Abstract
T cell development depends on the coordinated interplay between receptor signaling and transcriptional activation/repression. We performed a genetic complementation screen, and identified a novel transcriptional repressor, NKAP. NKAP associates with HDAC3 and is part of a DNA-binding complex, as shown by chromatin immunoprecipitation. NKAP also associates with CIR, a part of the Notch corepressor complex. The expression of NKAP during T cell development inversely correlates with the expression of Notch target genes, implying that NKAP may modulate Notch-mediated transcription. To examine the function of NKAP in T cell development, Lck-cre NKAP conditional knockout mice were generated. Interestingly, loss of NKAP blocks development of αβ but not γδ T cells. In addition, Lck-cre NKAP cKO DP T cells express 8- to 20-fold higher levels of Hes1, Deltex1 and CD25, providing in vivo evidence that NKAP functions as a transcriptional repressor, acting at least in part, by repression of the Notch pathway.
Introduction
T cells develop in the thymus through a series of tightly regulated steps, whereby uncommitted progenitors become restricted to the T cell pathway, rearrange a functional TCR and undergo positive/negative selection based on their TCR specificity (reviewed in Taghon and Rothenberg, 2008). During this process, signals emanating from cell surface receptors lead to alterations in gene expression. Loss of any of the components of these pathways results in arrested development. One of the critical checkpoints encountered by developing T cells is the DN3 to DP transition, also known as the β-selection checkpoint. At the DN3 stage of αβ T cell development, a successfully rearranged TCRβ chain pairs with the pre-TCRα chain and is expressed on the cell surface, producing a signal that leads to increased proliferation, expression of CD4 and CD8 on the cell surface, and subsequent rearrangement of the TCRα chain. Failure to productively rearrange TCRβ, or to transmit pre-TCR signals (such as in the absence of Lck or SLP-76), leads to developmental arrest at the DN3 stage.
Although pre-TCR signalling is necessary for progression through the β-selection checkpoint, it is not sufficient for T cell development to progress. Signals through Notch are also required. Notch comprises a family (Notch 1–4 in mammals) of transmembrane receptors (reviewed in Kadesch, 2004 and Maillard et al, 2005). Upon interaction with a ligand (members of the Delta-like and Jagged families), two separate proteolytic cleavage events liberate the cytoplasmic tail of Notch (intracellular Notch, or ICN) which translocates to the nucleus. ICN displaces a co-repressor complex from the ubiquitously expressed transcription factor CSL (CBF1/Su(H)/LAG-1, also known as RBP-Jκ). The corepressor complex functions, in part, through the recruitment of histone deacetylases (HDACs), resulting in an altered chromatin structure that is not amenable to active transcription. When ICN associates with CSL, it recruits the histone acetyltransferases GCN4 and PCAF, which results in chromatin remodeling. ICN also recruits transcriptional co-activators, including members of the mastermind-like family (MAML-1, 2 and 3), allowing for Notch target gene expression. Conditional loss of Notch1 using Lck-cre transgenics, or inhibition of Notch signaling using an inducible dominant negative MAML1, results in arrest of T cell development at the DN3 to DP transition (Maillard et al., 2006; Wolfer et al., 2002).
Signaling through the pre-TCR and Notch leads to epigenetic changes and alterations in transcription, which also play a critical role in T cell development. In particular, transcriptional corepressors and chromatin modifiers are also required at the DN3 to DP transition. Mice deficient in the common components of corepressor complexes, NCoR and mSin3a, arrest T cell development at the DN3 stage (Cowley et al., 2005; Jepsen et al., 2000). Conditional deletion of Brg1, the ATPase subunit of the SWI-SNF chromatin remodeling complexes, during T cell development (using Lck-cre) also results in a block at the DN3 stage (Gebuhr et al., 2003). Similarly, loss of Mi-2β, a component of the NuRD chromatin remoding complex, also leads to a block at the DN3 to DP transition (Williams et al., 2004). DNA methylation is associated with repressed transcription, and loss of the DNA methyltransferase Dnmt1 using Lck-cre also disrupts T cell development at the DN3 stage (Lee et al., 2001). Therefore, T cell development past the β-selection checkpoint requires pre-TCR and Notch signaling, as well as epigenetic chromatin alterations, to proceed to the DP stage.
Insights into the biochemical basis of TCR signal transduction arose from the generation and analysis of Jurkat mutant T cell lines (reviewed in Abraham and Weiss, 2004). The central role of the signaling molecules identified in Jurkat T cells was ultimately confirmed when knockout mice were generated. To gain a greater understanding of the pathways that regulate T cell function, we performed a genetic complementation screen using a retroviral cDNA library to rescue the defect in a Jurkat T cell mutant cell line. Using this approach, we describe the isolation of a novel transcriptional repressor, NKAP, and demonstrate that NKAP regulates the Notch signaling pathway in vivo and is required for T cell development.
Results
Genetic complementation of a Jurkat signaling mutant cell line by NKAP
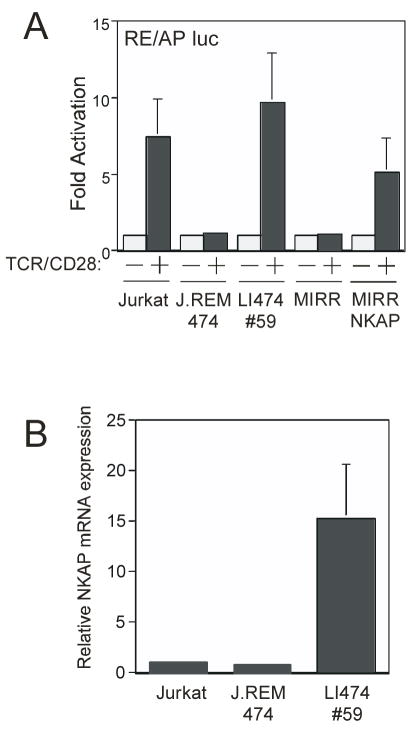
Previously, we generated a panel of Jurkat T cell mutant cell lines (the J.REMs) that fail to upregulate a GFP reporter under the control of the RE/AP element from the IL-2 promoter upon TCR activation (Greene and Shapiro, 2003). We were unable to identify the molecular defect in any of these cell lines using a candidate approach. Consequently, we initiated a genetic complementation screen to rescue the defect and identify novel regulators of T cell function. Using a retroviral leukocyte cDNA library (Clontech), we transduced one of the J.REM mutant cell lines (#474) and screened for clones in which the upregulation of the RE/AP GFP reporter was restored. One appropriate clone was identified from the initial screen, LI474-59 (“Library Infected J.REM 474 – clone 59, figure 1A). Using PCR primers flanking the site of cDNA insertion in the retrovirus, we determined that the integrated retrovirus contained the entire coding region (in the correct orientation for expression) of NKAP (Chen et al., 2003). To verify that NKAP was responsible for the reversal of the T cell activation defect, we generated a MIRR-NKAP retrovirus and used it to re-infect the original J.REM cell line. RE/AP activation was restored upon infection with MIRR-NKAP retrovirus, and not with an empty MIRR retrovirus (figure 1A). To determine whether NKAP deficiency was the cause of the molecular defect in the J.REM cell line, NKAP expression was examined. Using Q-PCR, it was determined that NKAP mRNA expression was equivalent between the parental Jurkat cell line and J.REM 474 (figure 1B). Additionally, sequencing of the entire NKAP mRNA coding region in J.REM 474 by RT-PCR did not reveal any mutations (data not shown). Therefore, alterations in NKAP are not responsible for the defect in the J.REM 474 cell line, and ectopic overexpression of NKAP in LI474-59 (figure 1B) must compensate for a defect in a different gene in J.REM 474.
Figure 1. Genetic complementation of a T cell activation defect by NKAP.
(A) Wild-type Jurkat T cells, J.REM 474, LI474-59, MIRR or MIRR NKAP transduced J.REM474 cells were transfected with an RE/AP luciferase reporter. The following day, cells were left unstimulated or stimulated with anti-TCR and anti-CD28. Results are shown as fold activation of stimulated relative to unstimulated controls (=1). Error bars reflect SEM from three independent transfections.
(B) RNA was generated from wild-type Jurkat, J.REM 474 and LI474-59 cell lines and analyzed by Q-PCR using NKAP mRNA expression.
NKAP is a 415-amino acid, widely expressed nuclear protein which is highly conserved from fly to human, with no known functional domains (Chen et al., 2003). Overexpression of NKAP in 293 cells enhanced NF-κB reporter activity 3-fold, while having minimal effect on the activation of an AP-1 reporter, leading to the conclusion that it is involved in NF-κB activation (Chen et al., 2003). Since the RE/AP reporter contains an NF-κB site, increased activation of NF-κB by NKAP could conceivably rescue the T cell activation defect in J.REM 474. To test this, J.REM 474 and the rescued LI474-59 cell lines were stimulated through TCR/CD28, and NF-κB activation examined. However, no differences were observed in either the timing or extent of signaling events downstream of TCR/CD28 leading to NF-κB activation. Specifically, TCR/CD28-mediated phosphorylation of Akt (S473 and T308), IKKα/β and IkBα were unaltered (Supplemental Figure 1A). In addition, no enhancement of the activation of an NF-κB reporter was observed in the LI474-59 cell line compared to the J.REM 474 cell line (Supplemental Figure 1B). Since no differences were observed in NF-κB activation between J.REM 474 and LI474-59, we considered that NKAP may have functions outside of the NF-κB pathway.
Genetic evidence that NKAP is a transcriptional repressor and component of the Notch corepressor complex
NKAP is evolutionarily conserved, and NKAP orthologs have been isolated in genetic screens in both Drosophila and C. elegans, although the function of NKAP in either of these organisms is uncharacterized. In Drosophila, a large scale yeast two hybrid screen was performed to define a proteome-wide protein interaction map (Giot et al., 2003), and three potential binding partners for CG6066 (the Drosophila NKAP ortholog) were found. Two of these potential binding partners are uncharacterized. However, a third fly protein, CG6843, is an ortholog of the human protein CSL/CBF1 interacting corepressor (CIR), a component of the Notch co-repressor complex (Hsieh et al., 1999). This suggested that NKAP may also be part of the Notch corepressor complex, through an association with CIR. Notch signaling regulates many aspects of T cell development, activation and effector function, including IL-2 expression (reviewed in Maillard et al., 2005). Since our complementation screen centered around transcriptional regulation of the IL-2 gene, the isolation of Notch regulators is possible. Additional evidence that NKAP functions as a negative regulator of Notch signaling arises from a C. elegans genetic screen to identify genes that modify vulval development (Poulin et al., 2005). Within vulval precursors, signals through Ras and Notch antagonize each other, causing differential cell fates (reviewed in Sundaram, 2004). In general, mutations that decrease Ras signaling or increase Notch signaling result in a ‘multi-vulval’ phenotype. A genome-wide RNAi screen identified novel genes that could synergize with lin-15 to produce a multi-vulval fate (Poulin et al., 2005). One of the genes was the C. elegans homolog of NKAP, E01A2.4. As knockdown of a negative regulator of Notch would be expected to produce a multi-vulval phenotype, the isolation of NKAP in this screen is consistent with a role for NKAP in the Notch corepressor complex. Thus, genetic evidence from Drosophila and C. elegans suggest that NKAP may function as a transcriptional repressor and negative regulator of Notch signaling through the corepressor complex.
NKAP interacts with CIR, a component of the Notch corepressor complex
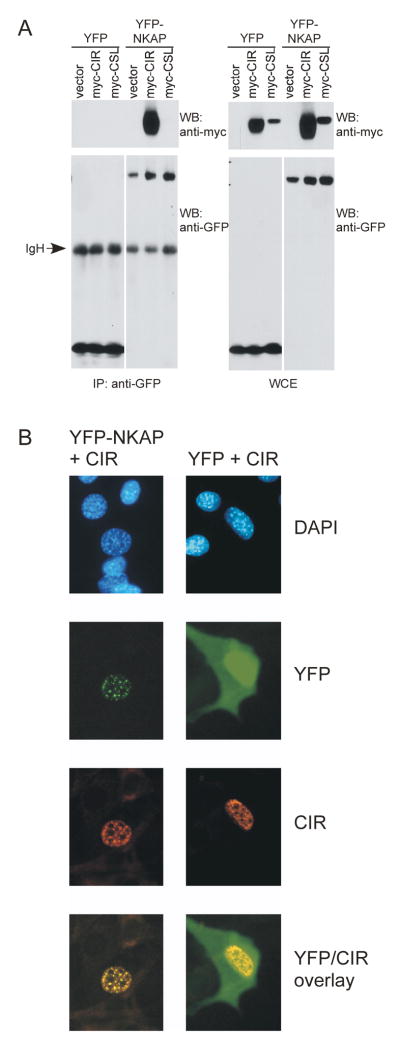
To confirm the Drosophila yeast-two-hybrid results demonstrating that NKAP and CIR associate, 293T cells were transfected with myc-tagged human CIR, YFP-tagged human NKAP or vector controls. As shown in figure 2A, CIR coimmunoprecipitated with NKAP, but not with YFP. We also examined whether NKAP could directly associate with CSL, the transcription factor to which the Notch corepressor associates, but no interaction was found (figure 2A). In their initial description, both NKAP and CIR were found to localize to nuclei in a punctate pattern (Chen et al., 2003; Hsieh et al., 1999). To determine whether NKAP and CIR colocalize, NIH3T3 cells were transfected with YFP or YFP-NKAP expression plasmids, along with myc-tagged CIR, and their subcellular localization was analyzed by fluorescent microscopy. DAPI staining was used to delineate nuclei. As shown in figure 2B, YFP-NKAP and CIR colocalize in a punctate nuclear pattern, while YFP is predominantly cytoplasmic and does not localize with CIR. Similar results were observed in Jurkat T cells (data not shown). Interestingly, ICN and many Notch regulators including CSL, SKIP, SMRT and MAML are also distributed in a punctate nuclear pattern, and CIR has been previously shown to colocalize with CSL (Hsieh et al., 1999). Therefore, these data show that NKAP can associate with CIR and may function as part of the Notch corepressor complex.
Figure 2. Association and colocalization of NKAP with CIR.
(A) 293T cells were transfected with YFP or YFP-NKAP expression plasmids, with either empty vector, myc-CIR or myc-CSL. The next day, cell extracts were generated and subject to immunoprecipitation with anti-GFP. Samples of extracts (WCE) and immunoprecipitates (IP) were analyzed by Western blotting (WB) with either anti-myc to examine CIR and CSL coprecipitation or anti-GFP to assess immunoprecipitation. Secondary antibody cross-reactivity to the immunoglobulin heavy chain (IgH) used in IP is denoted in the figure.
(B) NIH 3T3 cells were transfected with either YFP or YFP-NKAP, in addition to myc-CIR expression plasmids. The next day, cells were fixed, permeabilized and stained with rabbit anti-myc. DAPI was used to delineate nuclei. In the bottom panel, myc and YFP are overlaid.
NKAP associates specifically with HDAC3 via its c-terminal DUF926 domain
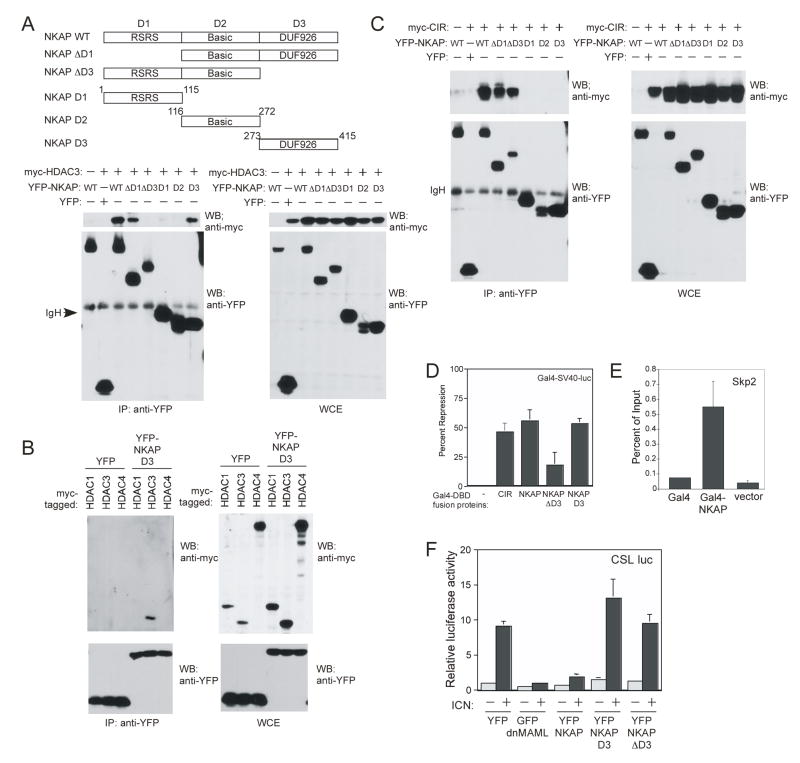
The function of a corepressor complex is to recruit HDACs, resulting in alterations in chromatin structure and inhibition of transcription. If NKAP functions as a transcriptional repressor, it would be expected to associate with HDACs. To test this, YFP-tagged NKAP and myc-tagged HDAC3 were co-transfected into 293T cells. As shown in figure 3A, HDAC3 co-immunoprecipitated with NKAP, but not with YFP. This interaction could be either direct, or mediated indirectly by CIR, which was previously shown to associate with HDAC2 (Zhou et al., 2000). To differentiate between these possibilities, we mapped the sites of interaction of NKAP with HDAC3 and CIR to determine if they associate with distinct regions of NKAP. Since NKAP has no previously characterized domains, the protein was divided approximately into thirds (D1, D2 and D3), as shown schematically in figure 3A. The D1 domain (aa 1-178) contains RSRS (Arg-Ser-Arg-Ser) tetrapeptide repeats, which have been previously shown to be important for nuclear localization to speckles (Hedley et al., 1995). The D2 domain (aa 179-272) is highly basic, containing repetitive sequences. The D3 domain (aa 273-415) contains a “DUF926” (Domain of Unknown Function #926, in the NCBI Conserved Domain Database). The DUF926 domain is shared between NKAP and a hypothetical open reading frame on chromosome 6, c6orf194, but the function of this domain in either protein is unknown. As shown in figure 3A, deletion of the D1 domain (NKAP ΔD1) did not alter the ability of NKAP to associate with HDAC3, however deletion of the D3 domain (NKAP ΔD3) completely abrogated HDAC3 binding. HDAC3 also associated with the D3 domain alone, showing that it is both necessary and sufficient for HDAC3 association. To determine whether this domain could associate with additional HDACs, 293T cells were transfected with YFP NKAP D3 and myc-tagged HDAC1, HDAC3 or HDAC4. As shown in figure 3B, the NKAP D3 domain bound specifically to HDAC3, but not to either HDAC1 or HDAC4. Similar results were found when full-length NKAP was used (data not shown). In addition, no association was found between NKAP D3 and either HDAC2, HDAC5 or HDAC7 (data not shown). Therefore, we have shown that the NKAP DUF926 (D3) domain can mediate an association specifically with HDAC3.
Figure 3. NKAP independently associates with CIR and HDAC3.
(A) Co-immunoprecipitation experiments were performed as in figure 2A, using myc-HDAC3 with full length YFP-NKAP or truncations as denoted in schematic to map the site of NKAP with HDAC3.
(B) Co-immunoprecipitation experiments were performed as in figure 2A, using YFP-tagged NKAP D3 cotransfected into 293T cells with myc-tagged HDAC1, HDAC3 and HDAC4.
(C) Co-immunoprecipitation experiments were performed as in figure 2A, to map the site of NKAP association with myc-tagged CIR.
(D) Gal4DBD-fusion proteins were generated with full-length CIR, full-length NKAP or NKAP ΔD3 and D3 truncations (as outlined in figure 3A). NIH3T3 cells were transfected with 0.1 μg of a Gal3-SV40-luciferase reporter, alone with 0.1 μg of either Gal4DBD alone or Gal4DBD-CIR, Gal4DBD-NKAP, Gal4DBD-NKAP ΔD3 or Gal4DBD-NKAP D3 expression plasmids. Results shown are the average of three independent experiments, shown as percent repression compared to the relative luciferase activity of the reporter transfected with Gal4DBD alone. Error bars show SEM.
(E) Gal4-TK-293T cells were transfected with Gal4DBD, Gal4DBD-NKAP or empty vector, and ChIP were performed using anti-Gal4. Input DNA from each transfection and IP was analyzed for Skp2 promoter genomic sequence. Data shown is from three independent IPs for each sample, and is shown as percent of input recovered in the ChIPs. Error bars show SEM.
(F) NIH3T3 cells were transfected with CSL-luciferase reporter, in the presence or absence of ICN, with either YFP, GFP-dnMAML, YFP-NKAP, YFP NKAP D3 or YPF-NKAPΔD3 expression plasmids. The next day, luciferase activity was quantified. Data from triplicate experiments were internally standardized to the amount of luciferase activity from YFP-transfected alone (=1). Error bars reflect SEM.
NKAP associates independently with CIR and HDAC3
To assess whether NKAP might bind HDAC3 indirectly through CIR, we also mapped the association of NKAP with CIR using the NKAP truncations described above. As shown in figure 3C, deletion of either the D1 domain (NKAP ΔD1) or the D3 domain (NKAP ΔD3) did not alter the ability of NKAP to associate with CIR, implying that CIR associates with NKAP through the D2 domain. Surprisingly, no association of NKAP D2 with CIR was found. This cannot be attributed to altered subcellular localization, as the NKAP D2 domain was found to colocalize to nuclear speckles with CIR, similar to full-length NKAP (data not shown). It may be that in isolation the NKAP D2 domain does not fold into its normal conformation. However, it is clear that the association of NKAP with CIR does not require the DUF926 domain. Since CIR and HDAC3 bind to different portions of NKAP, the association of NKAP with HDAC3 occurs independently of its association with CIR.
NKAP functions as a transcriptional repressor
Since NKAP has been shown to associate with CIR and HDAC3, it may function directly as a transcriptional repressor. This was tested by examining the ability of Gal4DBD-NKAP-fusion proteins to repress transcription from a luciferase reporter containing four copies of a GAL4-binding sequence upstream of the SV40 promoter/enhancer in NIH3T3 cells. As a positive control, Gal4DBD-CIR was utilized (Hsieh et al., 1999), resulting in a 50% repression of reporter activity as compared to Gal4DBD alone (figure 3D). Expression of full-length NKAP fused to the Gal4DBD also resulted in a similar level of transcriptional repression in this system, demonstrating that NKAP also functions as a transcriptional repressor (figure 3D, p<0.01). This repression is dependent on the HDAC3-binding NKAP D3 domain, since NKAP-mediated repression was reduced when the domain was deleted. In addition, the isolated NKAP D3 domain was sufficient for transcriptional repression. Therefore, NKAP, functions as a transcriptional repressor, with the majority of its repressive function mapping to the HDAC3-binding DUF926 domain.
NKAP is part of a DNA-binding complex
If NKAP is transcriptional repressor, it should be found as part of a DNA-binding complex. To examine whether NKAP associates with DNA, chromatin immunoprecipitations (ChIP) were performed. Gal4DBD-NKAP, Gal4DBD or empty vector were transfected into a 293T cell line, in which a Gal4-TK-reporter was stably integrated (Nguyen et al., 2008), and precipitated with anti-Gal4. Gal4DBD and Gal4DBD-NKAP associated with the integrated Gal4 reporter to similar extents (data not shown), demonstrating equivalent precipitation. To examine specific NKAP association with DNA, we used previously characterized chIP primers for the Notch binding site in the human Skp2 promoter (Dohda et al., 2007). As shown in figure 3E, Gal4DBD-NKAP associated with the Skp2 promoter. This was specific to NKAP, since Skp2 ChIP with Gal4DBD alone was similar to empty vector control (Gal4DBD-NKAP was statistically significantly increased as compared to these two controls, p<0.05). Thus, NKAP functions as a transcriptional repressor and is found associated with DNA at a Notch-regulated promoter.
NKAP is a repressor of Notch-mediated transcription
Since CIR is a component of the Notch corepressor complex, we next examined whether NKAP overexpression could inhibit Notch-mediated transcriptional activation. NIH3T3 cells were transfected with constitutively-active intracellular Notch1 (ICN) and either YFP vector, YFP-NKAP or GFP-dnMAML (dominant negative Mastermind-like) with a CSL luciferase reporter. DnMAML is a potent inhibitor of Notch-mediated transcription (Weng et al., 2003). As shown in figure 3F, substantial activation of CSL luciferase was observed upon expression of ICN. As expected, cotransfection of GFP-dnMAML inhibited the effects of ICN. NKAP overexpression similarly inhibited the ICN-mediated transcriptional activation of the CSL luciferase reporter (p<0.001), while having minimal effect on CSL reporter activity in the absence of ICN. In addition, we examined the ability of two NKAP truncations to inhibit Notch-mediated transcription: NKAP D3 which contains the HDAC3 binding domain and is required for transcriptional repression, and NKAP ΔD3 which binds CIR and could recruit NKAP to the Notch corepressor complex. As shown in figure 3F, neither of these truncations repressed ICN-mediated upregulation of the CSL luciferase reporter, demonstrating that neither the HDAC3-binding nor the CIR-binding activities alone are sufficient for NKAP to function as a repressor of Notch-mediated transcription.
Inverse correlation between NKAP expression and Notch target gene expression during T cell development
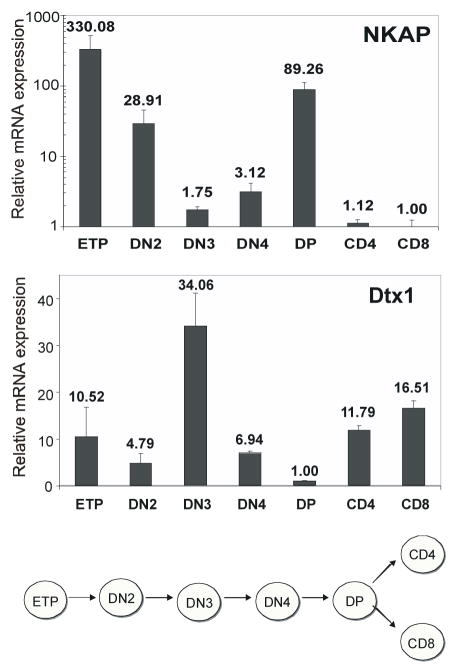
To examine the expression of NKAP during T cell development, we isolated ETP, DN2, DN3, DN4, CD4+CD8+(DP), CD4+ SP, and CD8+ SP T cells by FACS sorting and examined NKAP expression by QPCR. There is a remarkably dynamic modulation of NKAP expression, varying by two orders of magnitude between development stages (figure 4). Interestingly, the sharp decline in NKAP at the DN3 stage correlated with the highest expression levels of three Notch target genes, Deltex-1 (Dtx1), Notch-1 and Notch-3 (Taghon et al., 2006). Therefore, we examined Deltex1 mRNA gene expression. This analysis demonstrated an inverse relationship between NKAP and Deltex1 gene expression throughout T cell development, which is consistent with the model that NKAP functions as a transcriptional repressor and negative regulator of Notch signaling.
Figure 4. NKAP expression during T cell development inversely correlates with the expression of the Notch target gene Deltex1.
Thymocytes at different stages of development were isolated by FACS sorting. mRNA was generated and examined by quantitative PCR for NKAP and Dtx1 mRNA expression. The level of mRNA is normalized relative to the sorted population with the lowest expression of either NKAP or Dtx1 (=1). The average mRNA level from triplicate Q-PCRs is indicated above the bar. Relative NKAP mRNA levels are shown on a logarithmic scale, while Dtx1 is shown on a linear scale.
NKAP-deficiency blocks αβ T cell development at the DN3 stage
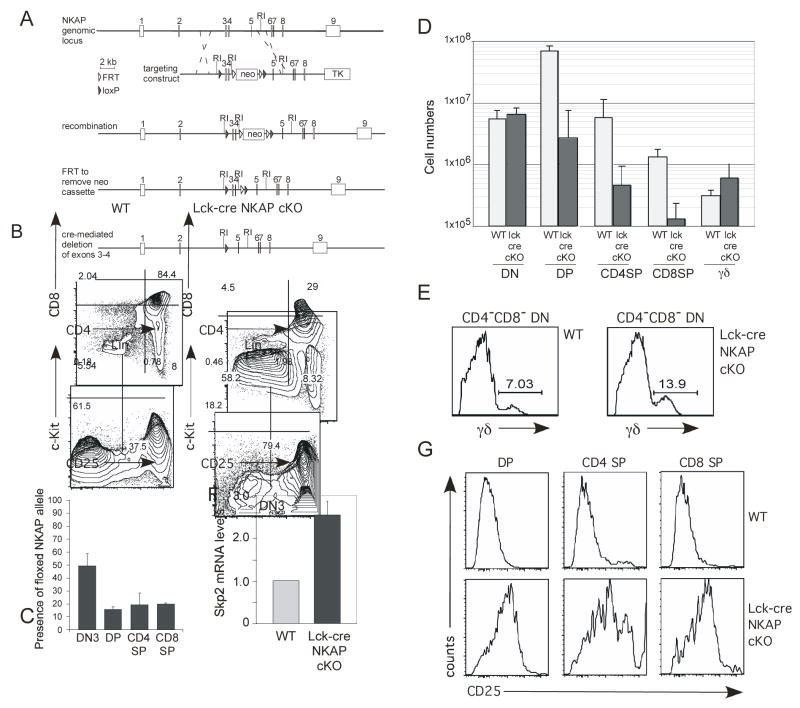
Since many mice deficient in Notch or its regulators are embryonic lethal (reviewed in Maillard et al., 2005), we designed a strategy for the generation of conditional NKAP knockout mice to examine the role of NKAP in T cell development (figure 5A). Exons three and four were floxed using recombineering (Liu et al., 2003), so that cre-mediated deletion would produce a frame shift resulting in deletion of both the CIR and HDAC3 binding sites. The floxed NKAP mice were bred to Lck-cre transgenic mice (Lee et al., 2001), which initiate deletion as early as the DN2 stage of T cell development. Lck-cre NKAP cKO mice had greatly reduced thymic cellularity; while wild-type mice had on average 83.0 × 106 ± 16.8 × 106 thymocytes, Lck-cre NKAP cKO mice had almost 10-fold fewer thymocytes, 9.87 × 106 ± 4.48 × 106. As shown in figure 5B, the majority of Lck-cre NKAP cKO thymocytes are within the CD4/CD8 DN gate, with a disproportionate increase in the DN3 (lin−CD44−CD25+) population, indicative of a block in T cell development at that stage. The incomplete block at the DN3 stage could be due to incomplete deletion of NKAP. To test this, PCR primers were generated that flank the upstream loxP site in the NKAP locus; upon cre-mediated deletion, one of the primer binding sites would be lost, allowing for direct quantification of deletion efficiency by QPCR using (cre-negative) NKAP floxed mice for normalization. DN3, DP and SP thymocytes were isolated from Lck-cre NKAP cKO mice, and analyzed for deletion efficiency. At the DN3 stage, the deletion of NKAP was inefficient, with approximately 50% deletion observed (figure 5C). DP and SP thymocytes exhibited 85–90% deletion, indicating that the majority of thymocytes which do develop past the DN3 stage in the Lck-cre NKAP cKO have not escaped deletion altogether, although it remains possible that loss of NKAP protein expression did not occur until after the requirement for NKAP at the DN3 to DP transition. Although αβ T cell development was disrupted, γδ T cell development was not altered in the absence of NKAP (figure 5D and E). Quantification of the various thymocyte populations (figure 5D) demonstrates that the absolute number of cells in the DN and γδ T cell populations are similar between wild-type and Lck-cre NKAP cKO mice, but that the number of DP, CD4 SP and CD8 SP thymocytes were severely decreased in the absence of NKAP (p<0.01 for DP and SP thymocytes). In particular, the absolute number of DP thymocytes decreased approximately 25-fold. Therefore, in the absence of NKAP, there is a block in αβ T cell development at the DN3 to DP transition, although γδ T cells develop in normal numbers.
Figure 5. NKAP is required for T cell development.
(A) Schematic of the targeting construct used to generated floxed NKAP mice
(B, E) Thymocytes from lck-cre NKAP cKO and a wild-type control mouse was examined for T cell populations by flow cytometry as denoted in the figure.
(C) Genomic DNA from sorted wt and lck-cre NKAP cKO thymocyte populations were examined by Q-PCR for the presence of the floxed NKAP allele, using PCR primers which flank the upstream loxP site. Within each population, signal from lck-cre NKAP cKO populations were normalized to floxed, cre-negative wildtype populations. Error bars reflect SEM from triplicate samples.
(D) Absolute numbers of total DN, DP, CD4 SP, and CD8 SP cells from wild-type and lck-cre NKAP cKO mice were calculated. Results shown are the average absolute numbers, from between 3–5 lck-cre NKAP cKO and 3–5 wild-type mice, at 8–10 weeks of age. Error bars reflect SEM.
(F) DN3 thymocytes from wild-type and lck-cre NKAP cKO mice were isolated by FACS sorting. mRNA was generated and examined by Q-PCR for Skp2 mRNA expression, and normalized relative to wt Skp2 expression (=1). The data shown is from two wild-type and two cKO mice, each from independent sorts and independent Q-PCR. Error bars reflect SEM.
(G) Thymocyte populations from wild-type and cKO mice were analyzed for expression of CD25 as denoted in the figure.
Loss of NKAP repression leads to increases in Notch target gene expression
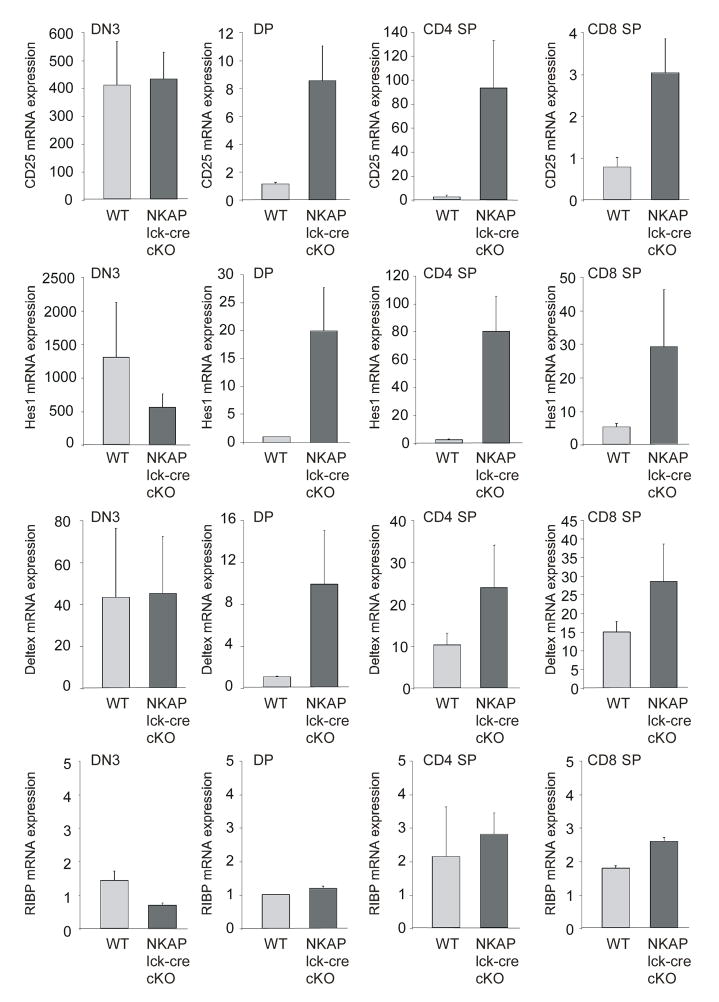
Based upon our initial biochemical characterization of NKAP as a transcriptional repressor that inhibits Notch-mediated transcription, it was predicted that Notch target gene expression will be increased upon NKAP deletion. NKAP was found to associate with the Skp2 promoter via ChIP (figure 3E). As shown in figure 5F, Skp2 mRNA QPCR demonstrates that Lck-cre NKAP cKO have significantly higher levels of Skp2mRNA than wt (p<0.01). In addition, DN4 cells from NKAP Lck-cre cKO mice had higher levels of expression of CD25, which is positively regulated by Notch activation (figure 5B). DP, CD4 SP and CD8 SP thymocytes from NKAP Lck-cre cKO mice also exhibited higher levels of CD25 expression (figure 5G), indicating that in the absence of NKAP, the mRNA expression of additional Notch target genes may be upregulated. To examine this, DN3, DP, CD4 SP and CD8 SP were sorted, and examined by QPCR for the expression of the Notch target genes CD25, Hes1 and Deltex1. As shown in figure 6, in DP thymocytes, the expression of CD25 increased 8-fold in the absence of NKAP, the expression of Deltex1 increased 10-fold and the expression of Hes1 increased 20-fold (all p<0.01). Increases in Notch target gene expression were also observed in the CD4 SP and CD8 SP populations as well (all but Dtx1 expression in CD8 SP T cells were statistically significant with at least p<0.01). RIBP expression, which remains constant throughout T cell development was examined as a control (Perchonock et al., 2007). Deletion of NKAP did not alter RIBP expression in DP, CD4 SP or CD8 SP populations, demonstrating that loss of NKAP does not generally increase transcription. The lack of effect on DN3 for Hes1, Deltex1 and CD25 is likely due to the combination of maximal expression of these genes at this stage of T cell development, coupled with low NKAP expression and incomplete deletion. Thus, NKAP functions as a transcriptional repressor in vivo, as demonstrated by the potentiation of Notch target gene expression upon NKAP deletion.
Figure 6. Potentiation of Notch target gene expression in the absence of NKAP.
DN3, DP, CD4 SP and CD8 SP thymocytes from wild-type and lck-cre NKAP cKO mice were isolated by FACS sorting. mRNA was generated and examined by quantitative PCR for CD25, Hes1, Deltex1 and RIBP mRNA expression levels, and normalized relative to the sorted wild-type population with the lowest expression of each gene (=1). The data shown is from two wild-type and three lck-cre NKAP cKO mice, each from independent sorts and independent Q-PCR. Error bars reflect SEM.
The block at the DN3 to DP transition is not due to loss of pre-TCR signaling
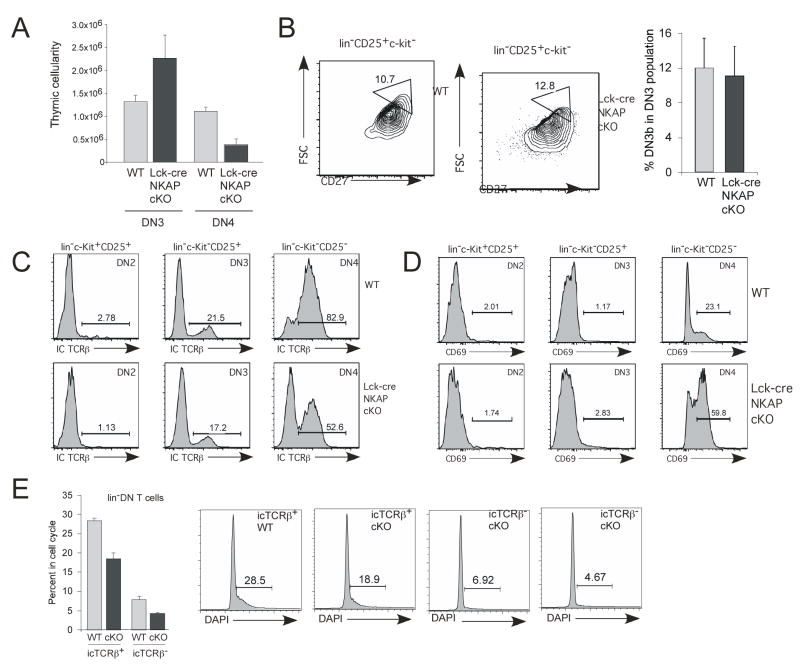
To further characterize the point at which NKAP deletion leads to T cell loss, the absolute number of DN3 (lin−CD44−CD25+) and DN4 (lin−CD44−CD25−) T cells were analyzed (figure 7A). Interestingly, Lck-cre NKAP cKO mice have approximately two-fold greater absolute number of DN3 T cells (p<0.1), and two-fold fewer DN4 T cells as compared to (Lck-cre+) littermate controls (p<0.01), indicating that the block initiates as cells transit from DN3 to DN4. Since Notch signaling is required at the DN3/β-selection checkpoint, it was surprising that NKAP deletion in lck-cre NKAP cKO mice, which potentiates Notch signaling, also resulted in a DN3 block. Transition through the β-selection checkpoint also depends on pre-TCR signaling, and DN3 T cells can be separated into pre-selection DN3a and post-selection DN3b cells based upon forward scatter and upregulation of CD27 (Taghon et al., 2006). As shown in figure 7B, no defect was observed in the proportion of DN3a and DN3b cells in the absence of NKAP, implying that pre-TCR signaling is intact. Pre-TCR signaling also leads to the upregulation of intracellular TCRβ expression, as well as expression of the activation marker CD69. As shown in figure 7C, similar levels of intracellular TCRβ chain was expressed in DN3 cells from wt and Lck-cre NKAP cKO mice. While Lck-cre NKAP cKO DN4 cells did upregulate intracellular TCRβ expression, the percentage is lower than that observed in wt DN4 cells (figure 7C). Further, Lck-cre NKAP cKO DN4 cells have a greater proportion of cells expressing CD69 compared to wt (figure 7D). Examination of cell cycle status by DAPI demonstrated that there is a partial defect in proliferation in DN thymocytes from Lck-cre NKAP cKO mice as compared to wt (p<0.01, figure 7E). Therefore, the block at the DN3 to DP transition upon loss of NKAP is not due to abrogation of pre-TCR signaling, but rather may involve the interpretation of the pre-TCR signals, since differences are observed in proliferation and in the expression of intracellular TCRβ and CD69 in the DN4 population.
Figure 7. pre-TCR signaling is intact in Lck-cre NKAP cKO mice.
(A) Absolute numbers of total DN3 (lin−CD25+c-kit−, or CD4−CD8−CD25+CD44−) and DN4 (lin−CD25+c-kit−) cells from five wild-type and three cKO mice were calculated, as in figure 5D above. Error bars reflect SEM.
(B) Flow cytometry was performed to analyze the percent of cells in DN3a (lin−CD25+c-kit−CD27medFSCmed) and DN3b populations (triangular gate, lin−CD25+c-kit−CD27hiFSChi). Shown is a representative FACS plot to show gating strategy, with average DN3b percentage calculated from three wt and four cKO mice also shown. Error bars reflect SEM.
(C, D) FACS was performed to analyze expression of intracellular TCRβ (IC TCRβ) and CD69 expression on the cell surface of DN2, DN3 and DN4 cells. Shown is representative data from two independent experiments.
(E) FACS was performed to analyze cell cycle status of IC TCRβ+ and IC TCRβ− lin−DN thymocytes. Shown is a representative FACS plot to indicate gating strategy, with average % cell cycle (S+G2M) calculated from analysis of three wt and three cKO mice. Error bars reflect SEM.
Discussion
Lck-cre NKAP cKO mice have a block at the DN3 to DP transition
We have identified a novel transcriptional repressor, NKAP, that is required for T cell development. NKAP overexpression inhibits Notch-mediated transcriptional activation. NKAP associates with the Skp2 promoter as demonstrated by chromatin immunoprecipitation, and loss of NKAP in lck-cre cKO mice leads to increased expression of Skp2, as well as the Notch target genes CD25, Deltex1 and Hes1. Lck-cre NKAP cKO mice have a block at the DN3 to DP transition, although this block is not due to a failure of either Notch or pre-TCR to function, as demonstrated by potentiation of Notch target gene expression (figure 6) and a normal transition from pre-β-selection DN3a to post-β-selection DN3b (figure 7B). However, it has been previously shown that this stage in T cell development requires the function of transcriptional repressors and chromatin modifiers. DN3 T cells that lack either of two generic components of corepressor complexes, NCoR or mSin3A, or the chromatin remodelers BRG1 or Mi-2β, have blocks at this stage of development (Cowley et al., 2005; Gebuhr et al., 2003; Jepsen et al., 2000; Williams et al., 2004), implying that epigenetic alterations play a critical role in creating a permissive environment for the transition to DP T cells. Although the developmental block is characterized in all four of these knockouts, the specific dysregulated gene(s) responsible for the phenotype has not yet been identified in any of these mice, and therefore we cannot determine whether a similar mechanism is responsible for the block in T cell development in the lck-cre NKAP cKO mice. However, although Notch signaling is required for T cell to progress past the DN3/β-selection checkpoint, there is one report that high levels of Notch signaling can also block T cell development (Michie et al., 2007). Rag2-deficient DN3 cells transduced with an ICN1-expressing retrovirus that co-expresses GFP from an internal IRES site develop into DP T cells, but only when low/medium levels of GFP/ICN1 are expressed. In contrast, thymocytes expressing high levels of GFP/ICN1 are developmentally arrested and do not develop into DP thymocytes (Michie et al., 2007), indicating that potentiation of Notch signaling could be in part responsible for the defect in lck-cre NKAP cKO mice. NKAP could also regulate genes outside of the Notch pathway that contribute to the developmental block. Current work is focused on identifying the cause of the block in T cell development in lck-cre NKAP cKO mice.
Non-redundant roles for NKAP and MINT in T cell development – implication for more than a single Notch corepressor complex
Our data demonstrate that NKAP is a novel component of the Notch corepressor complex, and that loss of NKAP leads to the potentiation of Notch target gene expression. Interestingly, mice deficient in a different component of the Notch corepressor complex, MINT (Msx2-interacting nuclear target protein, also known as SHARP) have a different phenotype (Kuroda et al., 2003; Oswald et al., 2002). MINT was originally identified as a Notch repressor, since overexpression of MINT inhibited Notch-mediated transcription, and MINT was shown to compete with Notch for binding to CSL (Kuroda et al., 2003; Oswald et al., 2002). However, loss of MINT has a limited effect on T cell development, with minimal alteration on Hes1 and Deltex1 expression (Kuroda et al., 2003; Tsuji et al., 2007). In contrast, loss of NKAP increased Hes1 and Deltex1 expression by 10- to 20-fold while decreasing the absolute number of DP T cells by 25-fold. MINT expression was unchanged in the lck-cre NKAP cKO mice (data not shown), indicating that MINT cannot compensate for loss of NKAP in developing T cells. While the effect on T cell development is limited, MINT-deficiency in B cells leads to an increase in marginal zone B cells (Kuroda et al., 2003). Marginal zone B cell development requires Notch2 signaling (Saito et al., 2003), thus the effect of MINT-deficiency in lymphocytes is consistent with enhanced Notch signaling. One possible explanation for the different phenotypes between MINT-deficient and Lck-cre NKAP cKO mice may be that these two proteins are components of separate and distinct Notch corepressor complexes. The enhancement in marginal zone B cells with minimal effect on T cell development caused by MINT deficiency is consistent with it being a specific negative regulator of Notch2. Similarly, the strong potentiation of Notch target gene expression during T cell development caused by NKAP-deficiency is consistent with it being a negative regulator of Notch1 signaling. The effect of NKAP-deficiency on B cell development in general, and marginal zone B cell development in particular, is being investigated.
Potential for NKAP-deficiency to lead to leukemic transformation
Activating mutations in Notch-1 are a primary factor in the development of T-cell acute lymphoblastic leukemia (T-ALL). Initially, a role for Notch-1 was identified in TALL tumor cells in which a translocation resulted in the generation of a truncated, constitutively intracellular Notch (Ellisen et al., 1991). Subsequently, other activating Notch-1 point mutants were isolated in over 50% of human primary T-ALL (Weng et al., 2004). The importance of these Notch mutations was demonstrated by treatment of these T-ALL with a γ-secretase inhibitor to block one of the proteolytic cleavage events that produces transcriptionally active ICN, resulting in growth suppression (Weng et al., 2004). We have shown that Notch target genes are potentiated in Lck-cre NKAP cKO mice, including Skp2, and that NKAP associates with the Skp2 promoter by ChIP. Skp2, an F-box containing protein which is part of the SCF E3 ubiquitin-ligase complex, is commonly overexpressed in many human cancers, including leukemias and lymphomas where Skp2 overexpression correlates with a poorer prognosis (reviewed in Hershko, 2008). Therefore, lck-cre NKAP cKO mice, may develop leukemias with age, as a result of upregulation of Notch target genes including Skp2, which is being examined.
Materials and Methods
Retroviral library transduction and selection
A human leukocyte cDNA retroviral library was purchased from BD Biosciences (Cat# HL8007BB) and retroviruses were generated according to the manufacturer’s instructions. Ecotropic-receptor expressing J.REM 474 Jurkat mutant cell lines were tranduced by spinfection for two hours in the presence of 8 μg/ml polybrene. Stimulations, FACS sorting and clonal cell isolation were performed as in (Greene and Shapiro, 2003). The NKAP cDNA retroviral insert was identified by amplication of LI474-59 genomic DNA using pLIB primers (BD Clontech).
Flow cytometry
Analysis of T cell development and the isolation of thymic T cell subsets were performed as previously described (Perchonock et al., 2007).
Immunofluorescence Microscopy
Transfected NIH3T3 cells were cultured on glass coverslips. Cells were then fixed, permeabilized and incubated with rabbit anti-myc (Cell Signaling Technologies), rinsed then incubated with AlexaFluor594 Goat anti-Rabbit IgG at 1:1000 dilution. Slides were mounted using prolong Gold reagent with DAPI (Molecular Probes). Microscopy was performed using a Leica DM IRB/E microscope, and analyzed using OpenLab.
Plasmids and Recombinant DNA
Epitope tagged HDACs, Gal4-DBD, and Gal4-luciferase constructs were a gift of Dr. Mitchell Lazar. MIRR (dsRED MIGR), dnMAML, and ICN constructs were a gift of Dr. Warren Pear. CSL-luciferase construct was a gift of Dr. Tom Kadesch. Full-length human NKAP and full-length human CIR cDNA were obtained from OpenBiosystems. NKAP truncation mutants D1 (aa 1-178), D2 (aa 179-272), D3 (aa 273-415), ΔD1 (aa 178-415) and ΔD3 (aa 1-272) were generated by PCR amplification which also introduced restriction sites for subcloning into pEYFPC.1, pEF6 myc/his, and pCMX-Gal.
Reporter assays
Transient transfections in Jurkat, 293T, and NIH-3T3 cells were performed by cotransfecting expression plasmids and SV40-Gal4-Luc for Gal4 repression assays or CSL-Luc for Notch reporter assays. Jurkat cells were transfected via electroporation and stimulated as previously described (Greene and Shapiro, 2003), while NIH-3T3 and 293T cells transfections were carried out using Fugene6 (Roche) according to the manufacturer’s instructions.
Real-Time quantitative PCR analysis
DNA or mRNA was isolated from sorted T cell populations. cDNA generated with Superscript II was amplified and detected using TaqMan probes (Applied Biosystems) for NKAP, Skp2, Hes1, CD25, RIBP and Deltex1, as well as 18S rRNA as an internal control. Primers to detect NKAP deletion were designed flanking the upstream loxP site, and standardized using either primers that amplified genomic GAPDH or Deltex1 (sequence available upon request). An ABI RT-PCR System was used, and error was calcucated via the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Transient transfection and immunoprecipitation assays
293T cells were transfected with Fugene6 reagent (Roche Applied Science) according to the manufacturer’s protocol. Cells were lysed in 1 ml lysis buffer consisting of PBS supplemented with 2 mM EDTA, 50 mM NaF, 1% NP40 detergent, phosphatase and protease inhibitor cocktails (Sigma), 0.2% SDS and 0.5% sodium deoxycholate. Lysates were incubated on ice for 15′, and clarified in a microcentrifuge prior to adding mouse anti-GFP antibody for IP (Roche Applied Science or Invitrogen). The anti-myc and anti-GFP antibodies used in Western blotting were from Cell Signaling Technologies.
Chromatin IPs
293T cells were transfected as described above with a plasmid encoding full length NKAP fused to the GAL4 DNA binding domain or a plasmid encoding the isolated GAL4-DBD or an empty vector. ChIPs were performed using the EZ Chip kit (Millipore) according to the manufacturer’s protocol, using 2 μg of anti-Gal4 (RK5C1, Santa Cruz Biotechnology). Precipitates were analyzed by QPCR using primers flanking the Notch binding site in the human Skp2 promoter (Dohda et al., 2007). QPCR was also performed using input DNA so that the percent recovery of the input target sequence by ChIP could be calculated.
Generation of floxed NKAP mice
A targeting vector was designed using recombineering (Liu et al., 2003), to flox exons 3 and 4 of NKAP (figure 5A). The construct was electroporated into R1 ES cells (a gift of Reka Nagy, Andras Nagy, Janet Rossant and Wanda Abramow-Newerly). After selection with G418 and gancyclovir, DNA from resistant clones was digested with EcoRV and screened by Southern blotting. C57Bl/6 blastocysts were injected with a targeted clone. Chimeric males were bred to FLP1-expressing females to generate mice carrying a floxed NKAP allele, but lacking the Frt’d neomycin expression cassette. Female progeny, which carry one wild-type and one floxed NKAP allele, were bred to lck-cre transgenic male mice (Jackson laboratories, Lee et al., 2001). Since NKAP is expressed on the X chromosome, only male progeny could be lck-cre NKAP conditional knockouts and were examined. Lck-cre NKAP cKO mice, and either littermates or age-matched controls, were examined at approximately 8 weeks of age. All animal work was performed with the approval of the IACUC committees at the University of Pennsylvania and the Mayo Clinic, in accordance with their guidelines.
Supplementary Material
Acknowledgments
We thank Warren Pear, Ivan Mailliard, Tom Kadesch, Avinash Bhandoola, David Schultz and Mitch Lazar for reagents and thoughtful discussions. We thank members of the Bhandoola and Allman laboratories for flow cytometry assistance, and Jean Richa for ES cell injection. This work was supported by an Arthritis Foundation Investigator Award and an NIH R21 grant to V.S.S.
Abbreviations
- CSL
CBF1/Su(H)/Lag-1
- CIR
CBF1-interacting corepressor
- ICN
intracellular Notch
- dnMAML
dominant negative Mastermind-like
- Gal4DBD
Gal4-DNA binding domain
- Dtx1
Deltex1
- HDAC
histone deacetylase
- HAT
histone acetyltransfease
Footnotes
The authors state that they have no financial conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham RT, Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat Rev Immunol. 2004;4:301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- Chen D, Li Z, Yang Q, Zhang J, Zhai Z, Shu HB. Identification of a nuclear protein that promotes NF-κB activation. Biochem Biophys Res Commun. 2003;310:720–724. doi: 10.1016/j.bbrc.2003.09.074. [DOI] [PubMed] [Google Scholar]
- Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohda T, Maljukova A, Liu L, Heyman M, Grander D, Brodin D, Sangfelt O, Lendahl U. Notch signaling induces Skp2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res. 2007;313:3141–3142. doi: 10.1016/j.yexcr.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, the catalytic subunit of mammalian chromatin-remoding complexes, in T cell development. J Exp Med. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brower C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Greene TA, Shapiro VS. Genetic analysis of CD28 signaling. Immunol Res. 2003;27:513–520. doi: 10.1385/IR:27:2-3:513. [DOI] [PubMed] [Google Scholar]
- Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–1425. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a copressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, et al. Combinatorial roles for the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Kadesch T. Notch signaling: the demise of elegant simplicity. Curr Op Gen Dev. 2004;14:506–512. doi: 10.1016/j.gde.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, survival and function. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation and function by the Notch pathway. Ann Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland JM, Keeshan K, Shestove Xu, Bhandoola LA, Pear WS. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie AM, Chan AC, Ciofani M, Carleton M, Lefebvre JM, He Y, Allman DM, Wiest DL, Zuniga-Pflucker JC, Izon DJ. Constitutive Notch signaling promotes CD4−CD8− thymocyte differentiation in the absence of the pre-TCR complex, by mimicking pre-TCR signals. Int Immunol. 2007;19:1421–1430. doi: 10.1093/intimm/dxm113. [DOI] [PubMed] [Google Scholar]
- Nguyen J, Yamada D, Schultz DC, Defossez PA. Assessment of sera for chromatin-immunoprecipitation. Biotechniques. 2008;44:66–68. doi: 10.2144/000112681. [DOI] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. SHARP is a novel component of the Notch/RBP-Jκ signaling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchonock CE, Pajerowski AG, Nguyen C, Shapiro MJ, Shapiro VS. The related adaptors ALX and RIBP have nonredundant function in lymphocytes. J Immunol. 2007;179:1768–1775. doi: 10.4049/jimmunol.179.3.1768. [DOI] [PubMed] [Google Scholar]
- Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 2005;24:2613–2623. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- Shapiro MJ, Chen YY, Shapiro VS. The carboxyl-terminal segment of the adaptor protein ALX directs its nuclear export during T cell activation. J Biol Chem. 2005;280:38242–38246. doi: 10.1074/jbc.M507441200. [DOI] [PubMed] [Google Scholar]
- Shapiro VS, Mollenauer MN, Greene WC, Weiss A. c-Rel Regulation of IL-2 Gene Expression may be Mediated through Activation of AP-1. J Exp Med. 1996;184:1663–1670. doi: 10.1084/jem.184.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro VS, Truitt KE, Imboden JB, Weiss A. CD28 mediates transcriptional upregualtion of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram MV. Vulval development: the battle between Ras and Notch. Curr Biol. 2004;14:R311–313. doi: 10.1016/j.cub.2004.03.052. [DOI] [PubMed] [Google Scholar]
- Taghon T, Rothenberg EV. Molecular mechanisms that control mouse and human TCR-αβ and TCR-γδ T cell development. Semin Immunopathol. 2008;30:383–398. doi: 10.1007/s00281-008-0134-3. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Development and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear taret protein (MINT) deficiency reveals negative regulation of early thymocyte differentiation in Notch/RBP-J signaling. Proc Natl Acad Sci USA. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early alpha/beta lineage thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Hayward SD. A role for SKIP is EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74:1939–1947. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.