Abstract
Knowledge of the genetic variability, population structure, and evolutionary history of Triatoma infestans may be useful for developing rational vector control strategies. A 661-bp fragment of the mitochondrial gene cytochrome oxidase I (COI) was sequenced and analyzed in bugs from Argentina, Uruguay, Peru, and Bolivia, including peridomestic, domestic, Andean, and Chaco sylvatic bugs. A total of 48 polymorphic sites among 37 haplotypes were described. Nucleotide variation fluctuated among samples, with the highest nucleotide diversity observed in seven Argentinean provinces. Within this group, some populations showed patterns of variability compatible with population expansions and/or fine-scale population structure, whereas others suggested population bottlenecks and/or population admixture processes. A maximum parsimony analysis of the haplotypes showed the presence of a Bolivian/Peruvian and an Argentinean/Uruguayan clade. Bolivian sequences were further divided in Chaco sylvatic and Andean domestic and sylvatic. Two different nested clades were found within the Argentinean/Uruguayan cluster. Analysis of molecular variance (AMOVA) and KST* analysis supported a strong population structure in Argentina, where genetic differentiation was correlated with geographic distance. Departures from neutrality expectations and a nested cladistic analysis suggest a recent population expansion of T. infestans in Argentina, followed by restricted gene flow and patterns of isolation by distance. This expansion could have taken place as a two-wave process, as was shown by the phylogenetic analysis and signatures of population admixture in the southernmost Argentinean populations.
Keywords: Triatoma infestans, cytochrome oxidase I, population structure, population expansion, population admixture
Triatoma infestans is the most widespread domestic vector of Trypanosoma cruzi, the causative agent of Chagas disease, in South America, with a broad distribution range including Argentina, Bolivia, Brazil, Chile, Paraguay, Peru, and Uruguay (Schofield 1988). Knowledge of the evolutionary history, population structure, and patterns of dispersion of T. infestans lags far behind data on its ecology and physiology. Because these data may be relevant for developing and implementing vector control strategies, intraspecific studies in T. infestans populations have been considered a high priority (Dujardin 1996). For instance, T. infestans populations reproduce relatively slowly and were believed to have low levels of genetic variability from which insecticide resistance alleles would be unlikely to appear (Schofield 1988, Guhl and Schofield 1996, Monteiro et al. 2001). However, the recent detection of T. infestans populations resistant to pyrethroid insecticides in Argentina and Bolivia (Picollo et al. 2005, Toloza et al. 2008) suggests that the actual levels of genetic variability of the species should be reassessed. Another important issue is to distinguish between sylvatic and domestic T. infestans bugs to understand the putative links between both populations and their role in house reinfestation after insecticide spraying (Monteiro et al. 2001, Noireau et al. 2005).
Genetic studies based on isoenzymatic loci provided valuable information on genetic flow and population structure in T. infestans (Dujardin et al. 1987, 1998; García et al. 1995). Bolivian and Peruvian T. infestans populations seem to follow the isolation by distance model, with each village representing the smallest panmictic unit (Dujardin et al. 1998). In some cases, however, the deme was represented by single houses, chicken coops, or goat or pig corrals (Breniére et al. 1998).
More recently, DNA sequencing has proven to be a useful tool for studying the systematics and evolutionary trends within Triatominae (reviewed in Abad-Franch and Monteiro 2005). Monteiro et al. (1999) were the first to analyze nucleotide variation in T. infestans with the mitochondrial gene cyt b. Despite using only one to six bugs per population, they detected two main clusters: one with haplotypes from Bolivia and another with sequences from Argentina and Brazil. The Bolivian cluster was further subdivided into two clusters: one including the three Andean populations analyzed and the other the sylvatic “dark morph” insects from the Bolivian Chaco (Noireau et al. 1997). Two haplotypes were detected in Argentina, and one of them was also present in Brazil. This division into two allopatric groups (one Andean, including Bolivia and Peru and another non-Andean including the remainder) was supported by another study based on cyt b variability in the Bolivian Department of Chuquisaca (Giordano et al. 2005), by patterns of C-banding in chromosomes and DNA content (Panzera et al. 2004) and by nuclear rDNA (Bargues et al. 2006). In two other population genetic studies, mitochondrial 12S and 16S rRNA genes showed a low degree of differentiation between populations from four Argentinean provinces (García et al. 2003), whereas microsatellite loci showed a substantial degree of genetic differentiation caused by genetic drift and limited gene flow in 19 Argentinean localities from nine provinces (Pérez de Rosas et al. 2007). Microsatellites also showed substantial differentiation and strong population structure in Bolivian populations (Pizarro et al. 2008).
As part of a broader project on the eco-epidemiology of Chagas disease in northern Argentina, the main aims of this study were (1) to estimate levels of nucleotide variation, haplotype diversity, and population structure in populations of T. infestans; (2) to search for signs of demographic processes such as population expansions and bottlenecks; and (3) to analyze the genealogic and phylogeographical relationships between haplotypes from Argentina, Bolivia, Peru, and Uruguay.
Materials and Methods
DNA Sequencing
A total of 244 insects was used for DNA sequencing, including 207 from Argentina (domestic and peridomestic), 25 from Bolivia (14 domestic, 10 Andean sylvatic and 1 Chaco dark morph), 7 from Peru (domestic), and 5 from Uruguay (domestic) (Table 1). Argentinean bugs were grouped according to the nine provinces where they were collected.
Table 1.
Geographical localization of the T. infestans samples
| Country | Province | Department | Locality | Habitat | Year | Latitude (°S) | Longitude (°W) | N |
|---|---|---|---|---|---|---|---|---|
| Argentina | Salta | San Martín | El Chorro | U (1) | 2003 | 22.13 | 63.91 | 2 |
| Rivadavia | El Mistel | U (1) | 2000 | 23.91 | 62.54 | 1 | ||
| La Bolsa | U (2) | 2000 | 23.91 | 62.54 | 2 | |||
| El Hogar Viejo | U (1) | 2000 | 23.91 | 62.54 | 1 | |||
| Unknown | U (U) | 2000 | 23.91 | 62.54 | 9 | |||
| Formosa | Patińo | El Ceibal | P (5) D (3) | 2002 | 24.53 | 59.65 | 12 | |
| San Martín 1 | P (1) U (3) | 2002 | 24.59 | 59.87 | 5 | |||
| Sgo del Estero | Moreno | Quilumpa | P (8) | 2003 | 26.95 | 62.92 | 15 | |
| Amamá | P (14) | 2001/2002 | 27.21 | 63.04 | 22 | |||
| Pampa Pozo | P (1) | 2002 | 27.27 | 63.03 | 1 | |||
| Mercedes | P (1) | 2002 | 27.25 | 62.97 | 1 | |||
| Villa Isabel | P (1) | 2002 | 27.07 | 63.07 | 1 | |||
| Figueroa | Km 40 | P (1) | 2004 | 27.45 | 63.52 | 2 | ||
| La Loma | P (1) | 2004 | 27.47 | 63.45 | 1 | |||
| Invernada Norte | P (2) | 2004 | 27.37 | 63.48 | 9 | |||
| Tucumán | Tafí del Valle | Anjuana | U (U) | 2005 | 26.44 | 66.00 | 9 | |
| Quilmes de | U (U) | 2005 | 26.44 | 66.00 | 5 | |||
| Abajo | ||||||||
| El Paso | U (U) | 2005 | 26.56 | 65.97 | 5 | |||
| Catamarca | Tinogasta | Bo. Estación | D (1) | 2005 | 28.42 | 65.73 | 7 | |
| Norte | ||||||||
| Zona Centro | P (1) | 2005 | 28.42 | 65.73 | 5 | |||
| Capital | Capital | P (1) | 2005 | 28.48 | 65.78 | 3 | ||
| Santa Fé | Vera | Caraguay | P (2) | 2005 | 29.40 | 60.13 | 3 | |
| 9 de Julio | Las Gamas | P (1) | 2005 | 29.41 | 60.38 | 2 | ||
| Fortín Seis | P (1) | 2005 | 28.35 | 61.21 | 2 | |||
| Santa Margarita | P (3) | 2005 | 28.32 | 61.55 | 4 | |||
| Las Palmas | P (4) | 2005 | 29.35 | 60.20 | 5 | |||
| Córdoba | Cruz del Eje | Río Seco | P (2) | 2006 | 29.90 | 63.72 | 14 | |
| La Rioja | Belgrano | Baldes de | P (7) | 2000 | 30.63 | 66.05 | 7 | |
| Pacheco | ||||||||
| Tala Verde | P (4) | 2000 | 30.50 | 66.13 | 4 | |||
| Saladillo | P (1) | 2000 | 30.57 | 66.27 | 1 | |||
| La Argentina | P (1) | 1999 | 30.48 | 66.10 | 2 | |||
| La Selva | P (1) | 2000 | 30.48 | 66.18 | 1 | |||
| Loma Alta | P (1) | 1999 | 30.53 | 66.10 | 1 | |||
| Los Chańaditos | P (1) | 2000 | 30.53 | 66.95 | 1 | |||
| Bajo Hondo | P (1) | 2000 | 30.68 | 66.17 | 1 | |||
| Chamical | Los Bordos | P (4) | 2000 | 30.37 | 66.32 | 4 | ||
| San Luis | Gral | Santa Rosa | P (3) | 2003 | 32.32 | 66.93 | 8 | |
| Belgrano | ||||||||
| San Juan | Angaco | Unknown | D (1) | 2005 | 31.37 | 68.33 | 2 | |
| Caucete | La Planta | D (1) | 2005 | 31.47 | 67.40 | 4 | ||
| Marayes | ||||||||
| Pocito | Lote Hogar N° 39 | P (1) | 2005 | 31.68 | 68.58 | 3 | ||
| Villa San Martín | P (1) | 2005 | 31.68 | 68.58 | 1 | |||
| Rawson | Villa Novoa | P (1) | 2005 | 31.68 | 68.27 | 2 | ||
| Valle Fértil | Usno | P (1) | 2005 | 30.58 | 67.61 | 3 | ||
| Río Negro | Avellaneda | Choele Choel | P (3) D (1) | 2005 | 39.37 | 65.73 | 10 | |
| Lamarque | P (1) | 2005 | 39.40 | 65.73 | 5 | |||
| Perú | Arequipa | Sachaca | Unknown | D (2) | 2003 | 16.42 | 71.57 | 7 |
| Bolivia | Cochabamba | Campero | Mataral | D (5) P (3) | 2005 | 18.60 | 65.12 | 14 |
| Quillacollo | Cotapachi | S (5) | 2004 | 17.43 | 66.28 | 10 | ||
| Santa Cruz | Cordillera | Izozog | S (1) | 2005 | 19.40 | 62.75 | 1 | |
| Uruguay | Rivera | Seccional 4 | Zona 1 | P (1) | 2005 | 30.88 | 55.53 | 5 |
No. of domestic, peridomestic, or sylvatic structures sampled are shown in parentheses.
Year, collection year; N, no. of bugs sequenced; P, peridomestic; D, domestic; S, sylvatic; U, unknown.
Adult or nymph legs were used for DNA extraction with the Wizard Genomic DNA kit (Promega, Madison, WI), following the manufacturer’s protocol with slight modifications. A 692-bp fragment of the mitochondrial gene cytochrome oxidase I (COI) was polymerase chain reaction (PCR)-amplified with primers S1718 and A2442 (Normark 1996). This fragment was chosen because it was useful to resolve intrapopulation variability or phylogenetic relationships between closely related species in other insect groups (Lunt et al. 1996, Jordal et al. 2002, Scataglini et al. 2006).
Reactions were performed in a final volume of 50µl containing 8µl of dNTPs (1.25 mM each), 5 µl of 10×reaction buffer, 2.5 µl of MgCl2 (50 mM), 0.2 µl of TaqDNA polymerase (Invitrogen, Carlsbad, CA) 5 U/µl, 100 ng of sense and antisense primers, and 50–100 ng of DNA. Amplifications were carried out in a BIO-RAD thermal cycler (BIO-RAD, Hercules, CA) with 1 cycle of 1 min at 94°C, 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 46°C), and extension (1.5 min at 72°C), and a final extension step of 3.5 min at 72°C. The amplified samples were run on a 1% agarose gel and purified with Wizard SV gel and PCR Clean-Up System (Promega). The purified PCR product was used as a template for direct sequencing of both strands using the same primers as in the amplification. Sequences were all performed with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) in an ABI PRISM 3100 or 3700 DNA Analyzer automated sequencer (Applied Biosystems). The 661-bp obtained sequences were aligned manually or with MEGA 3.1 software (Kumar et al. 2004). Newly reported sequences are available at GenBank under accession numbers EF451005–EF451040.
Data Analyses
Nucleotide diversity was estimated according to Watterson’s estimator (θw) based on the number of segregating sites (Watterson 1975) and Tajima’s estimator (π) based on the average number of observed pairwise differences (Tajima 1983).
To study whether levels of variability were significantly different among all the populations, we used the approach taken by Schmid et al. (2005), based on likelihood-ratio tests (LRTs). The analysis was originally created for comparing loci, but as in this case, it can also be applied to populations. We tested whether the variability in the 14 populations of the dataset (11 from Argentina, 2 from Bolivia, and 1 from Uruguay) was better explained by models based on 1 (M1), 2 (M2), 3 (M3), 4 (M4), or 14 θw values (M14).
DT (Tajima 1989), FS (Fu 1997), and R2 (Ramos-Onsins and Rozas 2002) tests were applied to determine whether populations of T. infestans were in mutation-drift equilibrium, according to expectations of the neutral theory (Kimura 1983).
For the total Argentinian sample, we also estimated the levels of variability and applied the tests taking only one random sequence per sampled structure (house, corral, etc.) and one random sequence per locality, to consider the possible effects of population structure and sampling. The rationale underlying this procedure is that a simple coalescent process may take place under several models of population subdivision, such as metapopulations and general finite island models (Wakeley 2001, Wakeley and Aliacar 2001). According to these models, if only one sequence is sampled per deme, the behavior of a sample during most of the coalescent process is similar to a nonsubdivided population, but with an effective population size that depends on the number and sizes of demes and migration rates (Wakeley 2001, Wakeley and Aliacar 2001). In addition, Tajima’s DT does not depart from neutrality under a wide range of migration and extinction rates when sequences are taken from different demes (Pannell 2003). The significance of the tests was calculated by performing 1,000 coalescent simulations based on a Monte Carlo process with no recombination (Hudson 1990).
To confirm that departures from neutrality are caused by demography and not by natural selection, we also used the MK test (McDonald and Kreitman 1991). T. brasiliensis (GenBank accession number AF021184) and T. garciabesi (GenBank accession number EF451041) were used alternatively as out-groups.
Population structure within Argentina was inspected by means of analysis of molecular variance (AMOVA) (Excoffier et al. 1992) with two hierarchical levels (locality and province) and KST* statistics (Hudson et al. 1992). The significance of these tests was obtained by means of 10,000 permutations of haplotypes and corrected following Bonferroni criteria for multiple comparisons (Weir 1996). KST* values were used to construct a phenogram with the unweighted pair-group method with arithmetic average (UPGMA) and neighbor-joining (NJ) methods, and the fitting of the genetic data in the trees was calculated with the coefficient of determination R2. Genetic distances between populations were also correlated with geographic distance through a Mantel test (Sokal and Rohlf 1981).
Phylogenetic analyses were performed with a maximum parsimony approach. The shortest trees were found with the implicit enumeration search option. All characters were regarded as unordered and un-weighted because all parsimony informative nucleotide changes were synonymous transitions. Statistical support for clades was assessed by Bremer support values (Bremer 1994) and bootstrapping (Felsenstein 1985) based on 1,000 replicates. A T. delpontei (accession number FJ439768) was also sequenced and used as outgroup.
Because of the observation of low bootstrap values in the tree (probably reflecting the low divergence between conspecific individuals) and many zero length branches (most likely because of persistant ancestral haplotypes in the populations, Posada and Crandall 2001), a nested cladistic analysis (Templeton et al. 1995) was performed to better understand the phylogenetic history of the haplotypes combined with the information about the geographic origin of the bugs.
Intraspecific variability, KST* values, and all neutrality tests were calculated with DnaSP 4.1 (Rozas et al. 2003), whereas the LRTs were performed with MANVa 0.98BETA (Ramos-Onsins et al. 2008). AMOVA and Mantel tests were made with Arlequin 3.11 (Excoffier et al. 2005). The UPGMA and NJ trees were created with MEGA 3.1 (Kumar et al. 2004), and the representation of the distances in the trees was evaluated with Treefit (Kalinowski 2008). Maximum parsimony trees were built with TNT (Goloboff et al. 2008), and the nested cladistic analysis was carried out with ANeCPA 1.1 (Panchal 2007).
Results
Nucleotide Variation
The 661-bp COI fragment showed 48 variable sites among the T. infestans sampled (Table 2). All but three of the changes were synonymous. The exceptions were in positions 203, 239, and 527, which cause the changes valine-isoleucine, glutamic acid-lysine, and alanine-threonine, respectively. The first and third changes were conservative in the sense that they affected neither size nor hydrophobicity (Taylor 1986). The second change did not affect polarity but changed the amino acid size (small to large) and charge (acidic to basic).
Table 2.
Variable sites and haplotypes detected in T. infestans from Argentina (Arg), Bolivia (Bol), Peru (Per), and Uruguay (Uru)
| n | n | n | |||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 6 | |||||||
| 1 | 3 | 5 | 8 | 9 | 0 | 0 | 1 | 1 | 2 | 3 | 3 | 4 | 4 | 5 | 6 | 7 | 8 | 0 | 0 | 3 | 3 | 3 | 6 | 6 | 0 | 0 | 4 | 4 | 5 | 6 | 0 | 1 | 7 | 8 | 9 | 1 | 1 | 2 | 2 | 2 | 4 | 5 | 5 | 5 | 6 | 8 | 6 | ||
| 9 | 4 | 0 | 8 | 7 | 3 | 6 | 2 | 3 | 4 | 3 | 9 | 2 | 5 | 1 | 3 | 3 | 7 | 2 | 3 | 3 | 5 | 9 | 2 | 6 | 7 | 8 | 0 | 6 | 5 | 7 | 0 | 9 | 8 | 7 | 6 | 1 | 4 | 6 | 7 | 9 | 7 | 0 | 3 | 9 | 5 | 6 | 1 | ||
| Haplo a | A | C | T | G | A | A | C | T | C | T | C | A | G | T | T | C | T | A | C | G | A | G | G | T | C | T | T | T | A | T | C | T | T | A | C | T | G | C | G | A | A | A | A | A | T | T | C | A | Arg(SE) |
| Haplo c | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(LR, SE, SF, CA, FO, SA, TU, SJ), Uru |
| Haplo d | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SF, SE, CA), Uru |
| Haplo e | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(LR, SE, CO) |
| Haplo f | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | Arg(LR) |
| Haplo g | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | C | . | . | . | . | . | . | A | . | . | G | . | . | . | . | . | . | . | . | Arg(LR) |
| Haplo i | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(LR) |
| Haplo j | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(LR) |
| Haplo k | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | T | . | G | . | . | . | . | . | . | . | . | Arg(SE) |
| Haplo l | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | G | . | . | . | . | . | . | T | . | Arg(SE) |
| Haplo m | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SA) |
| Haplo n | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SA, FO) |
| Haplo o | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(CA) |
| Haplo p | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | T | . | . | . | . | . | . | . | Arg (SE) |
| Haplo q | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | C | . | . | . | Arg(LR, SF) |
| Haplo s | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SE, CA) |
| Haplo t | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SF, SE) |
| Haplo u | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(FO) |
| Haplo v | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(TU) |
| Haplo w | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SE) |
| Haplo z | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | . | . | . | . | . | . | . | Arg(RN) |
| Haplo af | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SF) |
| Haplo ag | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SF) |
| Haplo ah | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | Arg(SF) |
| Haplo ai | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | G | . | . | . | . | . | . | . | . | Arg(SF) |
| Haplo b | . | . | . | . | . | . | T | C | . | C | T | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | G | . | . | G | . | . | . | . | . | Arg (SE) |
| Haplo h | . | . | . | . | . | . | T | C | . | C | . | . | A | . | C | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | A | G | . | . | . | . | . | . | . | . | Arg(LR) |
| Haplo r | . | . | . | . | . | . | . | C | . | C | . | G | A | . | . | T | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | G | . | . | . | . | . | . | Arg (SE) |
| Haplo x | . | . | . | . | . | . | T | C | . | C | . | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | C | . | . | A | G | . | . | . | . | . | . | . | . | Arg(SJ, RN, SL) |
| Haplo y | . | . | . | . | . | . | . | C | . | C | . | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | A | G | . | . | . | . | . | . | . | . | Arg(SJ) |
| Haplo av | . | . | . | . | . | . | T | C | . | C | . | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | . | . | . | . | . | . | . | Arg(CO) |
| Haplo ay | . | . | C | . | . | . | T | C | . | C | . | . | A | . | . | T | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | G | . | . | . | . | Arg (SL) |
| Haplo ab | G | . | . | A | . | . | . | C | . | C | . | . | A | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | G | . | . | . | . | T | . | Bol (dom, syl) |
| Haplo ac | G | . | . | . | . | . | . | C | . | C | . | . | A | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | G | . | . | . | . | T | . | Bol (dom, syl) |
| Haplo ad | G | . | . | A | . | . | . | C | . | C | . | . | A | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | T | C | . | . | . | . | . | . | A | G | . | G | . | . | . | . | T | . | Bol (dom) |
| Haplo ae | G | . | . | A | . | . | . | C | . | C | . | . | A | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | A | G | . | G | . | . | . | . | T | . | Bol (dom, syl), Perú |
| Haplo aa | . | T | . | . | . | . | T | C | T | C | . | . | A | . | . | T | C | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | G | . | . | . | C | T | G | Bol (dm) |
Dot, identity with nucleotide in first haplotype; n, nonsynonymous change; SA, Salta; FO, Formosa; CA, Catamarca; TU, Tucumán; SE, Santiago del Estero; SF, Santa Fé; LR, La Rioja; CO, Córdoba; SJ, San Juan; SL, San Luis; RN, Río Negro; dom, domestic; syl, sylvatic; dm, dark morph.
Forty polymorphic sites were found in domestic or peridomestic bugs from Argentina (Table 2 and Table 3), three in domestic bugs from Bolivia, two in sylvatic nonmelanic Bolivian bugs, and one in the Uruguayan sample, whereas the Peruvian sample was monomorphic (Table 3).
Table 3.
Estimates of COI variability and results of neutrality tests in Argentinean, Bolivian, Peruvian, and Uruguayan T. infestans
| N | S | k | θw | π | h | Hd | DT | FS | R2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Arg | 207 | 40 | 3.864 | 0.0102 | 0.0059 | 32 | 0.812 | −1.248 | −10.732a | 0.048b |
| Arg (1 per structure) | 104 | 39 | 3.525 | 0.0113 | 0.0053 | 31 | 0.830 | −1.640a | −16.741c | 0.044a |
| Arg (1 per locality) | 39 | 19 | 3.714 | 0.0068 | 0.0056 | 15 | 0.780 | −0.573 | −3.784b | 0.092 |
| SE | 52 | 23 | 2.748 | 0.0077 | 0.0042 | 12 | 0.805 | −1.482a | −1.980 | 0.060b |
| LR | 22 | 16 | 2.563 | 0.0067 | 0.0039 | 8 | 0.771 | −1.512a | −1.121 | 0.115 |
| SJ | 15 | 9 | 4.400 | 0.0042 | 0.0067 | 3 | 0.676 | 2.211a | 5.847 | 0.244 |
| SF | 15 | 9 | 2.152 | 0.0042 | 0.0033 | 8 | 0.838 | −0.834 | −2.811 | 0.103 |
| RN | 15 | 8 | 3.352 | 0.0037 | 0.0051 | 2 | 0.419 | 1.335 | 7.074 | 0.210 |
| SL | 8 | 6 | 2.571 | 0.0035 | 0.0039 | 2 | 0.429 | 0.518 | 4.286 | 0.214 |
| CO | 14 | 7 | 2.201 | 0.0033 | 0.0056 | 2 | 0.527 | 2.500c | 7.349 | 0.264 |
| CA | 15 | 3 | 1.067 | 0.0014 | 0.0016 | 4 | 0.533 | 0.464 | 1.069 | 0.178 |
| FO | 17 | 3 | 0.353 | 0.0013 | 0.0005 | 3 | 0.228 | −1.706b | −0.963 | 0.171 |
| SA | 15 | 2 | 0.686 | 0.0009 | 0.0014 | 3 | 0.600 | 0.302 | 0.160 | 0.171 |
| TU | 19 | 2 | 0.526 | 0.0004 | 0.0008 | 2 | 0.526 | 1.547 | 1.452 | 0.263 |
| Bol (dom) | 14 | 3 | 1.242 | 0.0014 | 0.0019 | 4 | 0.780 | 0.968 | 0.057 | 0.168 |
| Bol (syl) | 10 | 2 | 0.667 | 0.0011 | 0.0010 | 3 | 0.511 | −0.184 | −0.272 | 0.190 |
| Uru | 5 | 1 | 0.400 | 0.0007 | 0.0006 | 2 | 0.400 | −0.817 | 0.090 | 0.400 |
| Per | 7 | 0 | 0.000 | 0.0000 | 0.0000 | 1 | 0.000 | — | — | — |
P < 0.05
0.09 < P; and
P < 0.01.
N, no. of sequences; S, no. of segregating sites; k, mean no. of pairwise differences per sequence; θw, Watterson’s estimator; π, Tajima’s estimator; h, no. of haplotypes; Hd, haplotype diversity; DT, Tajima’s test; FS, Fu’s test; R2, Ramos-Onsins and Rozas’ test.
In Argentina, the observed total nucleotide variability was of 0.010 and 0.006 according to θw and π, respectively (Table 3). Values were rather similar when random sequences were taken from each locality or structure, suggesting that the estimation of total variability in Argentina was not severely affected by the sampling.
Before comparing variability among Argentinean Provinces (Table 3; Fig. 1), and given the different number of localities clustered within each province (Table 1), we performed an AMOVA to look for structuring in our data. The analysis showed that genetic differentiation among bugs from different provinces was highly significant (σa2 = 1.115, ΦCT = 0.458, df = 10, P < 0.0001) and explained almost 46% of the total variation among sequences, whereas differentiation between localities within provinces was low and non-significant (σb 2 = 0.084, ΦSC = 0.064, df = 28, P = 0.359).
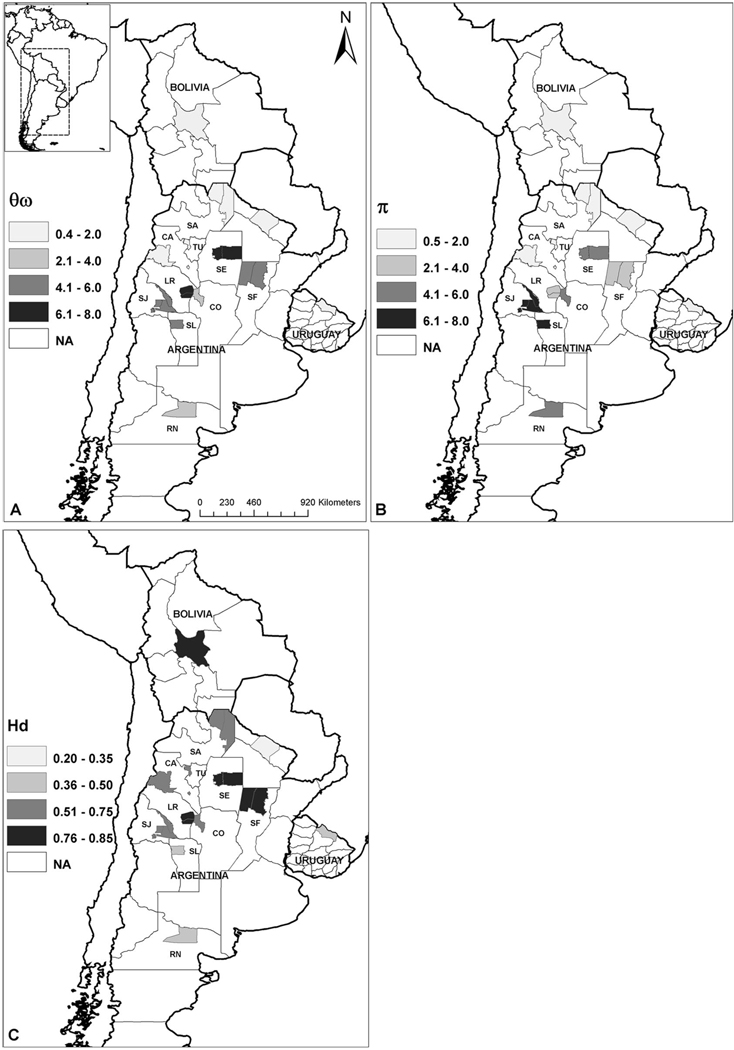
Fig. 1.
Geographical distribution of COI variability in Argentinean, Uruguayan, and Bolivian domestic and peridomestic T. infestans populations. (A) θw, Watterson’s estimator multiplied by 10−3. (B) π, Tajima’s estimator multiplied by 10−3. (C) Hd, haplotype diversity. Shaded areas in the map are the departments where the bugs were collected. Colors correspond to the four categories in which levels of variability were classified. NA, not available.
The LRTs showed that a model with two different θw values better explained the differences in COI variability among populations (Table 4). La Rioja, San Juan, Santa Fé, Río Negro, Santiago del Estero, San Luis, and Córdoba had higher genetic variability (θw 1 = 0.0052), whereas Catamarca, Salta, Tucumán, Formosa, Bolivian domestic, Bolivian sylvatic, and Uruguay were the least variable populations (θw 2 = 0.0012).
Table 4.
Maximum-likelihood estimates of θw per nucleotide and likelihood ratio tests
| Model | θw1 (no. pop) | θw2 (no. pop) | θw3 (no. pop) | θw4 (no. pop) | ML | LRT | P |
|---|---|---|---|---|---|---|---|
| M1 | 0.0031 (14) | −40.7985 | |||||
| M2a | 0.0012 (7) | 0.0052 (7) | −30.015 | 21.567 | 0.009 (M1 versus M2) | ||
| M3b | 0.0012 (7) | 0.0039 (5) | 0.0076 (2) | −28.355 | 3.320 | 0.810 (M2 versus M3) | |
| M4c | 0.0009 (4) | 0.0014 (3) | 0.0039 (5) | 0.0076 (2) | −28.0884 | 0.533 | 0.991 (M3 versus M4) |
| M14 | −27.9271 | 0.323 | 1.000 (M4 versus M14) |
θw1: CA, FO, SA, TU, Bol dom, Bol syl, Uru; θw2: SE, LR, SJ, SF, RN, SL, CO.
θw1: CA, FO, SA, TU, Bol dom, Bol syl, Uru; θw2: SJ, SF, RN, SL, CO; θw3: SE, LR.
θw1: TU, Bol dom, Bol syl, Uru; θw2: CA, FO, SA, SJ; θw3: SJ, SF, RN, SL, CO; θw4: SE, LR.
Haplotype Diversity
A total of 37 haplotypes were detected, 32 of them in Argentina, 2 in Uruguay, 1 in Peru, and 5 in Bolivian sylvatic and domestic bugs. Haplotype diversity ranged between 0.84 and 0.23 (Table 3; Fig. 1). Most Argentinian haplotypes were differentiated by only two or three nucleotide changes (Table 2), but there were also seven highly divergent haplotypes separated by five to eight mutational steps (haplotypes b, h, r, x, y, av, and ay). These distinct haplotypes were located in different provinces including La Rioja, Santiago del Estero, San Juan, Coórdoba, San Luis, and Río Negro (Table 2). The most frequent haplotype c was found in all Argentinian populations except Córdoba, San Luis, and Río Negro. Haplotypes d, e, n, q, s, t, and × were shared between two or more populations, whereas the remaining variants were observed exclusively in one population (Table 2). The highest number of haplotypes was detected in Santiago del Estero (12) followed by La Rioja and Santa Fé (8), Catamarca (4), Salta, Formosa and San Juan (3), and Tucumán, Córdoba, San Luis, and Río Negro (2). Insects from Uruguay and Argentina shared two haplotypes, including the most frequent haplotype c.
Bolivian insects showed five haplotypes not found in Argentina or Uruguay (Table 2). One was present in the domestic sample (ad), three were found in both the domestic and the sylvatic Andean samples (ab, ac, and ae), and haplotype aa was found in the darkmorph from the Bolivian Chaco (Table 2 and Table 3). The haplotype ae was also the only one present in the Peruvian sample.
Population Structure of Argentinean T. infestans
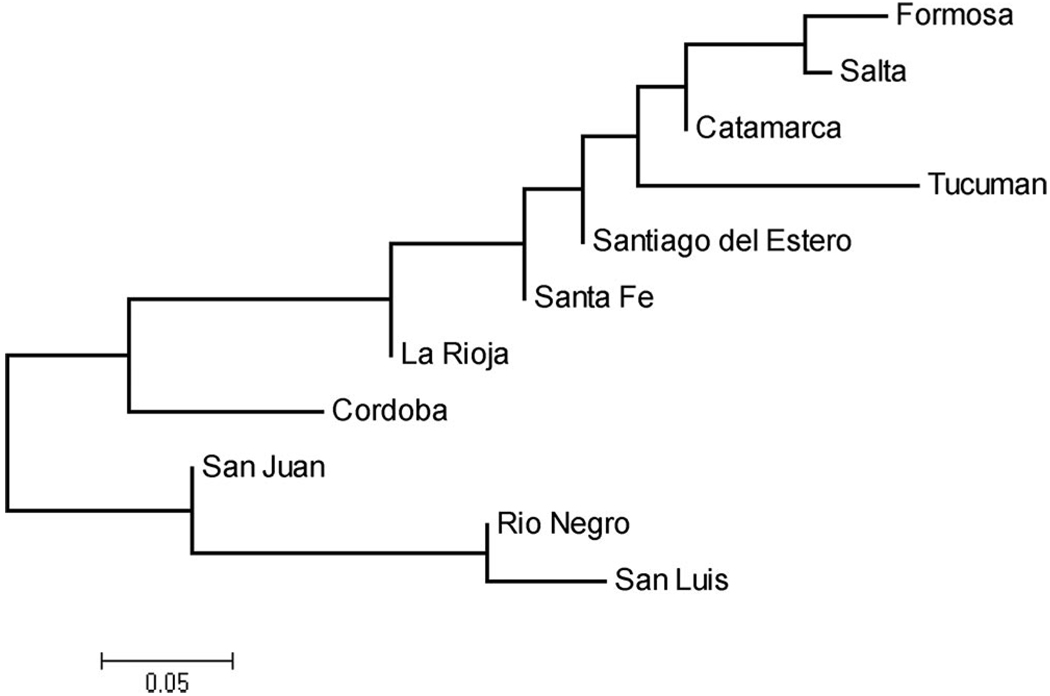
Pairwise comparison between Argentinian provinces gave highly significantKST* values, and several of them remained as such even after correcting for multiple comparisons. Differentiation was extremely variable, ranging from values close to zero (0.006, 0.008) to 0.683 (Table 5). When the KST* values were represented in a phenogram, the NJ showed a better representation of the distances (R2 = 0.93, P < 0.0001) than the UPGMA tree (R2 = 0.58, P < 0.0001); thus, the first methodology was chosen (Fig. 2). The tree showed that genetic differentiation had a good correspondence with geographical distance (Fig. 1 and Fig. 2). For example, the northernmost populations of Salta and Formosa were the more similar, and genetic distance from other populations increased continuously toward the south. This pattern was corroborated by a Mantel test, which showed a significant correlation betweenKST* values and geographic distance (r = 0.578, P = 0.0002) even when the geographically distant and highly differentiated population of Río Negro was removed from the matrices (r = 0.476, P = 0.0091).
Table 5.
Pairwise KSTa (below the diagonal) and P values (above the diagonal) in Argentinian populations of T. infestans
| SA | FO | TU | CA | SE | SF | LR | CO | SJ | SL | RN | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SA | 0.064 | <0.0001 | 0.01 | 0.001 | <0.0001 | 0.003 | <0.0001 | 0.002 | <0.0001 | <0.0001 | |
| FO | 0.04 | 0.003 | 0.03 | 0.002 | <0.0001 | 0.002 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| TU | 0.181a | 0.221 | 0.008 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| CA | 0.066 | 0.061 | 0.113 | 0.035 | 0.09 | 0.022 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| SE | 0.054 | 0.06 | 0.084a | 0.022 | 0.177 | 0.007 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| SF | 0.086a | 0.108a | 0.144a | 0.031 | 0.006 | 0.197 | <0.0001 | 0.001 | <0.0001 | <0.0001 | |
| LR | 0.087 | 0.104 | 0.155a | 0.049 | 0.025 | 0.008 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| CO | 0.362a | 0.431a | 0.397a | 0.297a | 0.119a | 0.179a | 0.134a | 0.004 | <0.0001 | <0.0001 | |
| SJ | 0.307 | 0.366a | 0.358a | 0.273a | 0.146a | 0.206a | 0.200a | 0.172 | 0.085 | 0.064 | |
| SL | 0.589a | 0.683a | 0.619a | 0.535a | 0.218a | 0.404a | 0.354a | 0.292a | 0.065 | 0.218 | |
| RN | 0.518a | 0.590a | 0.556a | 0.473a | 0.254a | 0.367a | 0.340a | 0.245a | 0.054 | 0.037 |
Significant after Bonferroni’s correction (P < 0.0009).
Fig. 2.
Neighbor-joining tree based on KST* values between Argentinean populations of T. infestans. The tree was arbitrarily rooted in the longest branch. The scale bar shows the length that equals a 0.05 distance value.
Neutrality Tests
Tajima’s DT was negative and significant for the total Argentinian sample (complete and when one sequence was taken per structure), La Rioja, and Santiago del Estero, whereas it was positive and significant for San Juan and Córdoba. FS was significant for the total Argentinian sample and when one sequence was randomly chosen per structure and marginally significant when one sequence was sampled from each locality. R2 was marginally significant for the total sample and Santiago del Estero and significant when one sequence was taken per structure (Table 3). The remaining Argentinian samples and the Bolivian domestic and sylvatic populations did not depart from neutral expectations.
The MK test was not applied to the Bolivian samples because of the lack of nonsynonymous polymorphism, and it was not significant for Argentinian bugs either when T. brasiliensis (total sample: G-value = 1.860, P = 0.173; one sequence per structure: G-value: 2.233, P = 0.135) or T. garciabesi (total sample: G-value = 0.271, P = 0.603; one sequence per structure: G-value = 0.459, P = 0.498) was used as the outgroup. The closer related species T. delpontei could not be used for this test because of the lack of nonsynymous fixed differences with T. infestans in the sequenced gene region.
Phylogeny of Haplotypes
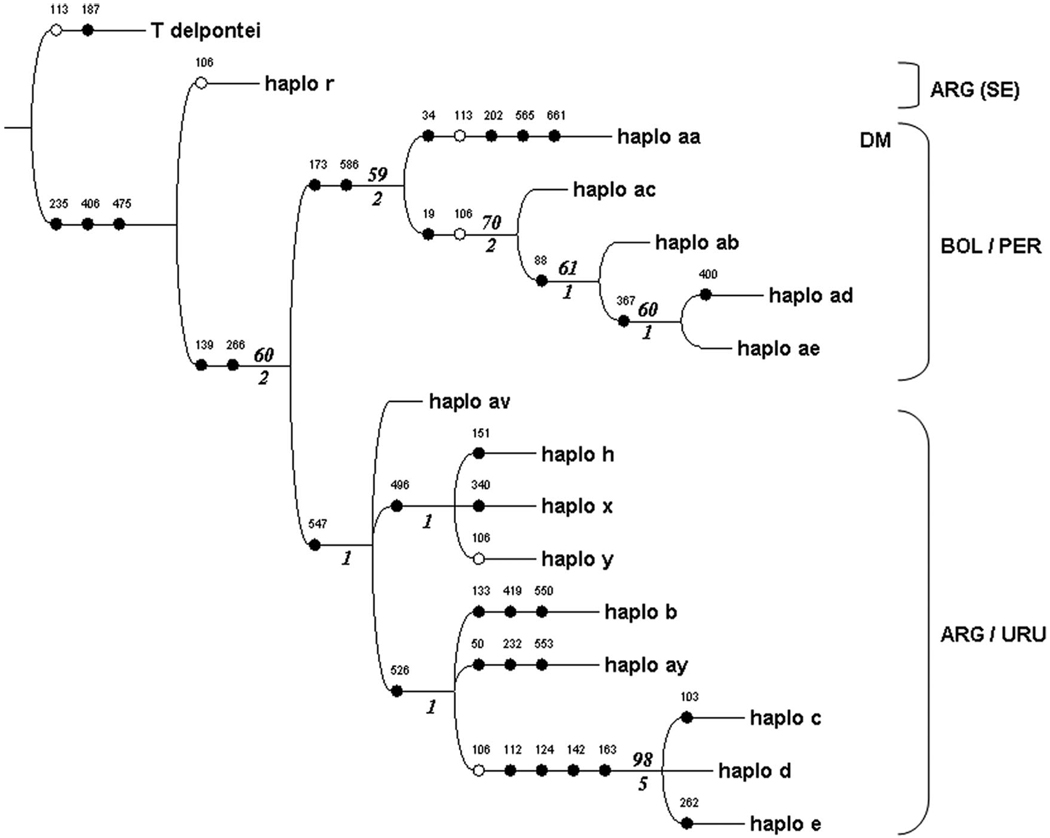
We obtained >1,000 equally parsimonious trees using a parsimony approach. The differences in the trees were mainly caused by minor changes in the relationships among haplotypes within the Argentinian clade, where most haplotypes were related alternatively to c, d, or e haplotypes (data not shown). Thus, data were reanalyzed with a subset of the Argentinean haplotypes, including only the seven highly divergent haplotypes and the c, d, and e haplotypes, which were the most frequent and shared by two or more populations. Only one most parsimonious tree of length 39 was found (Fig. 3). The most basal lineage was the Argentinean haplotype r and then two main clades appeared. One contained all the sequences from Bolivia and Peru and was further subdivided into two clusters: one including domestic and sylvatic Andean haplotypes and the other the sylvatic dark morph from the Bolivian Chaco.
Fig. 3.
Most parsimonious tree of COI haplotypes. Filled and empty circles are nonhomplasious and homoplasious changes, respectively. Numbers above circles are nucleotide positions. Bold italic numbers above and below the branches are 50% or higher bootstrap values and Bremer support values. ARG, Argentina; URU, Uruguay; BOL, Bolivia; PER, Perú; SE, Santiago del Estero; DM, “dark morph.”
The second main clade included all the Argentinean and Uruguayan haplotypes (Fig. 3), but within Argentina, the tree was not well resolved, with several branches of zero length and poor support. However, it showed the existence of two clades, one with haplotypes h, x, and y and the other with haplotypes b, ay, c, d, and e. Within this last group, haplotypes c, d, and e formed a well-supported cluster.
Nested Cladistic Analysis
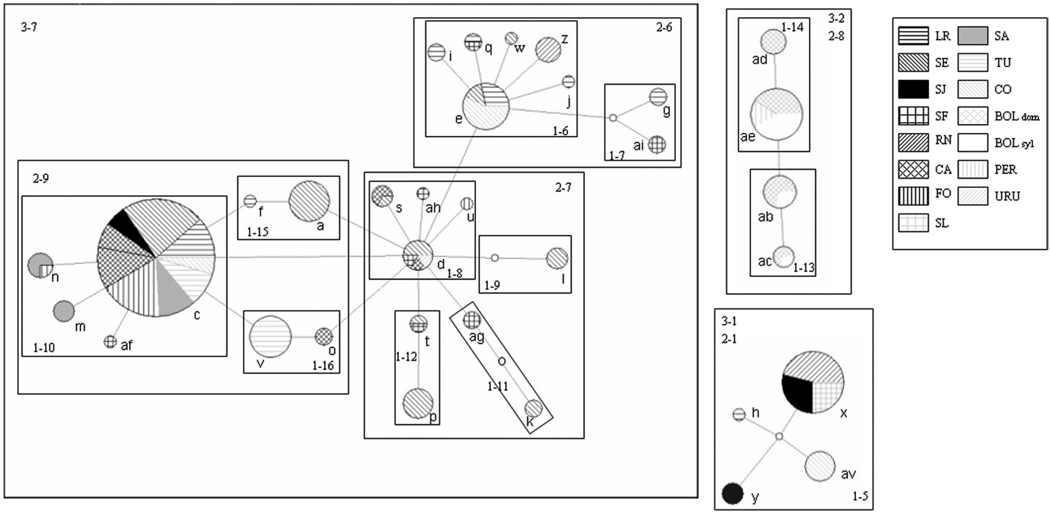
Four haplotypes (aa, b, r, and ay) were excluded from the analysis because their connections steps were beyond the 95% confidence interval. The remaining variants depicted a similar pattern as shown by the phylogenetic analysis, showing the existence of three discrete lineages (represented by three unlinked networks): one with Bolivian and Peruvian haplotypes, another with four divergent Argentinian haplotypes, and the third with Argentinean and Uruguayan haplotypes (Fig. 4). The presence of two loops in the main Argentinean/Uruguayan clade was analyzed with all the possible resolutions, and all of them reached the same outcome.
Fig. 4.
Nested cladistic haplotype network for T. infestans COI. Colored circles are recorded haplotypes and circle sizes are proportional to haplotypes frequencies. Each line in the network is a single mutational change; open circles are inferred intermediate haplotypes not sampled in the study. Boxes enclose haplotypes at each nested level.
Seven nested groups in the main Argentinean network, two in the Bolivian network and the total cladogram, showed geographic distributions that were significantly different from random expectations (Table 6). In Argentina, three clades suggested isolation by distance, one allopatric fragmentation, two were inconclusive, and the whole main network was compatible with a contiguous range expansion. In Bolivia, a clear phylogeographic pattern was not found.
Table 6.
Demographic inferences from nested geographical distance analyses (after Templeton 2005)
| Clade | Chain of inference | Population inference |
|---|---|---|
| 1–5 | 1–2 IO | I-T status undetermined: inconclusive outcome |
| 1–6 | 1–2-3–5-6–7-8 NO | Insufficient genetic resolution to discriminate between range expansion/colonization and restricted dispersal/gene flow |
| Sampling design inadequate to discriminate between isolation by distance (short distance movements) versus long distance dispersal | ||
| 1–10 | 1–2-3–4 NO | Restricted gene flow with isolation by distance |
| 1–12 | 1–19-20–2-3–4 NO | Restricted gene flow with isolation by distance |
| 1–13 | 1–2-3–5-6–7-8 NO | Insufficient genetic resolution to discriminate between range expansion/colonization and restricted dispersal/gene flow |
| Sampling design inadequate to discriminate between isolation by distance (short distance movements) versus long distance dispersal | ||
| 1–16 | 1–19 NO | Allopatric fragmentation |
| 2–7 | 1–2-3–4 NO | Restricted gene flow with isolation by distance |
| 2–8 | 1–2 IO | I-T status undetermined: inconclusive outcome |
| 2–9 | 1–2 IO | I-T status undetermined: inconclusive outcome |
| 3–7 | 1–2-11–12 NO | Contiguous range expansion |
| Total cladogram | 1–2 IO | I-T status undetermined: inconclusive outcome |
Discussion
COI Variability in T. infestans
Our study showed high levels of genetic variation of COI in T. infestans populations, with 48 variable sites distributed in 37 haplotypes. Within Argentina, nucleotide variation fluctuated greatly among provinces (Fig. 1), with a general pattern of populations either with higher (>0.0052) or with lower (<0.0012) variability.
Four of the most variable populations showed levels of polymorphism that departed from neutral expectations (Table 3). Two different patterns were found; Santiago del Estero and La Rioja populations had negative DT values because of a higher estimation of variability based on θw, whereas San Juan and Córdoba had positive DT values because of higher π values. This illustrates the different nature of the genetic variability and demographic history in T. infestans populations. In Santiago del Estero and La Rioja, the negative DT points out to an excess of low-frequency nucleotide changes, which in this case translates into the presence of many low-frequency haplotypes with small nucleotide changes between them. This pattern is in agreement with the presence of local population expansions and/or fine-scale population structure (Ptak and Przeworski 2002). In contrast, in Córdoba and San Juan, the positive DT values showed an excess of intermediate allele frequencies tht are compatible with a recent bottleneck or processes of population admixture (see below).
Levels of nucleotide variation remained high in samples from localities with recent and recurrent insecticide spraying, as recorded with microsatellite loci (Pérez de Rosas et al. 2007, Pizarro et al. 2008). For example, within the Santiago del Estero sample, the Amamá locality was subjected to community-wide deltamethrin sprays in 1985, 1992, and 2004, with multiple selective sprays between 1992 and 2004 (Gürtler et al. 2007), and the local bug population remained genetically highly variable (N = 25, θw = 0.0056, π = 0.0043, h = 8, Hd = 0.84). This finding was largely unexpected because severe population bottlenecks after repeated full-coverage insecticide spraying are expected to reduce the variability present in the population. Several hypotheses may help explain such a pattern: (1) Amamá bugs may belong to an ancient population of T. infestans and therefore may have accumulated a larger number of mutations than other populations; (2) the occurrence of bug sources outside the community could act as reservoirs of genetic variability during the reinfestation process; and (3) populations of T. infestans are subdivided within localities, and in such a case, a population bottleneck would result in several independent genetic drift effects that could randomly preserve different genetic combinations in each subpopulation. Recent studies in Amamá based on spatio-temporal patterns of reinfestation (Cecere et al. 2004), geometric morphometry (Schachter-Broide et al. 2004), and microsatellite markers (Marcet et al. 2008; Ceballos et al., unpublished data) support hypotheses 2 and 3 rather than hypothesis 1.
According to the LTR tests, Bolivian domestic bugs showed four haplotypes and high haplotype diversity, whereas nucleotide variation was comparable to those of the least variable Argentinian populations. Because the Cochabamba valley has long been proposed as the center of dispersal of T. infestans (Schofield 1988), Bolivian populations were expected to be the most ancient and the most genetically variable. However, there are at least two explanations for such a pattern. In the Cochabamba valley, indoor insecticide spraying against malaria vectors has been conducted regularly several times a year for at least two decades; therefore, this population could have undergone repeated bottlenecks. This result is partially supported by the positive, although nonsignificant, value of the Tajima’s statistic. Second, studies based on allozymes (Dujardin et al. 1998) and mitochondrial cyt b (Giordano et al. 2005) have pointed to the region encompassing Sucre, Vallegrande, and Potosi as the most probable area of origin of T. infestans, in contrast with the traditional hypothesis of Cochabamba. As in Argentina, the genetic diversity of T. infestans in different regions of Bolivia is probably strongly influenced by local geographic, ecological, and evolutionary histories (Giordano et al. 2005).
Sylvatic and domestic T. infestans bugs from Bolivia shared haplotypes, although haplotype diversity was higher in domestic than in sylvatic bugs. Previous studies using allozymes (Dujardin et al. 1987) and mitochondrial cyt b (Monteiro et al. 1999) were also unable to detect differences between sylvatic and domestic Bolivian populations of T. infestans, suggesting that the sylvatic foci could be derivatives from nearby peridomestic or domestic populations.
Phylogenetic Relationships Between Argentinean and Bolivian Haplotypes
The phylogeny of COI haplotypes validates the hypothesis of an Andean and a non-Andean T. infestans allopatric group, as shown by other nuclear and mitochondrial genes (Monteiro et al. 1999, Bargues et al. 2006). The maximum parsimony tree showed that the Bolivian/Peruvian haplotypes were clustered in one group, whereas almost all the Argentinean/Uruguayan haplotypes were clustered in another group.
However, the early divergence of haplotype r from Santiago del Estero, which was not grouped with any of the remaining variants in the tree is intriguing. One hypothesis to explain such a pattern is the presence of ancient haplotypes in the Argentinean Chaco, suggesting that the colonization of this area by T. infestans is older than suspected. The second hypothesis is that this mitocondrial sequence is the result of an old introgression event. Field observations and experimental studies have shown that T. infestans can form hybrids capable of backcrossing with the parental species with other species of the infestans complex such as T. platensis, T. delpontei, and T. rubrovaria (Abalos 1948, Pérez et al. 2005). The assumption that this bug is a hybrid itself can be dismissed, because it was included in a microgeographical-scale study based on 10 microsatellites (Marcet et al. 2008), and it showed the most common alleles of T. infestans.
The detection of sylvatic dark morphs in the Bolivian Chaco challenged the traditional view that Andean mesothermic valleys in Bolivia were the area of origin of T. infestans (Noireau et al. 2000, 2005). Carcavallo (1998) suggested that the dark morph populations from the dry subtropical Chaco forest may be the most ancient ones, whereas other authors proposed that this melanic bug could represent a direct expansion from the Andean population (Monteiro et al. 1999, Giordano et al. 2005) or the recolonization of sylvatic habitats by domestic bug populations from the Chaco (Noireau et al. 2000, Panzera et al. 2004, Bargues et al. 2006). The COI haplotype phylogeny is in agreement with the hypothesis of a direct Andean expansion and also shows several autapomorphies in the dark morph lineage, suggesting a considerable degree of isolation between melanic sylvatic bugs in the Chaco and the domestic and sylvatic Andean populations of T. infestans.
Population Structure in Argentina
Our data indicate the existence of a strong population structure in Argentinian T. infestans. AMOVA showed that differences among Provinces accounted for almost one half of the total observed variability. This observation contrasts with the rather low (6%) degree of differentiation shown by 12S and 16S mtDNA genes in populations from four Argentinian Provinces (García et al. 2003), even though these localities were rather close to the localities sampled in the same provinces in this study. Moreover, an AMOVA including only these four provinces also shows high variability between provinces when the localities sampled within provinces were take into account (σa2 = 0.547, ΦCT = 0.349, df = 3, P <0.05) or not (σa2 = 0.476, ΦST = 0.304, df = 3, P < 0.0001). Despite that sampling effects could not be completely ruled out, the contrasting results observed with both kinds of genes may be a consequence of the higher variability of COI, which makes it a more sensitive mitochondrial molecular marker for studying population structure.
The isolation by distance model was originally conceived by Wright (1943). He stated that distant populations may become differentiated when there is an extensive population range, but interbreeding is restricted by relatively short dispersal distances. This is precisely the pattern observed with KST* values in the neighbor-joining tree, also supported by the positive and significant correlation between geographic distance and COI variability. Our results are in agreement with previous studies based on microsatellites in Argentina (Pérez de Rosas et al. 2007) and on allozymes in Bolivia and Peru (Dujardin et al. 1997). Passive transportation associated with human activities has been proposed as the main dispersal mechanism in T. infestans (Schofield 1988, Bargues et al. 2006), and the isolation-by-distance pattern is readily explained by frequent movement of people and exchange of goods mainly restricted to close localities. However, active dispersal by flight has been estimated in a range of 0.2–2 km (Schweigmann et al. 1988, Schofield et al. 1992, Vazquez-Prokopec et al. 2004), values much smaller than the minimal distance between the study populations (≈100 km).
Demographic History of T. infestans in Argentina
Neutrality tests showed that Argentinian populations of T. infestans are not in mutation-drift equilibrium. The rejection of neutrality for these tests, either based on frequency spectrum or haplotype structure, has two possible explanations, involving random and nonrandom changes. Random changes are caused by demography (i.e., changes in population size, population structure), whereas nonrandom changes involve natural selection (Kreitman 2000). As a consequence, the observed negative values of DT and the departures from neutrality in FS and R2 are consistent with two hypotheses: (1) a population growth in Argentinian T. infestans and (b) the analyzed COI fragment has recently experienced a hitchhiking event caused by directional selection over a linked gene region (Tajima 1989, Braverman et al. 1995, Fu 1997, Ramos-Onsins and Rozas 2002). However, the MK test (which is less sensitive to demographic effects because it compares the relationship between polymorphism and divergence; Kreitman 2000) was nonsignificant. This strongly suggests that the observed departures from the mutation-drift equilibrium for the total Argentinian T. infestans sample are not caused by natural selection but most likely result from a recent population expansion. This hypothesis is also supported by the nested cladistic analysis, indicating that the patterns of geographic association for Argentinean haplotypes are compatible with a contiguous range expansion. Moreover, we infer from the relatively high-level clade at which the association was significant that this process may have occurred early in the radiation of T. infestans and was likely an important determinant of the genetic differentiation among populations. More recently, restricted gene flow, allopatric fragmentation, and isolation-by-distance all contributed to the current population structure in Argentina, as is suggested by the low-level clades associations and the Mantel test.
The phylogenetic and phylogeographic analyses are consistent with the presence of at least two clades within Argentinean haplotypes: one comprising variants h, x, and y and the other comprising haplotypes c, d, e, and similar. Unfortunately, the connections of variants av, b, and ay were not resolved because of the lack of synapomorphies in the parsimony tree or the high divergence in the nested cladistic analysis. The first clade seems to be restricted to populations of San Juan, San Luis, and Rio Negro, which are in the southernmost part of the sampled area, whereas the second clade is widespread. These data suggest a two-wave dispersal of T. infestans in Argentina. The clade with haplotypes h, x, and y may represent an older colonization event (possibly associated with the pre-Incan human migrations), whereas the remaining haplotypes may be the result of the more recent human migrations after the Spanish conquest, followed by a rapid population expansion process. In addition, the presence of haplotypes from the two different groups within the same populations (e.g., in San Juan, haplotypes x and y were found in the same house as haplotype c, and in Río Negro, haplotypes x and z coexisted) creates an unusual haplotype structure in these populations. This structure suggests a process of secondary contact after the two independent episodes of dispersal, leading to a current pattern of population admixture. This hypothesis is in agreement with the positive DT values found in San Juan, whereas in Río Negro, the test was consistent in sign but nonsignificant.
Mitochondrial genes have been used extensively as tools for inferring the evolutionary history and demographic past of both populations and species, and our work shows that COI is a useful marker to understand the historical population dynamics of T. infestans at a macrogeographical scale. However, these data must be considered with caution, because mitochondrial DNA can undergo particular evolutionary processes such as natural selection, high degree of homoplasy caused by an elevated A/T transition bias, and introgression, which may obscure the true species history (Ballard and Whitlock 2004, Lin and Danforth 2004). Further studies combining information from other independent gene regions will allow us to determine whether the patterns and evolutionary processes emerging from our COI data are particular for this gene or constitute distinctive features of this species.
Acknowledgments
We thank Drs. Y. Basmadjián and J. G. Cornejo del Carpio for providing bug samples; L. A. Jones for sequencing a subset of the samples; E. Hasson for sharing the facilities of his laboratory; S. Ramos-Onsins for useful suggestions about comparing θw values; C. Cecere and G. Vazquez-Prokopec for help in the design of Fig. 1; and F. Monteiro, C. Schofield, and three anonymous reviewers for valuable comments on earlier versions of this manuscript. This study benefited from international collaboration through the European Community–Latin America Triatomine Research (ECLAT) network. R.V.P. was supported by Fundación Antorchas and CONICET postdoctoral fellowships. R.V.P. and R.E.G. are members of Carrera de Investigador Científico (CONICET, Argentina). This project was financed by National Institutes of Health Research Grant R01 TW05836 funded by the Fogarty International Center and the National Institute of Environmental Health Sciences to U.K. and R.E.G. and by grants from the University of Buenos Aires and Agencia Nacional de Promoción Científica y Técnica (Argentina) to R.E.G.
References Cited
- Abad-Franch F, Monteiro FA. Molecular research and the control of Chagas disease vectors. An. Acad. Bras. Cienc. 2005;77:437–454. doi: 10.1590/s0001-37652005000300007. [DOI] [PubMed] [Google Scholar]
- Abalos JW. Sobre híbridos naturales y experimentales de Triatoma. An. Inst. Med. Regional. 1948;2:209–223. [Google Scholar]
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Bargues MD, Klisiowicz DR, Panzera F, Noireau F, Marcilla A, Perez R, Rojas MG, O’Connor JE, Gonzalez-Candelas F, Galvaõ C, et al. Origin and phylogeography of the Chagas disease main vector Triatoma infestans based on nuclear rDNA sequences and genome size. Infect. Genet. Evol. 2006;6:46–62. doi: 10.1016/j.meegid.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- Breniére SF, Bosseno MF, Vargas F, Yacsik N, Noireau F, Noel S, Dujardin JP, Tibayrenc M. Smallness of the panmictic unit of Triatoma infestans (Hemiptera: Reduviidae) J. Med. Entomol. 1998;35:911–917. doi: 10.1093/jmedent/35.6.911. [DOI] [PubMed] [Google Scholar]
- Carcavallo RU. Phylogeny of Triatominae. The Triatoma infestans complex. Mem. Inst. Oswaldo Cruz. 1998;93 Suppl 2:68–70. [Google Scholar]
- Cecere MC, Vazquez-Prokopec GM, Gürtler RE, Kitron U. Spatio-temporal analysis of reinfestation by Triatoma infestans (Hemiptera: Reduviidae) following insecticide spraying in a rural community in northwestern Argentina. Am. J. Trop. Med. Hyg. 2004;71:803–810. [PMC free article] [PubMed] [Google Scholar]
- Dujardin JP. Biosistemática, tendencias adaptativas y definición de poblaciones blanco. In: Schofield CJ, Dujardin JP, Jurberg J, editors. Proceedings of the International Workshop on Population Biology and Control of Triatominae. Mexico: INDRE, Cdad. de Mexico; 1996. pp. 97–98. [Google Scholar]
- Dujardin JP, Tibayrenc M, Venegas E, Maldonado L, Desjeux P, Ayala FJ. Isozyme evidence of lack of speciation between wild and domestic Triatoma infestans (Heteroptera: Reduviidae) in Bolivia. J. Med. Entomol. 1987;24:40–45. doi: 10.1093/jmedent/24.1.40. [DOI] [PubMed] [Google Scholar]
- Dujardin JP, Schofield CJ, Tibayrenc M. Population structure of Andean Triatoma infestans: allozyme frequencies and their epidemiological relevance. Med. Vet. Entomol. 1998;12:20–29. doi: 10.1046/j.1365-2915.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of the molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu Y-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García BA, Manfredi C, Fichera L, Segura EL. Short report: variation in mitochondrial 12S and 16S ribosomal DNA sequences in natural populations of Triatoma infestans (Hemiptera: Reduviidae) Am. J. Trop. Med. Hyg. 2003;68:692–694. [PubMed] [Google Scholar]
- García BA, Soares Barata JM, Blanco A. Enzyme polymorphism among Triatoma infestans (Hemiptera: Reduviidae) colonies. J. Med. Entomol. 1995;32:126–133. doi: 10.1093/jmedent/32.2.126. [DOI] [PubMed] [Google Scholar]
- Giordano R, Pizarro Cortez JC, Paulk S, Stevens L. Genetic diversity of Triatoma infestans (Hemiptera: Reduviidae) in Chuquisaca, Bolivia based on the mitochondrial cytochromoe b gene. Mem. Inst. Oswaldo Cruz. 2005;100:753–760. doi: 10.1590/s0074-02762005000700014. [DOI] [PubMed] [Google Scholar]
- Goloboff PA, Farris J, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:1–13. [Google Scholar]
- Guhl F, Schofield CJ. Population genetics and control of Triatominae. Parasitol. Today. 1996;12:169–170. [Google Scholar]
- Gürtler RE, Kitron U, Cecere MC, Segura EL, Cohen JE. Sustainable vector control and management of Chagas disease in the Gran Chaco, Argentina. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16194–16199. doi: 10.1073/pnas.0700863104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Gene genealogies and the coalescent process. Oxf. Surv. Evol. Biol. 1990;7:1–44. [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographical subdivision. Mol. Biol. Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Jordal BH, Normark BB, Farrell BD, Kirkendall LR. Extraordinary haplotype diversity in haplodiploid inbreeders: phylogenetics and evolution of the bark beetle genus Coccotrypes. Mol. Phylogenet. Evol. 2002;23:171–188. doi: 10.1016/S1055-7903(02)00013-1. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. Treefit. A computer program for evaluating how well evolutionary trees fit genetic distance data. 2008 http://www.montana.edu/kalinowski/Software/TreeFit.htm.
- Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- Kreitman M. Methods to detect selection in populations with applications to the human. Annu. Rev. Genomics Hum. Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3:integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lin C-P, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol. Phylog. Evol. 2004;30:686–702. doi: 10.1016/S1055-7903(03)00241-0. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Zhand DX, Szymura JM, Hewitt GM. The insect cytochrome oxidase I gene evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol. Biol. 1996;5:153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Marcet PL, Mora MS, Cutrera AP, Jones L, Gurtler RE, Kitron U, Dotson EM. Genetic structure of Triatoma infestans populations in rural communities of Santiago Del Estero, northern Argentina. Infect. Genet. Evol. 2008;8:835–846. doi: 10.1016/j.meegid.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptative protein evolution at the Adh locus in Drosophila. Nature (Lond.) 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Monteiro FA, Escalante AA, Beard CB. Molecular tools and triatomine systematics: a public health perspective. Trends Parasitol. 2001;17:344–347. doi: 10.1016/s1471-4922(01)01921-3. [DOI] [PubMed] [Google Scholar]
- Monteiro FA, Perez R, Panzera F, Dujardin JP, Galvaõ C, Rocha D, Noireau F, Schofield C, Beard CB. Mitochondrial DNA variation of Triatoma infestans populations and its implication on the specific status of T. melanosoma. Mem. Inst. Oswaldo Cruz. 1999;94 Suppl 1:229–238. doi: 10.1590/s0074-02761999000700037. [DOI] [PubMed] [Google Scholar]
- Noireau F, Flores R, Gutierrez T, Dujardin JP. Detection of sylvatic dark morphs of Triatoma infestans in the Bolivian Chaco. Mem.Inst. Oswaldo Cruz. 1997;92:583–584. doi: 10.1590/s0074-02761997000500003. [DOI] [PubMed] [Google Scholar]
- Noireau F, Bastrenta B, Catalá S, Dujardin JP, Panzera F, Torres M, Perez R, Galvaõ C, Jurberg J. Sylvatic population of Triatoma infestans from the Bolivian Chaco: from field collection to characterization. Mem. Inst. Oswaldo Cruz. 2000;95 Suppl 1:119–122. doi: 10.1590/s0074-02762000000700020. [DOI] [PubMed] [Google Scholar]
- Noireau F, Cortez MG, Monteiro FA, Jansen AM, Torrico F. Can wild Triatoma infestans foci in Bolivia jeopardize Chagas disease control efforts? Trends Parasitol. 2005;21:7–10. doi: 10.1016/j.pt.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Normark BB. Phylogeny and evolution of parthenogenetic weevils of the Aramigus tessellatus species complex (Coleoptera: Curculionidae: Naupactini): Evidence from mitochondrial DNA sequences. Evolution. 1996;50:734–745. doi: 10.1111/j.1558-5646.1996.tb03883.x. [DOI] [PubMed] [Google Scholar]
- Panchal M. The automation of nested clade phylogeographic analysis. Bioinformatics. 2007;23:509–510. doi: 10.1093/bioinformatics/btl614. [DOI] [PubMed] [Google Scholar]
- Pannell JR. Coalescence in a metapopulation with recurrent local extinction and recolonization. Evolution. 2003;57:949–961. doi: 10.1111/j.0014-3820.2003.tb00307.x. [DOI] [PubMed] [Google Scholar]
- Panzera F, Dujardin JP, Nicolini P, Caraccio MN, Rose V, Tellez T, Bermudez H, Bargues MD, Mas-Coma S, O’Connor JE, Perez R. Genomic changes of Chagas disease vector, South America. Emerg. Infect. Dis. 2004;10:438–446. doi: 10.3201/eid1003.020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez R, Hernández M, Quintero O, Canale ED, Méndez L, Cohanoff C, Martino M, Panzera F. Cytogenetic analysis of experimental hybrids in species of Triatominae (Hemiptera-Reduviidae) Genetica. 2005;125:261–270. doi: 10.1007/s10709-005-0369-z. [DOI] [PubMed] [Google Scholar]
- Pérez de Rosas AR, Segura EL, García BA. Microsatellite analysis of genetic structure in natural Triatoma infestans (Hemiptera: Reduviidae) populations from Argentina: its implications in assessing the effectiveness of chagas’ disease vector control programmes. Mol. Ecol. 2007;16:1401–1412. doi: 10.1111/j.1365-294X.2007.03251.x. [DOI] [PubMed] [Google Scholar]
- Picollo MI, Vassena C, Santo Orihuela P, Barrios S, Zaidemberg M, Zerba E. High resistance to pyrethroid insecticides associated with ineffective field treatments in Triatoma infestans (Hemiptera: Reduviidae) from Northern Argentina. J. Med. Entomol. 2005;42:637–642. doi: 10.1093/jmedent/42.4.637. [DOI] [PubMed] [Google Scholar]
- Pizarro JC, Gilligan LM, Stevens L. Microsatellites reveal a high population structure in Triatoma infestans from Chuquisaca, Bolivia. PLoS Negl. Trop. Dis. 2008;2:e202. doi: 10.1371/journal.pntd.0000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol. Evol. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Przeworski M. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 2002;18:559–563. doi: 10.1016/s0168-9525(02)02781-6. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins SE, Windsor A, Mitchell-Olds T. MANVa software: multilocus analysis of nucleotide variation. 2008 http://www.ub.edu/softevol/manva.
- Rozas J, Sánchez-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Scataglini MA, Lanteri AA, Confalonieri VA. Diversity of boll weevil populations in South America: a phylogeographic approach. Genetica. 2006;126:353–368. doi: 10.1007/s10709-005-1399-2. [DOI] [PubMed] [Google Scholar]
- Schachter-Broide J, Dujardin JP, Kitron U, Gürtler RE. Spatial structuring of Triatoma infestans (Hemiptera, Reduviidae) populations from northwestern Argentina using wing geometric morphometry. J. Med. Entomol. 2004;41:643–649. doi: 10.1603/0022-2585-41.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics. 2005;169:1601–1615. doi: 10.1534/genetics.104.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ. Biosystematics of the Triatominae. In: Service MW, editor. Biosystematics of Haematophagous insects. Oxford, United Kingdom: Clarendon Press; 1988. pp. 284–312. [Google Scholar]
- Schofield CJ, Lehane MJ, McEwen P, Catala SS, Gorla DE. Dispersive flight by Triatoma infestans under natural conditions in Argentina. Med. Vet. Entomol. 1992;6:51–56. doi: 10.1111/j.1365-2915.1992.tb00035.x. [DOI] [PubMed] [Google Scholar]
- Schweigmann N, Vallvé S, Muscio O, Ghillini M, Alberti A, Wisnivesky-Colli C. Dispersal flight by Triatoma infestans in an arid area of Argentina. Med. Vet. Entomol. 1988;2:401–402. doi: 10.1111/j.1365-2915.1988.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 2nd ed. San Francisco, CA: Freeman; 1981. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR. The classification of amino acid conservation. J. Theor. Biol. 1986;119:205–218. doi: 10.1016/s0022-5193(86)80075-3. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips CA. Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toloza AC, Germano M, Cueto GM, Vassena C, Zerba E, Picollo MI. Differential patterns of insecticide resistance in eggs and first instars of Triatoma infestans (Hemiptera: Reduviidae) from Argentina and Bolivia. J. Med. Entomol. 2008;45:421–426. doi: 10.1603/0022-2585(2008)45[421:dpoiri]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Vazquez-Prokopec G, Ceballos L, Kitron U, Güprtler RE. Active dispersal of natural populations of Triatoma infestans (Hemiptera: Triatominae) in rural northwestern Argentina. J. Med. Entomol. 2004;41:614–621. doi: 10.1603/0022-2585-41.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. The coalescent in an island model of population subdivision with variation among demes. Theor. Popul. Biol. 2001;59:133–144. doi: 10.1006/tpbi.2000.1495. [DOI] [PubMed] [Google Scholar]
- Wakeley J, Aliacar N. Gene genealogies in a metapopulation. Genetics. 2001;159:893–905. doi: 10.1093/genetics/159.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]