Abstract
Many sarcomas and leukemias carry non-random chromosomal translocations encoding mutant fusion transcription factors that are essential to their molecular pathogenesis. These novel, tumor-specific proteins provides a unique opportunity for the development of highly selective anticancer drugs that has yet to be exploited. A particularly clear example is provided by Ewing's Sarcoma Family Tumors (ESFT) which contain a characteristic t(11;22) translocation leading to expression of the oncogenic fusion protein EWS-FLI1. EWS-FLI1 is a disordered protein that precluded standard structure-based small molecule inhibitor design. Using surface plasmon resonance screening, we discovered a lead compound, NSC635437. A derivative compound, YK-4-279, blocks RHA binding to EWS-FLI1, induces apoptosis in ESFT cells, and reduces the growth of ESFT orthotopic xenografts. These findings provide proof of principle that inhibiting the interaction of mutant cancer-specific transcription factors with the normal cellular binding partners required for their oncogenic activity provides a promising strategy for the development of uniquely effective, tumor-specific anticancer agents.
There is a significant need for new cancer therapies that enhance efficacy and reduce long-term morbidity. Protein products of tumor-specific chromosomal translocations, which are present only in cancer cells, provide unique targets for anti-tumor therapies1. These translocations span a broad range of malignancies, including carcinomas, hematopoietic malignancies, and sarcomas2-4. In many cancers, these translocations lead to novel fusion proteins that both initiate and maintain oncogenesis. While some of these translocations, such as BCR-ABL5, lead to constitutively activated kinases, the majority lead to fusion proteins that function as transcription factors and lack intrinsic enzymatic activity. These translocation-generated transcription factor fusion proteins are ideal targets of anti-cancer therapies, yet no pharmaceuticals have been developed towards these targets.
The Ewing's sarcoma family of tumors (ESFT) can occur anywhere in the body and most often in the 2nd and 3rd decades. ESFT often respond well to initial chemotherapy, yet 40% of patients will develop recurrent disease. The majority of patients with recurrent disease will die from ESFT, while 75 - 80% of patients who present with metastatic ESFT will die within 5 years despite high-dose chemotherapy6. ESFT contain a well-characterized chromosomal translocation that fuses the amino-half of EWS to the carboxy-half of an ets-family DNA binding protein7. The most common fusion protein is the oncogenic transcription factor EWS-FLI1. Elimination of EWS-FLI1 using antisense and siRNA approaches results in the prolonged survival of ESFT xenograft-bearing animals8, but this approach currently lacks translation to clinical therapy9,10. Small-molecule targeting would be directed towards the disruption of EWS-FLI1 from established transcriptional complexes, since EWS-FLI1 lacks intrinsic enzymatic activity. The EWS-FLI1 transcriptional complex includes: RNA polymerase II, CREB-binding protein (CBP), and RNA Helicase A (RHA)11-13. Our previous investigations showed RHA augments EWS-FLI1 modulated oncogenesis, suggesting that this protein-protein complex is particularly important for tumor maintenance13. Small molecule inhibitors that block RHA interaction by targeting the oncogenic fusion protein EWS-FLI1 would be the first in a new class of anti-tumor therapy.
RHA has a critical role in embryogenesis and thus seems a reasonable partner for an oncogene in a highly undifferentiated tumor. RHA is indispensable for ectoderm survival in gastrulation of mammals14 and is required beyond embryogenesis because RHA null mouse fibroblast cells are not viable (personal communication, Dr. Chee-Gun Lee). However, transient reduction of RHA protein levels in COS cells did not affect the viability15. RHA provides a transcriptional coactivator role in models of tumorigenesis including NFkappaB16 and STAT617 transcriptomes. RHA binds to DNA in a sequence specific manner upon the promoters of p16INK4a 18 and MDR119. The amino-terminal region of RHA is most often the site for protein-protein interactions. CBP binds aa 1-250 of RHA 20, while additional partners for RHA bind in the amino-terminal region including RNA polymerase II21 and BRCA121. In modulating RNA interference in the RISC complex, RHA and Dicer, TRBP and Ago2 interact in the region of aa 1-272 of RHA22. EWS-FLI1 binds to RHA in a unique region that is not occupied by other transcriptional nor RNA metabolism proteins13, thus increasing the attractiveness of this protein-protein target.
Disruption of protein-protein interactions by small molecules is a rapidly evolving field. Proteins with more flexible structures, in some cases disordered proteins, have a greater potential for small molecule binding than rigid proteins because of higher induced fit sampling probabilities23. A disordered protein is defined, in part, by increased intrinsic movement and the inability to form rigid 3-dimensional structure (for review see24). EWS-FLI1 is a disordered protein and requires the disorder for maximal transactivation of transcription25,26. Based on these observations, EWS-FLI1, along with its binding to RHA, may provide a unique drug target.
RESULTS
RHA is a validated target in ESFT
A region of RHA that binds to EWS-FLI1 was identified based upon phage-display epitope screening13 (Fig. 1a). To validate RHA as critical in ESFT cells, RHA levels were reduced using shRNA and ESFT cell viability was reduced by 90% (Fig. 1b, c). A pancreatic cell line, PANC1 cells that do not express EWS-FLI1, were stably transfected with the same shRNA vectors with similar reduction in RHA levels (Supplementary Fig. 1a), but with no decrease in cell viability (Supplementary Fig. 1b). In order to further validate the protein-protein interaction of RHA with EWS-FLI1 as a therapeutic target for ESFT patients, we performed site-directed mutagenesis on the GST-RHA(647-1075) protein fragment. GST-RHA(647-1075) mutants were expressed and co-immunoprecipitated with full length recombinant EWS-FLI1. Mutants P824A and D827A showed a significant decrease in binding compared to wild-type control (Fig. 1d). The full length RHA mutant D827A maintained wild-type ATPase activity (Supplementary Fig. 2); therefore, we chose the D827A mutant to test whether RHA binding to EWS-FLI1 was required for neoplastic transformation.
Figure 1. RHA is necessary for optimal transformation by EWS-FLI1.
a. A schematic representation of RHA including the region that binds to EWS-FLI1. The E9R peptide corresponds to amino acids 823 to 832, located in the proximal of HA2 region of RHA. b. An shRNA expression vector was transfected into TC71 (ESFT) cells to reduce RHA levels. c. TC71 viability was reduced, as measured by WST reduction, following RHA shRNA expression. d. Alanine mutagenesis within E9R sequence was followed by in vitro immunoprecipitation with EWS-FLI1. The density of the GST-RHA band was measured and this graph is the average of three experiments. RHA P824A and D827A mutants have significantly lower binding to EWS-FLI1 (*p= 0.0129 and **p = 0.0034 respectively). e. Murine fibroblasts were placed in soft-agar for anchorage-independent growth assays (empty vector (W), EWS-FLI1 alone (WEF1)). f. The graph enumerates the colonies counted in three separate experiments; the difference between wild-type and mutant RHA was significant (*p= 0.0028). g. Protein expression for the fibroblasts is shown, detected with anti-FLAG (top) or anti-FLI1 (bottom). h. Densitometry of the EWS-FLI1 blot was performed using MultiGauge software.
RHA is required for EWS-FLI1 modulated transformation
Murine embryonic fibroblasts (W) that express low levels of endogenous RHA13 were stably transfected with EWS-FLI1 (WEF1) and either full-length wild-type RHA or full-length RHA(D827A). We observed a greater than additive effect when comparing the colony numbers from W + RHA (227±66) and WEF1 (115±8) to those of WEF1 + RHA (582±30) (Fig. 1e, f). The RHA(D827A) expressing cells demonstrated 3-fold lower anchorage-independent growth (p = 0.0028) than the wild-type (Fig. 1e, f). Similar protein expression levels of EWS-FLI1 and RHA were obtained in the fibroblasts (Fig. 1g). The EWS-FLI1 immunoblot was evaluated by densitometry and demonstrated reasonably similar protein levels amongst derived cell populations (Fig. 1h). The significant reduction of colony formation by the RHA(D827A) expressing cells suggests a critical role in anchorage-independent growth that is abrogated by RHA not binding to EWS-FLI1.
E9R peptide blocks RHA binding to EWS-FLI1
We developed reagents to block RHA binding to EWS-FLI1 since RHA is necessary for optimal EWS-FLI1 activity. The E9R peptide corresponds to amino acids 823 to 832, located in the proximal HA2 region of RHA (Fig. 1a). A cell-free protein interaction assay was developed to test whether E9R peptide inhibits RHA binding to EWS-FLI1. This immunoprecipitation assay demonstrated binding between bacterially expressed GST-RHA(647-1075) and full-length purified recombinant EWS-FLI1 (Fig. 2a, lane 3). Titration of E9R demonstrated a dose-dependent reduction in the binding of GST-RHA(647-1075) and full-length EWS-FLI1 with a decreased association to 50% with 0.1 μM E9R (Fig. 2a, lane 6). We thus sought to determine if disrupted EWS-FLI1/RHA binding inhibited cell growth.
Figure 2. E9R peptide prevents EWS-FLI1 binding to RHA with specific detrimental effects upon ESFT growth and transformation.
a. Immunoprecipitation of GST-RHA(647-1075) using recombinant full-length EWS-FLI1 bound to a FLI1 antibody. b. Growth reduction upon E9R-P (Antennapedia-E9R) treatment (10 μM) was observed in TC32 cells but not SKNAS cells. c. E9R-P peptide uptake was tracked with FITC label (upper panels). Merged images of DAPI nuclear counter-stain (middle panels) and FITC-Ant-E9R (lower panels) showed peptide was distributed throughout the cytoplasm and nucleus of both TC32 and SKNAS cells. Scale bar equals 20 μm. d. Neither Antennapedia alone (Antp), nor a mutant of an aspartic acid residue important for RHA binding to EWS-FLI1 (E9R-D5A-P) reduced growth of TC32 cells while E9R-P reduced cell growth. e. TC71 and SKNAS cells expressed EGFP empty vector (pG), EGFP-E9R (pGE9R), EGFP with nuclear export sequence (pGC) or EGFP-E9R with nuclear export sequence (pGCE9R). Only expression of EGFP-E9R in TC71 reduced anchorage-independent growth. f. Colony numbers of three experiments are averaged with a significant reduction in only TC71 cells when expressing E9R throughout the cell (*p = 0.0012). Scale bar equals 20 μm.
E9R peptide specifically inhibits ESFT growth
Peptide delivery to growing cells is greatly facilitated by cell permeable peptides (CPP)27. Antennapedia (Antp) is a CPP that was synthesized on the amino-terminus of E9R with or without the D827A mutation (E9R-P and E9R(D5A)-P, respectively) (Supplementary Table 1). Monolayer cultures of the EWS-FLI1-positive ESFT cell line, TC32, or a control EWS-FLI1-negative cell line, SKNAS (neuroblastoma), were treated with fluorescein conjugated peptides. Only the EWS-FLI1 containing cells showed reduced growth (Fig. 2b) and the SKNAS cells showed mild stimulation from the peptide based on an unknown mechanism. Confocal microscopy demonstrated uptake throughout the cell, including nuclei (shown by DAPI overlay, Fig. 2c). E9R-P significantly reduced ESFT cell growth (p = 0.048) while neither the D5A mutated control nor antennapedia peptides alone reduced ESFT cell growth (Fig. 2d). Neuroblastoma cells treated with the same peptides did not have a statistically significant alteration in growth, although a slight increase was observed with E9R(D5A) treated cells (p = 0.175). To determine the effect of E9R upon anchorage-independent growth, we stably transfected E9R as an EGFP fusion protein into TC71 (ESFT) or SKNAS (neuroblastoma) cells. An in-frame expression of LQLPPLERLTL excluded E9R from the nucleus28. Transfected cells showed E9R peptide expression either throughout the cell or excluded from the nucleus as predicted based on the intended targeting (Fig. 2e). TC71 colony formation was significantly reduced by 95% due to the expression of E9R, except when the peptide was excluded from the nucleus (Fig. 2e, f). The anchorage-independent growth of SKNAS was not affected by the E9R peptide (Fig. 2e, f). To further support specificity of the E9R peptide, a second small round blue cell tumor, embryonal rhabdomyosarcoma (RD cells), expressing pGE9R did not show reduced anchorage-independent growth (data not shown).
Small molecule binds to EWS-FLI1
A library of 3000 small molecules (NCI, DTP) was screened for EWS-FLI1 binding using surface plasmon resonance (SPR). We selected compounds that exhibited a binding level (actual resonance units, RUactual) to RUtheor ratio between 0.7 and 2.0 suggesting monomeric binding to EWS-FLI1 and also had favorable drug-like properties29. NSC635437 had an RUactual:RUtheor of 0.9 and was chosen for further evaluation based upon potential for chemical derivatization. We synthesized 1.0 gram of NSC635437 to complete our studies and for use as a standard during compound optimization (Fig. 3a).
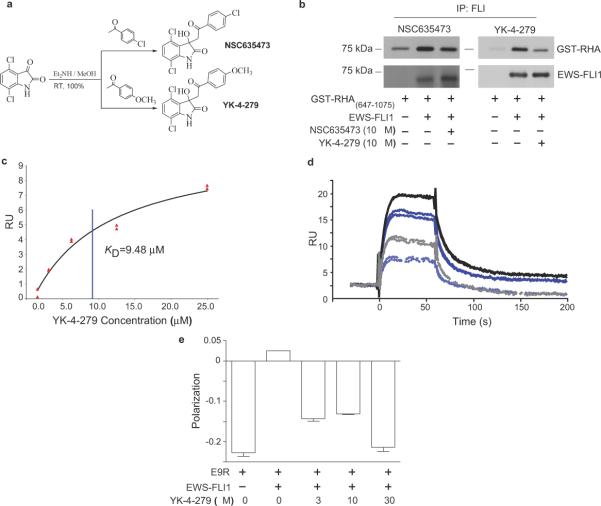
Figure 3. Small molecule binds to EWS-FLI1 and displaces E9R from EWS-FLI1.
a. NSC635437, 3-hydroxy-3-(2-oxo-2-phenyl-ethyl)-1,3-dihydro-indol-2-one was synthesized with 100% yield. Aromatic optimization produced YK-4-279 a para-methoxy derivative of NSC635437. b. EWS-FLI1 was incubated with NSC635437 (left panel) or YK-4-279 (right panel) followed by the addition of GST-RHA(647-1075). A FLI1 antibody complexed EWS-FLI1 and precipitated it from the solution. c. YK-4-279 steady state kinetics for binding to recombinant EWS-FLI1 that was immobilized on a CM5 Biacore chip. d. SPR displacement assay of 64 μM E9R alone (Black solid line) and with addition of YK-4-279 (Grey dashed line); 32 μM E9R alone (Dark blue solid line) and with addition of YK-4-279 (Light blue dashed line). e. Fluorescent polarization indicated the binding of 3.2 μM of FITC-E9R to EWS-FLI1, which was competitively inhibited by increasing concentrations of YK-4-279.
Methoxy-derivative is a more potent inhibitor of EWS-FLI1 binding to RHA
In a cell-free assay, NSC635437 reduced the direct binding of GST-RHA(647-1075) to full-length recombinant EWS-FLI1 (Fig. 3b, left panel). We used an aromatic optimization strategy to design analogs to improve upon the activity of NSC635437. One of these compounds (YK-4-279), substituted with a methoxy group at the para position (p-methoxy) of the aromatic ring (Fig. 3a) significantly reduced the protein-protein interaction of EWS-FLI1 with GST-RHA(647-1075) in vitro (Fig. 3b, right panel). We calculated the KD of 9.48 μM for the affinity of YK-4-279 with EWS-FLI1 using surface plasmon resonance (Fig. 3c). To support a model of YK-4-279 as having similar interaction qualities to E9R, SPR displacement assay shows 10 μM YK-4-279 reducing the binding of 64 μM E9R from 17 R.U. to 7 R.U. and 32 μM E9R from 13 R.U. to 5 R.U. (Fig. 3d). Fluorescence polarization further demonstrated E9R displacement of E9R when YK-4-279 was titrated into the experiment, showing complete displacement at 30 μM YK-4-279 (Fig. 3e).
YK-4-279 demonstrates functional inhibition of EWS-FLI1
ESFT cells treated with YK-4-279 demonstrated a dissociation of EWS-FLI1 from RHA by 10 μM, consistent with the KD value (Fig. 4a, top panel). YK-4-279 did not directly affect EWS-FLI1 nor RHA levels (Fig. 4a, middle and lower panels and Supplementary Fig. 3). To further support YK-4-279 as a functional inhibitor of EWS-FLI1, COS7 cells were cotransfected with EWS-FLI1 and NR0B1 reporter-luciferase plasmid (containing EWS-FLI1 regulatory GGAA elements31). The EWS-FLI1 transfected cells demonstrated a dose-dependent decrease in promoter activity when treated for 18 hours with 3 and 10 μM YK-4-279 (Fig. 4b). As an additional control for non-specific promoter effects, an NFκB responsive reporter was transfected into COS7 cells and activated with PMA. YK-4-279 did not affect the NFκB responsive promoter (Supplementary Fig. 4a). In a recent publication, EWS-FLI1 was shown to modulate cyclin D protein levels by altering a cyclin D splice site32. Blocking the interaction of EWS-FLI1 with RHA using YK-4-279 nearly eliminated cyclin D levels in TC32 cells treated for 14 hours (Fig. 4d), but did not affect cyclin D levels in four non-EWS-FLI1 containing cell lines (Supplementary Fig. 4b, c).
Figure 4. YK-4-279 reduces EWS-FLI1 functional activity.

a. TC32 cells were treated with YK-4-279 and resolved protein lysates were immunoblotted for co-precipitated RHA (top), EWS-FLI1 (middle), or total RHA (bottom). b. Luciferase reporter assay of EWS-FLI1 responsive NR0B1 promoter showed YK-4-279 dose-dependent (18-hour treatment) reduction in the promoter activity in COS7 cells. c. Protein lysates from transfected cells showed expression of EWS-FLI1. d. YK-4-279 treated TC32 cell lysates (treated for 14 hours) were blotted for cyclin D1 and actin.
YK-4-279 specifically inhibits ESFT cell growth and induces apoptosis
The compound identified from screening, NSC635437, was found to have an IC50 of 20 μM for TC32 cells growing in monolayer; however, YK-4-279 reduced the IC50 to 900 nM (Fig. 5a). YK-4-279 was relatively specific for ESFT cells as compared to the non-transformed HEK293 cells, demonstrating a 10-fold difference in IC50 (Fig. 5b). Primary cell lines, ES925 and GUES1, established from ESFT patients with recurrent tumors demonstrated sensitivity to YK-4-279 with anti-proliferative IC50 values of 1 and 8 μM, respectively (Fig. 5c). A panel of ESFT cell lines demonstrated IC50 values between 0.5 to 2 μM for YK-4-279 while cell lines that lack EWS-FLI1 have IC50 values in excess of 25 μM (Fig. 5d). An additional panel of non-transformed keratinocytes (HFK) and ectocervical cells (HEC) treated for 3 days with 30 μM YK-4-279 showed an IC50 that exceeded 30 μM (Supplementary Fig. 5a).
Figure 5. YK-4-279 is potent and specific inhibitor of ESFT.
a. TC32 cells were treated with a dose range of YK-4-279 and NSC635437. Cell number measured by MTT or WST reduction after seven days in culture. b. TC32 and HEK-293 (non-transformed, lacking EWS-FLI1) were treated similarly to (a). c. Primary ESFT explant cell lines GUES1 and ES925 were treated for 3 days with YK-4-279. d. Cell lines expressing EWS-FLI1 were compared to non-EWS-FLI1 malignant cell lines following 3 days in culture to establish the IC50 using WST assay. e. Caspase 3 activity of a panel of ESFT (TC32, TC71, A4573, and ES925), malignant non-EWS-FLI1 expressing (MCF-7, MDA-MB-231, PC3, ASPC1, COLO-PL), and non-transformed cells (HEK-293, HFK, and HEC) were measured. Graph plots level of fluorescence in treated divided by untreated lysate. f. Arrows indicated apoptotic nuclear fragmentation after 50 μM YK-4-279 treated ESFT (TC32) and non-transformed cells (HEK-293, HFK, and HEC). Scale bar equals 200 μm.
Apoptosis leads to tumor cell death through the activation of sequential caspase enzymes, with caspase-3 demonstrating a commitment to cellular suicide33. Caspase-3 activity rose in a dose-dependent fashion in TC32 cells treated with YK-4-279 for 24 hours (Supplementary Fig. 5b). Caspase activity was similar to 1 μM doxorubicin, a standard agent in the treatment of patients with ESFT6. Additional malignant and non-malignant cell lines were evaluated for caspase-3 activation in response to YK-4-279. While YK-4-279 induced caspase-3 activity in four ESFT cell lines (TC32, A4573, TC71, and ES925), none of the 5 non-EWS-FLI1 cancer cell lines nor 3 non-transformed cell lines (HFK, HEC, HEK293) treated with YK-4-279 resulted in apoptosis (Fig. 5e). Treatment of TC32, HEK293, HFK, and HEC with short-term (6 hours) high dose (50 μM) YK-4-279 resulted in significant apoptosis of the ESFT cells but no cell death in the non-transformed cells (Fig. 5f). Together, these results support the specific toxicity of YK-4-279 upon cell lines containing EWS-FLI1 compared with other tumor and non-transformed cells.
In order to further support for the target specificity of YK-4-279 toxicity upon ESFT cells, we reduced the levels of each of the critical proteins by using shRNA in A673 cells42. The RHA reduced cells demonstrated a YK-4-279 IC50 of >10 μM, while control shRNA (targeting luciferase) IC50 was less than 1 μM (Supplementary Fig. 5c). When EWS-FLI1 was reduced using shRNA, the IC50 increased 10-fold from 0.5 μM to approximately 5 μM (Supplementary Fig. 5d, e).
ESFT xenograft growth is inhibited by YK-4-279
ESFT (orthotopic) or prostate cancer cell xenograft tumors were established in SCID/bg mice. Tumor growth rate was reduced for CHP-100, ESFT, (Fig. 6a), but not the PC3, prostate tumors (Fig. 6b). Five independent experiments were performed with the ESFT xenografts (TC71 and CHP-100) and the cumulative data for these experiments shows a marked overall tumor reduction (p < 0.0001) in the YK-4-279 treated animals (Fig. 6c). Pathological analysis of animals treated with YK-4-279 did not show any signs of toxicity except changes related to IP injection. Tumors from animals treated with YK-4-279 were compared with DMSO treatment using immunohistochemistry to identify caspase-3 activity (Fig. 6d). The CHP-100 xenograft tumors from treated animals had a 3-fold increase in caspase-3 activity compared to control animals (Fig. 6e). These results show inhibition of two ESFT tumor models and concomitant increased apoptosis following YK-4-279 treatment.
Figure 6. YK-4-279 inhibited the growth of ESFT xenograft tumors.

Xenografts were established with injection of either ESFT (CHP-100 or TC71) or Prostate cancer (PC3) cells. a. CHP-100 intramuscular xenografts (arrow indicates when tumors were palpable) received DMSO (n=4; open circles) or 1.5 mg YK-4-279 (n=5; triangles) (p=0.016, by t-Test comparison). Single experiment growth curves depicted are representative of five independent experiments. b. PC3 subcutaneous xenografts (arrow indicates when tumors were palpable) were treated as CHP-100 cells (a) (n=5 per group, representative of 3 independent experiments). c. Overall response of ESFT xenografts (TC71, open symbols, and CHP-100, closed symbols) to YK-4-279 (1.5 mg/dose). Tumor volumes at day 14 after treatment initiation compared across 5 experiments (DMSO n = 19, YK-4-279 n = 25, p < 0.0001, by Mann-Whitney test). d. Tumors from the mice in (a) were analyzed for activation of caspase-3 activity using immunohistochemistry. e. Caspase-3 positive cells were counted (n>500 in 3 high-power-fields) in 4 separately stained slides for each group (p = 0.041).
DISCUSSION
EWS-FLI1 is a unique, cancer specific, molecule that is a potential therapeutic target in ESFT cells. RHA is critical to the function of EWS-FLI1. A peptide (E9R) that blocks RHA binding to EWS-FLI1 specifically reduced anchorage-independent growth. We also identified a small molecule lead compound that binds to EWS-FLI1. The lead compound derivative YK-4-279, along with E9R peptide, demonstrates that the EWS-FLI1/RHA interaction can be blocked with a detrimental effect on ESFT cells both in vitro and in vivo. These findings validate a highly specific cancer target, the interaction of EWS-FLI1 with RHA.
These are the first experiments that evaluate a small molecule inhibitor of EWS-FLI1 function. We demonstrate that the small molecule YK-4-279 binds to EWS-FLI1 and blocks the binding of RHA. A series of xenograft experiments demonstrates that 60 - 75 mg/kg YK-4-279 significantly reduced tumor growth. The small molecule not only inhibits RHA binding to EWS-FLI1, but also reduces EWS-FLI1 modulated transcription. An additional putative function of EWS-FLI1 is splice-site modification34, which was recently supported by the EWS-FLI1 altered splicing of cyclin D132. Treatment of ESFT cells with YK-4-279 led to decreased cyclin D1 levels. Additional investigations of the splicing complex are necessary to determine if this effect is due to the disruption of an EWS-FLI1/RHA complex or allosteric interference with EWS-FLI1. Small molecule inhibitors have great therapeutic potential, but will be immediately useful as functional probes.
EWS-FLI1 was recognized as a potential therapeutic target over 15 years ago, almost immediately after the protein was identified as a product of the breakpoint region t(11;22)35. The disordered biophysical nature of EWS-FLI1 precludes standard structure-based small molecule design26. Therefore, we pursued development of small molecule protein-protein inhibitors based upon the assumption that EWS-FLI1 would have a binding partner critical for its oncogenic function, which we previously identified as RHA13, and validated in the current study. The exact nature of the requirement of RHA for EWS-FLI1 is currently under investigation, however we speculate that RHA could be involved in EWS-FLI1 function, synthesis or stability. Our data support multiple mechanisms and therefore require further enzymatic and structural studies of EWS-FLI1 bound RHA for resolution. The fact that YK-4-279 is still toxic to A673 cells with reduced EWS-FLI1 could be due to residual EWS-FLI1 or suggest broader action of the compound. In addition, while our data suggest that YK-4-279 is specific for ESFT cell toxicity, we recognize that as additional cell and tumor models are tested, YK-4-279 may reveal other protein interactions.
Inhibitory peptides offer a greater likelihood of specificity to validate protein-protein interaction targets and to evaluate protein-complex disruption; however, peptides are problematic for clinical development. Our investigations use peptides to compare the effects of disrupting protein-protein interactions with our small molecules. While small peptides are currently being developed as therapeutic agents36,37, 10 - 20 aa peptides present formidable pharmacokinetic stability and delivery challenges. The E9R peptide may compete with full-length RHA binding to EWS-FLI1 and our data support a functional displacement of RHA by E9R. We demonstrate that using surface plasmon resonance and fluorescence polarization that YK-4-279 can `displace' E9R from EWS-FLI1. While our results support E9R and YK-4-279 binding to the same site on EWS-FLI1, allosteric interference cannot be excluded. Therefore, a structural model of EWS-FLI1 is required to both fully prove this interaction and YK-4-279 binding site, but is yet unavailable due to the challenges of disordered proteins23.
The interaction of RHA with EWS-FLI1 presents an ideal opportunity for the development of small molecule protein-protein interaction inhibitors (SMPPII). Both evidence and prevailing opinion support disordered proteins as potential targets of small molecule therapeutics38. Our data also support EWS-FLI1 protein interaction targeting to modulate oncogene function and potentially lead to novel therapeutics. Additional experiments to evaluate multi-species specificity, toxicity and ADME (absorption, distribution, metabolism, and excretion) are required to advance a further optimized derivative of YK-4-279 into clinical trials. Small molecules that disable EWS-FLI1 function with minimal toxicity, in particular sparing of hematopoetic stem cells, could potentially provide a valuable adjuvant therapy for patients with ESFT. In addition, this paradigm for drug discovery could be applied to many related sarcomas that share similar oncogenic fusion proteins.
METHODS
Materials
E9R peptide was obtained from Bio-synthesis Inc, Lewisville, TX. Protein G beads (Invitrogen, Carlsbad, CA), anti-GST, anti-FLI1, anti-Cyclin D1 antibodies (Santa Cruz, CA), Ac-DEVDAMC, caspase-3 fluorogenic substrate (BD Biosciences Pharmingen), and anti-Cleaved Caspase 3(Asp175) (Cell Signaling) were commercially obtained.
Site directed mutagenesis
Every non-alanine amino acid in aa 823-832 region of RHA changed to alanine by site directed mutagenesis by using QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX) according to manufacturer's protocol.
Cell cultures
Established TC32, TC71, A4573, CHP-100 and primary ES925 and GUES1 ESFT cell lines were maintained in RPMI (Invitrogen) media supplemented with 10% FBS (Gemini Bioproducts). HEC and HFK cell lines, previously described39. Stably EWS-FLI1 expressing subclones of these cells were tested in anchorage independent growth assay as described previously13.
Protein immunoprecipitation assays
Protein lysates and immunoprecipitations were performed as previously published13. Recombinant GST-RHA(647-1075) was prepared from crude bacterial extracts without further purification.
Small molecule library screening and selection of lead compound
A surface plasmon resonance assay using the Biacore T100 was established with EWS-FLI1, prepared in our laboratory as previously published26. DNA oligonucleotides were used to quality control the proper conformation of EWS-FLI1 on the surface of a CM5 chip. Small molecules obtained from the Developmental Therapeutics Program of the National Cancer Institute, NIH (http://dtp.nci.nih.gov/branches/dscb/repo_open.html) were prioritized based upon their molecular weight and solubility. An initial screening of molecules was performed at 1 or 10 μM compound, based on solubility. We used a model that compares the actual binding maximum (actual RU) with the theoretical binding maximum (RUtheor). If the RUactual to RUtheor is 0.9 - 1.0, this suggests a binding, and a compound was considered a `hit'. Hits (RUactual to RUtheor of 0.7 -2.0) were then reviewed by a team of medicinal chemists and those with structural potentials were selected for further study. Selected molecules were tested in vitro in a solution co-immunoprecipitation assay using recombinant EWS-FLI1 and GST-RHA(647-1075).
Synthesis and analysis of small molecule compounds
Fluorescence polarization assay
Increasing concentrations of FITC-E9R were added to a fixed concentration of EWS-FLI1 (4.8 μM) to obtain a saturated binding curve. The assay was performed in 20mM Tris, 500mM NaCl, 0.67M imidazole, pH 7.4. The fluorescence polarization was analyzed in a QuantaMaster fluorimeter (Photon Technology International, Ford, West Sussex, UK) equipped with polymer sheet polarizers at an excitation wavelength of 495 nm and emission wavelength of 517 nm. Increasing concentrations of YK-4-279 were added to a fixed concentration of EWS-FLI1 and FITC-E9R (3.2 μM, as determined from saturated binding curve) with the same buffer and instrumental settings as described above.
Plasmids and Reporter assay
EGFP-E9R fusion constructs prepared as published43. We transiently transfect the NR0B131 luciferase reporter and full-length EWS-FLI1 into COS-7 cells with Fugene-6 (Roche) and luciferase assay performed per manufacturer's protocol (Dual Luciferase Kit, Promega). Six hours following transfection, cells were treated with either 3 or 10 μM YK-4-279. Cell lysates luciferase activity levels were standardized to renilla activity from a non-affected promoter and plotted as relative luciferase activity (RLA).
Caspase-3 activity measurement and nuclear fragmentation
Cells were treated for 24 hours with 10 μM YK-4-279. The Caspase-3 substrate DEVD-AMC was incubated with equal amounts of protein lysate and fluorescence from cleaved substrate measured in a fluorimeter. TC32 and non-transformed cells HEK-293, HFK, and HEC were treated for 6 hours with high dose (50 μM) YK-4-279. DAPI stained cells were photographed at 600X using inverted fluorescence microscope.
Mouse strains and in vivo small molecule testing
One million TC71 or CHP-100 cells in 100 μL HBSS were injected orthotopically into the gastrocnemius muscle of 4-8 week old SCID/bg mice (Taconic, Germantown, New York). Prostate cancer xenografts were established by subcutaneous injection of 5 million PC3 cells into the flanks of 4-8week old nude mice (Taconic). Mice were randomized to receive three times per week intraperitoneal injections of DMSO, YK-4-279 at 1.5mg/dose when tumors were palpable. Each of the animal experiments was begun with 10 mice that were randomized into treatment and control groups when the tumors reached palpable size. In the control groups some tumors exceeded the IACUC maximal size (2 cm in any dimension) and were euthanized prior to day 14 and thus not included in the day 14 analysis (Fig. 6c). Tumor length and width were measured every 2-4 days and volume was calculated using the formula v = D × d2 × π/6 where D is the longest diameter and d is the shorter diameter. Xenograft studies were approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee.
Statistical Analysis
Statistical analyses performed using GraphPad Prism (http://www.graphpad.com/help/Prism5/Prism5Help.html?using_regression_guide_2.htm).
Supplementary Material
Acknowledgments
This work was generously supported by the Children's Cancer Foundation of Baltimore, MD (J.T. and A.Ü.), Go4theGoal Foundation (J.T.), Dani's Foundation of Denver, CO (J.T.), the Liddy Shriver Sarcoma Initiative (J.T.), the Amschwand Sarcoma Cancer Foundation (J.T.), Burroughs-Wellcome Clinical Scientist Award in Translational Research (J.T.), NIH R01CA138212 (J.T.), R01CA133662 (J.T.), and the Georgetown University Medical Center Drug Discovery Program. NIH support through the following grants Cancer Center Support Grant P30 CA051008 for use of FCCS and Microscopy core facilities, P01 CA47179 (M.M.). The authors would like to thank Dr. Steve Metallo for training in fluorescence polarization and Ms. Linshan Yuan for SPR technical assistance. Also, Drs. Tim Cripe and Luke Whitesell provided critical review of our manuscript. We also thank Drs. Stephen Lessnick, JV Frangioni, Olivier Delattre, Richard Schlegel for providing reagents, and the Developmental Therapeutics Program of the NCI, NIH for providing the Diversity set of compounds for screening. This article is dedicated to our patients who have fought, but succumbed to ESFT.
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.French CA, et al. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22:4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 3.Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- 4.Poppe B, et al. Expression analyses identify MLL as a prominent target of 11q23 amplification and support an etiologic role for MLL gain of function in myeloid malignancies. Blood. 2004;103:229–235. doi: 10.1182/blood-2003-06-2163. [DOI] [PubMed] [Google Scholar]
- 5.Carroll M, et al. CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood. 1997;90:4947–4952. [PubMed] [Google Scholar]
- 6.Grier HE, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing's sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003;348:694–701. doi: 10.1056/NEJMoa020890. [DOI] [PubMed] [Google Scholar]
- 7.Delattre O, et al. The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331:294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- 8.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequencespecific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 9.Kovar H, Ban J, Pospisilova S. Potentials for RNAi in sarcoma research and therapy: Ewing's sarcoma as a model. Semin Cancer Biol. 2003;13:275–281. doi: 10.1016/s1044-579x(03)00041-5. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, Iwakuma T, Harimaya K, Sato H, Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing's sarcoma and primitive neuroectodermal tumor cells. J Clin Invest. 1997;99:239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petermann R, et al. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 12.Nakatani F, et al. Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein. J Biol Chem. 2003;278:15105–15115. doi: 10.1074/jbc.M211470200. [DOI] [PubMed] [Google Scholar]
- 13.Toretsky JA, et al. Oncoprotein EWS-FLI1 activity is enhanced by RNA helicase A. Cancer Res. 2006;66:5574–5581. doi: 10.1158/0008-5472.CAN-05-3293. [DOI] [PubMed] [Google Scholar]
- 14.Lee CG, et al. RNA helicase A is essential for normal gastrulation. Proc Natl Acad Sci U S A. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman TR, et al. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 16.Tetsuka T, et al. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur J Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 17.Valineva T, Yang J, Silvennoinen O. Characterization of RNA helicase A as component of STAT6-dependent enhanceosome. Nucleic Acids Res. 2006;34:3938–3946. doi: 10.1093/nar/gkl539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myohanen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 19.Zhong X, Safa AR. RNA helicase A in the MEF1 transcription factor complex up-regulates the MDR1 gene in multidrug-resistant cancer cells. J Biol Chem. 2004;279:17134–17141. doi: 10.1074/jbc.M311057200. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima T, et al. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 21.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 22.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Bhalla J, Storchan GB, MacCarthy CM, Uversky VN, Tcherkasskaya O. Local flexibility in molecular function paradigm. Mol Cell Proteomics. 2006;5:1212–1223. doi: 10.1074/mcp.M500315-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Xie H, et al. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. Journal of proteome research. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng KP, et al. Multiple aromatic side chains within a disordered structure are critical for transcription and transforming activity of EWS family oncoproteins. Proc Natl Acad Sci U S A. 2007;104:479–484. doi: 10.1073/pnas.0607007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uren A, Tcherkasskaya O, Toretsky JA. Recombinant EWS-FLI1 oncoprotein activates transcription. Biochemistry. 2004;43:13579–13589. doi: 10.1021/bi048776q. [DOI] [PubMed] [Google Scholar]
- 27.Terrone D, Sang SL, Roudaia L, Silvius JR. Penetratin and related cell-penetrating cationic peptides can translocate across lipid bilayers in the presence of a transbilayer potential. Biochemistry. 2003;42:13787–13799. doi: 10.1021/bi035293y. [DOI] [PubMed] [Google Scholar]
- 28.Voss SD, DeGrand AM, Romeo GR, Cantley LC, Frangioni JV. An integrated vector system for cellular studies of phage display-derived peptides. Anal Biochem. 2002;308:364–372. doi: 10.1016/s0003-2697(02)00268-3. [DOI] [PubMed] [Google Scholar]
- 29.Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nature reviews. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 31.Gangwal K, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez G, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 34.Knoop LL, Baker SJ. EWS/FLI alters 5'-splice site selection. J Biol Chem. 2001;276:22317–22322. doi: 10.1074/jbc.M008950200. [DOI] [PubMed] [Google Scholar]
- 35.Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 36.Plescia J, et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7:457–468. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 37.Palermo CM, Bennett CA, Winters AC, Hemenway CS. The AF4-mimetic peptide, PFWT, induces necrotic cell death in MV4-11 leukemia cells. Leuk Res. 2007 doi: 10.1016/j.leukres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Y, et al. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Uren A, et al. Activation of the Canonical Wnt Pathway during Genital Keratinocyte Transformation: A Model for Cervical Cancer Progression. Cancer Res. 2005;65:6199–6206. doi: 10.1158/0008-5472.CAN-05-0455. [DOI] [PubMed] [Google Scholar]
- 40.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case DA, et al. The Amber biomolecular simulation programs. Journal of computational chemistry. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tirode F, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 43.Frangioni JV, Neel BG. Use of a general purpose mammalian expression vector for studying intracellular protein targeting: identification of critical residues in the nuclear lamin A/C nuclear localization signal. J Cell Sci. 1993;105(Pt 2):481–488. doi: 10.1242/jcs.105.2.481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.