Abstract
ACTIN-RELATED PROTEIN5 (ARP5) is a conserved subunit of the INO80 chromatin-remodeling complex in yeast and mammals. We have characterized the expression and subcellular distribution of Arabidopsis thaliana ARP5 and explored its role in the epigenetic control of multicellular development and DNA repair. ARP5-specific monoclonal antibodies localized ARP5 protein to the nucleoplasm of interphase cells in Arabidopsis and Nicotiana tabacum. ARP5 promoter-reporter fusions and the ARP5 protein are ubiquitously expressed. A null mutant and a severe knockdown allele produced moderately dwarfed plants with all organs smaller than wild type. The small and slightly deformed organs such as leaves and hypocotyls were composed of small sized cells. The ratio of leaf stomata to epidermal cells was high in the mutant, which also exhibited a delayed stomatal development compared to wild type. Mutant plants were hypersensitive to DNA damaging reagents including hydroxyurea, methylmethane sulfonate and bleocin, demonstrating a role for ARP5 in DNA repair. Interestingly, the hypersensitivity phenotype of ARP5 null allele arp5-1 is stronger than the severe knockdown allele arp5-2. Moreover, a wild type transgene fully complemented all developmental and DNA repair mutant phenotypes. Despite the common participation of both ARP4 and ARP5 in the INO80 complex, ARP4- and ARP5-deficient plants displayed only a small subset of common phenotypes and each displayed novel phenotypes suggesting that in Arabidopsis they have both shared and unique functions.
Keywords: Arabidopsis thaliana, nuclear ARPs, chromatin-remodeling, double stranded DNA break repair, DNA damaging agents, epigenetic control
Introduction
Nuclear ACTIN-RELATED PROTEINS (ARPs) are novel epigenetic factors that are distantly related in sequence and structure to conventional actin. Unlike the cytoplasmic cytoskeletal actin, however, the nuclear ARPs are involved in chromatin remodeling and gene regulation affecting development. The only known function of nuclear ARPs is as constituents of large multi-subunit machines that exert epigenetic control over chromatin structure (Meagher et al., 2005; Meagher et al., 2009; Olave et al., 2002). Mammalian and yeast nuclear ARPs bind members of the family of Swi2-related DNA Dependent ATPases to form nucleosome remodeling complexes. Fungal ARPs also bind the family of Vid21-related helicase-ATPases to form histone modifying complexes (Jonsson et al., 2004; Szerlong et al., 2008). The binding of ARP heterodimers or an ARP-actin heterodimer to a core ATPase appears essential to the assembly of complete and fully active chromatin remodeling or modifying complexes. In addition, yeast and/or mammalian ARP4, ARP5, and ARP8 bind nucleosomal histones, thereby targeting complexes to chromatin. Because no other family of subunit proteins participates in more complexes controlling chromatin dynamics than ARPs, ARP deficiencies have the potential to produce dramatic pleiotropic phenotypes. For example, defects in the expression of Arabidopsis ARP4, ARP6, and ARP7 severely affect apical stem cell development and organ initiation, cell proliferation and expansion, floral organ morphology and senescence, root and root hair morphology, leaf and trichome development, apical and lateral root growth, male and/or female fertility and the phase transition to flowering (Deal et al., 2005; Kandasamy et al., 2004; Kandasamy et al., 2005a; Meagher et al., 2005; Meagher et al., 2007). Because most of the studies in animals are done with cell lines, much less is known about the roles of mammalian nuclear ARPs in multicellular development.
In yeast and mammals, ARP5 is a subunit of the INO80 chromatin remodeling complex, which is best known for its role in DNA double strand break (DSB) repair (Kitayama et al., 2009; Shimada et al., 2008; van Attikum et al., 2004). Besides ARP5, the INO80 complex in yeast contains ARP4, ARP8, actin, the Swi2-related Ino80 and seven other subunits. ARP5 is essential to recruitment of INO80 to DSBs, to recombination repair, and to restarting stalled replication forks after repair (Meagher et al., 2009). HeLa cells partially silenced for ARP5 expression are very sensitive to bleomycin, a reagent that causes DSB (Kitayama et al., 2009). In Arabidopsis, loss of Ino80 function results in inefficient homologous recombination, presumably due to defects in DSB repair, although these plants were not hypersensitive to the DNA damaging agents, methyl methanesulfonate (MMS) or bleomycin (Fritsch et al., 2004).
The INO80 complex and its essential ARP5 subunit also have a less well-studied role in nucleosome remodeling that alters gene expression and development. For example, yeast mutants defective in ARP5 or Ino80 show an inositol requiring phenotype, for which the complex in named. This phenotype probably results from the requirement for INO80 binding to the INO1 promoter, altering its nucleosome structure and activating its expression (Ford et al., 2008). Furthermore, loss of Ino80 function results in a 2-fold misregulation of 3 to 8% of all yeast genes (Mizuguchi et al., 2004; van Attikum et al., 2004) and approximately 0.5% of Arabidopsis genes (Fritsch et al., 2004). Very little published data exist demonstrating roles for plant or animal ARP5 homologs in the epigenetic control of global gene expression and multicellular development, and nothing is known on the role of plant ARP5 in DNA repair.
In this study, we characterized the expression patterns and mutant phenotypes of Arabidopsis ARP5. Analysis of the organ and tissue specific expression of an ARP5 promoter reporter fusion and analysis of protein levels and subcellular localization with ARP5-specific monoclonal antibodies demonstrated that the plant ARP5 gene and its encoded protein are nearly ubiquitously expressed in the nuclei of most cell types. ARP5-defective plants showed dwarfed phenotypes with altered cell, tissue, and organ development. Moreover, the mutant plants were hypersensitive to DNA damaging agents that induce DNA single and double strand breaks. Our data suggest that in plants ARP5 protein participates in multicellular development and DNA repair, and may have roles outside of the conventional INO80 complex.
Materials and methods
ARP5 sequence annotation and plasmid construction
The sequence of the wild type Arabidopsis thaliana (Columbia) ARP5 locus At3g12380 at TAIR is incorrectly reported as encoding a 590 a.a. long protein, instead of the full length 726 a.a. polypeptide as described herein, due to the positioning of a stop codon interrupting the early part of the ARP5 reading frame. Perhaps this short form represents a rare allele. We have submitted to TAIR the correctly annotated Columbia ARP5 genomic sequence (bankit1199661, FJ850973) comprised of 11 exons (Fig. 3A) and a full-length transcript (bankit1199707, FJ850974) encoding the ARP5 protein sequence. We confirmed the size and coding sequence of the most common mature ARP5 cDNAs as encoding the longer protein and did not observe any cDNAs encoding the 590 a.a. form. Furthermore, our antibodies react with an approximately 80 kDa plant protein on SDS-PAGE western blots and not a 65 KDa protein as predicted at TAIR.
Fig. 3.
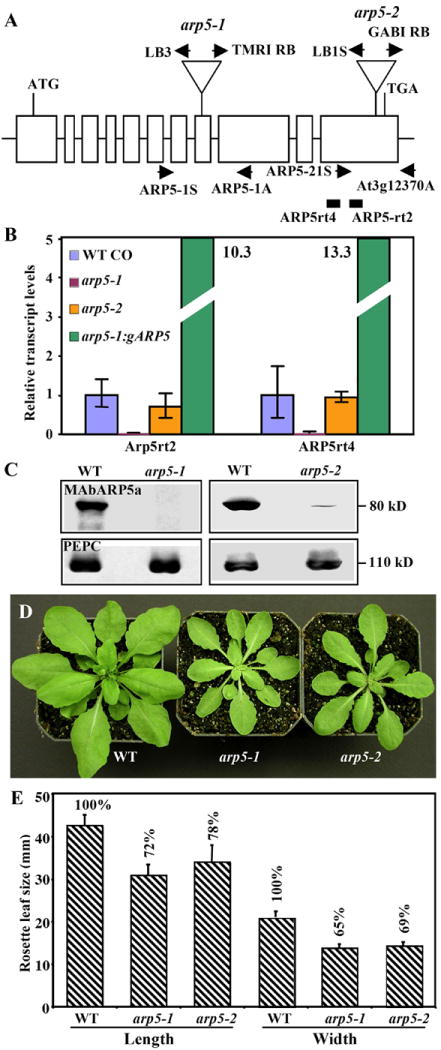
Characterization of molecular phenotypes of two ARP5-deficient mutants.
(A) A map of Arabidopsis ARP5 gene indicating the location of T-DNA insertions in the arp5-1 and arp5-2 mutant alleles. Open boxes indicate the 11 exons of ARP5 that encodes a protein of 726 a.a. T-DNA is inserted into the 8th and 11th exons in the arp5-1 and arp5-2 alleles, respectively. The locations of primers used for the genotyping of wild type and mutant alleles by PCR are marked with arrows. The solid rectangles indicate the RT-PCR products obtained while quantifying the transcript levels in (B) using the primers listed in Supplemental Table 1. (B) qRT-PCR analysis of ARP5 transcript levels in 15-day-old seedlings of the two mutant alleles and a complemented arp5-1 plant expressing gARP5. The data represent average values of two technical replicates and the bars correspond to standard deviation (SD). (C) Western blot analysis of ARP5 protein levels in the two mutants. The upper panels are probed with MAbARP5a and the lower panels are probed with anti-PEPC antibody. (D) Morphology of 32-day-old wild type and mutant plants. (E) A bar graph depicting a reduction in leaf size of arp5-1 and arp5-2 mutant plants compared to wild type (Bars indicate SD values).
For the complementation of the mutant plants, the Arabidopsis ARP5 genomic clone gARP5 was made in two steps. First, a 2080 bp fragment containing 562 bp promoter region and the 5′ half of the gene up to the first 5 amino acids of exon 7 was PCR amplified from the BAC clone T2E22 (ABRC) with Turbo Pfu (Stratagene). This PCR product was cloned into the KpnI and SalI sites of pCambia-Hyg vector (Hajdukiewicz et al., 1994). Second, a 2305 bp fragment containing the 3′ half of the ARP5 gene starting from intron 6 and including a 328 bp terminator region was PCR amplified as above and cloned into the SalI and EcoRI sites of the pCambia clone containing ARP5 5′fragment. The final gARP5 construct containing an ARP5 gene sequence totaling 4364 bp was mobilized into the Agrobacterium tumefaciens strain C58C1 and transformed into the mutant plants by infiltration of inflorescences and flower buds.
ARP5p:GUS construct was made by digesting the genomic clone gARP5 with HindIII and NcoI and swapping the resulting 562 bp ARP5 promoter and 5′UTR fragment with the corresponding ARP8 promoter and 5′UTR in the ARP8p:GUS construct (Kandasamy et al., 2008). ARP5p:GUS utilizes the 3′UTR and polyadenylation site of the Agrobacterium nopaline synthase gene.
For both GUS reporter analysis and complementation of the mutant phenotypes rather a small 562 bp promoter was used because this promoter spans into the ATG of the adjacent and divergently oriented locus At3g12390 (see Supplemental Figure 1). However, a genomic clone gARP5 directed from this promoter fully complements all the arp5-1 mutant phenotypes suggesting that this is a true ARP5 promoter sufficient for normal gene expression and function.
Plant material and growth conditions
Arabidopsis thaliana wild type (Columbia), arp5-1 (Torrey Mesa, Garlic_1185_A12) and arp5-2 (GABi-Kat, GK-386F02) mutant lines and ARP4RNAi plants (Kandasamy et al., 2005a), and Nicotiana tabacum seedlings were grown in growth chambers maintained at 22°C with 16-h-light/8-h-dark periods. The mutants were backcrossed twice with wild type and selfed to produce genetically clean lines. The homozygous F2 plants identified by PCR using the primers provided in Supplemental Table 1 and their progeny were used for phenotypic analysis. Low (15-20%) and stable ARP4 protein expression in the ARP4RNAi line #11 was described previously (Kandasamy et al., 2005a). By crossing homozygous arp5-1 and ARP4RNAi plants, the double ARP4- and ARP5-deficient plants (4Ri arp5-1) were generated. Seeds were germinated and seedlings were grown on MS agar medium (Murashige and Skoog, 1962) and then transferred to the soil for further growth or grown directly on soil depending on the nature of experiments.
Plant treatments with DNA damaging agents
To study the effect of genotoxic agents, surface sterilized seeds (50/plate) were plated on MS agar medium (~35 ml/plate) supplemented with 1% sucrose, and after 2-day incubation at 4°C, the plates were transferred to 22°C growth chambers for germination and further growth. We tested three chemicals that induce DNA damage: hydroxyurea (Sigma), MMS (Sigma) and bleocin (=bleomycin; Calbiochem). Because of their short aqueous half-lives, we used high concentrations of freshly prepared chemicals, which gave clear and consistent phenotypes. Four-day-old seedlings on a plate were treated with 1.5 ml of different concentrations of hydroxyurea (250 mM, 500 mM and 1M), MMS (0.2%, 0.25%, 0.5%) or bleocin (25, 50, 100, 250 and 500 μg/ml or 1mg/ml) in sterile water. These different solutions were applied directly on the seedings and then were spread uniformly on the plate and allowed to diffuse gradually into the agar medium. The plates were subsequently incubated in the growth chamber at 22°C and after 3 to 8 days of treatment, the seedlings were transferred to new MS agar plates and grown vertically to examine root growth or horizontally to check the effect on shoot growth.
Antibodies and protein gel blot analysis
Anti-ARP5 monoclonal antibodies were raised against a recombinant and truncated ARP5 protein, comprised of the C-terminal 281 a.a. tagged at its C-terminus with 6 histidine residues. To generate this recombinant protein, we cloned an 843 bp ARP5 cDNA fragment amplified from a mature flower cDNA library with the C-terminal His-tag encoding sequence and the stop codon into the NcoI and XhoI sites of the bacterial pET15b vector (Invitrogen). The 32 KDa protein was produced in the expression cell line BL21 DE3 and was purified following the manufacturer’s instructions for his-tagged proteins. The purified recombinant protein (50 mg) was injected three times into mice and anti-ARP5 antibody producing hybridoma cells were isolated by the Direct Selection of Hybridoma method as described in Price et al. (2009). Thus, two independent monoclonal antibodies (MAbAPR5a and MAbARP5b) were isolated and they were purified on a protein G column (Bio-Rad) as described previously (Harlow and Lane, 1999).
To prepare protein samples for western blot analysis, we ground various frozen plant organs and callus tissue in a high salt extraction buffer: 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 10% glycerol, 1 mM ß-mercaptoethanol, and protease inhibitor cocktail (Roche, Germany), and after centrifugation re-extracted the pellet with sample buffer (Laemmli, 1970), boiled, centrifuged and used the supernatant for SDS-PAGE analysis as described earlier (Kandasamy et al., 2003). We detected ARP5 bands on protein blots using MAbAPR5a or MAbARP5b primary antibody and horseradish peroxidase conjugated anti-mouse secondary antibody (GE Healthcare). Equal loading of samples and uniform transfer of proteins to the membrane were monitored by coomassie brilliant blue staining of gels, and probing of duplicate blots with anti-PEP carboxylase polyclonal antibody (Rockland), respectively. Quantification of the bands developed with the ECL kit (GE Healthcare) was done using films with short exposure time and NIH Sci Image program.
Quantitative real-time (qRT) PCR analysis
RNA was isolated from 15-day-old seedlings of wild-type, arp-5-1 and arp5-2 mutant and an arp5-1/gARP5 complemented line using the RNeasy plant mini kit (Qiagen) and treated with RQ1 RNase-free DNase (Promega) before RT. 1.5 μg of treated RNA was added to RT reactions using the Invitrogen SuperscriptIII first-strand synthesis kit (Invitrogen) with random hexamer primers to make cDNA. qRT-PCR was performed on an Applied Biosystems 7500 real-time PCR system using SYBR Green detection chemistry. The cDNA populations were analyzed using UBIQUITIN10 transcripts as the endogenous control as described earlier (Ruzicka et al., 2007).
Histochemical GUS assays
Complete young seedlings or different excised organs of adult plants carrying the ARP5pt:GUS construct were incubated overnight (~16 h) on multi-well plates containing the X-Gluc staining solution (Jefferson et al., 1987). The stained samples were cleared with ethanol and observed and photographed with a Leica stereomicroscope.
Microscopy
Scanning electron microscopic observations of the leaf surfaces of cryo-preserved samples were made using a Leo field emission scanning electron microscope (LEO Electron Microscopy) as described previously (Kandasamy et al., 2005b). Light microscopy observations of different plant organs were performed with a Leica stereomicroscope or compound microscope. For immunofluorescence microscopy observations of ARP5 localization, young seedlings were fixed with paraformaldehyde, and the cells were dissociated and labeled with the anti-ARP5 antibodies MAbARP5a or MAbARP5b as described earlier (Kandasamy et al., 2003). FITC-conjugated anti-mouse secondary antibody was used for visualization of ARP5 localization.
Results
Arabidopsis ARP5 is localized to the nucleus
In Arabidopsis thaliana, ARP5 (locus # At3g12380) is one of the divergent ARPs with a complex gene structure encoding a protein of 80 kDa. The 726 amino acid (a.a.) Arabidopsis ARP5 protein sequence is only 28% identical to yeast ARP5 (755 a.a.) and 36% identical to human ARP5 (607 a.a.) (See alignment in Supplemental Fig. 2). Despite its weak sequence conservation, several independently assembled protein sequence trees have placed Arabidopsis ARP5 in the same clade as the yeast, human, and protist ARP5 homologs, significantly separated from other clades of ARPs and conventional actin (Blessing et al., 2004; Kandasamy et al., 2004; Muller et al., 2005).
In order to compare the functions of Arabidopsis ARP5 with that of the divergent yeast and human ARP5 sequences, we raised two independent anti-ARP5 monoclonal antibodies using a C-terminal, 281 amino acid truncated recombinant protein as an antigen. Both antibodies (MAbARP5a and MAbARP5b) reacted strongly with the 32 kDa recombinant plant ARP5 protein, rARP5-C, and in addition they recognized the 80 kDa full-length ARP5 protein in Arabidopsis seedling extracts on western blots (Fig. 1A). When we examined the subcellular localization of Arabidopsis ARP5 using fixed and dissociated root apical cells, MAbARP5a as well as MAbARP5b revealed that this protein is predominantly concentrated in the nucleoplasm of interphase cells (Figs.1C-F). Only faint labeling was detected in the cytoplasm and there was usually weaker labeling in the nucleolus (See Figs.1I, J). In dividing cells at metaphase, ARP5 was not associated with the chromatin but was dispersed into the cytoplasm (not shown). Moreover, we tested whether ARP5 homologs in other plants also revealed similar subcellular localization patterns by immunolabeling root and leaf cells of the distant dicot tobacco with MAbARP5a. Tobacco cells also revealed nuclear localization of its ARP5 protein similar to Arabidopsis ARP5 (Figs.1K-N) and western blot analysis revealed that ARP5 from both species have identical molecular mass (Fig. 1B). Evidently the epitope detected by MAbARP5a has been well conserved over the 100 million years since these two plants had a common ancestor.
Fig. 1.
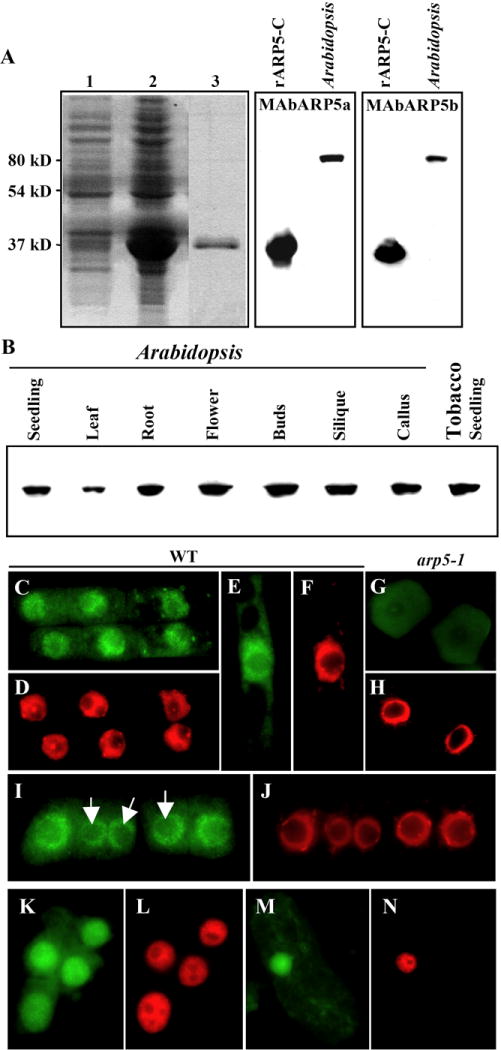
Expression and nuclear localization of ARP5 protein. (A) Reactivity of ARP5 antibodies MAbARP5a (middle panel) and MAbARP5b (right panel) with the truncated 32 KDa recombinant ARP5 (rARP5-C) and the native full length (80 KDa) ARP5 in the Arabidopsis seedling extract. An image of a coomassie brilliant blue stained gel showing the expression of the truncated ARP5 is depicted in the left panel. Lane 1, Control BL21 cells containing empty pET15b vector; lane 2, BL21 cells expressing ARP5 containing pET15b vector; Lane 3, Purified recombinant rARP5-C protein. (B) Western blot analysis of ARP5 in different organs and callus tissue of Arabidopsis with MAbARP5a. ARP5 homolog in tobacco seedling extract is shown in the last lane. (C-N) Immunocytochemical localization of ARP5 with MAbARP5a (C, G, I, K, M) and MAbARP5b (E) in root (C-L) and leaf cells (M, N). (C-J) Arabidopsis, (K-N) tobacco. (G, H) represent arp5-1 mutant cells and the rest are wild type (WT) cells. ARP5 staining with FITC-conjugated anti-mouse secondary antibody is shown in green and DNA staining with DAPI is shown in red. Arrows in I point to the nucleoli.
The Arabidopsis ARP5 promoter-reporter fusion and ARP5 protein are ubiquitously expressed
Previous microarray analyses of mRNA levels suggested that the ARP5 gene is expressed at low levels in all plant organs (Zimmermann et al., 2004). To refine our knowledge about the spatial and temporal pattern of expression of Arabidopsis ARP5, we studied the expression of ARP5 protein (Fig. 1B) and examined the activity of ARP5 regulatory sequences (Fig. 2). Western blot analysis of a variety of plant organs and callus tissue revealed that ARP5 protein is ubiquitously expressed in all major organs and callus (Fig. 1B). A reporter of ARP5 promoter activity was constructed with the β-glucuronidase (GUS) gene. When plants from different transgenic Arabidopsis lines expressing ARP5p:GUS fusion were incubated in X-glucuronide substrate, blue staining from the GUS activity was observed in all organs and most tissues (Fig. 2). The activity of the transgene was unexpectedly strong in some tissue such as the vascular tissue of differentiated mature roots and the connective tissue of anthers, and in different organs like hypocotyls, cotyledons, leaves, sepals, styles and filaments (Figs. 2A-K). Surprisingly, the GUS activity was relatively weak in the root apices at various stages of development (see Figs. 2A, E). However, microarray analyses of mRNA levels suggest that Arabidopsis ARP5 is moderately well expressed in the root tip (Zimmermann et al., 2004) and immunolocalization revealed the presence of ARP5 protein in the nuclei of root apical cells (Figs. 1C, E, I). More consistent with the GUS data, the intensity of ARP5 protein signal from the root apical cell nuclei shown in Figs. 1C, E and L was relatively poor compared to Arabidopsis ARP4, ARP6, or ARP7 (Deal et al., 2005; Kandasamy et al., 2003).
Fig. 2.

Expression of ARP5p:GUS reporter in Arabidopsis. (A) 28-h-old seedlings. (B) Two-day-old seedlings. (C) Cotyledon. (D) 20-day-old seedling. (E) Root. Note the lack of staining in the root tip and strong staining in the central vascular tissue. (F) Hypocotyl. (G) Cauline leaves from an adult plant. (H, I) Flower buds. (J) Flower. (K) Anthers and pistil. Co, cotyledon; Ct, connective tissue; Hy, hypocotyl; Fl, filament; Rt, root tip; Se, sepal; Sy, style; Vt, vascular tissue.
ARP5-deficient plants reveal defects in multicellular development
To study in more detail the role of Arabidopsis ARP5 during normal plant development, we analyzed the phenotypes of plants carrying T-DNA insertion mutations in the ARP5 gene. Two different T-DNA insertion alleles were examined (Fig. 3A). The arp5-1 mutant allele contained a T-DNA insertion in the 8th exon of the ARP5 gene. qRT-PCR analysis with independent pairs of ARP5-specific primers showed this insertion caused a complete knockout in the expression of ARP5 transcripts (Fig. 3B) and western blot analysis with MAbARP5a showed a complete knockout of ARP5 protein expression (Fig. 3C, left column, top row). Immunolabeling with MAbARP5a also revealed no detectable ARP5 protein in the nucleus of mutant root apical cells, confirming that arp5-1 is a null allele (see Figs. 1G, H). Likewise, arp5-2 allele had a T-DNA insertion in the terminal 11th exon that created a slightly truncated recombinant protein. The arp5-2 allele showed only a 20 to 30% reduction in transcript levels compared to wild type (Fig. 3B). However, western blot analysis revealed less than 5% of wild type levels of ARP5 protein in mutant seedling extracts, suggesting arp5-2 is a severe knockdown allele. Interestingly, both the alleles revealed an almost identical dwarf phenotype as shown for 32-day-old plants in Fig. 3D. The arp5-1 and arp5-2 mutant plants contained similar numbers of, but significantly smaller, leaves compared to the wild type. For instance, the wild type and the mutant plants all produced about 14 leaves before bolting, but the act5-1 mutant leaves were about 28% shorter in length and 35% smaller in width, whereas the arp5-2 plant leaves were 21% shorter in length and 31% smaller in width than wild type (Fig. 3E).
Because we characterized arp5-1 as a complete knockout, we selected this allele to pursue further analysis of multicellular phenotypes and the role of ARP5 in DNA repair. Our detailed analysis of the arp5-1 mutant suggested that the plants are smaller compared to wild type from the early seedling to the adult stage of development (see Figs. 4A, B), and all organs of the mutant plants are smaller than wild type (Figs. 4C-G, N). For example, the mutant plants produce slightly smaller flowers with narrower petals (Fig. 4C), and stunted hypocotyls (Figs. 4D, F) and shorter roots (Figs. 4E, N) with about 37% and 25% reduced length compared to wild type, respectively. The mutant leaves were often curled upwards, while wild type leaves never displayed such a curling phenotype during any stage of development (Figs. 4B, G).
Fig. 4.
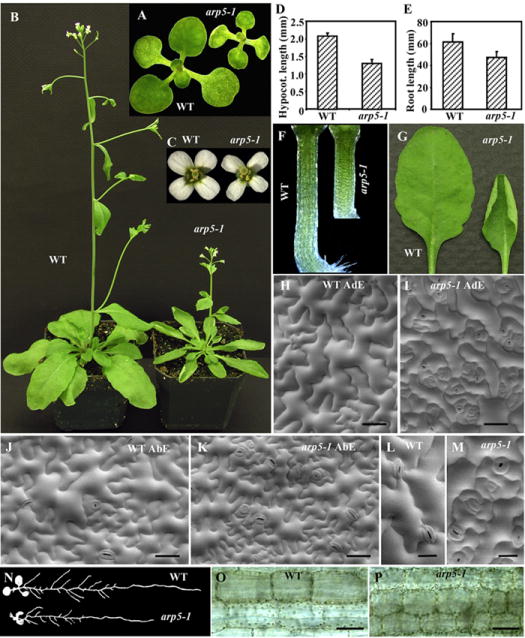
A detailed phenotypic analysis of the arp5-1 mutant allele. (A) 12-day-old wild type (WT) and arp5-1 seedlings. (B) ~40-day-old plants. (C) Flowers. (D) A bar graph depicting hypocotyl length of 6-day-old seedlings. (E) A bar graph showing root length of 12-day-old seedlings with SDs indicated. (F) Hypocotyls. (G) Rosette leaves. (H-M) SEM of adaxial (AdE; H, I) and abaxial (AbE; J, K) leaf epidermis. (L, M) Enlarged images of adaxial leaf epidermis showing uniform (wild type, L) vs delayed stomatal development in arp5-1 (M). (N) Root morphology of 12-day-old seedlings. (O, P) Enlarged images of wild type (O) and arp5-1 (P) mutant hypocotyls revealing difference in cell size. Bar = 50 μm in H-K, O and P; 20 μm in L and M.
To examine whether the smaller mutant plant organs are composed of smaller cells or fewer cells than wild type, we performed scanning electron microscopy (SEM) analysis of the leaf epidermis (Figs. 4H-M) and light microscopic observation of the hypocotyls (Figs. O, P). SEM of the adaxial epidermis suggested that the mutant leaf epidermal cells were much smaller than wild type and displayed poorly developed lobes (Figs. 4H, I). The abaxial epidermis of mutant leaves was also composed of relatively smaller cells than wild type, but unlike the adaxial mutant cells they displayed a similar lobe pattern as the wild type cells (Figs. 4J, K).
The difference in the development of lobes between the mutant adaxial and abaxial epidermal cells and the presence of small and some moderately sized cells in the abaxial epidermis versus mostly smaller cells in the adaxial epidermis may be the reason for the upward curling of mutant leaves. In addition, the mutant leaf epidermis contained 2-3 times more stomata per unit area than the wild type (see Figs. 4H-K). The development of a large percentage (40-50%) of stomata is also delayed in the mutant leaves, hence they exhibited highly crowded stomata at various stages of development as shown in Fig. 4M. The wild type leaves of identical age mostly contained all stomata at similar developmental stage (Fig. 4L). A close examination of the mutant hypocotyls suggested that they were also composed of smaller cells than wild type (Figs. 4O, P). Thus, the smaller organs of the dwarf mutant plants were composed of smaller cells than wild type.
To confirm that the different morphological phenotypes observed in the arp5-1 mutant were the result of deficiency in the expression of ARP5 protein, we transformed the mutant plants with a genomic clone (gARP5) containing the ARP5 coding sequence under the control of its endogenous 5′ and 3′ regulatory flanking sequences. Fifteen independent transgenic lines were isolated and the protein expression for five of them is shown in Fig. 5A (top panel). As revealed by western blot analyses all transgenic plants expressed the transgene, but the quantification of the ARP5 bands suggested that the levels of protein expression from the transgene varied from 55% to 160% compared to wild type (100%). Even after long exposure of protein blot to the film, there is still no ARP5 protein expression detected in the untransformed apr5-1 mutant (Fig. 5A, upper panel) and PEPC analysis revealed that all the lanes contained approximately equal levels of total protein (Fig. 5A, lower panel). All transformed plants, however, showed restoration of plant (Figs. 5B, G) and organ (Figs. 5C, F) size to the wild type levels. The plants with 50 to 60% higher levels of ARP5 than wild type often showed marginally (10-15%) improved root growth over wild type size (see Fig. 5F). SEM of leaf adaxial epidermis suggested that the cell size and morphology of the wild type and complemented plants were indistinguishable (Figs. 5D, E). Thus, the gARP5 transgene fully restored normal morphology to dwarf arp5-1 mutant plants.
Fig. 5.
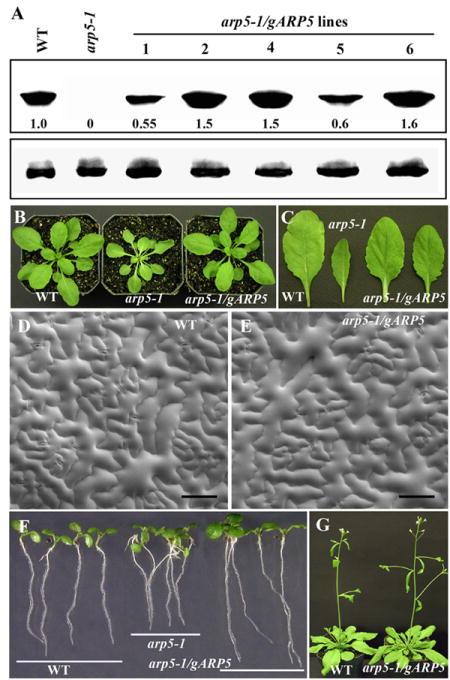
Complementation of arp5-1 mutant phenotypes with the expression of a genomic ARP5 transgene. (A) Western blot analysis of ARP5 protein levels in different arp5-1 complemented lines expressing transgenic gARP5 (arp5-1/gARP5 lines). WT, wild type. Upper panel is probed with MAbARP5a and the lower panel is probed with anti-PEPC antibody. The levels of ARP5 protein are indicated at the bottom the blot. (B) 30-day-old plants. (C) Rosette leaves. (D, E) SEM of adaxial leaf epidermis of wild type (D) and a complemented plant (E). (F) 6-day-old seedlings showing complementation of root growth phenotype. (G) 40-day-old plants. Bar = 100 μm in D and E.
ARP5 deficient plants are hypersensitive to DNA damaging agents
INO80 is a large chromatin remodeling complex, the activity of which in yeast and mammals requires Ino80, a DNA-dependent Snf2-like ATPase, ARP4, ARP5 and ARP8. Budding yeast mutants defective in Ino80 function are not only hypersensitive to reagents that induce DNA double-strand breaks (DSBs), but also to those that impair replication fork progression (Shen et al. 2000; Shimada et al. 2008). In Arabidopsis, plants deficient in Ino80 protein have reduced homologous recombination (HR) frequency, but they are not hypersensitive to genotoxic agents such as Mitomycin C, MMS, bleomycin and UV-C (Fritsch et al., 2004). In higher eukaryotes with larger genomes, such as Arabidopsis and humans, HR is usually rare and most DSBs are rejoined by mechanisms other than HR, such as the non-homologous end joining (NHEJ) pathway (Britt and May, 2003; West et al., 2002). Indeed, recent studies directly implicate the INO80 complex in DNA replication and DNA repair, most notably at DSBs (Shimada et al., 2008).
To understand whether Arabidopsis ARP5, a homolog of which in yeast and mammals is a subunit of the INO80 complex, has any role in the DSB repair or other DNA damage repair, we treated wild type and ARP5-defective plants with three distinct DNA damaging agents. Seeds were germinated and grown on MS agar germination medium (GM) for 4 days before exposing the seedlings to hydroxyurea (HU), methyl methanesulfonate (MMS), or bleocin. HU inhibits DNA synthesis by reducing the dNTP pool, whereas MMS alkylates nitrogen and oxygen atoms of the DNA bases inducing single- and double-stranded breaks in DNA. Bleocin cleaves double-stranded DNA as well as inhibiting DNA synthesis. Because all three chemicals have very short aqueous half-lives (only a few hours), freshly prepared stocks containing high concentrations of these agents were applied directly to seedlings on agar plates and the chemicals were allowed to diffuse into the medium of a defined volume (see Materials and Methods). After 3 to 8 days of treatment, plants were transferred to new GM plates and grown vertically for observation of root growth and horizontally for observation of shoot growth. For controls, wild-type and mutant plants were either treated with sterile water or allowed to grow further on GM without any chemical treatment.
The arp5-1 mutant plants treated for 3 days with HU (1.5 ml of 500 mM HU per 35 ml agar medium in a plate) showed about 72% inhibition of root growth compared to similarly HU-treated wild-type plants (Figs. 6A, B). At this concentration or other concentrations tested (see Methods), HU slows down the growth of both wild type and mutant plants, but root and shoot growth inhibition was always drastically higher in the mutant than wild type (Figs. 6A, C). On the other hand, the arp5-1 plants grown on control GM plates had roots that were only about 16% shorter than wild type (Figs. 6A, B). The HU hypersensitivity phenotype of arp5-1 plants was fully rescued in mutant plants complemented with the gARP5 transgene as shown in Fig. 6D. Thus, ARP5 is involved, either directly or indirectly, in repairing the DNA damage caused by HU.
Fig. 6.
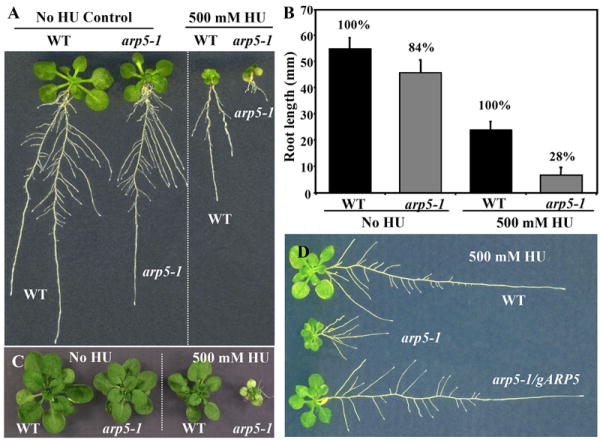
Hypersensitivity phenotype of arp5-1 to hydroxyurea. (A, B) Wild type and arp5-1 seedlings grown for 12 days throughout on MS plates (control) or 4-day-old seedlings treated with 500 mM hydroxyurea (HU) for three days and grown further for 5 days on MS plates. A bar graph representing root length phenotype is shown in (B). (C) 28-d-old arp5-1 plants showing hypersensitive HU shoot phenotype. (D) Rescue of the HU-sensitive root growth phenotype of arp5-1 by complementation with gARP5 (arp5-1/gARP5).
Similarly, arp5-1 mutant plants are more sensitive to MMS than wild type, and here too the gARP5 transgene fully reversed the hypersensitive phenotype of the mutant (Figs. 7A, B). When 4-day-old Arabidopsis seedlings were exposed to 0.2% MMS for 3 or 4 days and then grown further on GM medium, the root growth was inhibited 61% compared to wild-type. The mutant plants treated with MMS also showed poor lateral root formation (Fig. 7A). Surprisingly the roots of a complemented plant line (arp5-1/gARP5 line 2 in Fig. 5A) expressing 50% higher levels of ARP5 protein than wild-type plants grew slightly better than wild-type (see Figs. 7A, B). The shoot growth of mutant plants was also drastically affected after treatment with MMS compared to wild type or complemented plants (Figs. 7C-E).
Fig. 7.
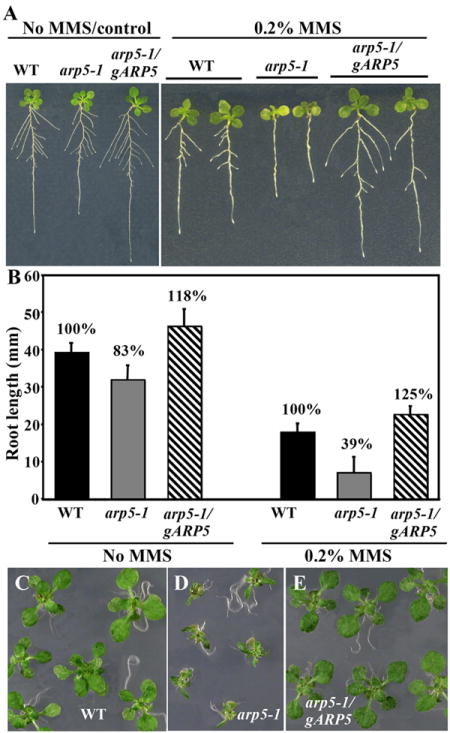
Hypersensitivity phenotype of arp5-1 to MMS. (A, B) Wild type, arp5-1 and complemented plants (arp5-1/gARP5) grown for 11 days throughout on MS plates (control, left panel) or 4-day-old seedlings treated with 0.2% MMS for three days and grown further for 5 days on MS plates (right panel). A bar graph representing the root length phenotype is shown in (B). (C-E) Shoot phenotype. 4-day-old seedlings grown on MS plates were treated with 0.2% MMS for 6 days and grown further on MS plates.
To further explore the participation of ARP5 in DSB repair pathway, we treated the mutant and wild type seedlings with 25μg/ml bleocin, a generic form of bleomycin. As shown in Figs. 8E and G, the arp5-1 mutant seedlings are more sensitive than wild type to bleocin. Even though longer treatment of bleocin (8-day) severely inhibits the growth of both the wild type and arp5-1 mutant plants, the mutant seedlings revealed chlorosis and eventual death much before wild type (Fig. 8G). After 5-day treatment of bleocin, the root growth of arp5-1 mutant seedlings was inhibited 80% more compared to wild type seedlings (Fig. 8E), whereas the complemented mutant plants (arp5-1/gARP5) showed almost identical root growth as wild type (Fig. 8F). Our results with all three DNA damaging agents tested, therefore, clearly suggest that ARP5 participates in the repair of DNA damage.
Fig. 8.

ARP4 and ARP5 deficient plants reveal common and unique phenotypes.
(A) Early flowering phenotype. WT, wild type; 4Ri, ARP4RNAi; 5-1, arp5-1 mutant, 4Ri arp5-1, ARP4 and ARP5 double deficient plant. Note the ARP4RNAi and ARP4 and ARP5 double deficient plants flower earlier than WT and arp5-1 plants. The arp5-1 and the double deficient plants have curled leaves. (B) Rosette leaves. The ARP4RNAi, and ARP4 and ARP5 double deficient plants have fewer leaves (8 or 9) compared to WT and arp5-1 plants (13 or 14). (C) Delayed floral senescence phenotype. Note in the inflorescences of ARP4RNAi and 4Ri arp5-1 double deficient plants, all the flowers shown have persistent sepals and petals. In wild type and arp5-1 plants only 4 or 5 flowers have sepals and petals. (D-G) Sensitivity of ARP4- and ARP5-deficient seedlings to bleocin. (D) Control plants treated with no bleocin. (E) 4-day-old WT, arp5-1, arp5-2 and ARP4RNAi seedlings treated with 0.25 μg/ml bleocin for 5 days, and grown further vertically on MS plates to monitor root growth. The numbers represent the average root length in mm of ten samples ± standard deviation. (F) Complementation of bleocin induced root growth phenotype of arp5-1 mutant with gARP5 transgene. (G) 4-day-old seedlings treated with 0.25 μg/ml bleocin for 8 days, and grown further horizontally on MS plates to monitor plant growth and survival. (H-I) Sensitivity of various ARP-deficient seedlings to MMS. 4-day-old seedlings treated with 0.2% (H) or 0.4% (I) MMS for 4 days, and grown further on MS plates to monitor root growth or plant survival. (J) Sensitivity of various ARP-deficient seedlings to hydroxyurea. 4-day-old seedlings were treated with 500 mM HU for 4 days and grown further for a week on MS plates.
ARP4- and ARP5-deficient plants revealed independent as well as common developmental phenotypes
The nuclear ARPs function only as subunits of chromatin remodeling complexes. ARP4 and ARP5 are both components of the INO80 complex in yeast and mammals (Kitayama et al., 2009; Shen et al., 2003). We therefore analyzed ARP4- and ARP5-deficient plants to see whether they share common phenotypes and if the ARP4 and ARP5 proteins function similarly in controlling different pathways of multicellular development and DNA repair. We used the arp5-1 null mutant allele and an ARP4RNAi epiallelic line in which RNA interference was used to silence ARP4 protein expression to 15-20% of wild type. Both ARP4- and ARP5-deficient plants were dwarf and had smaller organs such as leaves that were composed of smaller cells. Typically, the arp5-1 mutant plants revealed a delay in the development of a large number (~45%) of stomata and hence the leaves often contained stomata at various developmental stages (Figs. 4I, K, M). In addition, the mutant leaves had two to three times more stomata per unit area than wild type (see Figs. 4H and I). The ARP4-deficient plants have leaves with small cells, but they reveal almost normal development and even distribution of stomata similar to wild type (Meagher et al., 2007). The arp5-1 plants also had rosette leaves that were often curled upwards (Fig. 8A), but the ARP4-defective plants produced wavy but not such curly leaves (Fig. 8B).
ARP4RNAi plants specifically displayed early flowering and delayed floral senescence phenotypes. During long day photoperiod growth conditions, the arp5-1 plants flowered almost at the same time (~29 days after germination) as wild type with 13 to 14 rosette leaves, but the ARP4RNAi plants flowered a week earlier with only 7 to 8 leaves (Figs. 8A, B). Wild type and arp5-1 plant inflorescences have four to five flowers with intact sepals and petals, whereas ARP4RNAi plants have 15 or more flowers with intact sepals and/or petals. Surprisingly, even after full development of the siliques, the ARP4RNAi flowers retained the perianth organs (see Fig. 8C). Plants deficient in both ARP4 and ARP5 proteins revealed a combination of all the morphological phenotypes: smaller, fewer and curled rosette leaves, early flowering, and delayed floral senescence compared to wild type (Figs. 8A-C). In summary, the ARP4- and ARP5-deficient plants exemplified only a small subset of common developmental phenotypes and each displayed novel independent phenotypes (see Supplemental Table 2).
ARP4- and ARP5-deficient plants are both hypersensitive to DNA damaging agents
Treatment with genotoxic agents such as bleocin (Figs. 8D-G), MMS (Figs. 8H, I), and hydroxyurea (Fig. 8J) suggested that ARP4RNAi plants are more sensitive than wild type, but are not as sensitive as arp5-1 mutant plants. To verify whether this disparity is due to the presence of trace quantities of ARP4 protein in ARP4RNAi plants and no ARP5 protein in the arp5-1 null allele, we compared the slightly leaky ARP5 mutant allele arp5-2 with ARP4RNAi plants. For example, five days treatment with 0.25 μg/ml bleocin arrested almost all (80%) root growth of arp5-1 seedlings, but the root growth of arp5-2 mutant allele and ARP4RNAi plants was inhibited only 50 and 58% compared to wild type, respectively. Thus the leaky arp5-2 and ARP4Ri plants very much resembled each other in their response to Bleocin. Moreover, eight days treatment with 0.25 μg/ml bleocin killed almost all arp5-1 seedlings, but still, although sick, approximately 30% of ARP4RNAi and arp5-2 plants were alive (Fig. 8G). Eventually, however, all ARP-deficient plants became chlorotic and died before wild type (not shown). Similar results were observed with MMS (0.2%) and HU (500 mM) treatment, with the null arp5-1 showing stronger root growth inhibition phenotype compared to the severe knockdown arp5-2 and ARP4RNAi plants (Figs. 8H, J). However, at higher MMS concentration (0.4%) all the ARP deficient plants became chlorotic and eventually died while the wild type plants still remained green and healthy (Fig. 8I). Although arp5-1, arp5-2 and ARP4Ri plants are all sensitive to the genotoxic agents, the response is most severe with arp5-1 allele perhaps because it is a null allele. The arp5-2 and ARP4Ri plants may reveal moderate phenotypes because they still contain traces of respective ARP proteins.
Discussion
Arabidopsis ARP5 is a ubiquitously expressed nuclear protein. ARP5-deficient plants were severely altered in their multicellular development. The dwarfed mutant plants produced hypocotyls, leaves, stems, and floral organs that were all smaller than wild-type, being generally composed of smaller cells. The mutant leaves displayed small cells on their adaxial surfaces and stochastic clusters of small and moderately sized cells on their abaxial surfaces resulting in small upwardly curled leaves with serrated edges that were distinct from wild type. However, the overall plant architecture was similar to wild type. Two independent mutant alleles, a null arp5-1 and a severe knockdown allele arp5-2, produced the same morphological phenotypes. A genomic clone gARP5 fully complemented all morphological and cellular phenotypes of the arp5-1 mutant. Hence, all the mutant phenotypes described are the result of ARP5 deficiency and not due to differences in the genetic background of the mutant(s) from our wild type plant line.
Moreover, the arp5-1 mutant seedlings were hypersensitive to treatment with the DNA damaging agents HU, MMS, and bleomycin (bleocin) relative to wild type. Bleomycin is a DNA oxidizing agent that causes DSBs directly, resulting primarily in small deletions (Guttenplan et al., 2004). Repairing deletions in yeast and animal cells generally requires the ARP5-dependent activities of the INO80 complex in all three steps of recombination repair: recognizing damage, repairing the DNA, and restoring normal chromatin functions including transcription and DNA replication (Conaway and Conaway, 2009; Kitayama et al., 2009). Hence, it is not surprising that ARP5-defective Arabidopsis seedlings were hypersensitive to bleomycin, as are ARP5-defective animal cells (Kitayama et al., 2009). HU inhibits ribonucleotide reductase, causing DNA polymerase to be starved for nucleotide triphosphates, and the stalling of replication forks. MMS is a DNA base-modifying agent that generally causes base substitution mutations. If HU or MMS treatments are severe enough they may cause deletions that require recombination repair (Galli and Schiestl, 1999; Koc et al., 2004). The hypersensitivity of Arabidopsis ARP5-defective plants to HU and MMS may result from loss of chromatin remodeling required for recombination repair and/or restoration of normal DNA replication, paralleling the HU and MMS hypersensitivity reported for ARP5- or Ino80-defective yeast (Papamichos-Chronakis and Peterson, 2008; Shen et al., 2003; Shimada et al., 2008). In contrast, human ARP5 did not complement the HU and MMS hypersensitivity of yeast ARP5-defective mutants, while it partially complemented ultraviolet light and hydrogen peroxide hypersensitivity (Kitayama et al., 2009).
In yeast and animals, ARP5 has only been identified in INO80 complexes. If Arabidopsis ARP5 and the essential Swi2-related Ino80 subunit may only function together in classical INO80 complexes, the loss of either ARP5 or Ino80 function should produce the same epigenetic phenotypes. Therefore, it is surprising that our ARP5 defective plants showed hypersensitivity to chemical DNA damaging reagents, while Ino80 defective Arabidopsis plants did not (Fritsch et al., 2004). Furthermore, we reported more dramatic developmental defects for ARP5 deficiency than Fritsch et al. (2004) found for Ino80 mutants. Among a few possibilities, two seem most likely to explain this discrepancy. First, most studies including those examining Ino80 function in Arabidopsis (Fritsch et al., 2004) have incorporated low concentrations of drugs into aqueous media, where they will be quickly hydrolyzed and rendered ineffective. All three drugs have aqueous half-lives of only a few hours at room temperature, so we treated plants directly with high concentrations of freshly prepared HU, MMS, and bleomycin stocks (see Materials and Methods) to ensure that sufficient DNA damage occurred to require recombination repair. Perhaps the decay products of these chemicals can inhibit cell growth, as is observed in many studies, but by mechanisms independent from those inducing DSBs and requiring recombination repair and INO80 function. We observed the same, but much weaker phenotypes than we reported herein, when we incorporated these drugs into the media.
Second, Arabidopsis encodes 44 members of the Swi2-related DNA dependent ATPase family. Perhaps in addition to functioning with Ino80, ARP5 functions together with different Swi2-homologs in altered isoforms of the INO80 complex that control development and DNA repair. Although ARP5 may have an unknown interaction with any number of Swi2 homologs, there are two Swi2-related proteins that are particularly close sequence homologs of Arabidopsis Ino80 (1507 a.a.) and both are well characterized. The similarly large Pie1 (Swr1, 2055 a.a.) is 44% identical to Ino80 and the much smaller Ddm1 (DDM1, CHR1, 754 a.a.) is 41% identical in the sequence regions that may be aligned. Pie1 is a nucleosome remodeling factor known to function in a SWR1-related histone variant exchange complex. Loss of Pie1 activity causes many developmental phenotypes distinct from those we observed for ARP5, such as early flowering and serrated and elongated leaves (Deal et al., 2007; Noh and Amasino, 2003). Ddm1 is also a nucleosome remodeling factor, but its activity is essential to both the methylation of lysine 9 in histone H3 and methylation of C residue in CpG dinucleotides and both these activities maintain basal levels of gene silencing (Brzeski and Jerzmanowski, 2003). Ddm1 defective plants display a wide variety of developmental abnormalities affecting most organs including small leaves and late flowering (Kakutani et al., 1996; Kakutani et al., 1995). Loss of activity for Arabidopsis Pie1, Ddm1, and seven other Swi2-related genes produced defects in repair of gamma radiation DNA damage (Shaked et al., 2006), leaving open the possibility that one or more of these proteins may combine with ARP5 in novel remodeling and repair complexes.
Considering that nuclear ARPs are only found in chromatin remodeling complexes as heterodimers with other ARPs or actin, and yeast ARP5 is found along with ARP4, ARP8, and actin in the INO80 complex, it was reasonable to consider that ARP5-defective plants might share all their phenotypes with, for example, ARP4-deficient plants (Supplementary Table 2). ARP5- and ARP4-defective Arabidopsis plants have in common the small leaf morphology phenotype primarily due to small epidermal cells (Kandasamy et al., 2005a) and hypersensitivity to DNA damaging agents. Although the ARP4- and ARP5-deficient plants are both sensitive to HU, MMS and bleocin, the ARP4RNAi plants are relatively less sensitive than arp5-1 mutant plants. This may be due to the presence of very low levels of ARP4 protein (15 to 20% of wild type levels) in the knockdown RNAi lines compared to the lack of any ARP5 protein in the null arp5-1 mutant. This assumption is partly supported by the fact that the slightly leaky arp5-2 mutant allele, similar to ARP4RNAi plants, also revealed relatively moderate sensitivity phenotype to genotoxic agents. On the other hand, ARP5 may also function in additional complexes that do not contain ARP4 as a subunit and that are involved in DNA repair. ARP4-defective plants have many other independent phenotypes (e.g. early flowering and delayed floral senescence), which is unsurprising considering that ARP4 is found commonly in the vast majority of chromatin remodeling and modification complexes (Meagher et al., 2009; Olave et al., 2002). Thus, ARP4-deficient plants should have many other epigenetic defects not observed for ARP5-deficient plants. What is surprising, however, is that ARP5-defective plants have a few developmental phenotypes that are not found for ARP4-defective plants, including excess stomata and upward curling of leaves. These unique phenotypes again suggest that Arabidopsis ARP5 may function in some remodeling complexes outside of those that require ARP4, implying an activity for ARP5 independent of the classical INO80 complex.
Based primarily on work in yeast and to some extent on studies in mammals, ARP5-containing chromatin complexes are known to dynamically remodel nucleosomes. These temporary changes to chromatin structure lead to altered gene expression and development and to DNA repair, which may be inherited for a few hours or a few cell generations, but most are not inherited indefinitely by the organism’s offspring. The apparently random appearance of patches of small cells mingled among moderately sized cells in the abaxial epidermis and excessive numbers of stomatal complexes developing on both surfaces of ARP5-defective leaves resemble the stochastic phenotypes typically resulting from inappropriate epigenetic control of a Drosophila eye pigment gene (Csink and Henikoff, 1996). Our ARP5-defective leaf cell phenotypes also compare well with the first epigenetic phenotype reported for any ARP-deficiency, reversible switching of colony color from white to red and back again to white in ARP4-defective yeast expressing an epigenetically controlled reporter for color (Jiang and Stillman, 1996). In summary, our data on ARP5 gene and protein expression and the phenotypes of ARP5-deficient mutants suggest that Arabidopsis ARP5 functions are essential to normal epigenetic control in plants.
Supplementary Material
Acknowledgments
We acknowledge the Arabidopsis Biological Resource Center, USA and the GABI-Kat MPI for Plant Breeding Research, Germany for providing mutant seeds. We thank the UGA’s Center for Ultrastructural Research for the SEM facilities and Lori King-Reid for critically reading the manuscript. This work was supported by a grant from the National Institutes of Health (GM36397).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–442. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Britt AB, May GD. Re-engineering plant gene targeting. Trends Plant Sci. 2003;8:90–95. doi: 10.1016/S1360-1385(03)00002-5. [DOI] [PubMed] [Google Scholar]
- Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- Deal RB, Kandasamy MK, McKinney EC, Meagher RB. The Nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell. 2005;17:2633–2646. doi: 10.1105/tpc.105.035196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell. 2007;19:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Odeyale O, Shen CH. Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochem Biophys Res Commun. 2008;373:602–606. doi: 10.1016/j.bbrc.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B. The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol Cell. 2004;16:479–485. doi: 10.1016/j.molcel.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Galli A, Schiestl RH. Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat Res. 1999;429:13–26. doi: 10.1016/s0027-5107(99)00097-4. [DOI] [PubMed] [Google Scholar]
- Guttenplan JB, Khmelnitsky M, Haesevoets R, Kosinska W. Mutational spectrum of bleomycin in lacZ mouse kidney: a possible model for mutational spectrum of reactive oxygen species. Mutat Res. 2004;554:185–192. doi: 10.1016/j.mrfmmm.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane DP. Using Antibodies: A Laboratory Manual. Coldspring Harbor, NY: CSH Laboratory Press; 1999. p. 495. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Stillman DJ. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci U S A. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Richards EJ. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 1995;23:130–137. doi: 10.1093/nar/23.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB. Plant actin-related proteins. Trends Plant Sci. 2004;9:196–202. doi: 10.1016/j.tplants.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB. Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 2005a;41:845–858. doi: 10.1111/j.1365-313X.2005.02345.x. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Deal RB, Meagher RB. Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol. 2005b;138:2019–2032. doi: 10.1104/pp.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB. Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J. 2003;33:939–948. doi: 10.1046/j.1365-313x.2003.01691.x. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB. ACTIN-RELATED PROTEIN8 encodes an F-box protein localized to the nucleolus in Arabidopsis. Plant Cell Physiol. 2008;49:858–863. doi: 10.1093/pcp/pcn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama K, Kamo M, Oma Y, Matsuda R, Uchida T, Ikura T, Tashiro S, Ohyama T, Winsor B, Harata M. The human actin-related protein hArp5: nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp Cell Res. 2009;315:206–217. doi: 10.1016/j.yexcr.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Koc A, Wheeler LJ, Mathews CK, Merrill GF. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem. 2004;279:223–230. doi: 10.1074/jbc.M303952200. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Meagher RB, Deal RB, Kandasamy MK, McKinney EC. Nuclear actin-related proteins as epigenetic regulators of development. Plant Physiol. 2005;139:1576–1585. doi: 10.1104/pp.105.072447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, Kandasamy MK, Deal RB, McKinney EC. Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 2007;17:325–332. doi: 10.1016/j.tcb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Meagher RB, Kandasamy MK, McKinney EC. Actin-related proteins in epigenetic control. Intern Rev Cell Mol Biol. 2009 doi: 10.1016/S1937-6448(09)77005-4. In Preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005;16:5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 1962;15:473–497. [Google Scholar]
- Noh YS, Amasino RM. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell. 2003;15:1671–1682. doi: 10.1105/tpc.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- Price PW, McKinney EC, Wang Y, Sasser LE, Kandasamy MK, Matsuuchi L, Milcarek C, Deal RB, Culver DG, Meagher RB. Engineered cell surface expression of membrane immunoglobulin as a means to identify monoclonal antibody-secreting hybridomas. J Immunol Methods. 2009;343:28–41. doi: 10.1016/j.jim.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka DR, Kandasamy MK, McKinney EC, Burgos-Rivera B, Meagher RB. The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 2007;52:460–472. doi: 10.1111/j.1365-313X.2007.03257.x. [DOI] [PubMed] [Google Scholar]
- Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, Bray CM. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 2002;31:517–528. doi: 10.1046/j.1365-313x.2002.01370.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox ( www.genevestigator.ethz.ch) Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


