Abstract
Exposure to pollutant particles increased respiratory morbidity and mortality. The alveolar macrophages (AMs) are one cell type in the lung directly exposed to particles. Upon contact with particles, AMs are activated and produce reactive oxygen species, but the scope of this oxidative stress response remains poorly defined. In this study, we determined the gene expression profile in human AMs exposed to particles, and sought to characterize the global response of pro- and antioxidant genes. We exposed AMs obtained by bronchoscopy from normal individuals to Chapel Hill particulate matter of 2.5-μm diameter or smaller (PM2.5; 1 μg/ml) or vehicle for 4 hours (n = 6 independent samples). mRNAs were extracted, amplified, and hybridized to Agilent human 1A microarray. Significant genes were identified by significance analysis of microarrays (false discovery rate, 10%; P ≤ 0.05) and mapped with Gene Ontology in the Database for Annotation, Visualization, and Integrated Discovery. We found 34 and 41 up- and down-regulated genes, respectively; 22 genes (∼30%) were involved in metal binding, and 11 were linked to oxidative stress, including up-regulation of five metallothionein (MT)-1 isoforms. Exogenous MT1 attenuated PM2.5-induced H2O2 release. PM2.5 premixed with MT1 stimulated less H2O2 release. Knockdown of MT1F gene increased PM2.5-induced H2O2 release. PM2.5 at 1 μg/ml did not increase H2O2 release. Mount St. Helens PM2.5 and acid-extracted Chapel Hill PM2.5, both poor in metals, did not induce MT1F or H2O2 release. Our results show that PM2.5 induced a gene expression profile prevalent with genes related to metal binding and oxidative stress in human AMs, independent of oxidative stress. Metals associated with PM may play an important role in particle-induced gene changes.
Keywords: particulate matter, air pollution, microarray, metallothionein
Exposure to particulate matter (PM) is consistently associated with increased morbidity and mortality, attributable, in part, to respiratory illnesses (1, 2). These adverse effects include increased hospital admissions, asthma attacks, pulmonary infection, and mortality (3, 4). The mechanisms by which PM increases morbidity and mortality are not entirely clear, but pulmonary oxidative stress and inflammation induced by PM appear to play an important role (5–11).
The alveolar macrophage (AM) is one major cell type in the lung constantly exposed to ambient pollutants. Upon contact with environmental particulate pollutants, AMs are activated, and produce a large quantity of reactive oxygen species (ROS) from various enzymatic sources (5, 12–16). PM exposure may also increase or decrease antioxidant defense mechanisms in the lung, which further modulates oxidative stress and enhances pulmonary and systemic inflammation (13, 14, 17–25). The scope of this pro- and antioxidant response, however, remains poorly defined, in part because of the multiple enzymatic sources of ROS, and the numerous antioxidant enzymes that can be affected by PM.
To better understand how this complex network of pro- and antioxidant enzymes may be affected by PM, we examined the gene expression profile of human AM exposed to Chapel Hill fine particles of 2.5-μm diameter or smaller (PM2.5). The gene expression analysis highlighted the role of genes involved in metal binding and oxidative stress. The functional role of metallothionein (MT)-1 in response to PM was verified by RNA interference.
MATERIALS AND METHODS
Materials
All chemicals were purchased from Sigma, Inc. (St. Louis, MO) unless otherwise stated.
Particle Collection and Extraction
Particles used in this study were collected in April of 2002 in Chapel Hill, North Carolina, outside the U.S. Environmental Protection Agency Human Studies Facility using a ChemVol Model 2,400 high-volume cascade impactor (Rupprecht and Patashnick Co., Albany, NY) (5). PM2.5 on polyurethane foam (McMaster-Carr, Atlanta, GA), which were previously cleaned with methanol and water, and dried under sterile conditions, were retrieved weekly. The foam or filter was pre-etted with a small amount of 70% ethanol, and the endotoxin-free water was added to a total volume of 40 ml. The particles were removed from the foam or filter by sonication for 1 hour in a water bath (FS220, Fisher Scientific, Atlanta, GA). The foam was removed, and particles were then lyophilized. The metal constituents of PM2.5 of April 2002 were: Al, 930.0 ng/mg; As, 44.7 ng/mg; Cr, 10.2 ng/mg; Cu, 77.7 ng/mg; Fe, 1,713.0 ng/mg; Pb, 133.2 ng/mg; Ni, 26.7 ng/mg; Se, 70.8 ng/mg; Si, 729.0 ng/mg; Ti, 21.0 ng/mg; V, 66.0 ng/mg; and Zn, 522 (5).
Isolation of Human AMs and Bronchial Epithelial Cells
Human AMs and bronchial epithelial cells (BECs) were obtained by bronchoalveolar lavage and bronchial brushings from normal individuals according to procedures described previously (26, 27). Subjects were informed of the procedures and potential risks, and each signed an informed consent. The protocol was approved by the University of North Carolina School of Medicine Committee on Protection of the Rights of Human Subjects.
Procedures for human AM isolation and culture have been described previously (5, 13). Bronchoalveolar lavage samples were put on ice immediately and centrifuged at 300 × g for 10 minutes at 4°C. The lavaged cells were washed once with ice-cold RPMI 1640 medium with 20 mg/ml gentamicin (Life Technologies, Rockville, MD). Cell counts were performed with a hemacytometer. Cytocentrifuge slides were prepared and stained with Diff Quick (Leukostat Solution; Fisher Scientific) to check for AM purity. The cell preparation consisted of 85–95% AMs. The viability of AMs was determined by trypan blue exclusion. Viability exceeded 85% in all samples. AMs were suspended in 1 ml of RPMI 1,640 supplemented with 2.5% FBS (Life Technologies, Gaithersburg, MD) in 5-ml polypropylene tubes. The suspended cells were then incubated with vehicle or PM2.5.
Procedures for human BEC isolation and culture have been described previously (5). BECs adherent to the brushes were treated for 15 minutes at room temperature with 1 mM DTT in BEC growth medium (BEGM; Clonetics, San Diego CA) supplemented with bovine pituitary extract, 5 μg/ml insulin, 0.5 μg/ml hydrocortisone, 50 μg/ml gentamicin, 0.1 ng/ml retinoic acid, 10 μg/ml transferrin, 6.5 ng/ml triiodothyrodine, 0.5 μg/ml epinephrine, and 0.5 ng/ml human epidermal growth factor, and dislodged from the brushes by pipeting. The cells were then plated at a density of 1 × 105 viable cells/cm2 in six-well cluster plates (surface area, 9.6 cm2) that were precoated with 50 mg/ml human placental collagen type IV and used after passage 2 or 3.
Exposure to Chapel Hill PM2.5 and RNA Isolation
A suspension of AMs (1 × 106 cells) were incubated with control vehicle or Chapel Hill PM2.5 (1 μg/ml) for 4 hours in a static condition. At the end of the 4-hour incubation, total cellular RNA was extracted from AMs with Trizol reagent (GIBCO BRL/Life Technologies, Gaithersburg, MD), and further purified with phenol/chloroform. The RNA was amplified with a fluorescent linear amplification kit (no. 5184-3523; Agilent Technologies, Inc., Palo Alto, CA) according to the manufacturer's recommended procedures. The RNA integrity was assessed with an Agilent 2,100 bioanalyzer (Agilent Technologies, Inc.). The 260 nm:280 nm ratios for all RNAs were greater than 2.0.
Hybridization of RNA
Total RNAs (250 ng) were labeled using a Low Input Linear Amplification kit (Agilent Technologies) according to the manufacturer's protocol. Briefly, first- and second-strand cDNA synthesis was immediately followed by an in vitro transcription reaction, during which Cy3- or Cy5-UTP were incorporated. Labeled cRNA was purified on RNAEasy columns (Qiagen, Valencia, CA) and quantified using NanoDrop ND-1,000 UV-Vis spectrophotometer; 1 μg each of Cy3- and Cy5-cRNA were combined, defragmented, and hybridized on Agilent's In Situ Oligo Human 1A version 1 arrays per Agilent's protocol. Hybridizations were performed overnight (16–18 h) at 60°C in a rotisserie oven. Slides were washed using Agilent's sodium chloride–sodium citrate wash buffers and procedures, and scanned on an Axon 4000B confocal scanner (Molecular Devices, Sunnyvale, CA).
Microarray Analysis
The data discussed in this article have been deposited in the Gene Expression Omnibus (accession no. GSE10394; http://www.ncbi.nlm.nih.gov/geo/). All analyses were performed using TM4 software suite (28). Intensities were extracted from raw tiff images using TIGR Spotfinder. Cy3 and Cy5 intensities were lowess normalized for each individual hybridization using TIGR MIDAS. Dye flip replicas were filtered, merged to produce a single expression ratio measure for each gene, and log–base 2 transformed. Differentially expressed genes were identified using significance analysis of microarrays with 100 permutations (29). FDR was set at 10%.
Functional Classification
We also mapped the significant genes with Gene Ontology in the Database for Annotation, Visualization, and Integrated Discovery (DAVID Bioinformatic Resources 2007, National Institute of Allergy and Infectious Disease, http://apps1.niaid.nih.gov/david/) (30).
Quantitative PCR
Quantitative PCR (Q-PCR) was performed for selected genes. cDNAs were synthesized from 0.4 μg of total RNA in 100 μl of buffer containing 5 μM random hexaoligonucleotide primers (Pharmacia, Piscataway, NJ), 10 U/μl Moloney murine leukemia virus reverse transcriptase (GIBCO BRL/Life Technologies), 1 U/μl RNase inhibitor (RNasin; Promega, Madison, WI), 0.5 mM deoxynucleotide triphosphate solution (Pharmacia), 50 mM KCl, 3 mM MgCl2, and 10 mM Tris-HCl (pH 9.3) for 1 hour at 39°C. Reverse transcriptase was heat inactivated at 94°C for 4 minutes.
Q-PCR of specimen and standard cDNA was completed using TaqMan predeveloped assay reagents. Quantitative fluorogenic amplification of cDNA was performed using the ABI Prism 7,500 Sequence Detection System, primers and probes of interest, and TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA). The relative abundance of mRNA levels was determined from standard curves generated from a serially diluted standard pool of cDNA prepared from control human pulmonary artery endothelial cell cultures. The relative abundance of glyceraldehyde-3-phosphate dehydrogenase (Unigene accession no. 544,577) mRNA was used to normalize levels of the mRNAs of interest. For Q-PCR verification, RNA from six additional experiments was collected. RNA samples for microarray and Q-PCR were collected from different experiments, and do not represent the same sample.
Measurement of H2O2
Release of H2O2 into the medium was measured by the Amplex red reagent (10-acetyl-3,7-dihydroxyphenoxazine) (Molecular Probes/Invitrogen Corp., Carlsbad, CA). Amplex red reacts with H2O2 to produce highly fluorescent resorufin in the presence of horseradish peroxidase. The fluorescent signals were measured over 120 minutes with a Bioassay HTS7,000 plate reader (Perkin Elmer, Inc., Wellesley, MA) with HTSoft v1.0 software (PE Applied Biosystems, Weiterstadt, Germany). The excitation and emission wavelengths were 530 and 590 nm, respectively. Generation of H2O2 was calculated by subtracting the fluorescence signal at 0 minutes (baseline) from that at 120 minutes, and expressed as optical density of resorufin. Positive control samples using H2O2 (0.5–2.0 μM) were included for all Amplex red assays. Human AMs were also incubated with PM2.5 with or without rabbit liver zinc–containing MT1 (Alexis Biochemicals, San Diego, CA). MT was added 20 minutes before Amplex red.
Transfection of Human BECs with MT1F siRNA
Human BECs were grown to 60–70% confluency and then transfected with Gene Silencer transfecting agent plus (Gene Therapy System, Inc., San Diego, CA) with MT1F short interfering RNA (siRNA; 100 nM) (Ambion, Austin, TX) (cat. no. 16,708/siRNA, ID no. 44,734/lot no. ASO04QXP/Locus ID 4,494/RefSeq no. NM_005949) in serum-free basic growth medium-2 for 3 hours, according to the manufacturer's recommendations. All control cells were incubated with Gene Silencer select negative control no. 1 siRNA (part no. 4390844) (Ambion). Fresh endothelial growth medium-2 with 2% FBS was then added, and cells were cultured for an additional 48 hours. Cell lysates from some wells were collected for MT1F gene expression by RT-PCR. Other wells were used for measuring H2O2 release.
Statistical Analysis
Data are presented as mean values (±SE). Data from nonmicroarray experiments were analyzed by one-way ANOVA, followed by Scheffé's test for post hoc comparisons. The statistical analysis was performed using commercially available software (Statview version 5.0.1; SAS Institute Inc., Cary, NC). A P value of less than 0.05 was taken as statistically significant.
RESULTS
Genes Induced by PM2.5 in Cultured AMs
Using the above statistical algorithm, we identified 38 and 43 probes that were up- and down-regulated, respectively, at 10% FDR in AMs exposed to PM2.5. After eliminating probes with no corresponding genes, we reduced the list to 34 up-regulated genes (Table 1) and 41 down-regulated genes (Table 2). Expression of 13 up- and down-regulated genes was confirmed by Q-PCR (Figures 1 and 2). We further categorized these differentially expressed genes by their molecular functions using Gene Ontology. We found that 22 genes were involved in metal binding (Tables 1 and 2) (GOTERM_MF_3; P = 0.049).
TABLE 1.
GENES UP-REGULATED BY PARTICULATE MATTER OF 2.5-μM DIAMETER OR SMALLER
| UniGene | Symbol | Fold | Gene Description | Functions |
|---|---|---|---|---|
| HS.534330 | MT1F | 3.164 | Metallothionein 1F (functional) | Metal binding, antioxidant |
| HS.36102 | MT1E | 3.055 | Metallothionein 1E (functional) | Metal binding, antioxidant |
| HS.181768 | NCF1 | 2.908 | Neutrophil cytosolic factor 1, (chronic granulomatous disease, autosomal 1) | Pro-oxidant |
| HS.188518 | MT1H | 2.765 | Metallothionein 1H (functional) | Metal binding, antioxidant |
| HS.513626 | MT1G | 2.760 | Metallothionein 1G (functional) | Metal binding, antioxidant |
| HS.645456 | MT1B | 2.743 | Metallothionein 1B (functional) | Metal binding, antioxidant |
| HS.559477 | NINJ1 | 2.714 | Ninjurin 1 | |
| HS.441047 | ADM | 2.655 | Adrenomedullin | |
| HS.325978 | IL18BP | 2.573 | Interleukin 18 binding protein | |
| HS.127022 | RELB | 2.319 | V-rel reticuloendotheliosis viral oncogene homolog b, nuclear factor of kappa light polypeptide gene enhancer in b-cells 3 (avian) | |
| HS.154654 | CYP1B1 | 2.258 | Cytochrome p450, family 1, subfamily b, polypeptide 1 | Metal binding, pro-oxidant |
| HS.517145 | ENO1 | 2.133 | Enolase 1, (alpha) | Metal binding |
| HS.31210 | BCL3 | 2.119 | B-cell cll/lymphoma 3 | |
| HS.284491 | PFKP | 2.105 | Phosphofructokinase, platelet | Metal binding |
| HS.494457 | NQO1 | 2.007 | NAD(P)H dehydrogenase, quinone 1 | Antioxidant |
| NA | ENO1B | 1.989 | Enolase-α, lung specific | Metal binding |
| HS.297413 | MMP9 | 1.985 | Matrix metallopeptidase 9 | Metal binding |
| HS.76556 | PTPRE | 1.983 | Protein tyrosine phosphatase, receptor type, e | |
| HS.479728 | GAPDH | 1.953 | Glyceraldehyde 3-phosphate dehydrogenase | Antioxidant |
| HS.470126 | KYNU | 1.949 | Kynureninase (l-kynurenine hydrolase) | |
| HS.520708 | DFNA5 | 1.862 | Deafness, autosomal dominant 5 | |
| HS.117062 | AIFM2 | 1.817 | Apoptosis-inducing factor, mitochondrion-associated, 2 (AMID) | Pro-oxidant |
| HS.636480 | TUBB | 1.687 | Tubulin-β | |
| HS.493919 | MPZL1 | 1.683 | Myelin protein zero-like 1 | |
| HS.307905 | TFPI | 1.625 | Tissue factor pathway inhibitor | |
| HS.589369 | PDXK | 1.620 | Pyridoxal (pyridoxine, vitamin B6) kinase | Metal binding |
| HS.26010 | PPP1R15A | 1.602 | Protein phosphatase 1, regulatory (inhibitor) subunit 15a | |
| HS.125213 | ATOX1 | 1.578 | Atx1 antioxidant protein 1 homolog (yeast) | Metal binding, antioxidant |
| HS.406515 | NR1H3 | 1.548 | Nuclear receptor subfamily 1, group h, member 3 | Metal binding |
| HS.1422 | FGR | 1.512 | Gardner-rasheed feline sarcoma viral (v-fgr) oncogene homolog | |
| HS.438462 | MYOD1 | 1.496 | Myogenic differentiation 1 | |
| HS.556274 | CHST11 | 1.396 | Carbohydrate (chondroitin 4) sulfotransferase 11 | |
| HS.66542 | GPR3 | 1.392 | G protein-coupled receptor 3 | |
| HS.438863 |
NRBP |
1.368 |
Nuclear receptor binding protein 1 |
Genes that are involved in metal binding and oxidative stress are indicated under “Functions.”
TABLE 2.
GENES DOWN-REGULATED BY PARTICULATE MATTER OF 2.5-μM DIAMETER OR SMALLER
| UniGene | Symbol | Fold | Gene Description | Function |
|---|---|---|---|---|
| HS.567398 | CCL4 | −5.464 | Chemokine (C-C motif) ligand 4 | |
| HS.416073 | S100A8 | −4.016 | S100 calcium binding protein a8 (calgranulin a) | Metal binding |
| HS.51120 | CAMP | −3.610 | Cathelicidin antimicrobial peptide | |
| HS.647049 | LAT2 | −3.534 | Linker for activation of T cells family, member 2 | |
| HS.7884 | SLCO2B1 | −2.941 | Solute carrier organic anion transporter family, member 2b1 | |
| HS.233389 | CPVL | −2.849 | Carboxypeptidase, vitellogenic-like | |
| HS.153837 | MNDA | −2.725 | Myeloid cell nuclear differentiation antigen | |
| HS.75182 | MRC1 | −2.674 | Mannose receptor, c type 1 | Metal binding |
| HS.515061 | EDG6 | −2.577 | Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 6 | |
| HS.64016 | PROS1 | −2.481 | Protein S (α) | Metal binding |
| HS.20315 | IFIT1 | −2.463 | Interferon-induced protein with tetratricopeptide repeats 1 | |
| HS.377894 | GCA | −2.445 | Grancalcin, EF-hand calcium binding protein | Metal binding |
| HS.2551 | ADRB2 | −2.404 | Adrenergic,β-2-, receptor, surface | |
| HS.119192 | H2AFZ | −2.392 | H2a histone family, member z | |
| HS.75360 | CPE | −2.320 | Carboxypeptidase e | Metal binding |
| HS.504657 | CLEC4A | −2.315 | C-type lectin domain family 4, member a | |
| HS.479208 | FBXL5 | −2.309 | F-box and leucine-rich repeat protein 5 | |
| HS.75765 | CXCL2 | −2.198 | Chemokine (C-X-C motif) ligand 2 | |
| HS.12056 | ASGR1 | −2.183 | Asialoglycoprotein receptor 1 | Metal binding |
| HS.528055 | HIST1H4A | −2.183 | H4 histone, family 2 | |
| HS.202 | BZRP | −2.179 | Benzodiazapine receptor (peripheral) | |
| HS.12333 | RNF13 | −2.174 | Ring finger protein 13 | Metal binding |
| HS.25155 | NET1 | −2.160 | Neuroepithelial cell transforming gene 1 | |
| HS.518198 | CSTA | −2.146 | Cystatin a (stefin a) | |
| HS.499094 | PYCARD | −2.141 | PYD and CARD domain containing | |
| HS.520989 | FGL2 | −2.132 | Fibrinogen-like 2 | |
| HS.446123 | CAPZA2 | −2.105 | Capping protein (actin filament) muscle z-line, α 2 | |
| HS.516834 | C20ORF46 | −2.092 | Chromosome 20 open reading frame 46 | |
| HS.486410 | ECHDC1 | −2.088 | Enoyl coenzyme a hydratase domain containing 1 | |
| HS.26136 | PIGY | −2.079 | Phosphatidylinositol glycan anchor biosynthesis, class y | |
| HS.75367 | SLA | −2.079 | Src-like-adaptor | |
| HS.209983 | STMN1 | −2.075 | Stathmin 1/oncoprotein 18 | |
| HS.514412 | PECAM1 | −2.062 | Platelet/endothelial cell adhesion molecule (CD31 antigen) | |
| HS.70327 | CRIP1 | −2.058 | Cysteine-rich protein 1 (intestinal) | Metal binding |
| HS.523702 | MS4A6A | −2.058 | CD20-like precusor | |
| HS.484990 | HIST1H3H | −2.049 | Histone 1, h3a | |
| HS.325960 | MS4A4A | −2.041 | Membrane-spanning 4-domains, subfamily a, member 4 | |
| HS.8904 | VSIG4 | −2.020 | V-set and immunoglobulin domain containing 4 | |
| HS.558359 | BIRC1 | −2.016 | Baculoviral IAP repeat-containing 1 | |
| HS.282410 | CALM2 | −2.016 | Calmodulin 1 (phosphorylase kinase, δ) | Metal binding |
| HS.180878 |
LPL |
−2.016 |
Lipoprotein lipase |
Genes that are involved in metal binding are indicated under “Function.”
Figure 1.
PCR confirmation of selected up-regulated genes (n = 6 independent experiments for quantitative PCR [Q-PCR]). Solid bars, microarray; shaded bars, PCR.
Figure 2.
PCR confirmation of selected down-regulated genes (n = 6 independent experiments for Q-PCR). Solid bars, microarray; shaded bars, PCR.
Many of these genes, such as MT1F, MT1E, MT1H, MT1G, MT1B, ATOX1, CYP1B1, and MMP9, are also involved in oxidative stress (Tables 1 and 2). Other differentially expressed genes involved in oxidative stress included up-regulation of NCF1, NQO1, and GAPD (Tables 1 and 2). Hierarchical clustering analyses of significant genes are shown in Figure 3, which shows two main clusters in up-regulated genes. One cluster contains all MT1 isoform genes, NCF1, NQO1, and MMP9, and the other contains CYP1B1, GAPD, AMID, and ATOX1. Because of up-regulation of multiple MT1 isoform genes, we further investigated the role of MT1F, one of the main isoforms in the lung (31–33), in oxidative stress induced by PM2.5.
Figure 3.
Hierarchical clustering images for (A) up-regulated and (B) down-regulated genes. Hierarchical clustering on genes and samples was performed using Euclidean distance metric.
Effects of MT1 on H2O2 Production
Incubation of human AM with PM2.5 increased the extracellular H2O2 release detected by Amplex red in a dose-dependent manner, which was inhibited completely by catalase (200 U/ml) (Figure 4A). The enhanced release of H2O2 was also inhibited by the addition of MT1 protein in the medium (Figure 4B). These concentrations of PM2.5 were not cytotoxic, as there was no increase in lactic dehydrogenase release (data not shown). In addition, PM2.5 preincubated with 100 μM MT1 for 30 minutes before being added to the medium induced lower levels of H2O2 release compared with PM2.5 preincubated with BSA (10 μg/ml) in human AM (Figure 4C) or cell-free system (Figure 4D), indicating that metal chelation is one mechanism by which MT1 decreases PM2.5-induced H2O2 release.
Figure 4.
Effects of metallothionein (MT)-1 on H2O2 release in human alveolar macrophages (AMs). (A) Particulate matter (PM) of 2.5-μm diameter or smaller (PM2.5) dose-dependently increases H2O2 release. Catalase inhibited H2O2 release stimulated by PM2.5. The dose of PM2.5 used in the catalase experiments was 30 μg/ml. (B) Addition of MT1 into the medium decreases PM2.5-induced H2O2 release detected by increases in resorufin signals. PM2.5 preincubated with MT1 (100 μM) for 30 minutes induced lower levels of H2O2 release compared with PM2.5 preincubated with BSA (10 μg/ml) in human AMs (C) or cell-free medium (D). The concentration of PM2.5 was 30 μg/ml. *P < 0.05; n = 4–6 each.
Effects of MT1F Knockdown on H2O2 Release
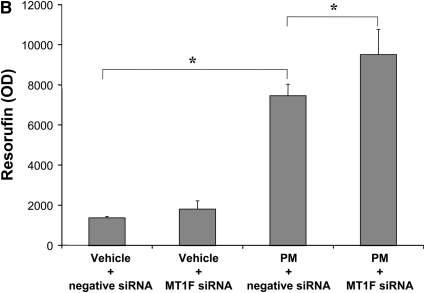
To determine how MT1F regulates H2O2 release, we knocked down MT1F gene expression using siRNA in human BECs. We used human BECs instead of human AMs for these siRNA experiments, because siRNA was ineffective in human AMs. We verified that human BECs, like human AMs, increased MT1F and CYP1B1 gene expression (measured by Q-PCR) by five- and sixfold, respectively, after PM2.5 exposure (100 μg/ml or 10 μg/cm2). Incubation with siRNA reduced MT1F gene expression by approximately 50% (Figure 5A) and increased PM2.5-induced H2O2 release by approximately 30% (Figure 5B).
Figure 5.
Effects of MT1F knockdown on H2O2 release in primary human bronchial epithelial cells. (A) Incubation of siRNA against MT1F inhibited gene expression of MT1F induced by PM2.5 by more than 50%. (B) Knockdown of MT1F enhanced PM2.5-induced H2O2 release. The concentration of PM2.5 was 100 μg/ml or 10 μg/cm2. *P < 0.05; n = 4–6 each.
Effects of PM Metal Components on H2O2 Release
To determine whether or not the metal components of PM are important in inducing MT1F, human AMs were incubated with untreated PM2.5 (1 μg/ml), metal-poor Chapel Hill PM2.5 and Mount St. Helens (MSH) PM2.5 (1 μg/ml), which contains little metal. To prepare metal-poor PM2.5, 1 μg/ml of Chapel Hill PM2.5 was washed with 1 N HCl three times, and the pellet was resuspended in sterile water (34, 35). Human AMs incubated with acid-extracted Chapel Hill PM2.5 and MSH PM2.5 showed no increase in MT1F gene expression (Figure 6A). Neither the acid-extracted Chapel Hill PM2.5 nor MSH PM2.5 at 1 μg/ml increased H2O2 release (Figure 6B). We also tested higher doses of these metal-poor particle preparations, and there was no increase in H2O2 release at doses up to 100 μg/ml (data not shown). These results indicate that metals associated with Chapel Hill PM2.5 play an important role in gene expression changes induced by the whole particles, independent of oxidative stress.
Figure 6.
Effects of PM2.5-associated metals on MT1F gene expression (A) and H2O2 release (B). Human AMs were treated with vehicle (control), untreated Chapel Hill PM2.5 (1 μg/ml), or Chapel Hill PM2.5 (1 μg/ml) extracted with 1 N HCl or Mount St. Helens PM2.5 (1 μg/ml). *P < 0.05; n = 4–6 each.
DISCUSSION
Our findings suggest that metal components associated with PM may be important in inducing gene changes, independent of oxidative stress. Using microarray analysis and gene ontology, we found that about one-third of the differentially expressed genes in AMs challenged with PM2.5 had molecular functions related to metal binding. We also noted that five MT-1 isoform genes (MT1F, MT1E, MT1H, MT1G, and MT1B) were among the top 10 up-regulated genes. The MT1 genes are known to be involved in heavy metal binding. The MT1 proteins are also important antioxidant enzymes. Three other metal-binding genes, ATOX1, CYP1B1, and MMP9, are also linked to oxidative stress. Furthermore, four other genes not in the metal-binding category, including NCF1, NQO1, AIFM2, and GAPD, are also known to be involved in oxidative stress. Thus, a total of 11 genes could be included in the pro- and antioxidant network. The 11 up-regulated oxidative stress genes fall into two main groups that include MT1 isoform genes and CYP1B1, respectively. Seven genes are involved in calcium binding (MMP9, GCA, PROS1, ASGR1, CALM2, MRC1, and S100A8), and may further modulate oxidant signaling by altering intracellular calcium (36–39).
PM is known to produce oxidative stress in lung cells. Our microarray study identified three pro-oxidant genes that were up-regulated: NCF1, CYP1B1, and AIFM2. NCF1 encodes a 47-kD cytosolic subunit of the membrane reduced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, an enzyme responsible for the respiratory burst. The membrane NADPH oxidase on AMs may be activated by PM exposure and produce ROS (5, 12, 16). CYP1B1 encodes a mono-oxygenase in the cytochrome p450 system localized in the endoplasmic reticulum. CYP1B1 is part of the phase-I xenobiotic enzyme system. It may be up-regulated by PM exposure, and is a major source of ROS in cells stimulated with PM (40–43). AIFM2 (AMID) encodes a mitochondrial protein with significant homology with reduced nicotinamide adenine dinucleotide oxidoreductase and apoptosis-inducing factor. Up-regulation of this gene by PM has not been reported. AIFM2 may be one of the important sources of mitochondrial ROS produced by AMs during PM exposure (13, 15, 44), and is probably linked to a caspase-independent mitochondrial effector of apoptotic cell death.
The up-regulation of multiple MT1 isoform genes is intriguing. MTs are low-molecular weight, metal and sulfur-rich proteins that are widely distributed in all organs, including the lung (32, 45). These intracellular proteins are thought to be involved in both heavy metal detoxification and the homeostasis of essential trace metals, such as zinc and copper. The metal-binding property and the abundance of cysteines (18–23 residues) in MTs have also been implicated as the mechanisms by which MTs protect cells from oxidative injury (46–49). We have shown that addition of exogenous MT1 protein attenuated the extracellular resorufin signals. In human BECs that also showed up-regulation of MT1F and CYP1B1 gene expression after PM2.5 exposure similar to human AMs, knocking down MT1F gene expression by more than 50% was associated with an approximately 30% increase in PM2.5-induced H2O2 release. In addition, particles preincubated with MT1 before being added to the cells attenuated the release of H2O2. These results confirm the capability of MT1 to scavenge ROS and bind metals in human lung cells during PM exposure. MT1 may bind to redox-active metals associated with PM via its cysteine residues, and decrease hydroxyl radicals generated from the Fenton reaction. Binding of MT1 to PM may also interfere with the engulfment of MT1–PM complex by the AMs and inhibit downstream oxidant pathways. The abundant thiol groups present in MT1 also may directly scavenge oxygen radicals, independent of its metal-binding capability (49).
We further noted that approximately 30% of the 75 differentially expressed genes have molecular functions related to metal ion binding. Of these, the five MT1 isoforms are known to be essential in detoxifying environmental heavy metals, such as arsenic, cadmium, and mercury (50–53). MT1 levels in various tissues may also increase in animals exposed to environmental zinc (54, 55). The MT1 genes also are involved in binding of intracellular metals, such as zinc and copper (32, 56–58), and, thus, may regulate signaling mediated by these trace metals. ATOX1 encodes a copper chaperone that binds and transports cytosolic copper to ATPase proteins in the trans-Golgi network for later incorporation to the ceruloplasmin (59–61). Thus, our gene profiling study indicates that PM exposure may induce host defense mechanisms that are normally invoked to combat heavy metal toxicity, and that molecular signaling pathways mediated by zinc and copper may be important in PM-induced health effects.
Metals associated with PM likely were the primary inducer for gene changes, as metal-poor PM preparations did not induce MT1. Oxidative stress was unlikely the inducer for changes in gene expression, because PM2.5 at 1 μg/ml, which altered gene expression, did not increase H2O2 release. The induction of MT1 genes further indicates that certain metal components in PM may be the biologically active constituents for pulmonary health effects. MT1 genes may be responsive specifically to some heavy metals that are normally present in PM, such as zinc and copper (32, 50, 62). The PM2.5 sample used in this study contains approximately 500 ng/mg of zinc and 70 ng/mg of copper (5). Zinc is a major metal element detected in traffic-derived PM (63–66), presumably from tire wear. Incubation of human BECs with zinc up-regulated MT1 isoform genes (33). Copper in PM has also been linked to traffic sources, as copper is an element in car brakes (65, 67, 68). Zinc and copper may also come from certain industrial sources (63, 65). In a previous study exposing healthy volunteers to Chapel Hill PM2.5, the principal component analysis identified a factor containing zinc and copper correlating with increased blood fibrinogen (26). Also, in an in vitro study of cultured human BECs, the principal component analysis identified a copper-containing factor that was associated with Chapel Hill PM–induced IL-6 release (5).
AMs have an important role in clearing particles from the lungs (69). Binding and subsequent engulfment of particles may activate signaling pathways, resulting in production of ROS by various pro-oxidant enzymes, such as NADPH (16). Peroxinitrite may also be formed from the interaction between ROS and nitric oxide (25). PM effects may be recognized via binding receptors on AM, which activates downstream inflammatory pathways (70). Thus, interactions between PM and cell membrane proteins of AMs may be a critical event that triggers subsequent oxidative stress and inflammation. Our results demonstrate changes in the expression of genes encoding membrane proteins, such as PTPRE, MRC1, ADRB2, CLEC4A, ASGR1, MS4A4A, and MS4A6A. All of these genes, except PTPRE, were down-regulated. How PM-induced changes in these novel membrane proteins affect oxidative stress and inflammation, and thus health effects, will need to be investigated in future studies.
In summary, our gene profiling experiments identified an array of gene expression response in human AMs exposed to PM2.5, independent of oxidative stress. This profile is most notable for genes unique to heavy metal binding and oxidative stress. Metals associated with PM may be important in inducing host defense mechanisms normally invoked to combat heavy metal toxicity, and molecular signaling pathways mediated by metals may be important in PM-induced health effects.
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute, and the U.S. Environmental Protection Agency.
Originally Published in Press as DOI: 10.1165/rcmb.2008-0064OC on February 27, 2009
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Franklin M, Zeka A, Schwartz J. Association between PM2.5 and all-cause and specific-cause mortality in 27 US communities. J Expo Sci Environ Epidemiol 2007;17:279–287. [DOI] [PubMed] [Google Scholar]
- 2.Pope CA III, Dockery DW, Spengler JD, Raizenne ME. Respiratory health and PM10 pollution: a daily time series analysis. Am Rev Respir Dis 1991;144:668–674. [DOI] [PubMed] [Google Scholar]
- 3.Committee of the Environmental and Occupational Health Assembly of the American Thoracic Society. Health effects of outdoor air pollution. Am J Respir Crit Care Med 1996;153:3–50. [DOI] [PubMed] [Google Scholar]
- 4.Brunekreef B, Forsberg B. Epidemiological evidence of effects of coarse airborne particles on health. Eur Respir J 2005;26:309–318. [DOI] [PubMed] [Google Scholar]
- 5.Becker S, Dailey LA, Soukup JM, Grambow SC, Devlin RB, Huang YC. Seasonal variations in air pollution particle–induced inflammatory mediator release and oxidative stress. Environ Health Perspect 2005;113:1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson K, Stone V, Borm PJ, Jimenez LA, Gilmour PS, Schins RP, Knaapen AM, Rahman I, Faux SP, Brown DM, et al. Oxidative stress and calcium signaling in the adverse effects of environmental particles (PM10). Free Radic Biol Med 2003;34:1369–1382. [DOI] [PubMed] [Google Scholar]
- 7.Sorensen M, Daneshvar B, Hansen M, Dragsted LO, Hertel O, Knudsen L, Loft S. Personal PM2.5 exposure and markers of oxidative stress in blood. Environ Health Perspect 2003;111:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao F, Gonzalez-Flecha B, Kobzik L. Reactive oxygen species in pulmonary inflammation by ambient particulates. Free Radic Biol Med 2003;35:327–340. [DOI] [PubMed] [Google Scholar]
- 9.Xiao GG, Wang M, Li N, Loo JA, Nel AE. Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J Biol Chem 2003;278:50781–50790. [DOI] [PubMed] [Google Scholar]
- 10.Alexis NE, Lay JC, Zeman K, Bennett WE, Peden DB, Soukup JM, Devlin RB, Becker S. Biological material on inhaled coarse fraction particulate matter activates airway phagocytes in vivo in healthy volunteers. J Allergy Clin Immunol 2006;117:1396–1403. [DOI] [PubMed] [Google Scholar]
- 11.Larsson BM, Sehlstedt M, Grunewald J, Skold CM, Lundin A, Blomberg A, Sandstrom T, Eklund A, Svartengren M. Road tunnel air pollution induces bronchoalveolar inflammation in healthy subjects. Eur Respir J 2007;29:699–705. [DOI] [PubMed] [Google Scholar]
- 12.Becker S, Soukup JM, Gilmour MI, Devlin RB. Stimulation of human and rat alveolar macrophages by urban air particulates: Effects on oxidant radical generation and cytokine production. Toxicol Appl Pharmacol 1996;141:637–648. [DOI] [PubMed] [Google Scholar]
- 13.Huang YC, Soukup J, Harder S, Becker S. Mitochondrial oxidant production by a pollutant dust and NO-mediated apoptosis in human alveolar macrophage. Am J Physiol Cell Physiol 2003;284:C24–C32. [DOI] [PubMed] [Google Scholar]
- 14.Aam BB, Fonnum F. Carbon black particles increase reactive oxygen species formation in rat alveolar macrophages in vitro. Arch Toxicol 2007;81:441–446. [DOI] [PubMed] [Google Scholar]
- 15.Becker S, Soukup JM, Gallagher JE. Differential particulate air pollution induced oxidant stress in human granulocytes, monocytes and alveolar macrophages. Toxicol In Vitro 2002;16:209–218. [DOI] [PubMed] [Google Scholar]
- 16.Park JB. Phagocytosis induces superoxide formation and apoptosis in macrophages. Exp Mol Med 2003;35:325–335. [DOI] [PubMed] [Google Scholar]
- 17.Pourazar J, Mudway IS, Samet JM, Helleday R, Blomberg A, Wilson SJ, Frew AJ, Kelly FJ, Sandstrom T. Diesel exhaust activates redox-sensitive transcription factors and kinases in human airways. Am J Physiol Lung Cell Mol Physiol 2005;289:L724–L730. [DOI] [PubMed] [Google Scholar]
- 18.Behndig AF, Mudway IS, Brown JL, Stenfors N, Helleday R, Duggan ST, Wilson SJ, Boman C, Cassee FR, Frew AJ, et al. Airway antioxidant and inflammatory responses to diesel exhaust exposure in healthy humans. Eur Respir J 2006;27:359–365. [DOI] [PubMed] [Google Scholar]
- 19.Barregard L, Sällsten G, Andersson L, Almstrand AC, Gustafson P, Andersson M, Olin AC. Experimental exposure to wood smoke: effects on airway inflammation and oxidative stress. Occup Environ Med 2008;65:319–324. [DOI] [PubMed] [Google Scholar]
- 20.Xia T, Kovochich M, Nel A. The role of reactive oxygen species and oxidative stress in mediating particulate matter injury. Clin Occup Environ Med 2006;5:817–836. [DOI] [PubMed] [Google Scholar]
- 21.Maier KL, Alessandrini F, Beck-Speier I, Hofer TP, Diabate S, Bitterle E, Stoger T, Jakob T, Behrendt H, Horsch M, et al. Health effects of ambient particulate matter–biological mechanisms and inflammatory responses to in vitro and in vivo particle exposures. Inhal Toxicol 2008;20:319–337. [DOI] [PubMed] [Google Scholar]
- 22.Sun Q, Yue P, Kirk RI, Wang A, Moatti D, Jin X, Lu B, Schecter AD, Lippmann M, Gordon T, et al. Ambient air particulate matter exposure and tissue factor expression in atherosclerosis. Inhal Toxicol 2008;20:127–137. [DOI] [PubMed] [Google Scholar]
- 23.Watterson TL, Sorensen J, Martin R, Coulombe RA Jr. Effects of PM2.5 collected from Cache Valley Utah on genes associated with the inflammatory response in human lung cells. J Toxicol Environ Health A 2007;70:1731–1744. [DOI] [PubMed] [Google Scholar]
- 24.Li N, Xia T, Nel AE. The role of oxidative stress in ambient particulate matter-induced lung diseases and its implications in the toxicity of engineered nanoparticles. Free Radic Biol Med 2008;44:1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hukkanen M, Corbett SA, Platts LA, Konttinen YT, Santavirta S, Hughes SP, Polak JM. Nitric oxide in the local host reaction to total hip replacement. Clin Orthop Relat Res 1998;53–65. [PubMed]
- 26.Huang YC, Ghio AJ, Stonehuerner J, McGee J, Carter JD, Grambow SC, Devlin RB. The role of soluble components in ambient fine particles-induced changes in human lungs and blood. Inhal Toxicol 2003;15:327–342. [DOI] [PubMed] [Google Scholar]
- 27.Ghio AJ, Kim C, Devlin RB. Concentrated ambient air particles induce mild pulmonary inflammation in healthy human volunteers. Am J Respir Crit Care Med 2000;162:981–988. [DOI] [PubMed] [Google Scholar]
- 28.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003;34:374–378. [DOI] [PubMed] [Google Scholar]
- 29.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol 2003;4:3. [PubMed] [Google Scholar]
- 31.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol 2007;8:R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coyle P, Philcox JC, Carey LC, Rofe AM. Metallothionein: the multipurpose protein. Cell Mol Life Sci 2002;59:627–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Stonehuerner J, Devlin RB, Huang YC. Discrimination of vanadium from zinc using gene profiling in human bronchial epithelial cells. Environ Health Perspect 2005;113:1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obot CJ, Morandi MT, Beebe TP, Hamilton RF, Holian A. Surface components of airborne particulate matter induce macrophage apoptosis through scavenger receptors. Toxicol Appl Pharmacol 2002;184:98–106. [PubMed] [Google Scholar]
- 35.Huang YC, Li Z, Harder SD, Soukup JM. Apoptotic and inflammatory effects induced by different particles in human alveolar macrophages. Inhal Toxicol 2004;16:863–878. [DOI] [PubMed] [Google Scholar]
- 36.Hoyal CR, Giron-Calle J, Forman HJ. The alveolar macrophage as a model of calcium signaling in oxidative stress. J Toxicol Environ Health B Crit Rev 1998;1:117–134. [DOI] [PubMed] [Google Scholar]
- 37.MacNee W, Donaldson K. Mechanism of lung injury caused by PM10 and ultrafine particles with special reference to COPD. Eur Respir J Suppl 2003;40:47s–51s. [DOI] [PubMed] [Google Scholar]
- 38.Brown DM, Hutchison L, Donaldson K, Stone V. The effects of PM10 particles and oxidative stress on macrophages and lung epithelial cells: modulating effects of calcium-signaling antagonists. Am J Physiol Lung Cell Mol Physiol 2007;292:L1444–L1451. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Duncan RF, Robison TW, Gao L, Forman HJ. Ca(2+)-dependent p47phox translocation in hydroperoxide modulation of the alveolar macrophage respiratory burst. Am J Physiol 1997;273:L1042–L1047. [DOI] [PubMed] [Google Scholar]
- 40.Kumagai Y, Arimoto T, Shinyashiki M, Shimojo N, Nakai Y, Yoshikawa T, Sagai M. Generation of reactive oxygen species during interaction of diesel exhaust particle components with NADPH–cytochrome p450 reductase and involvement of the bioactivation in the DNA damage. Free Radic Biol Med 1997;22:479–487. [DOI] [PubMed] [Google Scholar]
- 41.Yin XJ, Ma JY, Antonini JM, Castranova V, Ma JK. Roles of reactive oxygen species and heme oxygenase-1 in modulation of alveolar macrophage–mediated pulmonary immune responses to Listeria monocytogenes by diesel exhaust particles. Toxicol Sci 2004;82:143–153. [DOI] [PubMed] [Google Scholar]
- 42.Murphy G Jr, Rouse RL, Polk WW, Henk WG, Barker SA, Boudreaux MJ, Floyd ZE, Penn AL. Combustion-derived hydrocarbons localize to lipid droplets in respiratory cells. Am J Respir Cell Mol Biol 2008;38:532–540. [DOI] [PubMed] [Google Scholar]
- 43.Verheyen GR, Nuijten JM, Van Hummelen P, Schoeters GR. Microarray analysis of the effect of diesel exhaust particles on in vitro cultured macrophages. Toxicol In Vitro 2004;18:377–391. [DOI] [PubMed] [Google Scholar]
- 44.Li N, Sioutas C, Cho A, Schmitz D, Misra C, Sempf J, Wang M, Oberley T, Froines J, Nel A. Ultrafine particulate pollutants induce oxidative stress and mitochondrial damage. Environ Health Perspect 2003;111:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Courtade M, Carrera G, Paternain JL, Martel S, Carre PC, Folch J, Pipy B. Metallothionein expression in human lung and its varying levels after lung transplantation. Toulouse Lung Transplantation Group. Chest 1998;113:371–378. [DOI] [PubMed] [Google Scholar]
- 46.Irato P, Santovito G, Piccinni E, Albergoni V. Oxidative burst and metallothionein as a scavenger in macrophages. Immunol Cell Biol 2001;79:251–254. [DOI] [PubMed] [Google Scholar]
- 47.Kang YJ. The antioxidant function of metallothionein in the heart. Proc Soc Exp Biol Med 1999;222:263–273. [DOI] [PubMed] [Google Scholar]
- 48.Sato M, Bremner I. Oxygen free radicals and metallothionein. Free Radic Biol Med 1993;14:325–337. [DOI] [PubMed] [Google Scholar]
- 49.Sato M, Kondoh M. Recent studies on metallothionein: protection against toxicity of heavy metals and oxygen free radicals. Tohoku J Exp Med 2002;196:9–22. [DOI] [PubMed] [Google Scholar]
- 50.Peng Z, Peng L, Fan Y, Zandi E, Shertzer HG, Xia Y. A critical role for IkappaB kinase beta in metallothionein-1 expression and protection against arsenic toxicity. J Biol Chem 2007;282:21487–21496. [DOI] [PubMed] [Google Scholar]
- 51.Abe T, Yamamoto O, Gotoh S, Yan Y, Todaka N, Higashi K. Cadmium-induced mRNA expression of HSP32 is augmented in metallothionein-I and -II knock-out mice. Arch Biochem Biophys 2000;382:81–88. [DOI] [PubMed] [Google Scholar]
- 52.Park JD, Liu Y, Klaassen CD. Protective effect of metallothionein against the toxicity of cadmium and other metals. Toxicology 2001;163:93–100. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida M, Satoh M, Shimada A, Yasutake A, Sumi Y, Tohyama C. Pulmonary toxicity caused by acute exposure to mercury vapor is enhanced in metallothionein-null mice. Life Sci 1999;64:1861–1867. [DOI] [PubMed] [Google Scholar]
- 54.Onosaka S, Cherian MG. The induced synthesis of metallothionein in various tissues of rats in response to metals. II. Influence of zinc status and specific effect on pancreatic metallothionein. Toxicology 1982;23:11–20. [DOI] [PubMed] [Google Scholar]
- 55.Waalkes MP, Klaassen CD. Concentration of metallothionein in major organs of rats after administration of various metals. Fundam Appl Toxicol 1985;5:473–477. [DOI] [PubMed] [Google Scholar]
- 56.Wiseman DA, Wells SM, Hubbard M, Welker JE, Black SM. Alterations in zinc homeostasis underlie endothelial cell death induced by oxidative stress from acute exposure to hydrogen peroxide. Am J Physiol Lung Cell Mol Physiol 2007;292:L165–L177. [DOI] [PubMed] [Google Scholar]
- 57.Vasak M, Hasler DW. Metallothioneins: new functional and structural insights. Curr Opin Chem Biol 2000;4:177–183. [DOI] [PubMed] [Google Scholar]
- 58.Pauwels M, van Weyenbergh J, Soumillion A, Proost P, De Ley M. Induction by zinc of specific metallothionein isoforms in human monocytes. Eur J Biochem 1994;220:105–110. [DOI] [PubMed] [Google Scholar]
- 59.Prohaska JR, Gybina AA. Intracellular copper transport in mammals. J Nutr 2004;134:1003–1006. [DOI] [PubMed] [Google Scholar]
- 60.Lutsenko S, LeShane ES, Shinde U. Biochemical basis of regulation of human copper-transporting atpases. Arch Biochem Biophys 2007;463:134–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hussain F, Wittung-Stafshede P. Impact of cofactor on stability of bacterial (CopZ) and human (Atox1) copper chaperones. Biochim Biophys Acta 2007;1774:1316–1322. [DOI] [PubMed] [Google Scholar]
- 62.Cosma G, Fulton H, DeFeo T, Gordon T. Rat lung metallothionein and heme oxygenase gene expression following ozone and zinc oxide exposure. Toxicol Appl Pharmacol 1992;117:75–80. [DOI] [PubMed] [Google Scholar]
- 63.Buzcu-Guven B, Brown SG, Frankel A, Hafner HR, Roberts PT. Analysis and apportionment of organic carbon and fine particulate matter sources at multiple sites in the midwestern United States. J Air Waste Manag Assoc 2007;57:606–619. [DOI] [PubMed] [Google Scholar]
- 64.Chen LWA, Watson JG, Chow JC, Magliano KL. Quantifying PM2.5 source contributions for the San Joaquin Valley with multivariate receptor models. Environ Sci Technol 2007;41:2818–2826. [DOI] [PubMed] [Google Scholar]
- 65.Fang GC, Wu YS, Wen CC, Huang SH, Rau JY. Ambient air particulate concentrations and metallic elements principal component analysis at Taichung Harbor (TH) and Wuchi traffic (WT) near Taiwan Strait during 2004–2005. J Hazard Mater 2006;137:314–323. [DOI] [PubMed] [Google Scholar]
- 66.Kinoshita T, Yamaguchi K, Akita S, Nii S, Kawaizumi F, Takahashi K. Hydrometallurgical recovery of zinc from ashes of automobile tire wastes. Chemosphere 2005;59:1105–1111. [DOI] [PubMed] [Google Scholar]
- 67.Gerlofs-Nijland ME, Dormans JA, Bloemen HJ, Leseman DL, John A, Boere F, Kelly FJ, Mudway IS, Jimenez AA, Donaldson K, et al. Toxicity of coarse and fine particulate matter from sites with contrasting traffic profiles. Inhal Toxicol 2007;19:1055–1069. [DOI] [PubMed] [Google Scholar]
- 68.Laden F, Neas LM, Dockery DW, Schwartz J. Association of fine particulate matter from different sources with daily mortality in six U.S. cities. Environ Health Perspect 2000;108:941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palecanda A, Kobzik L. Receptors for unopsonized particles: the role of alveolar macrophage scavenger receptors. Curr Mol Med 2001;1:589–595. [DOI] [PubMed] [Google Scholar]
- 70.Becker S, Soukup JM, Sioutas C, Cassee FR. Response of human alveolar macrophages to ultrafine, fine, and coarse urban air pollution particles. Exp Lung Res 2003;29:29–44. [DOI] [PubMed] [Google Scholar]