology for HMG proteins analyses) and proposed that TFAM dimerization was likely mediated through the predicted coil–coil domain that resides within the second HMG-box domain (26). These disagreements prompted us to use equilibrium sedimentation analytical ultracentrifugation (AUC) to unequivocally determine the stoichiometry of TFAM and its deletion constructs. Our results showed that individual HMG-box domain (HMG1 or HMG2) of TFAM exists as monomer in solution (Table 1). The data of both HMG1 and HMG2 were fitted well to single-exponential models, using partial specific volume (νB) of 0.7319 (HMG1) or 0.7107 (HMG2) and solvent density (ρ) of 1.007 at 5°C (all values were calculated using Sednterp software). Averaged apparent molecular weights (Mw,app) of 8.80 and 8.34 kDa were obtained for HMG1 and HMG2, respectively. These values correspond to calculated molecular weights of HMG1 monomer (Mw = 8.63 kDa) and HMG2 monomer (Mw = 8.07 kDa). Attempt to fit the TFAM data to single-exponential model was not successful (Figure 2A). However, the data was fitted well to double-exponential model using νB = 0.7296 and ρ = 1.007 (Figure 2B), indicating that TFAM is neither a homogeneous solution of monomer nor dimer. Based on the averaged Mw,app found (24.31 and 47.15 kDa), TFAM exists as monomer (calculated Mw = 23.95 kDa) and dimer (calculated Mw = 47.90 kDa) in equilibrium. Surprisingly, TFAM-ΔC was a monomer from our AUC experiments. Upon fitting to single-exponential model (νB = 0.7296 and ρ = 1.007), an averaged Mw,app of 20.91 kDa was acquired, corresponding to monomeric size of TFAM-ΔC (calculated Mw = 20.69 kDa). To revalidate these findings, we derived an effective molecular weight of both TFAM and TFAM-ΔC at 5°C from NMR 1H T2 relaxation time constant estimates. Judging from the spectral profiles (number of peaks and peak positions), TFAM and TFAM-ΔC were structurally indistinguishable, as expected. Overlapping spectra at different τ clearly indicated that TFAM-ΔC, but not TFAM, was homogeneous solution (data not shown). Combining data from size-exclusion, AUC and NMR, we proposed that: (i) DNA-free TFAM exists as monomer–dimer in equilibrium; (ii) self-association of TFAM is mediated through its C-terminal tail; and (iii) high-mobility of TFAM observed in size-exclusion is contributed mainly by its first HMG-box domain.
Table 1.
Apparent molecular weights of various TFAM deletion constructs, obtained from equilibrium sedimentation AUC
| Single-exponential model |
Double-exponential model |
|||||
|---|---|---|---|---|---|---|
| Apparent Mw (kDa) | Fa | Apparent Mw (kDa) of first species | Fa | Apparent Mw (kDa) of second species | Fa | |
| HMG1 (Calculated Mw = 8.63 kDa) | ||||||
| 50.0 µM | 8.84 ± 0.04 | 1.02 | – | – | – | – |
| 200.0 µM | 8.76 ± 0.01 | 1.02 | – | – | – | – |
| HMG2 (Calculated Mw = 8.07 kDa) | ||||||
| 5.0 µM | 8.68 ± 0.05 | 1.08 | – | – | – | – |
| 50.0 µM | 7.41 ± 0.03 | 0.86 | – | – | – | – |
| 200.0 µM | 8.94 ± 0.01 | 1.11 | – | – | – | – |
| 400.0 µM | – | – | 7.47 ± 0.07 | 0.93 | 13.52 ± 0.15 | 1.68 |
| TFAM (Calculated Mw = 23.95 kDa) | ||||||
| 5.0 µM | – | – | 25.29 ± 0.37 | 1.06 | 54.46 ± 0.65 | 2.27 |
| 10.0 µM | – | – | 25.02 ± 0.75 | 1.04 | 43.75 ± 3.29 | 1.83 |
| 50.0 µM | – | – | 24.71 ± 0.14 | 1.03 | 44.49 ± 0.37 | 1.86 |
| 100.0 µM | – | – | 24.78 ± 0.08 | 1.03 | 53.02 ± 0.55 | 2.21 |
| 150.0 µM | – | – | 22.88 ± 0.30 | 0.96 | 42.45 ± 0.88 | 1.77 |
| 189.4 µM | – | – | 23.18 ± 0.11 | 0.97 | 44.71 ± 0.44 | 1.87 |
| TFAM-ΔC (Calculated Mw = 20.69 kDa) | ||||||
| 10.0 µM | 20.76 ± 0.08 | 1.00 | – | – | – | – |
| 50.0 µM | 20.91 ± 0.03 | 1.01 | – | – | – | – |
| 100.0 µM | 21.07 ± 0.03 | 1.02 | – | – | – | – |
aF = Apprent Mw/calculated Mw.
Data were fitted to either a single-exponential model or a double-exponential model.
Figure 2.
(Upper panels) Equilibrium sedimentation AUC of TFAM (5 µM) at speeds of 25 000 r.p.m. (open circle), 30 000 r.p.m. (time) and 35 000 r.p.m (open square). Experiments were conducted at 5°C in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 1 mM DTT. Data were fitted to either a single-exponential model (A) or a double-exponential model (B). Curve fits were shown as smooth lines. (Lower panels) Residuals of curve fits.
TFAM is thermally unstable
We next examined the thermal stability of TFAM through circular dichroism. Since TFAM is rich in helical content from PSIPRED prediction [Supplementary Figure S2; (27)], we followed the change of ellipticity at 222 nm as temperature was raised from 2°C to 98°C. The reversibility of thermal denaturation was checked by reheating the sample (rescan), after cooling the sample to its starting temperature. Supplementary Figure S3 illustrates the thermal melting curves of various TFAM deletion constructs. The data were fitted to a 2-state protein unfolding model (28). The fit returned the melting temperatures (Tm, midpoints of transition) of 308.9 K (HMG1), 296.1 K (HMG2), 310.9 K (TFAM) and 311.3 K (TFAM-ΔC) (Table 2). The thermal denaturation of individual HMG-box domain (HMG1 or HMG2) of TFAM is fully reversible, although Tm of HMG2 is 12.8 K lower than that of HMG1. In the recently published X-ray crystal structure of HMG2 [PDB 3fgh; (23)], the hydrophobic core between helices α1 and α2 contained two amino acids (Y162 and Y200) that had multiple conformations suggesting a highly dynamic core that has not been seen in the non-sequence-specific HMG structures. The lower Tm of HMG2 thus reflects this less rigid hydrophobic core that is also observed in sequence-specific transcription factors in the absence of DNA (e.g. Sox5 and SRY). Since N- and C-terminal tails (usually unstructured) of all HMG-boxes lie close together, we hypothesized that an extension to N-terminal end of HMG2 would help stabilizing the protein fold (Supplementary Figure S4). To test this hypothesis, we created a longer construct of HMG2 (denoted as NEx-HMG2), in which a partial linker (11 residues) was added to its N-terminus. In accordance with our prediction, NEx-HMG2 displayed a Tm of 302.6 K that was 6.5 K higher than Tm of HMG2. Therefore, the linker region of TFAM plays a role in maintaining the overall fold, and thus stability, of the second HMG-box domain. Both TFAM and TFAM-ΔC was slightly more thermal stable than HMG1. Upon thermal denaturation, we noticed slight protein aggregation in TFAM and TFAM-ΔC samples. This would explain why thermal denaturations of these proteins were not fully reversible, in contrast to HMG1 and HMG2 (Supplementary Figure S3). Since Tm of TFAM was similar to those of HMG1 and NEx-HMG2, we believe that the two HMG-box domains of TFAM unfold independently of one another.
Table 2.
Transition temperatures (Tm) of CD thermal melting curves for various TFAM deletion constructs
| Up-scan melting temperature (K) | Re-scan melting temperature (K) | |
|---|---|---|
| HMG1 | 308.9 ± 0.1 | 308.4 ± 0.1 |
| HMG2 | 296.1 ± 0.2 | 296.8 ± 0.2 |
| NEx-HMG2 | 302.6 ± 0.1 | 302.0 ± 0.1 |
| TFAM | 310.9 ± 0.3 | 310.5 ± 0.3 |
| TFAM-ΔC | 311.3 ± 0.8 | 310.2 ± 0.5 |
TFAM binds DNA co-operatively and with high affinity
Using fluorescence anisotropy, we performed solution-based DNA-binding experiments to measure the affinity of various TFAM deletion constructs to ssDNA and dsDNA (Table 3). In contrast to radioactivity-based electrophoretic mobility shift assay (EMSA) that is commonly employed to study DNA binding of TFAM qualitatively (23,25,29), fluorescence anisotropy allows quantitative measurement of DNA affinity. Three types of DNAs were tested for each protein construct: a 28-bp dsDNA consisting of TFAM-binding site on human mtDNA [MITOMAP map locus: MT-TFH (30); map position: 523–550 (31); denoted as 28-bp dsDNA herein], a 28-base ssDNA corresponding to the ‘heavy’-strand of the former dsDNA (28-base ssDNA) and a 50-bp dsDNA of random sequence (50-bp dsDNA).
Table 3.
Binding of DNAs (ssDNA or dsDNA) and p53N (residues 1–93) to various TFAM deletion constructs, measured using fluorescence anisotropy
| Kd (M) (simple one-state model)a | Hill equationa,b |
Number of titrations repeated | ||
|---|---|---|---|---|
| Kd (M) | Hill constant | |||
| HMG1 | ||||
| 28-bp dsDNA | 1.82 ± 0.06 (10−6) | 1.60 ± 0.14 (10−6) | 1.11 ± 0.09 | 4 |
| 28-base ssDNA | 3.81 ± 0.42 (10−6) | 3.33 ± 0.16 (10−6) | 1.08 ± 0.03 | 3 |
| 50-bp dsDNA | 3.58 ± 0.14 (10−6) | 2.60 ± 0.19 (10−6) | 1.23 ± 0.05 | 3 |
| p53N | 1.40 ± 0.18 (10−5) | 1.26 ± 0.21 (10−5) | 1.06 ± 0.07 | 4 |
| HMG2 | ||||
| 28-bp dsDNA | n.d. | |||
| 28-base ssDNA | n.d. | |||
| 50-bp dsDNA | n.d. | |||
| p53N | n.d. | |||
| TFAM | ||||
| 28-bp dsDNA | Poor fit | 6.76 ± 0.83 (10−8) | 1.61 ± 0.24 | 5 |
| 28-base ssDNA | Poor fit | 5.94 ± 0.57 (10−8) | 1.76 ± 0.18 | 3 |
| 50-bp dsDNA | Poor fit | 8.78 ± 0.40 (10−8) | 2.36 ± 0.09 | 3 |
| p53N | 1.95 ± 0.19 (10−6) | 2.05 ± 0.15 (10−6) | 0.96 ± 0.10 | 4 |
| TFAM-ΔC | ||||
| 28-bp dsDNA | 7.10 ± 0.40 (10−8) | 7.91 ± 0.31 (10−8) | 1.22 ± 0.04 | 3 |
| 28-base ssDNA | Poor fit | 11.72 ± 0.88 (10−8) | 1.24 ± 0.06 | 3 |
| 50-bp dsDNA | Poor fit | 11.63 ± 0.67 (10−8) | 1.38 ± 0.11 | 3 |
| p53N | 2.92 ± 0.42 (10−6) | 3.25 ± 0.71 (10−6) | 0.93 ± 0.05 | 4 |
aA linear drift term was added to the fitting equation.
bHill equation: [Protein]h/(Kdh + [Protein]h) where h is Hill constant.
DNAs and p53N were labeled with 5′-fluorescein or AlexaFluor 546, respectively. Data were fitted to either a simple 1-state model or Hill equation. All experiments were conducted at 37°C.
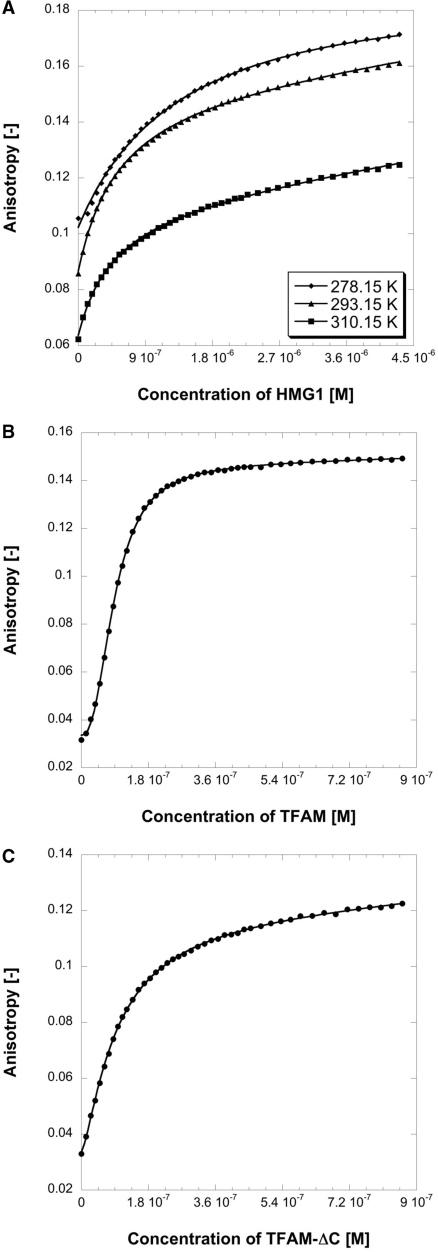
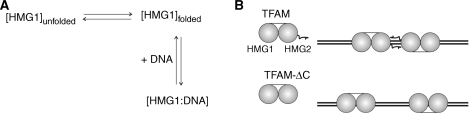
The dissociation constant (Kd) of HMG1 for DNA was temperature-dependent, which is reminiscent of some DNA-binding proteins for instance ORF56 (32). In the case of HMG1, Kd measured at three different temperatures followed the order of 37°C < 20°C < 5°C (Figure 3A), despite the low Tm of HMG1 (∼36°C, Table 2). We ascribed this observation to the nature of reversible thermal denaturation of HMG1 (Figure 4A). At body temperature (37°C), folded HMG1 plausibly exists in equilibrium with unfolded HMG1 and folded HMG1 is competent in DNA binding. The presence of DNA would, therefore, shift the equilibrium in favor of folded HMG1. For the purpose of comparing the contribution of each TFAM domain to DNA binding, we decided to conduct our DNA-binding experiments at physiological conditions namely 37°C and 150 mM ionic strength. In general, DNA-binding data of HMG1 were fitted well to a simple 1-state binding model. Kd for 28-bp dsDNA was 1.82 µM, which was ∼2-fold higher than Kd for 28-base ssDNA (3.81 µM) and for 50-bp dsDNA (3.58 µM). On the contrary, HMG2 displayed very weak DNA-binding capability (Supplementary Figure S5), corroborating findings from other research group (23). This unusual property of HMG2 is probably due to the presence of a polar residue (N163) in the position typically occupied by a non-polar residue capable of intercalating into the minor groove of DNA. Moreover, the region known to make DNA contacts is completely absent from the N-terminus of helix α2 in HMG2 (23). When we aligned and compared TFAMs from various species (Supplementary Table S3), all HMG-box domains showed basic pI values except the second HMG-box domains of Homo sapiens (pI = 7.31) and of Mus musculus (pI = 6.88). DNA binding of HMG proteins usually involves electrostatic interactions, hydrogen bonds, van der Waals contacts, as well as important intercalating non-polar residues reside in either helices α1 or α2 of HMG box. Near-neutral pI of HMG2, therefore, could possibly be another explanation of its low-DNA affinity. Nevertheless, crystal structure of HMG2 clearly revealed the presence of a positively charged concave surface area that is typically involved in DNA binding of HMG boxes (23). Hence, we tested DNA binding of NEx-HMG2, the longer and more stable HMG2 construct. NEx-HMG2 showed increased DNA binding compared with HMG2 (Supplementary Figure S5A; Table 4), although affinity was 10-fold lower than that of HMG1. We reason that linker enhances DNA binding of HMG2 by providing additional DNA contacts (33,34). Thus, the second HMG-box domain of TFAM is a potential DNA binder with its DNA-binding capacity being modulated by the linker region. A smaller isoform of TFAM mRNA was found in most human tissue with a 96-base deletion. Since the fifth exon is identical to the 96 bases skipped in this shorter mRNA, this isoform is concluded to be generated by alternative splicing (11). Interestingly, the region of deletion corresponds to the second HMG-box domain (E148→Q179, 32 residues; Figure 1A) that showed weak DNA binding.
Figure 3.
Binding titration profiles of various TFAM deletion constructs with fluorescein-labeled dsDNA. Experiments were conducted in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT. (A) Profiles of HMG1 at three different temperatures, 5°C (diamond), 20°C (triangle) and 37°C (square). (B) Profile of TFAM at 37°C. (C) Profile of TFAM-ΔC at 37°C.
Figure 4.
(A) Proposed DNA-binding mechanism of HMG1. Folded HMG1 exists in equilibrium with unfolded HMG1 and folded HMG1 is competent in DNA binding. The presence of DNA would shift the equilibrium in favor of folded HMG1. (B) Schematic DNA-binding models of TFAM and TFAM-ΔC. The C-tail modulates self-association of TFAM and promotes TFAM dimer formation upon DNA binding. This accounts for more positive co-operativity in DNA binding of TFAM, in comparison to TFAM-ΔC.
Table 4.
Binding of 5’-fluorescein-labeled DNAs (linear or bulged) to various TFAM deletion constructs, measured using fluorescence anisotropy
| Kd (M) (simple one-state model) | Hill equationa |
Number of titrations repeated | ||
|---|---|---|---|---|
| Kd (M) | Hill constant | |||
| TFAM | ||||
| Normal DNA-S | Poor fit | 4.13 ± 0.59 (10–7) | 1.26 ± 0.14 | 5 |
| Bulged DNA-S | Poor fit | 1.64 ± 0.17 (10–7) | 1.24 ± 0.12 | 3 |
| Normal DNA-Lb | Poor fit | 2.14 ± 0.33 (10–8) | 2.04 ± 0.19 | 3 |
| Bulged-DNA-Lb | Poor fit | 3.15 ± 1.08 (10–8) | 1.85 ± 0.21 | 3 |
| NEx-HMG2 | ||||
| Normal-DNA-L | 2.92 ± 0.34 (10–5) | 2.69 ± 0.32 (10–5) | 1.09 ± 0.16 | 3 |
| HMG2 | ||||
| Normal DNA-L | Estimated to be >40 µM | |||
aHill equation: [Protein]h/(Kdh + [Protein]h) where h is Hill constant.
bA linear drift term was added to the fitting equation.
Data were fitted to either a simple 1-state model or Hill equation. All experiments were conducted at 10°C.
To fit data from TFAM and TFAM-ΔC experiments, 1-state-binding model could not be applied. These data, however, were fitted well to the Hill equation indicating co-operativity in DNA binding (Figures 3B and 3C). Compared with HMG1, TFAM showed significantly higher affinity to 28-bp dsDNA (27-fold), 28-base ssDNA (64-fold) and 50-bp dsDNA (41-fold). When the C-terminal tail was truncated, DNA-binding affinity was reduced ∼1.2–2-folds. This result is consistent with findings from EMSA [2.2-fold reduction; (23)]. It is worth a note that positive co-operativity was reduced in all three DNAs tested, after removal of the TFAM C-tail (1.61→1.22 in 28-bp dsDNA, 1.76→1.24 in 28-base ssDNA and 2.36→1.38 in 50-bp dsDNA). Since the C-tail modulates self-association of TFAM, we postulate that TFAM binds as a dimer on DNA resulting in a more positive co-operativity (Figure 4B). The exact mechanism and interface of TFAM self-association, of course, requires further studies. TFAM dimerization upon DNA binding was also supported by evidences from non-contact atomic force microscopy (26) and from EMSA (23). Mutational analysis of TFAM revealed that the C-tail is important for specific DNA recognition and essential for transcriptional activation (35,36). The C-tail is necessary to bind mitochondrial transcription factor B (37) for eliciting transactivation. Here, we demonstrated that the C-tail facilitates TFAM dimerization on DNA. Ohgaki et al. reported that C-tail truncation decreased DNA-binding activity of TFAM by three orders of magnitude using surface plasmon resonance (SPR) kinetic analysis (38). We have observed non-specific interaction between TFAM and sensor chip (data not shown). The discrepancy between our data and those reported by Ohgaki et al. is likely contributed by various factors including different DNAs used (sequence and length), non-specific TFAM–sensorchip interaction and fitting model (1:1 Langmuir binding model was used in the report from Ohgaki et al.).
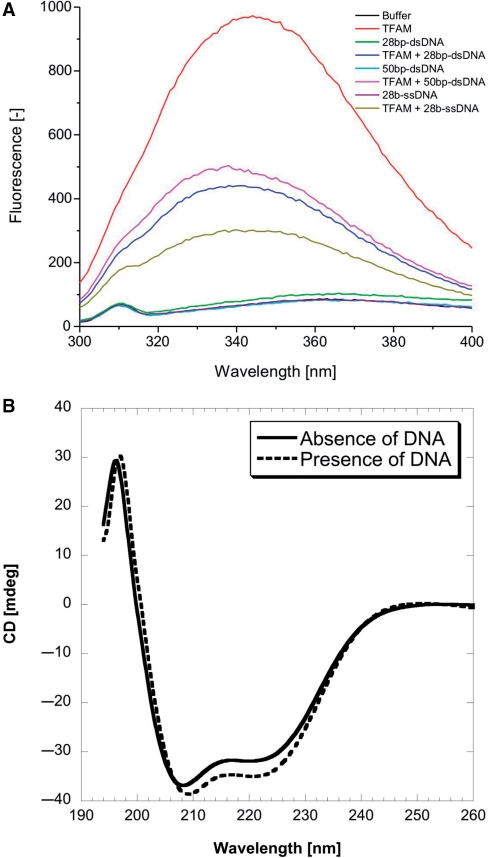
TFAM contains two tryptophan residues in each HMG-box domain (W88 and W107 in the first domain, W189 and W218 in the second domain). Of these, W88, W189 and W218 are strictly conserved among TFAMs from various species (Supplementary Figure S6). Figure 5A shows the intrinsic fluorescence spectra of TFAM, in the absence and presence of DNA. Excitation at 280 nm gave an emission maximum at 344 nm, implying that some of these tryptophan residues are solvent exposed. Upon binding DNA, emission maxima were blue-shifted to ∼338 nm and the fluorescence emissions were reduced 31–52%. Thus, these conserved tryptophan residues are involved in stabilizing protein–nucleic acid complexes, plausibly via stacking interactions with the nucleotide bases of DNA or intercalating into DNA grooves. The binding of TFAM to DNA has also an effect on its secondary structure, as evaluated by UV CD spectropolarimetry (Figure 5B). The CD-spectrum of TFAM–DNA complex shows a decrease in optical ellipticity in the spectral region 208–235 nm, compared with that of DNA-free TFAM. The decrease is indicative of formation of additional α-helical region or type-II β-turn. We did not observe significant change in the CD spectrum >240 nm, which would otherwise indicate DNA conformational change from B-DNA to either A-DNA or Z-DNA. Although HMG-box is widely recognized to induce DNA deformations (bending and unwinding) upon DNA binding, this feature is not apparent in our CD measurements using 28-bp dsDNA (Supplementary Table S1). The magnitude of DNA deformation induced by HMG-box is likely DNA-sequence or DNA-length dependent.
Figure 5.
(A) Intrinsic fluorescence emission spectra of TFAM in the absence (red) and presence of 28-bp dsDNA (blue), 28-bp ssDNA (dark yellow) and 50-bp dsDNA (magenta), at excitation wavelength of 280 nm. Experiments were conducted at 25°C in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT. Concentrations of both TFAM and DNAs were 500 nM. Control spectra without TFAM were also recorded: buffer (black), 28-bp dsDNA in buffer (green), 28-bp ssDNA in buffer (purple) and 50-bp dsDNA in buffer (cyan). (B) UV CD spectra of 10 µM DNA-free TFAM (continuous line) and TFAM complexed with 4 µM 28-bp dsDNA (broken line), corrected for the background of DNA alone in buffer. Experiments were conducted at 20°C, in 25 mM sodium phosphate pH 7.4, 100 mM KCl and 1 mM 1,4-dithioerythritol.
Taken together, our solution-based DNA-binding data conform to the reported biological roles of TFAM as a transcriptional activator (preference towards sequence-specific DNA) and a mtDNA-packaging factor (high affinity for non-sequence-specific DNA).
TFAM binds p53N
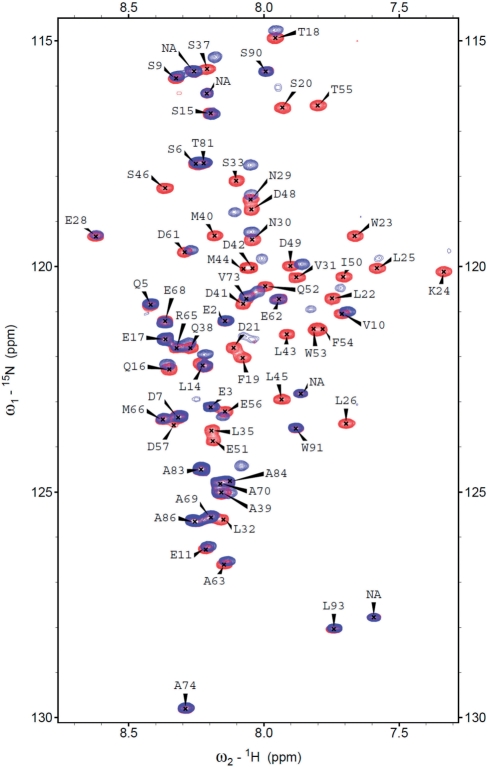
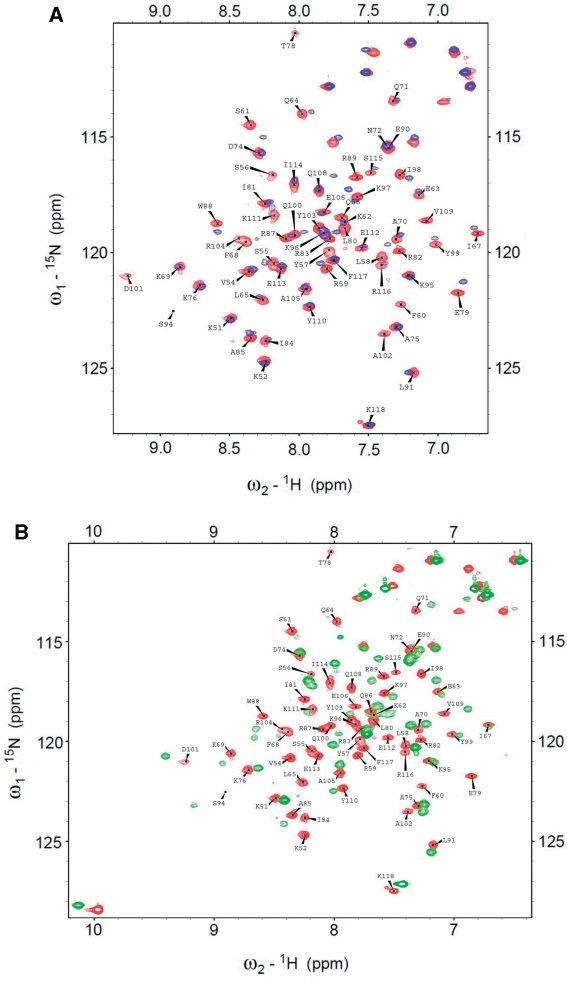
Tumor suppressor p53 has previously been demonstrated to interact with DNA-binding proteins. Salient examples include human mitochondrial single-stranded DNA-binding protein [HmtSSB; (13)], replication protein A [RPA; (39)] and transcriptional co-activator PC4 (40), via N-terminal transactivation domain of p53. It has also been reported that p53 interacts with HMG proteins, including HMGB1 (41) and TFAM (8). On the basis of these previous findings, we hypothesized that TFAM-binding site on p53 resides within its transactivation domain. By using fluorescence anisotropy, we assayed TFAM for binding to an AlexaFluor 546-labeled p53N (residues 1–93) at physiological ionic strength and our hypothesis was indeed verified (Figure 6). The binding data of all protein constructs tested were fitted well to a simple 1-state binding model (Table 3), and affinity towards p53N followed the order of TFAM (1.95 µM) > TFAM-ΔC (2.92 µM) > HMG1 (14 µM). To localize the binding interface on p53N, we recorded 2D 1H,15N-HSQC NMR spectra of 15N-labeled p53N (100 µM) in the absence and presence of unlabeled HMG1 (100 µM). In Figure 7, free 15N-labeled p53N spectrum (red) was overlaid with the HMG1-bound spectrum (blue). p53N interacts clearly with HMG1 through an extended interface. Large chemical shift perturbations were mainly localized in three regions: region I (residues 18–26), region II (residues 30–38) and region III (residues 40–56). The p53 transactivation domain is subdivided into two loosely defined sub-domains (42), TAD1 (residues 1–40) and TAD2 (residues 41–61). We found that HMG1 interacts simultaneously with TAD1 and TAD2, similar to HmtSSB (13) and transcriptional co-activator p300 (43). Both sub-domains contains a Φ-X-X-Φ-Φ motif (where Φ = hydrophobic amino acid and X = any amino acid) that been shown to be important in many protein–protein interactions [Supplementary Figure S7; (44)]. Our NMR experiment demonstrated that p53–HMG1 interaction involves these two motifs (F19-W23 in TAD1 and I50-F54 in TAD2), signifying the involvement of hydrophobic interaction in stabilizing p53–HMG1 complex. Further, HMG1-binding site identified in our NMR study coincides with p53N regions that were known to form helical structures upon complex formation [i.e. helix 1 (S15-P27), helix 2 (D41-M44) and helix 3 (P47-T55); Supplementary Figure S7] (39,45). We believe that electrostatic interactions contribute substantially to p53N–HMG1 complex formation, in view of acidic nature of p53N (pI = 3.47) and basic nature of HMG1 (pI = 9.99).
Figure 6.
Binding titration profiles of HMG1 (diamond), TFAM (triangle) and TFAM-ΔC (square) with AlexaFluor 546-labeled p53N. Experiments were conducted at 37°C, in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT.
Figure 7.
2D 1H,15N-HSQC NMR spectra of 15N-labeled p53N (residues 1–93, 100 µM) in the absence (red) and presence of 100 µM of HMG1 (blue). Spectra were recorded at 5°C in 50 mM MES pH 6.8, 100 mM NaCl, 5 mM DTT and 5% (v/v) D2O.
Yoshida et al. reported that interaction between TFAM and p53 required either one of the two HMG-box domains and residues 363–376 of p53 (8), based on pull-down assays. We, therefore, assayed TFAM for binding to a fluorescein-labeled p53C (residues 362–393) at physiological ionic strength, using fluorescent anisotropy. All protein constructs tested (TFAM, HMG2, NEx-HMG2) showed weak binding to p53C (Supplementary Figure S5B). We ascribe this low affinity to basic nature of p53C (pI = 9.61) and suggest that the C-tail of p53 is possibly a secondary binding site of TFAM. Protein partners having two different binding sites on p53 have previously been reported. These include S100 [binds transactivation and tetramerization domains of p53; (46)] and Mdm2 [binds transactivation and core domains of p53; (47)]
Both TFAM and p53 bind distorted DNA preferentially
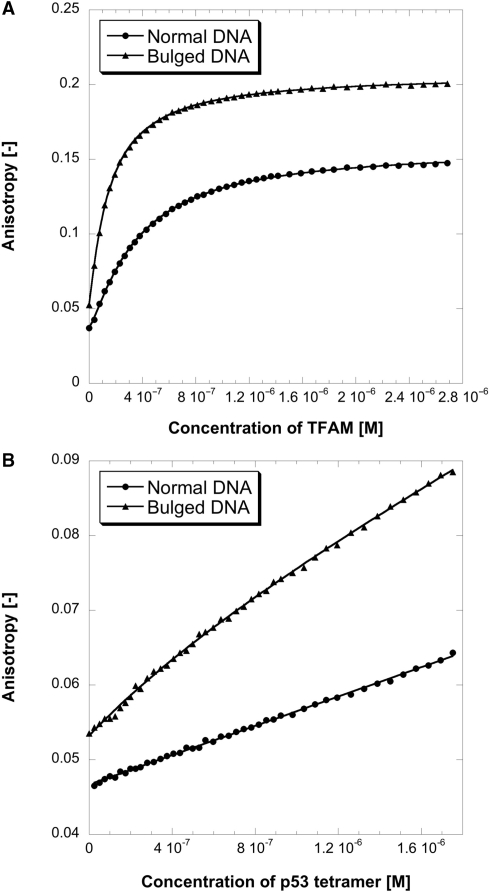
TFAM has previously been demonstrated to bind preferentially to oxidatively damaged DNA (e.g. 8-oxo-7,8-dihydroguanine), cisplatinated-DNA and DNA lesions induced by chemical carcinogens (e.g. N-acetoxy-acetylaminofluorene), using EMSA assays (48,49). To further investigate this preference quantitatively, we used fluorescence anisotropy to study binding of TFAM to two different DNAs. One of these was a linear DNA with 12 bp (denoted as Normal-DNA-S). The other was a bulged DNA containing a dA2 bulge, with its sequence being identical to the previous one (Bulged-DNA-S). We chose to study bulged DNA because it is structurally analogous to UV-induced pyrimidine dimers (naturally occurring DNA lesions) and cisplatinated-DNA (adducts induced in vivo by anticancer drug cisplatin). Both pyrimidine dimer and cisplatination induce DNA bends of magnitude comparable with the bulge-induced bends (50,51). Therefore, the study of bulged DNA is biologically relevant. As shown in Figure 8A and Table 4, TFAM bound distorted DNA preferentially. Kd for Bulged-DNA-S (164 nM) was 2.5-fold lower, compared with that for Normal-DNA-S (413 nM). Nevertheless, this preference was not evident when we used DNAs that were four times longer in length (Normal-DNA-L and Bulged-DNA-L). Interestingly, p53 displayed the same preference (Figure 8B), although Kd of p53 for short DNA was much higher. Taken together, p53–TFAM interaction may play a significant role in DNA damage and repair. Further, our data provided direct evidence that TFAM can indeed bind DNA as short as 12 bp, in agreement with estimation from Pellegrini et al. that TFAM was present at a ratio of one molecule per ∼10–12 bp of mtDNA (52). This estimation was based on quantitative western and Southern blot analyses, using protein–DNA isolated from mouse liver mitochondria.
Figure 8.
Binding titration profiles of TFAM (A) and p53 (B) with fluorescein-labeled linear DNA (circle) or bulged DNA (triangle). Experiments were conducted at 10°C, in 25 mM Tris–HCl pH 7.4, 150 mM NaCl and 5 mM DTT.
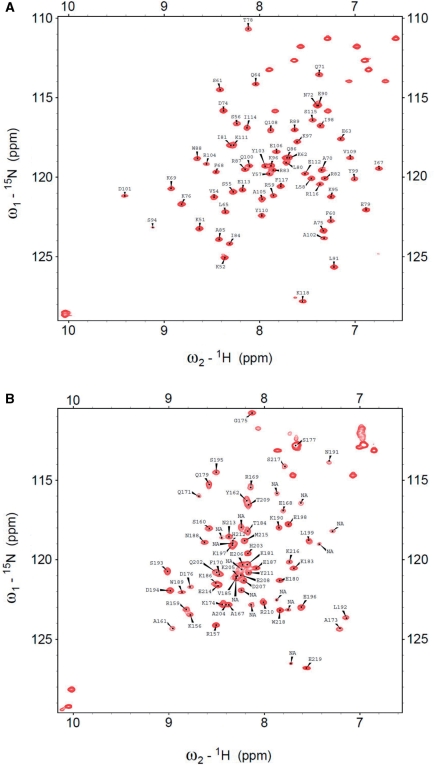
NMR backbone resonance assignments of HMG1 and HMG2
For the purpose of mapping p53N-binding site on HMG-box domain, we assigned the backbone resonances in the NMR spectra of both HMG1 and HMG2. 13C,15N-labeled HMG1 and 13C,15N-labeled HMG2 samples provided well-dispersed NMR spectra at 15°C and 5°C, respectively (Figure 9). Low temperatures were chosen to populate natively folded HMG1 and HMG2, in view of their low Tm (Supplementary Figure S3; Table 2). This allowed determination of >85% of the backbone resonance assignments from HMG1 (97%, Supplementary Table S4) and HMG2 (87%, Supplementary Table S5), as described in ‘Materials and Methods’ section. Even though the spectrum of HMG2 was recorded at 5°C, we noticed a large number of sharp overlapping resonances in the random coil region of the spectrum (1HN 8.0–8.5 ppm). Therefore, HMG2 is partially structured in the DNA-free protein, which explains its lower thermal stability (Supplementary Figure S3; Table 2). The intrinsic disorder in DNA-free HMG-box was also documented for SRY box (53) and LEF-1 box (54). To localize p53N-binding site, we acquired 2D 1H,15N-HSQC NMR spectra of 13C,15N-labeled HMG1 (100 µM) in the absence and presence of unlabeled p53N (100 µM). In Figure 10A, we presented the overlay spectra of free HMG1 (red) and p53N-bound HMG1 (blue). Large chemical shift perturbations were mapped to regions of high-helical propensity, as predicted by PSIPRED (Supplementary Figure S7). It appears that p53N interacts extensively with helices α1 and α2 (Supplementary Figure S7). The 1H,15N-HSQC spectrum of HMG1–DNA complex (1:1 ratio, 100 µM of each) is shown in Figure 10B (green). Drastic chemical shift changes have occurred for the HMG1 resonances, reflecting either extensive DNA-binding interface or conformational change. When we used a ratio of 2.5 HMG1 to 1 DNA, most peaks disappeared in the 1H,15N-HSQC spectrum (Supplementary Figure S8). This is a common phenomenon when high-molecular weight complexes are formed. In other words, there were more than one HMG-box bound to a single DNA molecule, when HMG-box was present in excess (a ‘pigeonhole principle’ applies in this case). Of note, we observed perturbation of tryptophan side chain resonances upon DNA binding (1HN 10–12 ppm, indole side-chain region). This indicates that tryptophan side chains were positioned in a different environment, corroborating our intrinsic fluorescence measurement (Figure 5A).
Figure 9.
2D 1H,15N-HSQC NMR spectra of HMG1 (A) and HMG2 (B), acquired at 15°C and 5°C, respectively. Non-assigned peaks were labeled with NA. Peaks corresponding to side chain amides were not labeled. Both spectra were recorded with 100 µM protein in 25 mM Tris–HCl, 150 mM NaCl, 1 mM DTT and 5% (v/v) D2O.
Figure 10.
(A) 2D 1H,15N-HSQC NMR spectra of 13C,15N-labeled HMG1 (100 µM) in the absence (red) and presence of 100 µM of p53N (blue). (B) 2D 1H,15N-HSQC NMR spectra of 13C,15N-labeled HMG1 (100 µM) in the absence (red) and presence of 100 µM of DNA (green). All spectra were recorded at 15°C in 25 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM DTT and 5% (v/v) D2O.
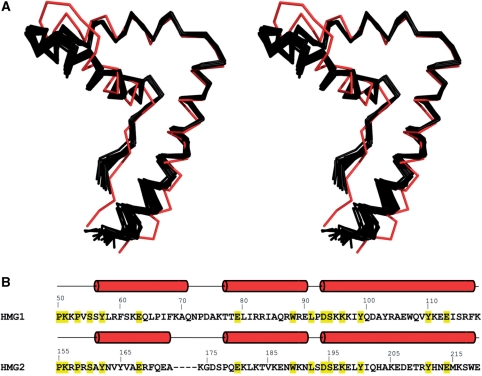
Comparison of HMG1 homology model with HMG2 structure
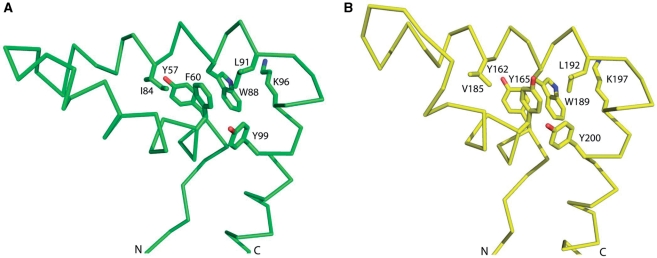
Using NMR chemical shift data of HMG1, we created a homology model of this HMG-box domain (see ‘Materials and Methods’ section). HMG1 adopts canonical HMG-box fold and consists of an extended region (residues 50–55) and three helices (residues 56–71, 77–90, 93–116) that are approximately orthogonal to one another (Figure 11A). The N-terminal extended region packs against the C-terminal helix α3. The core, formed at the intersection of the three helices, comprises residues Phe60, Trp88 and Tyr99 which are surrounded by Tyr57, Ile84, Leu91 and Lys96. In addition, the orientation of helix α1 toward helix α2 is stabilized by several hydrophobic contacts formed by residues Leu65, Phe68 and Leu80 (Figure 12A). Structural comparison of HMG1 model and HMG2 structure (3fgh) reveals that these two HMG-box domains are very similar and superpose with a root mean square deviation of 2.1 Å (Figure 11A). Most of the residues forming the hydrophobic core, at the intersection between the three helices, are invariant. The only exception is that Phe60 and Ile84 of HMG1 are replaced by Tyr165 and Val185 in HMG2, respectively (Figure 12). Despite the global similarity, several differences between HMG1 and HMG2 structures are apparent. These include the region connecting helices α1 and α2, and the length of helix α1 (Figure 11B). HMG2 contains a four-residue deletion resulting in a shorter helix α1. None of the residues forming contacts between helices α1 and α2 are conserved. Leu65, Phe68 and Leu80 of HMG1 are replaced by Phe170, Ala173 and Lys181 in HMG2, respectively. Thus, the orientation of helix α1 towards helix α2 in HMG2 is determined by the hydrophobic contacts between the aliphatic portion of Lys181 and the side-chains of Phe170 and Ala173. Although the large-to-small substitution of Phe68→Ala173 is partly compensated by the substitution of Leu65→Phe170, this rearrangement causes a small displacement of helix α1 relative to the axes of helices α2 and α3. The electrostatic potential surface comparison of HMG1 and HMG2 shows that the overall charge of the DNA-binding interface is preserved (Figure 13). The displacement of helix α1 in HMG2, however, widens the concave DNA-binding interface and could be a potential reason for the weaker DNA-binding property of this domain (Supplementary Figure S5A; Table 4). Modeling of the N-terminal linker of NEx-HMG2 (11 residues) suggests that some of the hydrophobic residues in this region might have a role in the stabilization of HMG-box, as observed experimentally (Supplementary Figure S3). In particular, Leu152 could form a hydrophobic contact with Trp218 and thus orient the main-chain carbonyl group of Gly153 in a position suitable for hydrogen bond formation (Supplementary Figure S4). Since this 11-residue linker is highly positively charged, we would expect that the linker contributes to DNA anchoring, as observed in other HMG-box containing proteins (33,34).
Figure 11.
Comparison of HMG-box domains (HMG1 and HMG2) of TFAM. (A) Stereo views of structural superposition of HMG2 (3fgh, red) overlaid with a bundle of 25 HMG1 models (black). Images were generated using PYMOL. (B) Structure-based sequence alignment of HMG1 and HMG2. Secondary structure elements were shown above the protein sequences. Invariant residues between HMG1 and HMG2 were highlighted in yellow.
Figure 12.
(A) Hydrophobic core of HMG1 model, with Y57, F60, I84, W88, L91, K96 and Y99 indicated. (B) Hydrophobic core of HMG2 (3fgh) with Y162, Y165, V185, W189, L192, K197 and Y200 indicated. Both images were generated using PYMOL.
Figure 13.
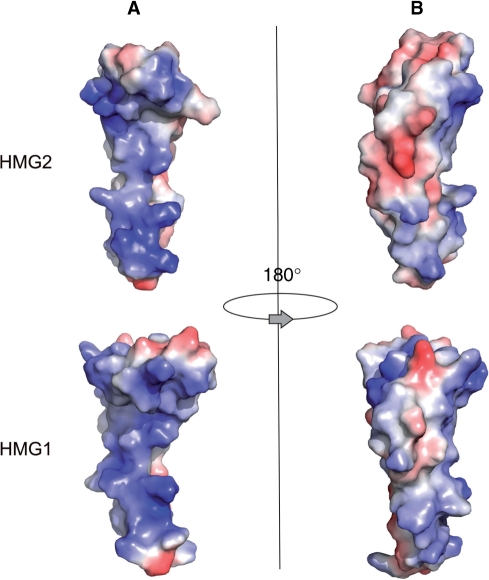
Electrostatic surface potential plots of HMG1 model and HMG2 (3fgh), generated with PYMOL. Regions of positive potential were colored in blue, and regions of negative potential were colored in red. (A) Front views showing DNA-binding interfaces. (B) Back views.
DISCUSSION
Structure–function relationships of TFAM
TFAM is a multi-functional protein, involved in various aspects of mtDNA integrity maintenance: transcription, replication, nucleoid formation, DNA protection, DNA repair and damage sensing (3). In part, its functional diversity is a result of TFAM’s domain architecture (Figure 1A). TFAM is constructed by two tandem HMG-box domains, which are connected by a basic 36-residue linker. To the C-terminus of the second HMG-box domain, a 27-residue tail is attached, that is also rich in basic amino acid residues. In order to investigate the DNA-binding propensity and function of each domain of TFAM, we adopted a ‘divide-and-conquer’ strategy by creating a series of TFAM deletion constructs as depicted in Figure 1A. Using equilibrium sedimentation AUC (Table 1), we showed that HMG1 and HMG2 were monomers in solution. On the contrary, TFAM exists as monomer–dimer in equilibrium (Figure 2) and its self-association is modulated by the basic C-tail. This finding was validated by 1D 1H-NMR spectra of TFAM and TFAM-ΔC (data not shown). The anomalous elution profile of TFAM in size-exclusion chromatography (Supplementary Figure S1) is contributed mainly by HMG1. This ‘high mobility’ is a consequence of asymmetric nature of TFAM and interaction between highly charged TFAM and chromatographic material. TFAM is thermally unstable, with a Tm close to body temperature (37.7°C) determined from CD spectropolarimetry (Supplementary Figure S3; Table 2). HMG1 displayed similar Tm (35.7°C), while HMG2 showed substantially lower Tm (23.0°C). The thermal instability of DNA-free HMG2 well reflects its less rigid hydrophobic core (23) and its intrinsic disorder that we observed in 2D 1H,15N-HSQC spectrum of HMG2 (Figure 9B). The linker region enhances stability of HMG2, possibly by providing additional contacts with C-end of HMG2 and thus maintaining the global fold of three helices in an L-shape configuration (23). From our solution-based DNA-binding experiments using fluorescent anisotropy, TFAM binds DNA co-operatively with nanomolar affinity and a preference towards sequence-specific DNA. Despite its low Tm, TFAM maintains high affinity towards DNA at 37°C. This is possible since thermal unfolding of TFAM is almost completely reversible (Supplementary Figure S3). The presence of DNA shifts the equilibrium towards folded HMG-box that is competent in DNA binding (Figure 4A). The C-tail contributes significantly to positive co-operativity of TFAM’s DNA binding, which is linked to its capability of inducing TFAM self-association (Figure 4B). The C-tail removal reduces DNA-binding affinity by a factor of 1.2–2, consistent with in vivo data reported (36). When TFAM-ΔC was expressed at levels comparable to those of endogenous TFAM in HeLa cells, mtDNA increased two folds, suggesting that TFAM-ΔC was as competent in maintaining mtDNA as endogenous TFAM. TFAM-ΔC, however, lost its transcriptional activity in vitro (36). TFAM dimerization at promoter region might be biologically significant to elicit its transcriptional activity. Interestingly, isolated HMG2 is a weak DNA binder as opposed to HMG1. This unusual behavior of HMG2 can be attributed to various reasons: the lack of DNA intercalating residue, missing region known to make DNA contacts, and near neutral pI (Table S3). Linker region compensates this inability of HMG2 (Figure S5A), likely by providing additional DNA contacts (33,34). It is worth noting that the highly conserved tryptophan residues (Supplementary Figure S6) are involved in DNA binding, as judged from intrinsic fluorescence measurement (Figure 5A) and 2D 1H,15N-HSQC spectrum of HMG1–DNA complex (Figure 10B). Finally, DNA binding induces conformational change of TFAM (Figure 5B). The decrease in optical ellipticity at spectral region 208–235 nm indicates formation of additional α-helical region or type-II β-turn, possibly at intrinsic disorder region resides within HMG2 or linker region.
Biological implications of TFAM–p53 interaction
Tumor suppressor p53 is involved in a multitude of cellular functions including cell-cycle arrest, apoptosis, senescence, DNA damage and repair (55). For the first time, we showed that TFAM interacts primarily with N-terminal transactivation domain of p53 (p53N, residues 1–93, Figures 6, 7 and 10A). C-terminal regulatory domain of p53 (p53C, residues 362–393) provides a secondary binding site for TFAM (Supplementary Figure S5B).
p53N is inherently devoid of any tertiary structure. Nevertheless, a pre-existing amphipathic helix (T18-L26) and two nascent turns (turn I: M40-M44 and turn II: D48-W53, both weakly amphipathic) were found in unbound p53N, using heteronuclear multi-dimensional NMR spectroscopy (45). These local secondary structures are selectively formed by functionally critical and positionally conserved hydrophobic residues (hydrophobic antennae, e.g. F19, L22, W23 and L26 in p53N) present in several acidic transactivation domains (AADs) (45). Such local structures are general features of AADs and may represent ‘specificity determinants’ that are important for transcriptional activity (56). We speculate that the structural transitions associated with p53N upon TFAM binding involve both (i) tightening of the pre-existing helix into a stable helix and (ii) conversion of the two nascent turns (turns I and II) into an amphipathic helix. The later coil-to-helix transition is either due to structural restraints imposed by TFAM or induced by hydrophobic cores of HMG-boxes. The fact that the overall helical content of several AADs increases upon addition of hydrophobic solvents that might mimic the hydrophobic environment of a target protein supports such a possibility (45). In p53N upon TFAM binding, we picture formation of two helices (S15-L32 and Q38-D57) with a symmetrical linker (S33-S37) between them. This would also explain the chemical shift perturbations we observed in p53N (Figure 7; Supplementary Figure S7). We believe that p53N docks into the positively charged DNA-binding cleft of HMG-box (Figure 13), mimicking single-stranded DNA structurally (39,40). Amphipathic helices of p53N could plausibly orient in a way that hydrophilic sides are exposed to solvent and hydrophobic sides are buried. In this orientation, HMG-box–p53 complex is stabilized by electrostatic interactions of the negatively charged side chains of p53N and the positively charged side chains of HMG-box, as well as hydrophobic interactions between the buried surface of p53N helix (I50, W53, F54) and the hydrophobic core of HMG-box (Y57, F60, W88). Therefore, HMG-box represents a structural entity endowing dual affinity (i) for acidic, amphipathic helices and (ii) for DNAs. The aforementioned speculations would certainly require experimental validations, e.g. by solving the crystal structure of HMG-box–p53 complex.
Similar to RPA–p53 interaction (57), TFAM–p53 interaction could possibly involve two distinct areas of p53 (p53N and p53C, Figure 6 and Supplementary Figure S5B) simultaneously. This is likely facilitated by the presence of DNA. In the absence of DNA, p53 displays an elongated cross-shaped structure with a pair of loosely coupled core domain dimers at the ends, which are accessible for binding to DNA or protein partners (58). The core domains in this ‘open conformation’ can close around a DNA to form a compact complex, resulting in a ‘close conformation’ (58). Further, DNA induces TFAM dimerization that is modulated by TFAM C-terminal tail (Figure 4B). In a possibly compact TFAM–p53–DNA ternary complex, one would envisage the feasibility of ‘two-point interaction’.
We believe that TFAM–p53 interaction is biologically relevant. The naturally oxidative environment of mitochondria and lack of an efficient DNA repair mechanism render mtDNA highly susceptible to mutations, and these mutations are frequently associated with a variety of human diseases (1). Since both TFAM and p53 binds preferentially distorted DNA (Figure 8, Table 4), TFAM–p53 interaction is implicated in DNA damage and repair. In this same vein, we recently reported that HmtSSB (a mtDNA packaging factor and sub-unit of mtDNA replisome) binds to p53N via an extended binding interface, with dissociation constant of 12.7 (±0.7 µM) (13). HmtSSB enhances intrinsic 3′-5′ exonuclease activity of p53 (59), particularly in hydrolysing 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) present at 3′-end of DNA (13). Our data suggest that p53 is involved in maintaining integrity of mitochondrial genome, in line with findings from other group (60). Remarkably, p53 null mouse and p53 knockdown human primary fibroblasts were shown to exhibit mtDNA depletion, decreased mitochondrial mass and membrane potential, reduced protein level of both ribonucleotide reductase p53R2 sub-unit and TFAM, as well as disruption of cellular ROS homeostasis (61). Altogether, p53 has a direct impact on mitochondrial homeostasis. Although Kd of TFAM to p53N falls in micromolar range (1.95 ± 0.19 µM), TFAM is a copious protein within mitochondria (62). In human HeLa cells, TFAM is 1700-fold more abundant than mtDNA (63). Pellegrini et al. estimated that TFAM was present at a ratio of one molecule per ∼10–12 bp of mtDNA (52). Further, TFAM was up-regulated in cisplastin-treated KB human epidermoid cancer cells (8). It is important to note that mitochondrial p53 isolated from ML1, HCT116 and RKO cells, after short term genotoxic stress, were phosphorylated on Ser 6, Ser 9, Ser 15, Ser 20, Ser 37 and Ser 46 (64). These serine residues were located within TAD1 and TAD2 sub-domains of p53, the identified binding interface between p53 and TFAM from NMR (Figure 7 and Supplementary Figure S7). Since phosphorylation increases negative charge of p53N, the affinity between TFAM–p53 in vivo is likely higher than value obtained from in vitro measurement. Relative affinities of the N-terminal transactivation domain of p53 for transcription co-activator p300 domains or PC4 were reported to be regulated by phosphorylation, and phosphorylation generally enhances binding affinity (40,65). Last but not least, mitochondrial translocation of p53 would result in its eventual compartmentalization within these organelles, leading to increased local p53 concentration in mitochondria.
Protein traffic upon DNA damage
As a predominantly nuclear protein, p53 lacks MTS. Nevertheless, mitochondrial localization of p53 was observed after cisplatin treatment [(8) and in present study; Supplementary Figure S9]. The mechanism by which p53 is transported into mitochondria remains, however, unclear. Being an important chemotherapeutics, cisplatin is best recognized as a DNA damaging agent and the cytotoxicity of cisplatin has generally been attributed to its ability to form inter- and intra-strand DNA crosslink. While most investigations of cisplatin pharmacology focussed on cisplatin/nuclear DNA adducts, only 1% of intracellular platinum is bound to nuclear DNA. The remaining majority of the intracellular drug is available for targeting nucleophilic sites on other molecules, including phospholipids, cytosolic, cytoskeletal and membrane proteins, RNA, or even mtDNA (66). The voltage-dependant anion channel (VDAC, also termed mitochondrial porin) contains two cysteines and two methionines, in total four potential targets for platinum binding. VDAC has been shown to form cisplatin/protein adducts, resulting in permeability alteration of mitochondrial membrane (66). Likewise, cisplatin could possibly change the permeability of nuclear envelope that was noticeable in Supplementary Figure S9. Since p53 was up-regulated after drug treatment, p53 observed in perinuclear mitochondria might be a consequence of passive diffusion (Supplementary Figure S9). Remarkably, it has been demonstrated that TFAM overexpression and impairment of its mitochondrial targeting could result in nuclear accumulation of this protein (24). A putative bipartite nuclear localization sequence (two clusters of positively charged residues separated by 11 amino acids) was found in HMG1. Nuclear TFAM was found to confer cytoprotection against chemotherapeutic drugs e.g. etoposide, camptothecin and cisplatin. Mitochondrial localization of p53 and nuclear accumulation of TFAM raise the possibility that p53 and TFAM were examples of ‘eclipsed distribution’ (67). Upon DNA damage, protein traffic is likely bi-directional (p53: nuclei → mitochondria, TFAM: mitochondria → nuclei).
CONCLUSION
In the present study, we characterized interaction between TFAM and p53. Biophysical measurements, in conjunction with cellular biology methods, provide a systematic platform to study the physical and functional interactions between these two proteins in vitro. The biological relevance of this binding in vivo would require more in-depth investigations, in view of the complex multi-functional roles of both TFAM and p53.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Union FP6 Proteomage (to A.R.F. and T.S.W.); MRC Career Development Fellowship (to T.S.W.). Funding for open access charge: Medical Research Council, UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Prof. Hans Spelbrink (University of Tampere, Finland) for providing the gene encoding TFAM, Dr. Robert Sade for useful discussion, Caroline M. Blair and Fiona Sait for technical assistance.
REFERENCE
- 1.Copeland WC. Inherited mitochondrial diseases of DNA replication. Ann. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Remmen H, Jones DP. Current thoughts on the role of mitochondria and free radicals in the biology of aging. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64:171–174. doi: 10.1093/gerona/gln058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang D, Hamasaki N. Mitochondrial transcription factor A in the maintenance of mitochondrial DNA: overview of its multiple roles. Ann. NY Acad. Sci. 2005;1042:101–108. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- 4.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Clayton DA. Mitochondrial DNA replication: what we know. IUBMB Life. 2003;55:213–217. doi: 10.1080/1521654031000134824. [DOI] [PubMed] [Google Scholar]
- 6.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Ann. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 7.Robberson DL, Clayton DA. Replication of mitochondrial DNA in mouse L cells and their thymidine kinase - derivatives: displacement replication on a covalently-closed circular template. Proc. Natl Acad. Sci. USA. 1972;69:3810–3814. doi: 10.1073/pnas.69.12.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 9.Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol. Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes A, Mezzina M, Gadaleta G. Human mitochondrial transcription factor A (mtTFA): gene structure and characterization of related pseudogenes. Gene. 2002;291:223–232. doi: 10.1016/s0378-1119(02)00600-5. [DOI] [PubMed] [Google Scholar]
- 11.Tominaga K, Hayashi J, Kagawa Y, Ohta S. Smaller isoform of human mitochondrial transcription factor 1: its wide distribution and production by alternative splicing. Biochem. Biophys. Res. Commun. 1993;194:544–551. doi: 10.1006/bbrc.1993.1854. [DOI] [PubMed] [Google Scholar]
- 12.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 13.Wong TS, Rajagopalan S, Townsley FM, Freund SM, Petrovich M, Loakes D, Fersht AR. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res. 2009;37:568–581. doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 15.Jung YS, Zweckstetter M. Mars – robust automatic backbone assignment of proteins. J. Biomol. NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 16.Anglister J, Grzesiek S, Ren H, Klee CB, Bax A. Isotope-edited multidimensional NMR of calcineurin B in the presence of the non-deuterated detergent CHAPS. J. Biomol. NMR. 1993;3:121–126. doi: 10.1007/BF00242480. [DOI] [PubMed] [Google Scholar]
- 17.Ginzinger SW, Coles M. SimShiftDB: local conformational restraints derived from chemical shift similarity searches on a large synthetic database. J. Biomol. NMR. 2009;43:179–185. doi: 10.1007/s10858-009-9301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginzinger SW, Graeupl T, Heun V. SimShiftDB: chemical-shift-based homology modeling. In: Hochreiter S, Wagner R, editors. Bioinformatics research and development. Vol. 4414. Springer Berlin: Heidelberg; 2007. pp. 357–370. [Google Scholar]
- 19.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 22.Sippl MJ, Wiederstein M. A note on difficult structure alignment problems. Bioinformatics. 2008;24:426–427. doi: 10.1093/bioinformatics/btm622. [DOI] [PubMed] [Google Scholar]
- 23.Gangelhoff TA, Mungalachetty PS, Nix JC, Churchill ME. Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A. Nucleic Acids Res. 2009;37:3153–3164. doi: 10.1093/nar/gkp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastukh V, Shokolenko I, Wang B, Wilson G, Alexeyev M. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007;274:6488–6499. doi: 10.1111/j.1742-4658.2007.06167.x. [DOI] [PubMed] [Google Scholar]
- 25.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 28.Fersht AR. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. New York: W. H. Freeman and Company; 1998. pp. 511–513. [Google Scholar]
- 29.Ikeda S, Sumiyoshi H, Oda T. DNA binding properties of recombinant human mitochondrial transcription factor 1. Cell Mol. Biol. (Noisy-le-grand) 1994;40:489–493. [PubMed] [Google Scholar]
- 30.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 32.Lipps G, Stegert M, Krauss G. Thermostable and site-specific DNA binding of the gene product ORF56 from the Sulfolobus islandicus plasmid pRN1, a putative archael plasmid copy control protein. Nucleic Acids Res. 2001;29:904–913. doi: 10.1093/nar/29.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lnenicek-Allen M, Read CM, Crane-Robinson C. The DNA bend angle and binding affinity of an HMG box increased by the presence of short terminal arms. Nucleic Acids Res. 1996;24:1047–1051. doi: 10.1093/nar/24.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stros M. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry. 2001;40:4769–4779. doi: 10.1021/bi002741i. [DOI] [PubMed] [Google Scholar]
- 35.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 36.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohgaki K, Kanki T, Fukuoh A, Kurisaki H, Aoki Y, Ikeuchi M, Kim SH, Hamasaki N, Kang D. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J. Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- 39.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl Acad. Sci. USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajagopalan S, Andreeva A, Teufel DP, Freund SM, Fersht AR. Interaction between the transactivation domain of p53 and PC4 exemplifies acidic activation domains as single-stranded DNA mimics. J. Biol. Chem. 2009;284:21728–21737. doi: 10.1074/jbc.M109.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang J, Kim DH, Lee SW, Choi KY, Sung YC. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J. Biol. Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 43.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl Acad. Sci. USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Mapping the interactions of the p53 transactivation domain with the KIX domain of CBP (dagger) Biochemistry. 2009;48:2115–2124. doi: 10.1021/bi802055v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 46.van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100family to p53 monomers and tetramers. J. Biol. Chem. 2009;284:13804–13811. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc. Natl Acad. Sci. USA. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietrowska M, Kolodziejczyk I, Widlak P. Mitochondrial transcription factor A is the major protein in rodent hepatocytes that recognizes DNA lesions induced by N-acetoxy-acetylaminofluorene. Acta. Biochim. Pol. 2006;53:777–782. [PubMed] [Google Scholar]
- 49.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, et al. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem. Biophys. Res. Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 50.Aboul-ela F, Murchie AI, Homans SW, Lilley DM. Nuclear magnetic resonance study of a deoxyoligonucleotide duplex containing a three base bulge. J. Mol. Biol. 1993;229:173–188. doi: 10.1006/jmbi.1993.1016. [DOI] [PubMed] [Google Scholar]
- 51.Dornberger U, Hillisch A, Gollmick FA, Fritzsche H, Diekmann S. Solution structure of a five-adenine bulge loop within a DNA duplex. Biochemistry. 1999;38:12860–12868. doi: 10.1021/bi9906874. [DOI] [PubMed] [Google Scholar]
- 52.Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim. Biophys. Acta. 2009;1787:296–302. doi: 10.1016/j.bbabio.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 53.Stott K, Tang GS, Lee KB, Thomas JO. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 2006;360:90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 54.Love JJ, Li X, Chung J, Dyson HJ, Wright PE. The LEF-1 high-mobility group domain undergoes a disorder-to-order transition upon formation of a complex with cognate DNA. Biochemistry. 2004;43:8725–8734. doi: 10.1021/bi049591m. [DOI] [PubMed] [Google Scholar]
- 55.Romer L, Klein C, Dehner A, Kessler H, Buchner J. p53–a natural cancer killer: structural insights and therapeutic concepts. Angew. Chem. Int. Ed. Engl. 2006;45:6440–6460. doi: 10.1002/anie.200600611. [DOI] [PubMed] [Google Scholar]
- 56.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 57.Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612–618. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc. Natl Acad. Sci. USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 Protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 60.Bakhanashvili M, Grinberg S, Bonda E, Simon AJ, Moshitch-Moshkovitz S, Rahav G. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15:1865–1874. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 61.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim. Biophys. Acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nemajerova A, Erster S, Moll UM. The post-translational phosphorylation and acetylation modification profile is not the determining factor in targeting endogenous stress-induced p53 to mitochondria. Cell Death Differ. 2005;12:197–200. doi: 10.1038/sj.cdd.4401526. [DOI] [PubMed] [Google Scholar]
- 65.Teufel DP, Bycroft M, Fersht AR. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene. 2009;28:2112–2118. doi: 10.1038/onc.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cullen KJ, Yang Z, Schumaker L, Guo Z. Mitochondria as a critical target of the chemotheraputic agent cisplatin in head and neck cancer. J. Bioenerg. Biomembr. 2007;39:43–50. doi: 10.1007/s10863-006-9059-5. [DOI] [PubMed] [Google Scholar]
- 67.Regev-Rudzki N, Pines O. Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. Bioessays. 2007;29:772–782. doi: 10.1002/bies.20609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.