Abstract
The proteasome degrades proteins modified by polyubiquitylation, so correctly controlled ubiquitylation is crucial to avoid unscheduled proteolysis of essential proteins. The mechanism regulating proteolysis of RNAPII has been controversial since two distinct ubiquitin ligases (E3s), Rsp5 (and its human homologue NEDD4) and Elongin-Cullin complex, have both been shown to be required for its DNA-damage-induced polyubiquitylation. Here we show that these E3s work sequentially in a two-step mechanism. First, Rsp5 adds mono-ubiquitin, or sometimes a ubiquitin chain linked via ubiquitin lysine 63 that does not trigger proteolysis. When produced, the K63 chain can be trimmed to mono-ubiquitylation by an Rsp5-associated ubiquitin protease, Ubp2. Based on this mono-ubiquitin moiety on RNAPII, an Elc1/Cul3 complex then produces a ubiquitin chain linked via lysine 48, which can trigger proteolysis. Likewise, for correct polyubiquitylation of human RNAPII, NEDD4 cooperates with the ElonginA/B/C-Cullin 5 complex. These data indicate that RNAPII polyubiquitylation requires cooperation between distinct, sequentially acting ubiquitin ligases, and raise the intriguing possibility that other members of the large and functionally diverse family of NEDD4-like ubiquitin ligases also require the assistance of a second E3 when targeting proteins for degradation.
Keywords: elongin, NEDD4, Rsp5, ubiquitylation
Protein ubiquitylation plays a crucial role in virtually all cell regulatory pathways. Mono-ubiquitylation commonly alters the activity of the target protein, or tags it for interaction with other factors, while the effect of polyubiquitylation depends on the type of ubiquitin chain being added. Ubiquitin lysine 48 (K48) chains most often result in degradation of the target protein by the proteasome, whereas other chains, such as those occurring through K63, are typically signals for proteolysis-independent pathways (1, 2).
One interesting substrate for protein ubiquitylation is RNAPII, which transcribes all protein-encoding genes in eukaryotes. Ubiquitylation and degradation of RNAPII was first thought to occur specifically in response to DNA damage (3–5), but more recent experiments have shown that RNAPII arrested during transcript elongation as a result of other transcription obstacles is also prone to ubiquitylation and degradation (6). Thus, degradation of RNAPII may be a “last resort,” used to clear active genes of persistently arrested RNAPII elongation complexes (6–9). Interestingly, the proteasome is nuclear and can be found on the coding region of genes by chromatin-immunoprecipitation (10), so RNAPII proteolysis may well occur on the DNA.
We have reconstituted RNAPII ubiquitylation in vitro with highly purified, physiologically relevant yeast, or human, ubiquitylation factors, respectively (6, 11, 12). The yeast HECT E3 Rsp5 binds RNAPII via the flexible C-terminal repeat domain (CTD) of the Rpb1 subunit (13), but modifies the polymerase in the main body of the Rpb1 subunit (6, 14). Mutation of RSP5 (rsp5-1; temperature-sensitive) results in a defect in ubiquitylating (and degrading) RNAPII in response to DNA damage at the restrictive temperature both in vivo (5) and in vitro (15). Human cells depleted for the Rsp5 homologue NEDD4 by RNA interference are similarly compromised for RNAPII ubiquitylation/degradation (11).
Surprisingly, however, yeast and human cells lacking Elongin C or some Elongin C-associated proteins also appear to be compromised for RNAPII ubiquitylation/degradation (16–19). This raises the question as to how ubiquitylation of RNAPII can be dependent on two different ubiquitin ligases. One possibility is that the role of one of the factors is indirect, or that these enzymes represent parallel pathways for RNAPII ubiquitylation. It is, however, also formally possible that both are required, and that their combined effort somehow brings about RNAPII polyubiquitylation and degradation.
As the cellular requirements and genetics of RNAPII ubiquitylation has already been extensively studied, we focused on trying to understand the underlying mechanism using a biochemical approach. Our results indicate that RNAPII polyubiquitylation and degradation both in the yeast and human system requires distinct, sequentially acting ubiquitin ligase complexes and thereby provide an explanation for several previous, apparently contradictory reports. Given that human NEDD4 is part of a large family of ubiquitin ligases, our data also open the possibility that two-step protein polyubiquitylation is much more widespread than presently realized.
Results
Rsp5 Forms K63-Linked Ubiquitin Chains on RNAPII.
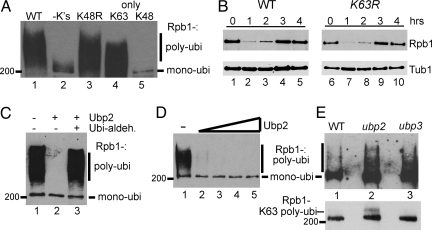
Highly purified Uba1, Ubc5, and Rsp5 can ubiquitylate yeast RNAPII in vitro, and the same factors are also required for the reaction in vivo. Ubiquitylation results in the appearance of a 200-kDa ubiquitylated RNAPII species, representing mono-ubiquitylated RNAPII, as well as a smear above it, representing polyubiquitylated RNAPII (5, 6) (Fig. 1 A and C). Various mutated forms of ubiquitin were used with the purified factors to investigate which type of ubiquitin chains are formed on RNAPII (Fig. 1A). Under the conditions of these experiments, wild-type (WT) ubiquitin mostly yielded polyubiquitylated RNAPII, whereas ubiquitin without lysines only supported mono-ubiquitylation, as expected (Fig. 1A, compare lanes 1 and 2). Efficient polyubiquitylation of RNAPII still occurred when lysine 48 was unavailable (Fig. 1A, lane 3), but not when it was the only position available for chain formation (Fig. 1A, lane 5). In contrast, polyubiquitylation occurred when only lysine 63 was available (Fig. 1A, lane 4), indicating that RNAPII polyubiquitylation by Rsp5 occurs via K63 links.
Fig. 1.
Rsp5-dependent polyubiquitylation of RNAPII is via K63-linked ubiquitin chains, which are hydrolyzed by Ubp2. (A) Western blot probed with anti-ubiquitin antibodies of RNAPII ubiquitylation reconstituted with Uba1, Ubc5, and Rsp5 (6) and various ubiquitin forms. (B) Western blot of total Rpb1 (Upper) after UV-irradiation (300 J/m2) in the indicated strains, with tubulin (Tub1) as loading control (Lower). (C) Purified Ubp2 (40 ng) was added to a reconstituted RNAPII ubiquitylation reaction (as in A) in the absence or presence of ubiquitin-aldehyde. (D) Purified Ubp2 (5, 10, 25, and 50 ng) was added to a reconstituted RNAPII ubiquitylation reaction as above. Numerous control experiments confirm that RNAPII is indeed the ubiquitylated protein in A, B, and D. Most notably, when it is left out of the reaction, the ubiquitylated species disappears (6). (E) Western blot of ubiquitylated RNAPII, isolated by affinity purification from the indicated strains, probed with anti-Rpb1 (4H8) antibody. Upper, RNAPII bound to GST-Dsk2 affinity matrix. Lower, RNAPII bound to a RAP80 UIM affinity matrix. The RAP80 matrix binds similar amounts of mono-ubiquitylated RNAPII from all strains, but it only binds polyubiquitylated RNAPII from the ubp2Δ strain.
Others have previously reported data suggesting that K63-chains can also result in proteasome-mediated protein degradation (20, 21). To address whether Rsp5-mediated RNAPII ubiquitylation and degradation occur via ubiquitin K63-linked chains in vivo, we used a yeast strain in which this lysine is mutated (K63R) (22). Proteasome-mediated RNAPII degradation can be observed as a temporary disappearance of the Rpb1 protein from cells upon a high dose of UV-irradiation (5, 7). The K63R strain showed no defect in this process (Fig. 1B, compare lanes 1–5 with 6–10), suggesting that polymerase degradation occurs via the traditional proteasomal pathway using K48-linked ubiquitin chains.
The Rsp5-Associated Ubiquitin Protease Ubp2 Can De-Ubiquitylate RNAPII.
Depending on the experimental conditions used, Rsp5 may predominantly add mono-ubiquitin or a polyubiquitin chain to RNAPII in vitro (6, 12). Therefore, it is formally possible that in vivo, Rsp5 normally only adds mono-ubiquitin. Interestingly, however, Rsp5 forms a complex with the ubiquitin protease Ubp2, which has been shown to affect certain other protein ubiquitylation pathways (23, 24). We purified TAP-tagged Ubp2 and found that substoichiometric amounts of Rsp5 co-purified, as previously observed by others (23, 24). To investigate Ubp2's effect on RNAPII ubiquitylation, a purified fraction was tested in the reconstituted RNAPII ubiquitylation system (Fig. 1C). As expected, Rsp5 efficiently polyubiquitylated RNAPII (Fig. 1C, lane 1), but when Ubp2 was added, only RNAPII mono-ubiquitylation was observed (Fig. 1C, lane 2). This was a result of Ubp2-mediated de-ubiquitylation of longer Rsp5-generated ubiquitin chains because adding the ubiquitin protease inhibitor ubiquitin aldehyde resulted in RNAPII polyubiquitylation (Fig. 1C, lane 3). Strikingly, although Ubp2 reversed polyubiquitylation, even a 10-fold excess of the enzyme amount required to disassemble chains failed to remove RNAPII mono-ubiquitylation (Fig. 1D, lanes 2–5), indicating that Ubp2 can efficiently hydrolyze the ubiquitin-ubiquitin links, but not the link between ubiquitin and RNAPII. This is in contrast to another RNAPII-targeting ubiquitin protease, Ubp3, which hydrolyzes this link (25).
Only a very small fraction of RNAPII is ubiquitylated at any given time in vivo, but this fraction can be dramatically enriched on an ubiquitin binding GST-Dsk2 affinity matrix (see Materials and Methods). Using this approach to isolate mono- and polyubiquitylated proteins, we noted that a ubp2Δ mutant had increased amounts of polyubiquitylated RNAPII (Fig. 1E Upper, compare lanes 1 and 2), supporting the idea that Ubp2 can also de-ubiquitylate RNAPII in vivo. As previously reported (24), ubp3 mutant cells also had an increased amount of ubiquitylated RNAPII (Fig. 1E Upper, lane 3). One possible explanation for the increase in polyubiquitylated RNAPII in ubp2Δ is that Ubp2 normally hydrolyzes non-degrading K63 chains produced by Rsp5. Indeed, using a RAP80 UIM affinity matrix that has a preference for binding K63 polyubiquitylated proteins (26), polyubiquitylated RNAPII could be isolated specifically from ubp2Δ cells (Fig. 1E Lower, compare lanes 1 and 2), while increased levels of polyubiquitylated RNAPII were not observed in ubp3Δ cells by this approach (Fig. 1E Lower, compare lanes 2 and 3), as predicted from its role in the rescue of RNAPII ‘marked’ for degradation by K48-linked polyubiquitin chains (24). Others have also shown that Ubp2 regulates K63-linked ubiquitin chain formation in vivo (27), so together these results suggest that Ubp2 has the ability to restrict RNAPII modification to mono-ubiquitylation by trimming K63-linked ubiquitin chains.
Evidence that Rsp5 Is Required Only for RNAPII Mono-Ubiquitylation.
Efficient RNAPII polyubiquitylation can also be reconstituted in cell-free yeast extracts, provided ubiquitin is added (15). Such ubiquitylation is dependent on Rsp5 and Def1 (15), as well as Elc1 (see below), strongly indicating that it faithfully reconstitutes the main aspects of the reaction as it occurs in vivo. Given that RNAPII polyubiquitylation in the system based on pure ubiquitylation factors occurred via ubiquitin K63 links, we investigated whether RNAPII ubiquitylation in the cell free system could occur with ubiquitin that only contains lysine 48; that is, whether other cellular factors might somehow facilitate correct chain formation for proteasomal degradation. RNAPII polyubiquitylation via K48 chains was indeed observed in extracts (heat-treated at 37 °C) from WT cells (Fig. 2A, lane 1). As expected, such ubiquitylation was dependent on Rsp5, as it did not occur in rsp5-1 extracts after heat-inactivation of Rsp5 (15) (lane 2), unless purified Rsp5 protein was added back (lane 3). This indicates that the crude extract either contains a factor capable of promoting an ability of Rsp5 to make K48-linked chains (for example an E4 activity), or that Rsp5 only adds the initial ubiquitin moiety (mono-ubiquitin), and that a distinct activity then extends it, producing RNAPII polyubiquitylated via K48-links.
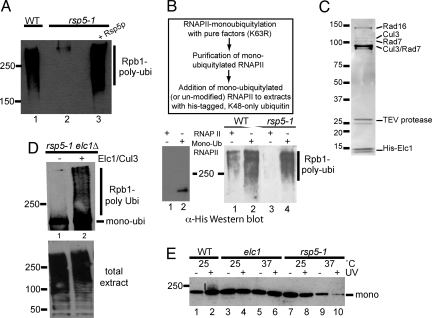
Fig. 2.
Ubiquitylation of RNAPII in crude extracts requires Rsp5, but only for mono-ubiquitylation. Elc1/Cul3 performs polyubiquitlyation based on that. (A) Western blot using anti-FLAG antibodies to detect RNAPII ubiquitylation (using FLAG- tagged K48-only ubiquitin) in heat-treated cell-free extracts. (B) (Boxed) Outline of experimental approach. (Lower Left) Un-modified and premono-ubiquitylated RNAPII purified/immobilized on beads. Only mono-ubiquitylated RNAPII cross-reacts with α-his antibody. (Lower Right) Polyubiquitylation of unmodified, or premono-ubiquitylated, RNAPII using K48-only, his-tagged ubiquitin. (C) Coomassie-stained SDS/PAGE gel showing purified Elc1 complex. The identity of proteins is indicated on the right. (D) Western blot using anti-FLAG antibodies to detect polyubiquitylation of immobilized premono-ubiquitylated RNAPII with FLAG-tagged K48-only ubiquitin in heat-treated rsp5-1 elc1Δ extract. (Upper) Signal from (bead-associated) RNAPII. No signal was detected without RNAPII on the beads. (Lower) Total ubiquitylated protein (supernatant). (E) Western blot of ubiquitylated RNAPII (isolated by affinity purification), probed with anti-Rpb1 (4H8) antibody.
To try and distinguish between these possibilities, we first tested the effect of deleting the gene encoding the E4 called Ufd2, which might conceivably catalyze the elongation of Rsp5-produced ubiquitin chains through K48 linkages on RNAPII (28). However, ufd2Δ cells still supported RNAPII ubiquitylation/degradation. We then investigated if an E3 distinct from Rsp5 is responsible for RNAPII polyubiquitylation. For this, RNAPII was first mono-ubiquitylated using highly purified ubiquitylation factors and K63R ubiquitin; the mono-ubiquitylated RNAPII was then purified, and added to the cell-free extract-based ubiquitylation system (Fig. 2B Upper). Un-modified and mono-ubiquitylated RNAPII were immobilized on beads (Fig. 2B Lower Left, compare lanes 1 and 2) and tested for polyubiquitylation in this system. In control experiments, heat-treated extract from WT, but not rsp5-1 cells, supported polyubiquitylation when un-modified RNAPII was used as substrate (Fig. 2B Lower Right, compare lanes 1 and 3), as observed previously. In contrast, both WT and rsp5-1 extract supported robust RNAPII polyubiquitylation if the purified, premono-ubiquitylated RNAPII was added as substrate (Fig. 2B, compare lanes 2 and 4).
Elc1 Is Required for Polyubiquitylating Premono-Ubiquitylated RNAPII.
The results presented above opened the possibility that a distinct E3 activity produces K48-linked ubiquitin chains on the basis of Rsp5-mediated RNAPII mono-ubiquitylation. Others have previously reported convincing data to support a role for Elongin proteins in RNAPII ubiquitylation in vivo, in both human and yeast cells (16, 17, 19). To investigate this possible connection, we Myc9-TEV2-His8-tagged (29) Elc1 at its genomic locus. Proteins co-purifying with Elc1 (Fig. 2C) were identified by mass spectrometric analysis. As expected from a previous report (30), Rad16 and Rad7 were identified, but neither of these proteins is required for RNAPII ubiquitylation (17). More importantly, Cul3, a member of the cullin family of ubiquitin ligases was also identified. In agreement with the idea that Elc1/Cul3 is involved in RNAPII ubiquitylation, others reported while this work was in progress that cells lacking CUL3 are indeed unable to ubiquitylate and degrade RNAPII in response to UV-induced DNA damage (18).
To address the possibility that Rsp5 mono-ubiquitylates RNAPII and that the Cul3/Elc1 complex then produces a polyubiquitin chain on this basis, we again reconstituted RNAPII ubiquitylation in cell-free extracts. Heat-treated extracts from rsp5-1 cells support polyubiquitylation of premono-ubiquitylated RNAPII (Fig. 2B), so we asked if Elc1 is required for this activity. Indeed, when extract from an rsp5-1 elc1Δ double mutant was used, no conversion of the added premono-ubiquitylated polymerase to the polyubiquitylated form was detected (Fig. 2D Upper, lane 1), even though efficient ubiquitylation of other proteins in the crude extract took place (Fig. 2D Lower, lane 1). However, addition of the purified Elc1/Cul3 complex restored RNAPII polyubiquitylation (Fig. 2D Upper, lane 2), indicating that Elc1/Cul3 complex polyubiquitylates RNAPII, which has first been mono-ubiquitylated by Rsp5. These biochemical results are consistent with the idea that Elc1/Cul3 plays a direct role in RNAPII ubiquitylation, as the second enzyme in a two-step mechanism.
One prediction from this model is that while deletion of ELC1 hinders polyubiquitylation, only inactivation of Rsp5 should affect the preceding RNAPII mono-ubiquitylation in vivo. To investigate this, ubiquitylated proteins were isolated from yeast before and after UV-irradiation using an affinity matrix containing a ubiquitin-binding domain (11), and then RNAPII ubiquitylation was assessed using anti-Rpb1 antibodies (Fig. 2E). Under the conditions of this particular experiment, the vast majority of detectable RNAPII ubiquitylation was mono-ubiquitylation; only a low level of polyubiquitylation after UV-irradiation in WT cells was detected (Fig. 2E, compare lanes 1 and 2). More importantly, however, the overall level of RNAPII mono-ubiquitylation was unaltered in elc1Δ cells (Fig. 2E, lanes 3–6), while it was significantly reduced in rsp5-1 cells at the restrictive temperature (Fig. 2E, lanes 9 and 10). These in vivo experiments demonstrate that while Rsp5 is required for the initial RNAPII mono-ubiquitylation, Elc1 is only required for subsequently building a polyubiquitin chain on this basis.
Reconstitution of Two-Step RNAPII Polyubiquitylation with Highly Purified Yeast and Human Factors, Respectively.
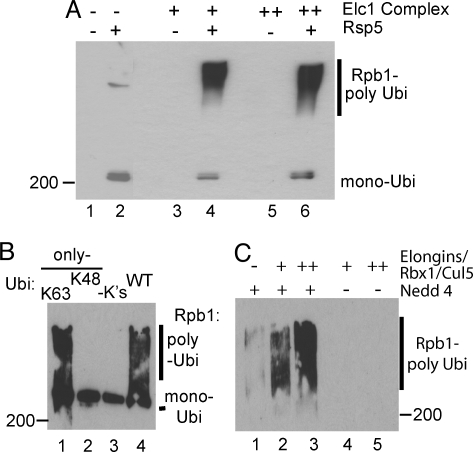
To investigate whether Rsp5 and Elc1/Cul3 are required and sufficient for RNAPII polyubiquitylation, we now attempted to reconstitute two-step polyubiquitylation of yeast RNAPII with highly purified factors (Fig. 3A). As a simple way of ensuring that the correct ubiquitin chains were indeed formed, K48-only ubiquitin was used. As expected, Rsp5 could only mono-ubiquitylate RNAPII with this form of ubiquitin (Fig. 3A, lane 2; and Fig. 1A). Strikingly, in the absence of Rsp5, the Elc1/Cul3 complex had little or no RNAPII ubiquitylation activity (Fig. 3A, lanes 3 and 5). However, in the presence of Rsp5, addition of the Elc/Cul3 complex resulted in efficient ubiquitylation (lanes 4 and 6), with mono-ubiquitylated Rpb1 being converted into poly ubiquitylated species (Fig. 3A, compare lane 2 with lanes 4 and 6).
Fig. 3.
Reconstitution of RNAPII polyubiquitylation with highly purified ubiquitylation factors. (A) Western blot probed with anti-FLAG antibodies of RNAPII ubiquitylated with highly purified Uba1, Ubc5, Rsp5, and the Elc1/Cul3 complex, using FLAG-tagged K48-only ubiquitin. Long ubiquitin chains have several epitopes per RNAPII molecule and show much stronger reactivity than mono-ubiquitylated RNAPII. (B) Anti-ubiquitin Western blot of human RNAPII ubiquitylation with highly purified factors [Uba1, UbcH7, and Nedd4 (11)], using WT or mutant ubiquitin. (C) Western blot of RNAPII ubiquitylation reconstituted with highly purified mammalian ElonginABC/Rbx1/Cullin5 complex in the absence or presence of limiting amounts of Nedd4.
In humans, NEDD4 and Elongin/cullin complex have also both been implicated in RNAPII ubiquitylation (11, 16, 19). To determine the nature of the chains formed by NEDD4, this E3 and mutated forms of ubiquitin were used in a reconstituted human RNAPII ubiquitylation system (11), with Uba1 and UbcH7 used as the E1 and E2, respectively. WT ubiquitin supported RNAPII polyubiquitylation, but ubiquitin without lysines only allowed mono-ubiquitylation (Fig. 3B, compare lanes 3 and 4). Similar to yeast, efficient polyubiquitylation occurred when only lysine K63, but not when only lysine K48 was available for chain formation (Fig. 3B, compare lanes 1 and 2).
We now expressed the Elongin ABC complex, Rbx1, and Cullin 5 in insect cells, purified, and combined them (31). This Elongin/Rbx1/Cullin 5 complex (with Uba1 and UbcH7) was unable to polyubiquitylate RNAPII under the conditions used (Fig. 3C, lanes 4 and 5). To try and reconstitute two-step polyubiquitylation of human RNAPII using wild-type ubiquitin, we used a limiting amount of NEDD4 that was in itself unable to support significant ubiquitylation of RNAPII in the assay. However, in contrast to when they were used in isolation, robust polyubiquitylation of human RNAPII occurred when the NEDD4 and Elongin/cullin E3 ligases were combined (Fig. 3C, lanes 2 and 3). These data indicate that Rsp5 (NEDD4) and an Elongin/cullin complex are required and sufficient to reconstitute polyubiquitylation of RNAPII.
Discussion
Rsp5 (NEDD4 in humans) and Elc1/Cul3 (Elongin/Rbx1/Cullin 5 complex) have previously been shown to be required for damage-induced RNAPII ubiquitylation and degradation in yeast and human cells, respectively (5, 6, 13, 15–19), but their relationship, if any, has been unclear. In previous studies the lack of ubiquitylation and degradation of yeast RNAPII in elc1Δ and cul3Δ cells was extrapolated to mean that these proteins were (solely) responsible for (directly) ubiquitylating RNAPII in response to DNA damage (17, 18). Prakash and coworkers even suggested that the effect of Rsp5 on RNAPII ubiquitylation was indirect and irrelevant (17), although previously reported data obtained in vitro pointed to a direct role for this protein (6, 13, 15). These points may now be moot as the results presented here explain why two distinct E3s are essential: they cooperate to bring about RNAPII polyubiquitylation for degradation via a two-step mechanism (Fig. 4). In the first step, Rsp5 adds mono-ubiquitin to RNAPII (Fig. 4, step 1). In the purified yeast ubiquitylation system, Rsp5 produces a mixture of mono- and polyubiquitylated RNAPII. However, the ubiquitin chains produced are K63-linked, and such chains are not required for RNAPII degradation in cells. Experiments with ubp2Δ cells support the idea that Rsp5 can add a K63 chain to RNAPII in vivo, but that this chain is trimmed by Ubp2. Rsp5 and Ubp2 interact (23, 24), and our data with purified factors show that Ubp2 is capable of hydrolyzing K63 chains to produce mono-ubiquitylated RNAPII, further supporting the idea that Ubp2-mediated ‘proofreading’ of Rsp5 action occurs. We previously reported that Ubp3 plays a role in ‘rescuing’ RNAPII from degradation by reversing RNAPII ubiquitylation, and that it can hydrolyze K48-linked ubiquitin chains (25). In contrast to Ubp2, Ubp3 can also efficiently cleave the ubiquitin chain off RNAPII to produce completely de-ubiquitylated polymerase. Ubp2 and Ubp3 thus appear to have temporally distinct roles in the polymerase ubiquitylation pathway.
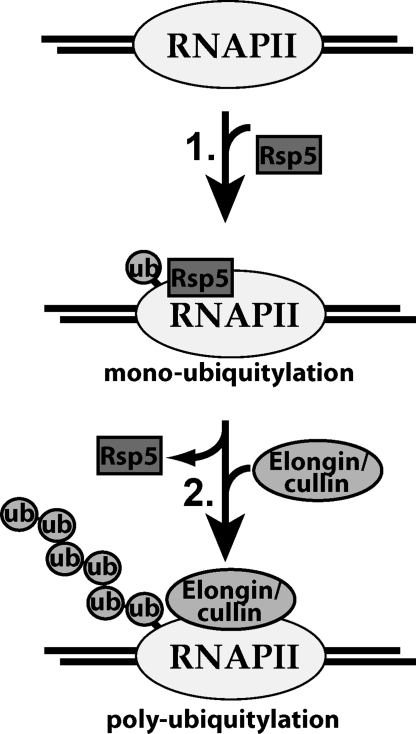
Fig. 4.
Simplified model for RNAPII polyubiquitylation by distinct, sequentially acting ubiquitin ligases. RNAPII is ubiquitylated via a two-step mechanism. First, yeast RNAPII is mono-ubiquitylated by Rsp5. Second, an Elongin/cullin complex uses the mono-ubiquitylated RNAPII as a basis for polyubiquitin chain formation, forming K48 ubiquitin linkages. This ubiquitin chain acts as a signal for RNAPII degradation by the proteasome. Likewise, in human cells, NEDD4 mono-ubiquitylates RNAPII, and an ElonginABC/Rbx1/Cullin 5 complex then polyubiquitylates it.
It is important to note that although Rsp5 is not directly involved in making a K48 polyubiquitin chain on RNAPII, there is no mono-ubiquitylation in its absence, and therefore no basis on which to produce polyubiquitylated RNAPII. This explains the previously observed requirement for Rsp5 in RNAPII polyubiquitylation and degradation (5). In the second step, a distinct E3 ligase, the Elc1/Cul3 complex, which in itself is unable to ubiquitylate RNAPII in vitro, then extends mono-ubiquitylated RNAPII to form K48 ubiquitin chains (Fig. 4, step 2). Elc1 is dispensable for mono-ubiquitylation of RNAPII in vivo, but it is absolutely required for polyubiquitylation, both in vivo (17), and in vitro. Future studies will focus on determining if the Elc1/Cul3 complex polyubiquitylates both RNAPII lysine residues ubiquitylated by Rsp5 in vitro (6, 13). The mechanism of sequential E3 ligases for ubiquitylation of RNAPII is almost certainly evolutionarily conserved, with NEDD4 and the Elongin ABC/Cullin 5 complex polyubiquitylating human RNAPII in a similar manner.
Besides providing an attractive explanation for why two distinct ubiquitin ligases are required for RNAPII polyubiquitylation, our data also raise interesting new questions. For example, although RSP5 function is clearly required for damage-induced RNAPII polyubiquitylation and degradation (5), Rsp5-mediated mono-ubiquitylation does not appear to change dramatically in response to DNA damage (Fig. 2E). Moreover, although Elc1 is also required for damage-induced RNAPII polyubiquitylation and degradation (17), the extent of co-immuno-precipitation of Elc1 and RNAPII does not appear to change significantly in response to UV-irradiation (Fig. S1). This raises the question, how DNA damage can induce the dramatic increase in RNAPII polyubiquitylation and degradation observed? Likewise, we previously showed that Def1 protein regulates DNA damage-induced RNAPII polyubiquitylation and degradation (7), and that it can increase Rsp5-mediated RNAPII polyubiquitylation in vitro (6). The precise mechanism of Def1 action will need to be re-evaluated in light of the findings reported here. These and several other questions raised by our findings are presently the focus of investigation.
It seems obvious that, in general, protein polyubiquitylation via K48 links needs to be very tightly regulated to avoid unscheduled degradation of the multiple essential proteins that are regulated this way. Splitting the process into two steps (mono-, then polyubiquitylation), and allowing constant ‘proof-reading’ by ubiquitin proteases would go far toward ensuring correctly targeted proteolysis. Indeed, other examples of splitting polyubiquitylation into multiple steps have been described (32–34). However, we propose that a two-step mechanism of protein ubiquitylation requiring distinct E3s for mono- and polyubiquitylation may potentially be much more widespread than presently realized. NEDD4 and its close homologues (a large family of nine enzymes involved in various cellular processes), such as Itch and Smurf, thus have numerous important targets that they mark for degradation, including, for example, PTEN (35), TGF-beta receptor (36, 37), and p73 (38). The possibility that polyubiquitylation and degradation of these and many other target proteins also require distinct, sequentially acting ubiquitin ligases certainly merits further investigation.
Materials and Methods
Yeast Strains.
Strains (sources) used in this study were: UbK63R (SUB413) and corresponding wild-type (SUB280) (D. Finley, Harvard University) (22), and UBP2-TAP (Open Biosystems). All other yeast strains were congenic with W303 (39). RPB1-3HA and ubp3Δ were described previously (15, 25). elc1Δ and ubp2Δ were constructed by deletion of the respective ORFs by homologous recombination using standard procedures. The rsp5-1 strain was created by homologous recombination of an integrating plasmid (40) containing the L733S RSP5 mutation (41) into W303–1a. rsp5-1 elc1Δ was made by mating the appropriate haploid strains described above, sporulating and dissecting tetrads. Elc1 was N-terminally tagged at the genomic locus to produce Myc9-TEV2-His8-tagged Elc1 (also called Myc-Elc1). (oligonucleotide sequences and other details are available upon request). Correct insertion was confirmed by genomic PCR, DNA sequencing, and Western blot analysis.
Plasmids.
A plasmid for bacterial expression of N-terminally FLAG-His6-tagged ubiquitin was constructed by amplifying a yeast ubiquitin ORF with the FLAG-His6 sequence added in-frame via the 5′ primer. The PCR product was cloned into vector pET-21b (Novagen). Standard mutagenesis techniques were then used to generate a K48-only ubiquitin expression plasmid, where all lysines in ubiquitin except K48 were mutated to arginine. A plasmid expressing Myc-tagged K63R ubiquitin was made by site-directed mutagenesis in a Myc-tagged ubiquitin construct (15). All constructs were confirmed by sequencing.
Proteins.
His-tagged ubiquitins, Uba1, and human UbcH7 protein were from Boston Biochemical. RNAPII, and ubiquitylation factors expressed in E. coli were purified as described (6, 11). Mammalian Elongin complex was purified as described (31). Protein mass spectrometric analysis was done as described (42). Antibodies used were anti-hexahistidine (Novagen), anti-Myc 9E10 (43), anti-Rpb1 4H8 (44), anti-tubulin (Abcam), anti-FLAG (Sigma), and anti-GST (Amersham Biosciences). Rsp5 without the GST-tag was generated by cleavage with PreScission Protease (Amersham Biosciences) in 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT (pH 7.0) for 16 h at 4 °C. Ubp2-TAP was purified from yeast using standard procedures (45). Expression and purification of Myc-ubiquitin was described previously (15). FLAG-His6 tagged ubiquitin was expressed in E. coli and purified by Ni-NTA agarose affinity chromatography using standard techniques. Tagged Elc1 was purified from yeast cells expressing Myc9-TEV2-His8-Elc1 essentially as described (29). The protein was further purified by MonoQ chromatography. Details are available on request.
RNAPII Ubiquitylation.
Ubiquitylation reactions (yeast proteins) were performed essentially as described (6), typically with 300–450 ng RNAPII, 40 ng rUba1, 40 ng rGST-Ubc5, 90 ng rGST-Rsp5 (or Rsp5), and 2 μg rUbiquitin. For de-ubiquitylation experiments, 40 ng purified Ubp2 (or 5–50 ng in Fig. 1D) was added. Ubiquitin-aldehyde (Boston Biochemical) was used at 4.9 μM. Samples were fractionated on a 4–12% SDS/PAGE gel and blotted and probed with an anti-His antibody to detect ubiquitylated RNAPII. Experiments to assess the ability of the purified Elc1 complex to ubiquitylate RNAPII were carried out the same way, except that K48-only FLAG-His6-ubiquitin was used.
To produce mono-ubiquitylated RNAPII for use in cell-free whole cell extracts (see below), the above assay was modified so that 3–4.5 μg RNAPII (Rpb1–3HA) was incubated with 1 μg rUba1, 0.6 μg rGST-Ubc5, 1.3 μg rGST-Rsp5, and 30 μg rUbiquitin (Myc-K63R, or FLAG-His6 K48-only). Samples were then incubated with glutathione Sepharose 4B beads to remove GST-Rsp5. Removal was confirmed by Western blotting using anti-GST antibodies (Amersham Biosciences). The Rsp5-free fraction was incubated with anti-HA affinity beads (Roche) for 4 h to overnight at 4 °C for further purification of RNAPII. RNAPII-beads were stored at 4 °C for up to 2 weeks before use.
Ubiquitylation in Yeast Extracts.
Preparation of whole cell extracts and its use in ubiquitylation reactions have been described (15, 46). To inactivate Rsp5–1, cells were incubated for 1 h at 37 °C before preparation of extracts. After incubation of approximately 1 μg immobilized HA-tagged RNAPII with 200 μg extract, 2 μg rUbiquitin, 1 mM ATP, and 20 μM lactocystine (Cayman Chemical Company), and 2 mM N-ethylmaleimide (Sigma), RNAPII was isolated by centrifugation (1,000 rpm in microcentrifuge), and the supernatant was removed to determine general protein ubiquitylation in the reaction. RNAPII-beads were washed thoroughly, and bound proteins were eluted by addition of SDS-sample buffer. Samples were loaded onto a 4–12% gel, blotted, and probed with the anti-His, or anti-Myc, antibody to reveal ubiquitylation.
In vitro ubiquitylation of human RNAPII with highly purified mammalian ubiquitylation factors was carried out essentially as described previously (11), except that the NEDD4 concentration was 6 nM and the Elongin complex [Elongin A/B/C, Rbx1, Cul5a (31)] was added as well, as indicated. Samples were fractionated by 4–12% SDS/PAGE and blotted and probed with anti-His antibody.
Ubiquitylation In Vivo.
Procedures for assessing the level of ubiquitylated RNAPII in vivo have been described (11, 25). Experiments with rsp5-1 cells included a shift to non-permissive temperature (37 °C) for 90 min. Two milligrams crude extract protein was mixed with ubiquitin affinity matrix (glutathione agarose beads complexed with GST fused to the UBA domain-containing Dsk2 protein (11), or Rap80 UIM-agarose (Boston Biochemical), as indicated) for 4 h at 4 °C to isolate ubiquitylated proteins. After washing, samples were eluted with SDS-sample buffer and fractionated on 3–8% SDS/PAGE or 4–12% SDS/PAGE gels, blotted, and probed with anti-Rpb1 antibody (4H8). Treatment of the beads with de-ubiquitylating enzyme [Rabbit isopeptidase (Calbiochem)] before washing resulted in a loss of RNAPII signal, confirming that the polymerases retained were indeed ubiquitylated.
Supplementary Material
Acknowledgments.
We thank Mark Skehel and coworkers (Protein Analysis and Proteomics Laboratory) for mass spectrometry analysis of the Elc1/Cul3 complex, Dan Finley for yeast strains, and Helle Ulrich and Barbara Dirac-Svejstrup for comments on the manuscript. This work was supported by an in-house grant from Cancer Research U.K. (to J.Q.S.) and by National Institute of General Medical Sciences Grant R37 GM41628 (to R.C.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907052106/DCSupplemental.
References
- 1.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 4.Bregman DB, et al. UV-induced ubiquitination of RNA polymerase II: A novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somesh BP, et al. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Woudstra EC, et al. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- 8.Svejstrup JQ. Rescue of arrested RNA polymerase II complexes. J Cell Sci. 2003;116:447–451. doi: 10.1242/jcs.00271. [DOI] [PubMed] [Google Scholar]
- 9.Svejstrup JQ. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci. 2007;32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Physical and functional association of RNA polymerase II and the proteasome. Proc Natl Acad Sci USA. 2004;101:5904–5909. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Somesh BP, et al. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007;129:57–68. doi: 10.1016/j.cell.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 13.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 15.Reid J, Svejstrup JQ. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J Biol Chem. 2004;279:29875–29878. doi: 10.1074/jbc.C400185200. [DOI] [PubMed] [Google Scholar]
- 16.Kuznetsova AV, et al. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci USA. 2003;100:2706–2711. doi: 10.1073/pnas.0436037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribar B, Prakash L, Prakash S. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3999–4005. doi: 10.1128/MCB.00293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribar B, Prakash L, Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol Cell Biol. 2007;27:3211–3216. doi: 10.1128/MCB.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasukawa T, et al. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 2008;27:3256–3266. doi: 10.1038/emboj.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 21.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam MH, et al. Interaction of the deubiquitinating enzyme Ubp2 and the e3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS ONE. 2009;4:e4259. doi: 10.1371/journal.pone.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvint K, et al. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol Cell. 2008;30:498–506. doi: 10.1016/j.molcel.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 27.Kee Y, Munoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- 28.Saeki Y, Tayama Y, Toh-e A, Yokosawa H. Definitive evidence for Ufd2-catalyzed elongation of the ubiquitin chain through Lys48 linkage. Biochem Biophys Res Commun. 2004;320:840–845. doi: 10.1016/j.bbrc.2004.05.216. [DOI] [PubMed] [Google Scholar]
- 29.Greenwood C, Selth LA, Dirac-Svejstrup AB, Svejstrup JQ. An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J Biol Chem. 2009;284:141–149. doi: 10.1074/jbc.M805312200. [DOI] [PubMed] [Google Scholar]
- 30.Ramsey KL, et al. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol Cell Biol. 2004;24:6362–6378. doi: 10.1128/MCB.24.14.6362-6378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamura T, et al. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem. 2001;276:29748–29753. doi: 10.1074/jbc.M103093200. [DOI] [PubMed] [Google Scholar]
- 32.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 33.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. NEDD4–1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebisawa T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 37.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 38.Rossi M, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otero G, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 45.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 46.Kong SE, Svejstrup JQ. Incision of a 1,3-intrastrand d(GpTpG)-cisplatin adduct by nucleotide excision repair proteins from yeast. DNA Repair (Amst) 2002;1:731–741. doi: 10.1016/s1568-7864(02)00080-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.