Abstract
Historical episodes of natural selection can skew the frequencies of genetic variants, leaving a signature that can persist for many tens or even hundreds of thousands of years. However, formal tests for selection based on allele frequency skew require strong assumptions about demographic history and mutation, which are rarely well understood. Here, we develop an empirical approach to test for signals of selection that compares patterns of genetic variation at a candidate locus with matched random regions of the genome collected in the same way. We apply this approach to four genes that have been implicated in syndromes of impaired neurological development, comparing the pattern of variation in our re-sequencing data with a large-scale, genomic data set that provides an empirical null distribution. We confirm a previously reported signal at FOXP2, and find a novel signal of selection centered at AHI1, a gene that is involved in motor and behavior abnormalities. The locus is marked by many high frequency derived alleles in non-Africans that are of low frequency in Africans, suggesting that selection at this or a closely neighboring gene occurred in the ancestral population of non-Africans. Our study also provides a prototype for how empirical scans for ancient selection can be carried out once many genomes are sequenced.
INTRODUCTION
Many of the most successful genome-wide scans for signals of natural selection to date have focused on the ‘long range haplotype test’ (LRH test), which searches for common haplotypes that extend much longer distances than would be expected under neutrality (1–3). Although this is a powerful method, it can only detect the signatures of natural selection that occurred within the last ∼10 000 years. An alternative approach to screening the genome for signatures of selection is to search for regions where the allele frequency distribution is distorted compared with the expectation in the absence of selection (4). Teshima et al. (5) showed that such signals are sometimes too subtle to be detected with statistical significance in a genome scan. However, screens for regions of the genome that have a distorted allele frequency spectrum do have power to detect a subset of real selection events (5), and are attractive because they can detect selection events that are much more ancient than the ones that are accessible to the LRH test. It has not been possible to carry out such studies on a large scale in a robust manner, however, because genome-wide data sets like the International Haplotype Map Project (HapMap) (6,7) are strongly affected by ascertainment bias: non-randomness in the polymorphisms chosen for analysis (8). Large numbers of genomes are currently being sequenced, and it should be possible to use these data to carry out robust genome-wide scans for distortions in allele frequencies.
The most straightforward way to search for signals of selection is to compare patterns of genetic variation at candidate loci to theoretical expectations from a neutral model of population history not involving selection. Most such analyses have assumed a constant-sized ancestral population, an ‘infinite sites’ model for mutation, a constant rate of recombination across the genome, and no sequencing error (9–12). However, deviations from a neutral model can produce false signals of selection both by skewing the expected values of test statistics, and by inflating the variance of the underlying distributions of test statistics (4,10,13–21). The variance effect is particularly problematic. For example, demographic histories like population bottlenecks are predicted to increase variability of test statistics across loci, even in the absence of natural selection. This can produce test statistics that appear to be multiple standard deviations from the mean under the theoretical expectation for a constant-sized population, but in fact are entirely consistent with the demographic history expected from a bottleneck.
Here, we focus on an ‘empirical’ approach to searching for signals of selection in humans. By comparing the pattern of genetic variation at candidate genes to patterns in random regions that are in principle all affected by the same history and the same processes of mutation, recombination and sequencing error, it should be possible to assess whether the pattern of genetic variation at tested loci is unusual (22,23).
Two approaches are possible in order to empirically test for selection: non-parametric and parametric. The most obvious approach, which is non-parametric, involves rank-ordering loci according to their value of a test statistic that is sensitive to selection, and designating significant loci as ones that are outliers from the distribution. The parametric approach recognizes that the distributions of some test statistics [such as Tajima's D (10)] are expected to have about the same shape whatever the demographic history, except that the parameters such as the mean and variance vary. By estimating the mean and variance from the genome wide distribution, it should be possible to extrapolate the tail, and to confidently infer P-values that are much more extreme than would be possible based on the rank ordering method. This is analogous to the ‘Genomic Control’ method that is commonly used to detect disease alleles in case–control association studies, where an ‘inflation factor’ of the chi-squared distribution is estimated from the genome-wide data, and the scaled chi-squared distribution is then used to determine P-values with more precision than would be possible with rank-ordering (24). Below, we report simulation experiments under different selection scenarios, which confirms that the ‘Genomic Control’ method for detecting selection results in overall higher sensitivities compared with the rank ordering method.
To test the Genomic Control approach for screening for selection, we chose four candidate genes. All four had previously shown evidence of positive selection in the last tens of millions of years on the primate lineage leading to humans, and all four have been associated with syndromes impaired neurological development, a category that in anecdotal studies has shown evidence of being unusually affected by selection during this period (25–33). In this study, we sought to test the distinct but related hypothesis that these genes have also been unusually subject to selection in the last few hundred thousand years since anatomically modern human arose in the fossil record. We chose one gene (FOXP2) as a ‘positive control’ since previous studies had found evidence of selective sweeps (possibly multiple rounds) at this locus (25,34,35). The other three genes had no evidence of selection within the last few hundred thousand years at the time when this study was designed. Mutations at FOXP2 have been shown to affect comprehension and production of human speech (36). Mutations at ASPM are associated with neurological impairment and reduced cortical size (37). Mutations at GPR56 are associated with reductions in the size of the frontal cortex (38). Mutations at AHI1 are associated with Joubert syndrome and motor and behavioral abnormalities (39).
We re-sequenced about 15–30 kilobases (kb) from each of the four genes using long-read Sanger sequencing, focusing on segments containing mutations that have been documented as medically important, or containing novel amino acid changes on the human lineage (Table 1). At FOXP2, we re-sequenced the region that was analyzed in a previous study (27) (exons 4–8), and also extended the sequencing to another region spanning exons 14–16 that had not previously been analyzed (Table 1). For all the regions we examined, we re-sequenced 16 samples of North European ancestry (CEU) and 16 West Africans from Nigeria (YRI), identified single nucleotide polymorphisms (SNPs) using automated software (Methods), and then genotyped the discovered SNPs within these segments in 90 CEU and 90 YRI HapMap samples (40,41), using a nearly identical protocol as was used by the ENCODE Project to sequence and then genotype about 2.5 Mb of the genome at the Broad Institute (7). We applied an array of statistical tests to detect significant deviations at the re-sequenced regions from the ENCODE comparison data, using four different summary statistics of genetic variation: Tajima's D (10), Fu and Li's F (12), and Fay and Wu's H (11) and FST between CEU and YRI.
Table 1.
Re-sequenced segments in this study
| Gene | Re-sequenced region | Span in base pairs | Physical coordinates (HG16) | Reason for ascertainment |
|---|---|---|---|---|
| AHI1 | Exon 5–12 | 23 127 | Chr6: 135,748,716-135,771,842 | Non-synonymous mutations, causal for Joubert Syndrome; elevated dN/dS (39). |
| Exon 15–17 | 4875 | Chr6: 135,731,090-135,735,964 | Functionally important coding region (39) | |
| ASPM | Exon 2–4 | 6334 | Chr1: 194,396,156-194,402,489 | Two frameshift deletions in exon 3 causing microcephaly (37); elevated dN/dS (31) |
| Exon 18 | 6755 | Chr1: 194,356,820-194,363,574 | One nonsense mutation causing microcephaly (37); elevated dN/dS (31). | |
| Exon 20–25 | 6251 | Chr1: 194,346,319-194,352,569 | One nonsense deletion in exon 21 causing microcephaly (37) | |
| FOXP2 | Exon 4–8 | 26 250 | Chr7: 113,818,098-113,844,347 | Two human lineage specific mutations (25), and evidence for more recent selection (25,29) |
| Exon 14–16 | 4372 | Chr7: 113,855,123-113,859,494 | Non-synonymous mutation putatively causal for a severe speech and language disorder (36) | |
| GPR56 | Exon 3–15 | 15 780 | Chr16: 57,458,562-57,474,341 | Numerous non-synonymous/missense mutations (38) |
Mekel-Bobrov et al. (42) previously reported an analysis of genetic variation at ASPM, suggesting that natural selection occurred at this gene within the last few 10 000 years. Our own empirical comparisons showed that the pattern at ASPM was not unusual compared with random regions of the genome (we published this result as a Technical Comment) (43). The fact that the patterns of genetic variation at ASPM stand out from simulations of a history that is meant to be similar to that of the analyzed populations, but do not stand out from empirical data, highlights the value of empirical comparisons and the difficulty of fully modeling the processes that produce patterns of genetic variation in real data. Here, we present the results for all four genes (which necessarily involves re-reporting the ASPM data), which allows us to more systematically evaluate the empirical approach. Our study serves as a proof-of-principle, showing how empirical comparisons can be used as the basis for robust tests of selection based on allele frequency skews once many human genomes are sequenced (44,45).
RESULTS
Genic regions and SNP ascertainment
Previous studies have suggested that AHI1, ASPM, FOXP2 and GPR56 are implicated in neurodevelopmental processes in the human brain (25,28,31,36–39,42,46) (Table 1). We re-sequenced eight segments from within the four genes, choosing the regions based on the fact that they co-localized either with clusters of non-synonymous sites that are thought to be responsible for neurological impairment in patients (36–39), or with amino acid changes that have arisen on the human lineage in the last tens of millions of years of evolution based on comparison with other primates (Table 1). These re-sequenced regions were chosen to span large contiguous regions when possible, and as a result, most of the sequenced data were in introns. Our tests for selection were based on skews in the allele frequency distribution rather than on analysis of amino-acid changes, and hence intronic sequence was just as valuable as exonic sequence for our analysis.
We designed a SNP ascertainment and genotyping strategy that closely matched what was used by the ENCODE Project, allowing us to use ∼2.5 Mb of the ENCODE data that had also been genotyped at the Broad Institute as a large-scale empirical control data set (Supplementary Material, Fig. S1). We matched the ascertainment strategy of SNPs with ENCODE in five respects: (a) we used the same set of 16 CEU and 16 YRI samples; (b) we used the same re-sequencing approach; (c) we used the same SNP calling software and algorithm; (d) we partially used the same SNP genotyping method [instead of a round of Illumina Golden Gate genotyping (47) followed by a round of Sequenom genotyping to fill in gaps (48), we used two rounds of Sequenom genotyping]; and (e) we used the same 30 CEU and 30 YRI trios in the follow-up genotyping (Supplementary Material, Fig. S1).
In total, we discovered 270 SNPs by re-sequencing 16 CEU and 16 YRI samples. Among the 222 SNPs for which we were able to design assays, 200 successfully genotyped and passed our quality control procedures. To ensure identical SNP ascertainments between our project and the ENCODE Project, we also re-curated the ENCODE genotype data set to include only the SNPs ascertained in the CEU or YRI (since our re-sequencing did not includes samples from East Asians). By matching our data collection to ENCODE, we ensured that any differences found between the four tested genes and the control data would be due to unusual patterns (potential natural selection) in the tested genes.
Qualitative signatures of natural selection at FOXP2 and AHI1
To determine whether there is any evidence of natural selection at the four genes, we first visually examined the derived allele frequency (DAF) spectrum of each gene in CEU and YRI (Fig. 1). After a selective sweep that fixes a newly arising advantageous mutation in the population, variants that are linked to it are expected to have skewed frequencies, with some having a high DAF (if they originally resided on the selected haplotype and hitchhiked to high frequency during the selective sweep), and some having a low DAF (suggesting that they were among the alleles that arose after the sweep) (4).
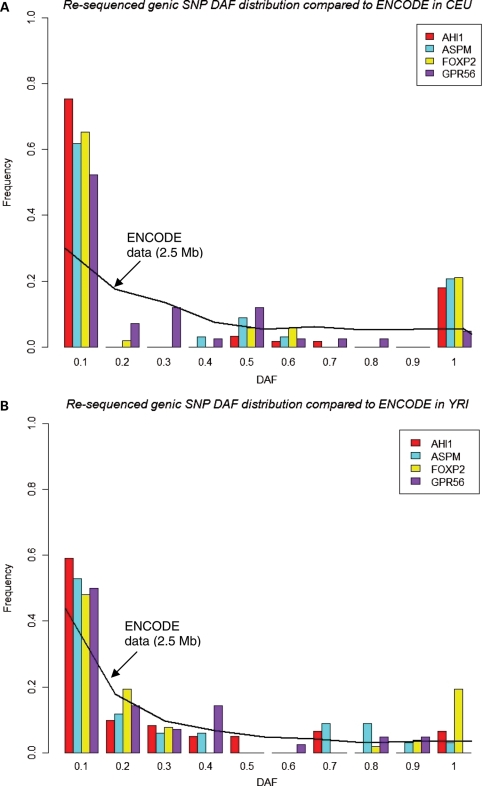
Figure 1.
DAF distribution by bin in SNPs from re-sequenced regions compared with SNPs from the ∼2.5 Mb of ENCODE data (black lines) in (A) CEU and (B) YRI. The DAF distribution is shown for each gene, merging the different interrogated segments. The derived allele is inferred by comparison with the chimpanzee allele.
Figure 1 compares the DAF distribution of SNPs in the four genes with ENCODE data. In CEU, three genes demonstrate greater density in both the high and low ends of the DAF distribution than is expected from ENCODE data (AHI1, ASPM and FOXP2; Fig. 1A). In YRI, one gene (FOXP2) has an evidently skewed DAF distribution (Fig. 1B).
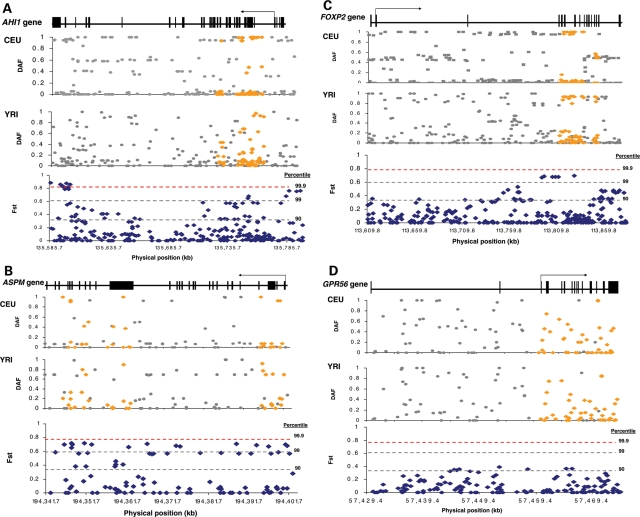
To provide a more comprehensive picture of the DAF distribution in the vicinity of the four genes, we also merged our re-sequencing data with data from HapMap, and visualized the data by plotting the DAF of each SNP against its physical distance (Fig. 2). Our tests for natural selection based on allele frequency patterns in the re-sequenced regions are based on matching only to ENCODE data (presented in the next section). However, the union with HapMap shows qualitatively that the same signals of natural selection that we found at FOXP2 and AHI1 are also present in the flanking regions that we did not re-sequence (Fig. 2).
Figure 2.
DAF and FST distributions of SNPs ascertained in the regions we re-sequenced (yellow) and HapMap SNPs (grey) within the physical genomic positions spanned by the genes, in (A) AHI1, (B) ASPM, (C) FOXP2 and (D) GPR56. The exon/intron map for the gene is shown at the top; the DAF plotted against physical position for CEU and YRI in the middle; and FST compared with percentiles from all of HapMap (90th, 99th and 99.9th) at the bottom. The scale for each of the genes is different (and hence the density of SNPs is different) because of their different physical distance spans.
Significant signals of selection at FOXP2 and AHI1 compared with empirical data
To assess whether the patterns of genetic variation at each of the re-sequenced segments are significantly different from what is expected based on empirical comparisons to data from random regions, we calculated three statistics that are designed to be sensitive to a history of selection at a locus: Tajima's D (10), Fu and Li's F (12) and Fay and Wu's H (11). Although all these statistics have known mathematical distributions in the case of a constant-sized population and an infinite sites model of mutation, human population history and genetic data are not well described by these models. To deal with this, we treated these quantities as statistical metrics with unknown distributions, and inferred the distributions empirically from data obtained from random regions of the genome. Unmodeled processes like demographic history should equally affect the entire genome, whereas selection should cause specific regions to stand out in a way that we can detect.
To collect empirical comparison data, we selected non-overlapping windows from the ENCODE data that matched the genetic distance and number of segregating sites in each of the tested regions that we analyzed, repeating this procedure for CEU and YRI separately. The statistical significance of each re-sequenced segment was then assessed by a direct rank-ordering technique compared with the empirical control data. The number of matching windows in the ENCODE data varied substantially depending on the genetic distance span and number of segregating sites in the re-sequenced segment, ranging from as few as 7 (for AHI1 exons 5–12 in YRI) to no more than 190 (for ASPM exon 18 in YRI; Tables 2 and 3). Many of the observed test statistics were more extreme than all windows of the empirical data from ENCODE (Tables 2 and 3).
Table 2.
Statistical tests in CEU for unusual allele frequencies in re-sequenced segments compared with ENCODE control data
| CEU | Re-sequenced region | Genetic distance span (cM) | #Segregating sites | Matches in empirical comparison | Tajima's D; empirical P-value, MBB P-valuea; (MBB σ) | Fu and Li's F; empirical P-valuea; (MBB σ) | Fay and Wu's H; empirical P-valuea;(MBB σ) |
|---|---|---|---|---|---|---|---|
| AHI1 | Exon 5–12 | 0.0017 | 27 | 28 | −1.6; 0.071*, 0.0062; (−2.74) | −1.5; 0.071*; (−4.17) | −9.6; 0.36; (−1.81) |
| Exon 15–17 | 0.00040 | 9 | 105 | −1.3; 0.019*, 0.020; (−2.33) | −1.1; 0.057; (−3.44) | −2.7; 0.29; (−1.03) | |
| ASPM | Exon 2–4 | 0.00024 | 8 | 97 | −1.3; 0.021*, 0.056; (−1.91) | −0.08; 0.16; (−1.72) | −2.3; 0.33; (−0.82) |
| Exon 18 | 0.0028 | 9 | 174 | 0.7; 0.97, 0.93; (−0.085) | 0.2; 0.33; (−1.16) | −3.4; 0.22; (−1.35) | |
| Exon 20–25 | 0.000097 | 9 | 56 | −0.2; 0.32, 0.16; (−1.41) | 1.0; 0.64; (−0.37) | −2.9; 0.36; (−0.96) | |
| FOXP2 | Exon 4–8 | 0.0093 | 14 | 93 | −1.9; 0.022*, 0.0025; (−3.03) | −0.4; 0.11; (−2.40) | −6.0; 0.15; (−1.70) |
| Exon 14–16 | 0.0043 | 6 | 186 | 3.4; 0.011*, 0.0052; (2.79) | 2.2; 0.011*; (1.90) | 0.1; 0.82; (0.55) | |
| GPR56 | Exon 3–15 | 0.093 | 25 | 22 | 1.9; 0.36, 0.18; (1.33) | 1.8; 0.64; (0.53) | 0.9; 0.36; (0.81) |
aIn each cell, the first value reports the value of the statistic. The second value reports the P-value based on rank-ordering compared with ENCODE data (for Tajima's D, this is followed by a P-value for a two-tailed z-test based on the MBB procedure). The third value in parenthesis gives the number of standard deviations (σ) from the mean.
*Indicates observed value that is more extreme than all empirical comparisons. Since P-values are 2-sided, the most extreme rank-ordering P-value that is possible is 2/n, where n is the number of windows in the control data.
Table 3.
Statistical tests in YRI for unusual allele frequencies in re-sequenced segments compared with ENCODE control data
| YRI | Re-sequenced region | Genetic distance span (cM) | #Segregating sites | Matches in empirical comparison | Tajima's D; empirical P-value, MBB P-valuea; (MBB σ) | Fu and Li's F; empirical P-valuea; (MBB σ) | Fay and Wu's H; empirical P-valuea; (MBB σ) |
|---|---|---|---|---|---|---|---|
| AHI1 | Exon 5–12 | 0.0017 | 45 | 7 | −0.3; 0.86, 0.73; (−0.35) | 1.2; 0.86; (0.38) | −1.9; 0.86; (−0.11) |
| Exon 15–17 | 0.00040 | 14 | 56 | 0.4; 0.96, 0.98; (0.020) | 1.0; 0.75; (−0.17) | 0.3; 0.75; (0.43) | |
| ASPM | Exon 2–4 | 0.00024 | 12 | 58 | 0.6; 0.97, 0.94; (0.080) | 1.4; 0.79; (0.51) | −1.6; 0.38; (−0.34) |
| Exon 18 | 0.0028 | 9 | 190 | −0.7; 0.32, 0.19; (−1.32) | 0.8; 0.89; (−0.45) | −4.8; 0.074; (−2.68) | |
| Exon 20–25 | 0.000097 | 10 | 53 | 0.8; 0.79, 0.74; (0.33) | 1.5; 0.68; (0.59) | 0.1; 0.98; (0.39) | |
| FOXP2 | Exon 4–8 | 0.0093 | 40 | 56 | −1.1; 0.036*, 0.056; (−1.91) | 0.5; 0.18; (−1.49) | −10.2; 0.18; (−1.87) |
| Exon 14–16 | 0.0043 | 9 | 165 | −0.8; 0.16, 0.15; (−1.45) | −0.4; 0.061 (−2.10) | −3.3; 0.12 (−1.79) | |
| GPR56 | Exon 3–15 | 0.093 | 41 | 21 | 0.1; 0.86, 0.44 (−0.78) | 0.4; 0.095*; (−2.28) | −2.3; 0.86; (−0.36) |
aIn each cell, the first value reports the value of the statistic. The second value reports the P-value based on rank-ordering compared with ENCODE data (for Tajima D, this is followed by a P-value for a two-tailed z-test based on the MBB procedure). The third value in parenthesis gives the number of standard deviations (σ) from the mean.
*Indicates observed value that is more extreme than all empirical comparisons. Since P-values are 2-sided, the most extreme rank-ordering P-value that is possible is 2/n, where n is the number of windows in the control data.
FOXP2 showed strong signals of selection, confirming previous reports of an unusual DAF distribution at the locus (25). In exons 4–8, Tajima's D in CEU was below all estimates from the empirical distribution of ENCODE regions (P < 0.022), a signal that was also seen in YRI (P < 0.036). These results are consistent with an episode of natural selection in the common history of CEU and YRI (35). In contrast, for exons 14–16 in CEU, the statistics are larger than the expectation for Tajima's D (P < 0.011) and Fu and Li's F (P < 0.011) (Table 2), which reflects an excess of alleles at intermediate frequency (Fig. 2C), a pattern that is also seen in East Asians (not shown). These results could be explained by a selective sweep followed by genetic drift associated with the out-of-Africa bottleneck.
The gene showing the strongest deviation from the ENCODE data is AHI1, particularly at exon 5–12 in CEU (Table 2), where Tajima's D and Fu and Li's F are both outside the distribution of matched windows from ENCODE (P < 0.071; Table 2). The re-sequencing data from AHI1 exons 15–17 in CEU provides further evidence of selection, with Tajima's D falling outside the distribution of windows from ENCODE (P < 0.019; Table 2). However, AHI1 does not show any evidence for selection in YRI, suggesting that the selection probably occurred in the ancestors of CEU after the split from YRI (Table 3).
We found almost no evidence for selection in ASPM or GPR56, in either CEU or YRI, by any test (Tables 2 and 3). The only exception was in ASPM exons 2–4 in CEU, where we observe an unusually low Tajima's D statistic that is more extreme than the values seen in all 97 empirical comparisons (P < 0.021) (Table 2); however, this is the only signal of selection at this gene that we observed by any test. We conclude that the gene does not show evidence of selection after correcting for multiple hypothesis testing, confirming our previous report about patterns of variation at this gene (43).
Genomic Control, an improvement on empirical methods to detect selection
A major limitation of non-parametric searchers for selection using empirical methods is that the P-values can only be as significant as the number of regions to which a test statistic is compared (at least when significance is assessed by rank-ordering comparison regions and assessing where a test statistic falls). This is particularly problematic for empirical comparison data sets that are small or based on contiguously re-sequenced regions (such as the ENCODE regions and whole-genome re-sequencing data), since correlation in genealogical histories among neighboring loci means that there are effectively fewer comparison regions than would be expected if all the windows were unlinked. As a result, even if a test statistic at a candidate gene for selection is more extreme than is observed in all N regions to which we empirically compare it, in the face of the correlation among regions we cannot confidently say that the P-value is <1/N.
To address these limitations, we used a Genomic Control method. The Genomic Control method is inspired by an idea from case–control association studies (24). The idea is that even if the distribution of a test statistic sensitive to selection has a mean and variance that is systematically different from what is expected under the constant-sized model due to the fact that the true demographic history is different, the parametric shape of the distribution may be conserved regardless of the demographic history. Thus, the Genomic Control strategy uses the empirical comparison data set to estimate quantities like the mean and variance of the distribution (taking into account uncertainty in these quantities), and then compares the test statistic for the candidate selected loci to these distributions.
We focused our exploration of the Genomic Control method on the Tajima's D statistic, which is expected to approximately conform to a normal distribution whether for a constant-sized or a non-constant-sized population, which we confirmed by examining the real ENCODE data (Supplementary Material, Fig. S2). We estimated the mean and variance of the Tajima's D distribution from the ENCODE data, while taking into account increased uncertainty in these parameters due to correlation in genealogical histories within an ENCODE region by a Moving Block Bootstrap (MBB) (Materials and Methods). It allowed us to assess the statistical significance of our observations, taking into account the limited data set size and linkage disequilibrium (LD) in the data set.
We carried out coalescent simulations to test the performance of our empirical methods for screening for selection—and in particular Genomic Control—under different selection and demographic scenarios. Eight different selection scenarios were simulated, involving the selected allele occurring within the tested region or at some distance from the tested region, and four different selection intensities (selection coefficients of 0.1, 0.05, 0.01 and 0.005) (Methods, Table 4). We compared the simulated selected loci to unselected regions 500 kb in size (chosen to match the size of the ENCODE regions), and analyzed all the data using the same procedure as our real data. The demography we explored for the simulations of selection was that of a constant-sized population (Table 4). For the simulations of non-selected regions, we also explored population bottlenecks and expansions (Methods). Simulating histories that are not that of a constant-sized population is important, as it allowed us to verify that demographies like expansions and bottlenecks do not inflate the false-positive rate in empirical scans for selection, as they do in traditional non-empirical scans that compare observed data to the theoretical expectation for a constant-sized population.
Table 4.
Simulations of empirical tests of selection
| Simulated selection scenario | Empirical approaches for detecting selection |
|
|---|---|---|
| Genomic Control power (%) | Rank method power (%) | |
| s = 0.1, causal mutant in the middle of a 30 kb region | 94 | 92.7 |
| s = 0.05, causal mutant in the middle of a 30 kb region | 91.9 | 88.5 |
| s = 0.01, causal mutant in the middle of a 30 kb region | 60.9 | 54.3 |
| s = 0.005, causal mutant in the middle of a 30 kb region | 35.8 | 29.1 |
| s = 0.1, causal mutant at 220 kb to the 30 kb region | 13 | 10 |
| s = 0.05, causal mutant at 220 kb to the 30 kb region | 3.4 | 2.7 |
| s = 0.01, causal mutant at 220 kb to the 30 kb region | 2 | 1 |
| s = 0.005, causal mutant at 220 kb to the 30 kb region | 1.4 | 0.8 |
| False positive rate for simulated neutral regions under a constant sized population model | 2.3 | 0.2 |
| False positive rate for simulated neutral regions under a population growth model | 0.5 | 1 |
| False positive rate for simulated neutral regions under a population bottleneck growth model | 2.8 | 0.8 |
| False positive rate for simulated neutral regions under a population growth model compared with control regions under a constant size model (this highlights the false-positives that arise under traditional tests of selection) | 41.0 | 35.3 |
The simulations confirmed that the empirical approach does not result in an inflated false-positive rate in screens for selection (Table 4). Interestingly, the Genomic Control method had a somewhat higher power for detecting selection (by ∼5–10%) compared with the non-parametric rank order approach. We believe that this reflects the fact that Genomic Control can more accurately estimate statistical significance for data points that fall outside the empirical rank order distribution; however, Genomic Control provides no increased power compared with rank ordering for data points that fall within the empirical distribution. Both empirical approaches had very little power to detect selection when selection occurred at some distance to the region of interest (Table 4).
Application of Genomic Control strengthens the selection signal at FOXP2 and AHI1
We applied the empirical approach to screening for selection to real data from the four genes. We found significant results at FOXP2 and AHI1, but not at the other genes. The P-values inferred by Genomic Control in CEU are highly significant for AHI1 exons 5–12 (P = 0.0062), FOXP2 exons 4–8 (P = 0.0025) and FOXP2 exons 14–16 (P = 0.0052) (Table 2). In YRI, the MBB indicates marginally significant signals at FOXP2 exons 4–8 (P = 0.056). The P-values at AHI1 and FOXP2 obtained by the Genomic Control approach are more extreme than those we obtained using the non-parametric rank-ordering approach, reflecting the fact that the Genomic Control approach allows us to extrapolate P-values further into the tail of the distribution.
The Genomic Control analysis finds no evidence for selection at ASPM, consistent with our published Technical Comment (43). The strongest signal is in CEU in exons 2–4 (P = 0.056), which is not significant after correcting for multiple hypothesis testing. At exon 18, the region previously highlighted as potentially containing a selected variant (42), there is no evidence for selection (P = 0.93), and this region is not in LD with exons 2–4. We also found no evidence for selection in the ASPM gene when it is treated as a whole (combining the three re-sequenced segments).
Analysis of allele frequency differentiation highlights alleles at AHI1
We also examined the allele frequency differentiation between CEU and YRI at all SNPs using the FST statistic (49,50). If selection occurred after the separation of North Europeans and West Africans, selected loci would be expected to be unusually differentiated in frequency across populations. On the other hand, selection that occurred before population separation is not expected to affect frequency differentiation.
We compared FST in the re-sequenced segments using the entire HapMap Phase II data set (41) as an empirical control. (The comparison with HapMap data is conservative for our analysis, and hence we do not need to restrict our analysis to comparison with ENCODE regions; Materials and Methods.) We observed a handful of highly differentiated SNPs (>99th percentile) in the re-sequenced segments, with the most striking signal at AHI1. When we extended the analysis to all HapMap Phase II SNPs (Fig. 2A), we observed even higher differentiation at the proximal end of AHI1. Here, the FST estimates of 10 different SNPs fall between the 99.9th and 99.99th percentile of HapMap.
The extraordinarily high SNP frequency differentiation at the proximal end of AHI1—about 220 kb away from the re-sequenced segments (Fig. 2A and Table 5)—suggests that if selection indeed occurred at AHI1, it may not have been in the re-sequenced segment, but could equally well have been centered elsewhere, with the speed of the selective sweep (due to a strong selective coefficient) being responsible for the large size of the affected locus. When we visually examined the DAF pattern around AHI1 in HapMap over a larger region than is shown in Supplementary Material, Fig. S4, it is in fact not clear whether the putative selective signal is due to variation at AHI1 at all. The DAF pattern is distorted in CEU over a region that extends to a couple of megabases, and this region contains many known genes in addition to AHI1 (Supplementary Material, Fig. S4). Thus, although AHI1 is an interesting candidate gene for selection, we cannot rule out the possibility that the signal we observe is due to hitchhiking from a powerful selective sweep at a neighboring gene.
Table 5.
Derived allele frequencies for SNPs in the vicinity of AHI1 that are highly differentiated between African and non-African populations
| SNP | Build34 | Region | HapMap |
HGDP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEU (%) | CHB+JPT (%) | YRI (%) | European (%) | West Asian (%) | Central and South Asian (%) | East Asian (%) | East Oceanian (%) | Native American (%) | African (%) | |||
| rs7453135 | 134,439,442 | proximal | 85 | 99 | 10 | 79 | 72 | 77 | 96 | 65 | 98 | 19 |
| rs9688660 | 134,445,499 | proximal | 85 | 99 | 10 | |||||||
| rs9321439 | 134,708,764 | proximal | 84 | 77 | 6 | |||||||
| rs6922545 | 134,709,544 | proximal | 84 | 77 | 8 | |||||||
| rs7775514 | 134,712,805 | proximal | 84 | 77 | 6 | |||||||
| rs9493942 | 134,713,104 | proximal | 85 | 77 | 7 | 77 | 60 | 83 | 85 | 100 | 98 | 18 |
| rs726948 | 134,714,168 | proximal | 85 | 77 | 13 | |||||||
| rs2327484 | 134,714,298 | proximal | 85 | 77 | 13 | 78 | 60 | 83 | 86 | 96 | 98 | 26 |
| rs1052502 | 135,587,135 | AHI1 | 95 | 86 | 6 | 93 | 85 | 90 | 89 | 30 | 96 | 25 |
| rs7741046 | 135,595,272 | AHI1 | 95 | 91 | 8 | |||||||
| rs2327612 | 135,597,189 | AHI1 | 97 | 97 | 13 | |||||||
| rs2142956 | 135,597,202 | AHI1 | 97 | 97 | 13 | |||||||
| rs7766656 | 135,598,171 | AHI1 | 92 | 91 | 8 | 92 | 87 | 90 | 92 | 30 | 97 | 30 |
| rs6933077 | 135,598,904 | AHI1 | 97 | 97 | 15 | |||||||
| rs9483826 | 135,600,086 | AHI1 | 97 | 97 | 8 | |||||||
| rs7765602 | 135,601,578 | AHI1 | 97 | 97 | 14 | |||||||
| rs7765971 | 135,601,742 | AHI1 | 97 | 97 | 14 | |||||||
| rs7756167 | 135,603,575 | AHI1 | 95 | 91 | 8 | |||||||
| rs9389294 | 135,787,775 | AHI1 | 94 | 92 | 17 | |||||||
| rs9402709 | 135,793,821 | AHI1 | 94 | 92 | 17 | |||||||
| rs4896149 | 135,797,073 | AHI1 | 95 | 92 | 17 | |||||||
| rs958072 | 135,808,136 | distal | 94 | 92 | 17 | |||||||
| rs9494266 | 135,832,143 | distal | 94 | 92 | 15 | 92 | 86 | 94 | 91 | 43 | 96 | 28 |
| rs7752627 | 135,856,515 | distal | 94 | 92 | 15 | |||||||
| rs9483910 | 136,461,542 | distal | 99 | 86 | 7 | |||||||
| rs9321552 | 136,462,182 | distal | 99 | 86 | 7 | |||||||
| rs3823159 | 136,463,297 | distal | 99 | 86 | 7 | 100 | 94 | 95 | 84 | 46 | 57 | 25 |
| rs6570067 | 136,477,401 | distal | 99 | 86 | 7 | |||||||
| rs1480642 | 136,480,098 | distal | 99 | 86 | 8 | |||||||
| rs3734548 | 136,488,969 | distal | 98 | 78 | 7 | |||||||
| rs3799396 | 136,492,042 | distal | 98 | 78 | 7 | |||||||
| rs7753890 | 136,496,827 | distal | 99 | 79 | 7 | 97 | 91 | 89 | 84 | 37 | 98 | 24 |
| rs11154872 | 136,778,327 | distal | 87 | 64 | 15 | |||||||
| rs3778308 | 136,786,352 | distal | 87 | 64 | 15 | |||||||
| rs9399183 | 136,798,058 | distal | 87 | 64 | 15 | 82 | 81 | 79 | 61 | 32 | 68 | 24 |
Note: This table reports all HapMap Phase II SNPs in the AHI1 region where we observe DAF < 17% in YRI and DAF > 83% in CEU. Most of these SNPs have a similarly elevated DAF in CHB+JPT, suggesting that the selective sweep at this locus occurred in the common ancestral population of North Europeans and East Asians after the split from West Africans. Where available, we also report data for the Human Genome Diversity Panel (HGDP) (51) for a wider range of populations, pooling samples into seven geographical regions following Ref. (61). The only non-African populations that do not consistently exhibit high derived allele frequencies across these regions are East Oceanians.
We also explored whether the evidence of high frequency derived alleles is unique to CEU, or whether it is also present in other non-African populations. Table 5 shows the frequencies for all SNPs across about 2 Mb centered at AHI1 that were highly differentiated in frequency between West Africans and North Europeans, with DAF < 17% in YRI and DAF > 83% in CEU. The observation of a high DAF in CEU always coincides with a high DAF in CHB and JPT. For 10 SNPs, we were also able to obtain data from Human Genome Diversity Panel (HGDP) samples (51), which showed the same high DAF pattern in all non-Africans with the possible exception of West Oceanians (Papuans and Melanesians). In contrast, all the HGDP African populations, including the San and Mbuti hunter-gatherers, had low derived allele frequencies at these sites (Table 5). These results indicate that the putative selection event near AHI1 probably occurred in the common history of non-Africans, around the time of the dispersal out of Africa.
In contrast with the pattern at AHI1, there is no similar signal of high FST between Africans and non-Africans at FOXP2, consistent with inference from previous studies that the selection event occurred more than a hundred thousand years ago (25). In particular, the fact that the selection signal is shared in CEU and YRI suggests that at least some of the selection at this locus occurred before these two populations diverged.
LRH tests do not detect recent positive selection at the four genes
We carried out LRH analyses (2,52,53) to test for evidence of more recent positive selection. The rationale for LRH is that a selective sweep can drive an advantageous allele to high frequency rapidly enough that the haplotype background on which the allele arises does not have much time to break down by recombination. Regions that have experienced recent positive selection can thus have a distinct signature of a high frequency allele associated with long-range LD. The LRH test (2,52,53) is particularly sensitive to selection that occurred in the past ∼10 000 years of history (53).
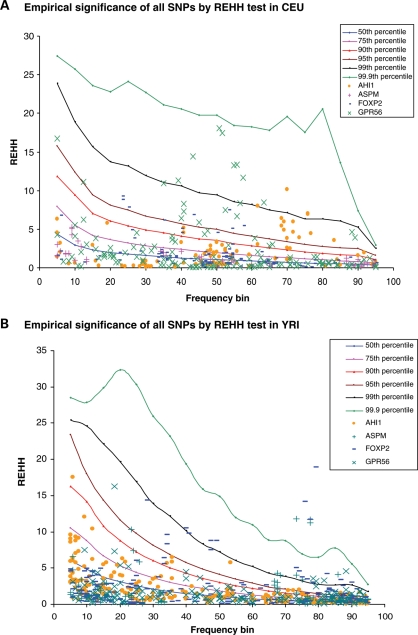
To carry out the LRH analyses, we integrated our data with HapMap to achieve greater marker density in the four genic regions. For each application of the LRH test, we used a single SNP as a core SNP (Materials and Methods). Comparing to the background of random genetic variants in CEU in all of HapMap Phase II, 4 SNPs in AHI1 and 8 in GPR56 exceeded the 99th percentile with nominally significant P-values (Fig. 3A and Supplementary Material, Table S1a). In YRI a handful of SNPs in ASPM (n = 11), FOXP2 (n = 18) and GPR56 (n = 4) exceeded the 99th percentile, and some SNPs in ASPM (n = 5) and FOXP2 (n = 3) even exceeded the 99.9th percentile. Although these results are intriguing, many SNPs were tested and thus there is a multiple hypothesis testing concern (1204 tests in CEU and 1804 in YRI, including tests on both sides of the core SNPs). It has previously been observed that SNPs with extreme relative extended haplotype homozygosity (REHH) signals can be observed in a genome scan without being convincingly associated with signals of natural selection (1). Frazer et al. (41) recommended an empirical rule of thumb, which was to identify regions as strong candidates for selection only if >10% of SNPs within a 100 kb region have an LRH score [ln(REHH) deviation from the genome-wide average] greater than 3.92. When we apply this criterion, none of the genes crossed the threshold, consistent with the failure of these four genes to emerge as strong candidates in previous genome-wide scans (1,3).
Figure 3.
Empirical significance of the LRH test for all SNPs in the four genes for (A) CEU and (B) YRI. We merged the SNPs discovered in the re-sequenced genic segments with the HapMap SNPs within each gene, and used each SNP across the span as a core to carry out a LRH test, separately reporting the scores at both sides of the core (2,52). LRH values were split into 20 bins (0–5, 5–10, 10–15, … , 95–100%) by their respective allele frequency, and compared to the empirical LRH distribution obtained from the HapMap SNPs using the same LRH methods. A few SNPs in ASPM and FOXP2 in YRI exceed the 99.9th percentile when compared with the HapMap. However, after accounting for multiple hypotheses testing, none of the genes stands out as showing an unusual LRH test compared with the genome-wide distribution (Supplementary Material, Table S1).
DISCUSSION
We have carried out a re-sequencing study of four genes relevant to neurological development, and applied an array of statistical tests to examine the hypothesis of selection during the last few hundred thousand years of human evolution at these loci. We confirmed the previously reported evidence of natural selection at FOXP2, and in particular showed that it likely occurred prior to the divergence of West African and North European populations, since the signal is shared in these two populations (35). We also highlighted a genomic region containing AHI1 as likely to have been affected by selection. At this locus, there is high differentiation between Africans and non-Africans throughout a region of a couple of megabases including AHI1, suggesting that the sweep occurred in a population that accounts for a larger proportion of ancestry in non-Africans than in Africans. The pattern also provides some intriguing hints about the duration of the putative selective sweep. Since the region of high frequency differentiation spans about 0.02 Morgans, we hypothesize that there was an intense period of selection during which the allele swept through the population in not much more than 1/0.02 = 50 generations. This calculation is intentionally approximate, as the goal of this study is to show how empirical comparisons can establish evidence of natural selection rather than to quantify the parameters of selection. A modeling analysis could obtain a more precise estimate.
Our confirmation of a previously detected signal of natural selection at FOXP2, and our detection of a novel signal near AHI1, provides some evidence for the hypothesis that genes that appear to be important in syndromes affecting neurological development, also are candidates for selection in the last few hundred thousand years. A gene's relevance to neurological impairment syndromes is no guarantee of selection, however, as we did not find a signal at either ASPM [contradicting the result of (42)] or GPR56 (5). Moreover, in the case of AHI1, the selection signal is so broad that our results do not distinguish between whether AHI1 itself or a neighboring locus was the subject of selection.
Our study is also significant as a proof-of-principle for how empirical comparisons can add rigor to studies of natural selection using the distribution of allele frequencies. Many past studies have been re-sequenced a gene, and then compared the observed patterns to expectations from computer simulations to assess whether the tested genes stand out (25,28,42,46). However, comparing a candidate gene to a large empirical control data set collected in the same way can provide evidence for selection that is more reliable than these comparisons with theoretical expectations (22,54,55). A novel feature of this study methodologically is the introduction of a Genomic Control approach to selection studies, where we empirically infer the parameters of a statistical distribution of a test statistic sensitive to selection (like Tajima's D)—taking into account LD in the data using a MBB analysis—and then assess statistical significance by comparing with this parametric distribution instead of using a simple rank-ordering approach. Genomic Control offers the special advantage—which will be important in future studies—that it can estimate the statistical significance of strong selection signals with much more precision than would be possible with a rank-ordering approach that can only measure a P-value that is as significant as 1 divided by the number of comparisons that are made. Comparisons to empirical data to establish evidence for selection will become all the more important once uniformly ascertained genetic variation data sets become available on a genome-wide scale (44), which will permit studies such as the one we report here to be carried out on all genes simultaneously.
MATERIALS AND METHODS
Human subjects
For the SNP discovery stage of this study, we re-sequenced the same CEU and YRI DNA samples used in the ENCODE Project, including 16 CEU (NA11829, NA11830, NA11831, NA11832, NA11992, NA11993, NA11994, NA11995, NA12003, NA12004, NA12005, NA12006, NA12154, NA12155, NA12156 and NA12236) and 16 YRI (NA18486, NA18489, NA18498, NA18499, NA18501, NA18502, NA18505, NA18507, NA18510, NA18511, NA18516, NA18517, NA18519, NA18520, NA18522 and NA18523). The ascertained SNPs were then genotyped in 90 CEU and 90 YRI samples from the HapMap Project (30 father–mother–child trios from each population, containing 120 independent chromosomes) to characterize these SNPs in larger numbers of samples. This study on de-identified human samples was approved by the Institutional Review Boards of Harvard Medical School and the Massachusetts Institutes of Technology.
Sequencing of the selected gene segments and genotyping of SNPs
We used an ABI 3730 DNA Analyzer (ABI, USA) to re-sequence eight segments selected from these four genes (AHI1, ASPM, FOXP2 and GPR56), ranging from about 15–30 kb in size. We could not sequence the entire intronic and exonic span of all these genes, and hence we prioritized the segments that seemed most likely to be important for human evolution or disease based on other lines of evidence. Segments were chosen based on two subjective criteria: first, they span nucleotides that have been medically documented to cause neurological impairment when mutated; and second, they contain amino acid changing substitutions that have arisen since divergence from other primates (Table 1). Within these constraints, we selected segments that were as contiguous as possible, which meant that in practice, our data set contained much more intronic than exonic sequence.
After the re-sequencing was complete, SNPs were identified by a combination of the PolyPhred and PolyDhan programs, using the same protocol as was used at the Broad Institute to identify SNPs in 2.5 Mb regions for the ENCODE Project. (The other 2.5 Mb of re-sequencing data in the ENCODE Project was gathered at the Human Genome Sequencing Center at the Baylor College of Medicine. We did not include the Baylor data in the present study as a somewhat different protocol was used to discover and genotype SNPs, and we wished our own data to be maximally comparable to the ENCODE data.)
We designed primers for follow-up genotyping using the Assay Design 3.0 software (Sequenom) and attempted to genotype the SNPs identified by re-sequencing in the 30 CEU and 30 YRI trios (father–mother–child) from HapMap using the mass-spectrometry-based MassArray platform (Sequenom) (48). A total of 200 SNPs passed our quality control criteria (polymorphic and >50% genotyping success rate). The average genotyping success rate across all samples (90 CEU and 90 YRI) was 81%. In all analyses except for phasing haplotypes, we only utilized data from the 120 unrelated chromosomes. The data are available at http://genetics.med.harvard.edu/~reich.
Physical and genetic maps
We used the July 2003 human reference genome sequence to determine physical positions (NCBI Build 34/hg16), and the Oxford high-resolution genetic map (56) to matching ENCODE regions to our tested regions in their genetic distance span. When no genetic position was available at a given physical position, it was interpolated based on the genetic and physical positions of the closest flanking SNPs for which data were available.
Generating an empirical data set for examining the DAF spectrum
To extract an empirical data set to which we could compare the regions we re-sequenced, we defined non-overlapping windows spanning each of the five ENCODE regions. Windows were sized to match the genetic distance span as well as the number of segregating sites in each corresponding interrogated segment (we randomly dropped segregating sites from a window in the ENCODE data when there were more segregating sites than in a test region). On the basis of these matched windows from the ENCODE data set, we assessed the statistical significance of the regions we analyzed by rank-ordering the statistics obtained from each of the interrogated segments within the empirical distribution obtained from all ENCODE windows. We used the publicly available Bioperl PopGen module (40) to compute the Tajima's D (10) and the Fu and Li's F (12) selection statistics. The Fay and Wu's H test was implemented based on the method described in Ref. (11). These statistics have different sensitivities. Tajima's D test searches for an excess of rare alleles and is sensitive to positive or negative selection. Fay and Wu's H statistic tests for an excess of high frequency derived alleles, which is a signature of a selective sweep. Fu and Li's F statistic focuses on singleton alleles, which can arise during positive selection. We inferred the ancestral and derived states of SNPs by aligning with the chimpanzee genome as in Ref. (43).
Using Genomic Control to assess statistical significance using a parametric distribution while accounting for correlation among ENCODE windows
We were concerned that our ability to detect statistically very significant loci was limited by the number of empirical comparison regions. We therefore wished to develop a method that would be able to use the fact that a test statistic was not just a moderate outlier, but quantitatively very separated from the values observed at other loci, to be able to detect more extreme signals in the face of limited empirical comparison data.
We call this approach to detecting loci affected by natural selection ‘Genomic Control’, based on the related method that was developed by Devlin and Roeder for genome-wide disease association studies (24). The idea is that the genomic distribution of a test statistic sensitive to natural selection may have a mean and variance that are skewed from the expectations of a simplistic model of demographic history. However, the Genomic Control idea assumes that once the mean and variance of the distribution are estimated, it should be possible to use the fact that the tail of the distribution can be inferred to assess the significance of even extreme observations. This makes it possible to obtain P-values that are much more accurate than are obtainable by a simple rank-ordering method, which can only produce a P-value as extreme as the number of comparisons that is made.
Genomic Control does not add information compared with the rank-ordering when a test statistic is within the distribution of the windows from the empirical comparison data.
To implement the Genomic Control idea we needed to first assess whether the test statistics that we analyzed could be well approximated by a parametric distribution. To assess this, we applied a leave-one-out cross-validation procedure in which we considered each window from the ENCODE control data set in turn and estimated the number of standard deviations by which it fell outside the distribution formed by applying the MBB procedure to the rest of the windows. We found that the normal distribution provided an excellent fit to the Tajima's D statistic (P = 0.61 for rejection of normality by a χ2 goodness-of-fit test; Supplementary Material, Fig. S1), with only 4% of cross-validations rejecting the null hypothesis of no selection at a 95% confidence level. Fu and Li's F and Fay and Wu's H did not fit a normal distribution (P < 10−6; Supplementary Material, Fig. S1), or any of the other parametric distributions we examined (not shown). As the number of windows in the leave-one-out procedure was small, we did not attempt to study the empirical distribution of these statistics in the cross validation, or to transform the distributions to a parametric distribution that we could handle.
In applying our empirical tests for selection (not only Genomic Control, but also the simple outlier approach), we were also concerned that when we observed values of test statistics that were more extreme than those in all windows of matched size in the ENCODE data, we would be overestimating statistical significance because this procedure assumes that all windows are independent whereas in fact they are correlated due to LD. The reason for this is that there are effectively fewer empirical comparisons than would be expected from the number of windows.
To account for LD among neighboring windows in the ENCODE data within our Genomic Control framework, we bootstrapped 10 000 random data sets of windows using the MBB (57,58). The MBB accounts for the correlation between SNPs in different windows by randomly re-sampling contiguous runs of windows from the data in each bootstrap, allowing us to derive estimates of the mean and standard deviation of each allele frequency spectrum selection statistic in a way that is not sensitive to the presence of LD.
Simulations to evaluate the performance of the empirical screens for selection
The simulations of selection were carried out with SelSim (59). Each simulated sample contained 100 haplotypes with a length of 30 kb, meant to approximately match the settings of our real data. We assumed a constant recombination rate of 1.25 cM/Mb and a mutation rate of 10−8 per nucleotide per generation. All selection scenarios assumed a constant population size. Selection was simulated via a two-deme structured-coalescent model, where we assumed random frequency trajectories and an additive model for the effect of the allele. For simplicity, we simulated completed selective sweeps that fixed in the current generation. For four of the simulated scenarios, we assumed that the advantageous mutant was located in the middle of the region, and for the other four scenarios, we assumed the advantageous mutants were 220 kb outside the 30 kb region. For both positions of the selected mutant, we simulated with selection coefficients of 0.1, 0.05, 0.01 and 0.005.
For the neutral simulations, we used the MS Software (60) to simulate 500 kb regions (mimicking the size of the ENCODE regions), and used the same number of haplotypes, and the same settings of recombination and mutation rate, as the selection scenarios. We simulated both a constant-sized demographic history (to match the selection simulations), and also a demographic history meant to mimic that of the North European population in our study and including another model in which the population experienced a severe ‘Out of Africa’-like population bottleneck. In the population growth model, the population began with a constant size of 3000, which reduced to 2000 at a time of 3500 generations ago, and then increased instantaneously to 100 000 at a time of 350 generations ago. In the population bottleneck model, the population began with a constant size of 10 000, and then experienced a bottleneck 1380 generations ago during which the population size was reduced instantaneously to a size of 330 for 100 generations [resulting in a bottleneck intensity T/2N = 0.151 (51)], and then increased instantaneously 1280 generations ago to a constant size of 10 000 and remained at that size until the present. Although these population growth and bottleneck models do not accurately represent the true population history of human populations, they produces patterns of genetic variation that are roughly similar to what we observe in real human populations from HapMap (51), and they allow us to qualitatively explore the performance of our method under such a wide range of demographic histories.
All the simulated data were analyzed using the same set of analysis software as our real data, and the null distribution of Tajima's D was generated by sampling sub-windows from the 500 kb regions. The threshold for statistical significance was set so that the false positive rate was 1%, and the cutoff of D for the 1% significance level was then determined and used to determine statistical power.
In the course of our simulations, we confirmed that ENCODE data set is of sufficient size to support robust application of the Genomic Control method. To show this, we randomly sampled five 500 kb simulated regions, and found that the inferred Tajima's D distribution is not significantly different from what was found when more regions were sampled (Student's t-test P-value = 0.33). The ability to build up a robust expectation for a statistic under the null hypothesis of no selection, whereas using only a limited amount of data, is an important advantage for our parametric Genomic Control approach compared with the non-parametric rank-order approach.
FST analysis to test for extreme allele frequency differentiation between populations
We estimated FST based on the reference and variant allele counts in CEU and YRI, using the formula in Refs. (49,50). To assess statistical significance, we rank-ordered the values observed in our gene segments and compared with the values of all SNPs in the HapMap Phase II data set (the percentile is then reported as a P-value). It is conservative to compare FST to the entire HapMap, since as we show in Supplementary Material, Fig. S2, it is only modestly different from ENCODE, and the difference is in the direction that the HapMap distribution has more SNPs with high FST values.
LRH test analysis
EHH is defined as ‘the probability that two randomly chosen chromosomes carrying a tested core allele are homozygous at all SNPs for the entire interval from the core allele to a given distance’ (52). REHH, which we use as our LRH statistic, is ‘the ratio of the EHH on the tested core allele compared with the EHH of the grouped set of core alleles at the region not including the core haplotype tested’ (52). The goal of the REHH statistic is to use the alleles at a site that are not hypothesized to be under selection as an internal control for local variation in recombination rates. To prepare data for LRH analysis, we merged our genotyping data with the HapMap Phase II data (41) for 500 kb flanking either side of the gene. We considered each SNP inside the re-sequenced regions as a ‘core’ for carrying out the LRH test, and compared with random loci as an empirical control data set. Several previous studies have reported that LRH statistics are not much affected by ascertainment bias (1,21,52), justifying the use of HapMap data for the LRH analysis. We assessed REHH in both directions from the core SNP at a distance where haplotype heterozygosity including all sites between the core and the chosen marker broke down to 4% of its value at the core SNP (this is recommended as a maximally powerful distance by the Sweep software package). Because we were analyzing multiple core SNPs in each genic region, we used the approach of Ref. (2) to correct for multiple hypothesis testing, requiring more than 10% of the SNPs within a 100 kb region to have ln(REHH) at least 3.92 standard deviations higher than the genome-wide distribution for SNPs of the same derived frequency.
SUPPLEMENTARY MATERIAL
FUNDING
F.Y., D.R. and A.K. were supported by NIH (U01-HG004168). R.F. was supported by NIH (K01-MH71801). C.A.W. is an Investigator of the Howard Hughes Medical Institute and D.R. holds a Burroughs Wellcome Career Development Award in the Biomedical Sciences.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mark Daly, Nick Patterson, Alkes Price and Pardis Sabeti for suggestions and critiques.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang E.T., Kodama G., Baldi P., Moyzis R.K. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teshima K.M., Coop G., Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 7.HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornton K.R., Jensen J.D. Controlling the false-positive rate in multilocus genome scans for selection. Genetics. 2007;175:737–750. doi: 10.1534/genetics.106.064642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith J.M., Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 2007;89:391–403. doi: 10.1017/S0016672308009579. [DOI] [PubMed] [Google Scholar]
- 10.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fay J.C., Wu C.I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y.X., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson A. Letters to the editors: remarks on the Lewontin–Krakauer test. Genetics. 1975;80:396. doi: 10.1093/genetics/80.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y.X. New statistical tests of neutrality for DNA samples from a population. Genetics. 1996;143:557–570. doi: 10.1093/genetics/143.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andolfatto P., Przeworski M. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics. 2000;156:257–268. doi: 10.1093/genetics/156.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- 17.Wall J.D., Andolfatto P., Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen R., Williamson S., Kim Y., Hubisz M.J., Clark A.G., Bustamante C.D. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haddrill P.R., Thornton K.R., Charlesworth B., Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen J.D., Kim Y., DuMont V.B., Aquadro C.F., Bustamante C.D. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics. 2005;170:1401–1410. doi: 10.1534/genetics.104.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A.G. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akey J.M., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamblin M.T., Thompson E.E., Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devlin B., Roeder K. Genomic Control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 25.Enard W., Przeworski M., Fisher S.E., Lai C.S., Wiebe V., Kitano T., Monaco A.P., Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert S.L., Dobyns W.B., Lahn B.T. Genetic links between brain development and brain evolution. Nat. Rev. Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- 27.Dorus S., Vallender E.J., Evans P.D., Anderson J.R., Gilbert S.L., Mahowald M., Wyckoff G.J., Malcom C.M., Lahn B.T. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Webb D.M., Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouprina N., Pavlicek A., Mochida G.H., Solomon G., Gersch W., Yoon Y.H., Collura R., Ruvolo M., Barrett J.C., Woods C.G., et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans P.D., Anderson J.R., Vallender E.J., Gilbert S.L., Malcom C.M., Dorus S., Lahn B.T. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum. Mol. Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 32.Evans P.D., Anderson J.R., Vallender E.J., Choi S.S., Lahn B.T. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- 33.Vallender E.J., Lahn B.T. Positive selection on the human genome. Hum. Mol. Genet. 2004;13(Spec no. 2):R245–R254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- 34.Krause J., Lalueza-Fox C., Orlando L., Enard W., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M., et al. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr. Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Coop G., Bullaughey K., Luca F., Przeworski M. The timing of selection at the human FOXP2 gene. Mol. Biol. Evol. 2008;25:1257–1259. doi: 10.1093/molbev/msn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai C.S., Fisher S.E., Hurst J.A., Vargha-Khadem F., Monaco A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 37.Bond J., Roberts E., Mochida G.H., Hampshire D.J., Scott S., Askham J.M., Springell K., Mahadevan M., Crow Y.J., Markham A.F., et al. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 38.Piao X., Hill R.S., Bodell A., Chang B.S., Basel-Vanagaite L., Straussberg R., Dobyns W.B., Qasrawi B., Winter R.M., Innes A.M., et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 39.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 40.Stajich J.E., Hahn M.W. Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 2005;22:63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]
- 41.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mekel-Bobrov N., Gilbert S.L., Evans P.D., Vallender E.J., Anderson J.R., Hudson R.R., Tishkoff S.A., Lahn B.T. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- 43.Yu F., Hill R.S., Schaffner S.F., Sabeti P.C., Wang E.T., Mignault A.A., Ferland R.J., Moyzis R.K., Walsh C.A., Reich D. Comment on ‘Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2007;316:370. doi: 10.1126/science.316.5823.370a. [DOI] [PubMed] [Google Scholar]
- 44.Kaiser J. DNA sequencing. A plan to capture human diversity in 1000 genomes. Science. 2008;319:395. doi: 10.1126/science.319.5862.395. [DOI] [PubMed] [Google Scholar]
- 45.Williamson S.H., Hubisz M.J., Clark A.G., Payseur B.A., Bustamante C.D., Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans P.D., Gilbert S.L., Mekel-Bobrov N., Vallender E.J., Anderson J.R., Vaez-Azizi L.M., Tishkoff S.A., Hudson R.R., Lahn B.T. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 47.Fan J.B., Oliphant A., Shen R., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 48.Tang K., Fu D.J., Julien D., Braun A., Cantor C.R., Koster H. Chip-based genotyping by mass spectrometry. Proc. Natl. Acad. Sci. USA. 1999;96:10016–10020. doi: 10.1073/pnas.96.18.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reynolds J., Weir B.S., Cockerham C.C. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cockerham C.C., Weir B.S. Estimation of gene flow from F-statistics. Evolution. 1993;47:855–863. doi: 10.1111/j.1558-5646.1993.tb01239.x. [DOI] [PubMed] [Google Scholar]
- 51.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 52.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J., et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 53.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 54.Wall J.D., Cox M.P., Mendez F.L., Woerner A., Severson T., Hammer M.F. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voight B.F., Adams A.M., Frisse L.A., Qian Y., Hudson R.R., Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. USA. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 57.Lahiri S.N. Resampling Methods for Dependent Data. New York: Springer; 2003. [Google Scholar]
- 58.Keinan A., Mullikin J.C., Patterson N., Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer C.C.A., Coop G. SelSim: a program to simulate population genetic data with natural selection and recombination. Bioinformatics. 2004;20:3673–3675. doi: 10.1093/bioinformatics/bth417. [DOI] [PubMed] [Google Scholar]
- 60.Hudson R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 61.Cavalli-Sforza L.L. The Human Genome Diversity Project: past, present and future. Nat. Rev. Genet. 2005;6:333–340. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.