Abstract
Cell-to-cell variability of gene expression in clonal populations of mammalian cells is ubiquitous. However, because molecular biologists habitually assume uniformity of the cell populations that serve as starting material for experimental analysis, attention to such non-genetic heterogeneity has been scant. As awareness of, and interest in, understanding its biological significance increases, this Primer attempts to clarify the confusing terminologies used in an emerging field that often conflates heterogeneity with noise, and provides a qualitative introduction to the fundamental dynamic principles that underlie heterogeneity. It thus aims to present a useful conceptual framework to organize, analyze and communicate observations made at the resolution of individual cells that indicate that heterogeneity of cell populations plays a biological role, such as in multipotency and cell fate decision.
Introduction
Hardly a phenomenon in metazoan biology is more ubiquitous, yet as neglected, as the inevitable heterogeneity of cell phenotypes within a population of cells of the same cell type. Nevertheless, the implications of such cell population heterogeneity are far reaching, and semantic questions are inevitable: if an apparently ‘clonal’ population of cells, which are, by definition, genetically identical, is heterogeneous and exhibits enduring cell-to-cell variations with distinct cell individuality, why then do we not talk about distinct cell types, or subtypes, in the population? Stem cell researchers will easily recognize the ontological (see Glossary, Box 1) problem (Orkin and Zon, 2002): if a culture of stem cells, even if clonal, is actually heterogeneous to start with, be it owing to impurities introduced by unavoidable technical limitations (Enver et al., 2009) or to intrinsic diversity, as discussed here, what then is the meaning of plasticity and multipotency, as defined as the ability of cells to generate a variety of cell types? Perhaps the process of generating and/or acquiring heterogeneity eo ipso could lie at the core of multipotency.
Box 1. Glossary
Attractor state. A stationary and stable network state into which a set of particular network states will eventually evolve (‘be attracted to’). This happens because the interactions between the network elements impose constraints so that the majority of theoretically possible network states is unstable and will move towards an attractor state.
Clone, or a clonal group of cells. A group of cells that contains only cells that share a common ancestry and hence are assumed to be genetically identical. According to this definition, however, entire organisms, being derived from a zygote, would be clones. Thus, in the context of phenotypic variability, ‘clonal’, which is often equated with ‘genetically identical’, must be more narrowly defined (see text).
Dichotomy. The partitioning of a whole entity X (a set, concept, or phenomenon) into two, and only two, subsets A and B, that are mutually exclusive and together cover all possible elements belonging to X.
Ergodicity. A property of a system or process that satisfies the ergodic hypothesis in statistical physics, according to which the average over time of a (fluctuating) quantity of the system is the same as the average at a given time over a large sample of replicates of that system (= ensemble).
Extrinsic heterogeneity. Cell-to-cell variability (typically at the level of micro-heterogeneity) in a population caused by non-uniform environmental factors that differentially affect individual cells.
Genetic heterogeneity. A property of a population (e.g. of cells) in which the genomes of the individual members (cells) are not identical for all members and which hence contains a mixture of distinct genomes. Frequently considered in tumor biology, where the genome sequence differences between the tumor cells could explain trait differences due to somatic mutations.
Intrinsic heterogeneity. Cell-to-cell variability (typically at the level of micro-heterogeneity) in the absence of inhomogeneities in the microenvironment. Most commonly explained by ‘gene expression noise’ (temporal noise), but applies also to population noise (see Fig. 3).
Macro-heterogeneity. Heterogeneity of a cell population due to the presence of a variety of discretely distinct cell types or of cells in obviously distinct states, such as progenitor versus differentiated cells. Macro-heterogeneity is manifest as a multi-modal distribution of a trait X in a histogram of the population (see Figs 2, 4).
Micro-heterogeneity. Heterogeneity within an apparently uniform cell population that is thought to consist of identical cells. Micro-heterogeneity is manifest as the variance (spread) of a single bell-shaped distribution (see Figs 2, 4).
Network state at a given time point. The state of a system (network) of interacting elements (genes) that is jointly defined by all values of activities of the elements (e.g. gene expression levels) at a given time. The network state of a gene regulatory network is thus reflected in the gene expression profile.
Non-genetic (or phenotypic) heterogeneity. A property of a population (e.g. of cells) that refers to the phenotypic variability between its members, which share the same genome. Hence, the trait differences are not due to genetic differences between the cells.
Onomasiology. A branch of semantics that deals with the question of what term to use to describe a (novel) concept or phenomenon, in contrast to semasiology, which deals with how to define a term that is already in use. If onomasiology is about naming, semasiology is about meaning (Baldinger, 1980).
Ontology. A domain of philosophy concerned with fundamental questions regarding the nature of being, such as the relationship between the very existence (‘to be’) of entities and their essence (‘to be such’). In the context of stem cell biology, ontological questions include: do stem cells exist as independent entities, and if so, what are their defining (essential) properties? Is ‘stemness’ an identity or just a state of a cell, or even a property of a group of cells? In informatics, ontology refers to the specification of a shared conceptualization, as in ‘gene ontology’.
Population noise. The variability between individuals in a nominally uniform population due to distinct stationary traits that differ between the individual members (see Fig. 3C).
Quasi-potential landscape. A conceptual construct, inspired by classical mechanics and the idea of potential energy, to help visualize forces that change the state of a system within the ‘state space’ (i.e. the abstract space of all theoretically possible system states). It uses the intuition that some form of ‘gravity’ pulls that system towards the lower points in the landscape (see Box 2).
Temporal noise. The change of a measurable quantity over time in a disordered pattern due to random fluctuations (Fig. 3A).
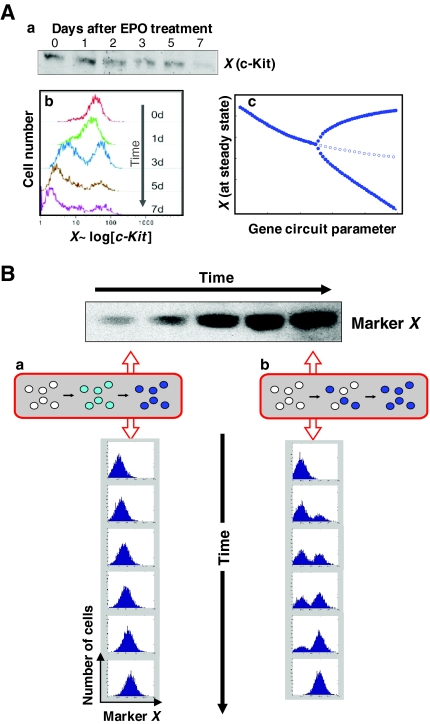
The problem of population heterogeneity is not purely academic. Just consider the experiment illustrated in Fig. 1. A western blot analysis of stem cell differentiation shows how c-Kit, a stem cell marker in many tissues, is downregulated upon the induction of differentiation (Fig. 1A) (H. Chang and S.H., unpublished). However, flow cytometry analysis for surface expression of c-Kit on individual cells, displayed as a histogram, reveals a different picture. As cells downregulate c-Kit, their distribution does not simply shift to the left of the histogram (to lower intensities of fluorescence signal) as one might expect. Instead, the initial population consistently splits into two subpopulations (‘peaks’). The majority of cells do indeed downregulate c-Kit surface expression, but a small subpopulation arises that contains cells with even higher c-Kit surface expression than the original median. The ‘average’ c-Kit expression (i.e. the normalized total) of the whole cell population, as measured in lysed cells by western blotting, still decreases. This splitting of the cell population reflects a heterogeneity of the population in terms of the response to the differentiating signal and is most aptly accounted for by what is termed ‘pitchfork bifurcation’. This is a general phenomenon that arises in some gene-regulatory circuits with feedback loops and forces cells to make a choice between two alternative stable states as their current state is destabilized (Huang et al., 2007).
Fig. 1.
Population-averaging versus individual-cell-based measurements of protein levels. (A) Examples of information masked when population-averaging methods, such as western blotting, are used to measure changes in protein levels. Hematopoietic progenitor cells were treated with erythropoietin (EPO) to induce erythroid differentiation, and c-Kit levels were monitored over time. (a) Western blotting shows a decrease in overall c-Kit levels. (b) Flow cytometry reveals the temporal progression of the population distribution of c-Kit surface expression. (c) A bifurcation diagram could explain the separation of the population into c-Kit-low and c-Kit-high subpopulations (solid lines), even though the overall levels decrease (dotted line). (B) Flow cytometry analysis at the individual cell level distinguishes between (a) a continuous increase in protein in each cell and (b) the non-synchronous near all-or-none (ON-OFF) switching of protein expression. Both give rise to the apparently gradual increase in the band intensity in the western blot shown. The gradual increase in band intensity in b arises from the statistical, non-synchronous switching on of expression, reflecting the fraction of cells in the population that are in the ON-state, as contained in the lysate used for the western blot. In reality, even a formal all-or-none response has a finite switch time, but the change in expression levels is still very steep, and true intermediate levels that are not due to mixing of asynchronously switching cells would be seen only at the single-cell level in dense-interval monitoring, observation at the population level being obscured by asynchrony.
This example illustrates how we have sidelined the notion of cell individuality in the quest to determine regulatory pathways by investigating which transcript or protein levels go up or down in response to a manipulation. Such investigations rely mostly on population-averaging lysate-based methods, such as immunoblots, PCR or microarrays. In using these methods, we tacitly assume cell populations to be uniform. However, heterogeneity in cell populations is not a new concept. It has been widely described in clonal populations of bacteria (Balaban et al., 2004; Novick and Weiner, 1957; Spudich and Koshland, 1976) and has also been referred to as ‘non-genetic individuality’. In the theory of evolutionary dynamics, the role of ‘phenotype variation’ at the organismal level that does not necessarily imply a genetic variation is becoming increasingly appreciated (Kaneko, 2007).
Studies of gene expression or protein levels in mammalian tissues using in situ hybridization or immunohistochemistry often reveal dramatic cell-to-cell variability within one cell type in the same tissue. Heterogeneity was more recently shown in single-cell analysis using fluorescently labeled probes for hybridization (Levsky et al., 2002) or visual gene-expression reporters (Sigal et al., 2006; Takasuka et al., 1998), as well as in single-cell PCR (Hayashi et al., 2008; Warren et al., 2006; Diercks et al., 2009). Apart from a few exceptions (Aird, 2004; Grundel and Rubin, 1988; Rubin, 1992), the biological significance of non-genetic cell population heterogeneity in mammals has not been explicitly articulated until recently, when it has been described in the context of stem cells and fate decision (Stockholm et al., 2007; Chambers et al., 2007; Chang et al., 2008; Dietrich and Hiiragi, 2007; Kalmar et al., 2009; Singh et al., 2007; Spencer et al., 2009; Kobayashi et al., 2009).
To facilitate the discourse on heterogeneity, we need clarity of terminology and a good grasp of the underlying physics. This Primer aims to provide an introduction to the phenomenon of heterogeneity through an exposition at three levels: first, terminology; second, how heterogeneity is measured experimentally; and third, the dynamics of heterogeneity (including its source). The goal is to equip experimental biologists with a solid set of vocabulary and with a conceptual framework for organizing observations, rather than with a comprehensive review of the literature. For in-depth discussions of the biological significance of non-genetic heterogeneity in cell fate control and for an alternative perspective from the related field of ‘gene expression noise’, we refer the reader to several excellent recent reviews (see Enver et al., 2009; Graf and Stadtfeld, 2008; Losick and Desplan, 2008; Raj and van Oudenaarden, 2008).
Terminology
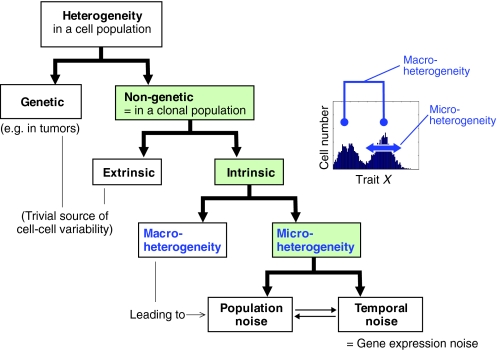
Heterogeneity is a property of a cell population, not of individual cells. Heterogeneity of a population implies the presence of cell-to-cell variability with respect to one (or more than one) measurable trait X, where X can be the cellular level of a given molecule, such as a protein, or any quantifiable morphological or functional parameter. A closer analysis of the term ‘heterogeneity’ reveals many facets and forms that are rarely explicitly articulated. Thus, an attempt at creating a taxonomy and at defining terms and naming newly described phenomena represents a linguistic problem that reaches into the field of onomasiology (see Glossary, Box 1). We propose the following scheme (shown in Fig. 2) that operationally organizes heterogeneity into a hierarchy of dichotomies (see Glossary, Box 1).
Fig. 2.
Cell population heterogeneity. A schematic representation of terminologies and concepts used in the study of cell population heterogeneity, organized into a hierarchy of dichotomies: genetic versus non-genetic heterogeneity; extrinsic versus intrinsic non-genetic heterogeneity; macro-versus micro-heterogeneity within intrinsic non-genetic heterogeneity; and population versus temporal noise within micro-heterogeneity (for details, see text and Box 1, Glossary). The inset represents a flow cytometry histogram that reveals a bimodal distribution, which reflects two distinct subpopulations.
Genetic versus non-genetic heterogeneity
Heterogeneity of cell populations is only non-trivial if one considers heterogeneity within a clonal, that is, genetically identical population of cells, for all too often in biology, any variation in phenotype is explained by a genetic variation. Such ‘genetic determinism’, which has dominated thinking in biology for decades (Strohman, 1997), leaves little room for non-genetic variations (Jablonka, 1994; Lewontin, 2000; Morange, 2001; Strohman, 1994). Genetic variability as a source of heterogeneity in cells is a straightforward, but nonetheless relevant, concept; in cancer biology, for instance, heterogeneity among tumor cells is commonly explained by genetic mutations. This genetic heterogeneity (see Glossary, Box 1), in turn, is thought to be promoted by genomic instability in cancer cells (Lengauer et al., 1998; Loeb, 2001; Tomlinson et al., 2002). In other words, the heterogeneity with respect to trait X is genetic in that the difference in X between individual cells is due to the presence of distinct genes that control trait X in these cells.
Conversely, non-genetic heterogeneity (see Glossary, Box 1) in clonal cell populations defies genetic determinism and raises a simple question, the answer to which we might take for granted: how can the same set of genes generate vastly distinct, stable, and often inherited gene expression profiles and, thereby, distinct phenotypes? This question is at the core of multipotency, as discussed below.
To avoid ambiguities in further discussion, two key terms need further clarification. First, the general term ‘non-genetic’ is to be distinguished from the more specific term ‘epigenetic’, which is used by physicists (Aurell and Sneppen, 2002; Walczak et al., 2005), organismal biologists and systems biologists (Huang, 2009; Jablonka and Lamb, 2002; Waddington, 1942) in a fashion that is different from the way molecular biologists use it.
In the former usage, increasingly encountered in systems biology, ‘epigenetic’ (as in ‘epigenetic landscape’) is employed, as originally coined by Conrad Waddington (Waddington, 1957), to refer to a multiplicity of stable gene expression states in systems that exhibit ‘multi-stability’, as explained below (see Box 1).
By contrast, molecular biologists use ‘epigenetic’ (as in ‘epigenetic mark’) to refer to DNA methylation and histone modifications (Goldberg et al., 2007; Kouzarides, 2007), which serve as a proximate explanation for relatively stable gene expression states. An ‘epigenetic mark’ is the conceptual cousin of a ‘genetic mutation’, both of which are used to explain lasting phenotypic changes by invoking a molecular event. However, epigenetic marks in the genome do not actually account for the existence of stable, inherited states, such as cell types, that originally inspired the term ‘epigenetic’, because covalent modifications are actually dynamic, reversible and lack locus specificity. Therefore, they are not, strictly speaking, explanatory, but represent a molecular mechanism for implementing stable gene expression patterns that first have to be orchestrated by the transcriptional network. It is this gene regulatory network that creates the ‘epigenetic landscape’ in Waddington's sense, as explained below (Bonifer et al., 2008; Huang, 2009; Ptashne, 2007; Slack, 2002).
Second, for a discourse on non-genetic heterogeneity in clonal (isogenic) populations (see Glossary, Box 1), a strict definition of what is a ‘clone’ is required. The term ‘clonal’ does not only mean ‘genetically identical’; if it were so, every developed multicellular organism being derived from a zygote would, in a trivial sense, be a clone. Yet, no one would call a mixture of (isogenic) bone marrow cells that include stem cells, white and red blood cell precursors and so forth, ‘clonal’. Thus, a stricter, non-trivial definition of clonality is that the cells of a clonal population (see Glossary, Box 1) are (1) recently derived from a single ancestor cell (‘colony’), (2) within the same uniform and constant micro-environment, and (3) have achieved, as a population, a certain stationarity.
Macro-heterogeneity versus micro-heterogeneity
In the trivial view of an entire organism as a clone, one could regard phenotypic differences between nominal cell types (e.g. liver cell versus neuron) as a sort of non-genetic heterogeneity. As the definition of what constitutes a cell type versus a subtype or a phenotypic variant is not at all clear, we propose the term macro-heterogeneity (see Glossary, Box 1) to describe evident variability between discrete (sub)populations, which can represent either cell types, subtypes, or just ‘variants’ with respect to a trait X. By contrast, we refer to the heterogeneity between cells within a nominally identical cell type in an apparently uniform population as micro-heterogeneity (see Glossary, Box 1). Crucially, despite the fuzziness that surrounds the definition of a ‘cell type’, a clear-cut delineation can be made based on the statistical distribution of a trait X observed in a population, such as in flow cytometry. Here, macro-heterogeneity is manifest as the presence of multiple discrete, but possibly overlapping, ‘peaks’ (i.e. a multi-modal distribution). By contrast, micro-heterogeneity is reflected in the spread (breadth) of one peak (see Fig. 2, inset). Multi-modality indicates multi-stability, i.e. the presence of multiple distinct attractor states, as discussed below.
Extrinsic versus intrinsic heterogeneity
Phenotype variation among cells of a single clonal population can be caused by extrinsic factors that do not act uniformly on the population and thus trigger a cellular response in only a fraction of the population. This is referred to as extrinsic heterogeneity (see Glossary, Box 1) and arises, for example, when a clonal population grows in a complex environment, such as tissue niches, where cells differ in their vicinity to blood vessels, other cells or structures, or when cells in culture differ with respect to their distance from neighboring cells and to the dish border, or with respect to oxygen gradients and so forth. The heterogeneity of the cell population would then simply mirror the heterogeneity of the environment. Such heterogeneity due to differential instruction from outside plays a key role in development. Notably, the very first diversifications of cell lineages in the early embryo, such as the lineage split into the trophectoderm and inner cell mass, appear to benefit from the physical asymmetries of the embryo (inside versus outside cells, polarity) (Zernicka-Goetz et al., 2009). Similarly, positional information, such as that provided by morphogen gradients, further drives (extrinsic) heterogeneity in later cell fate waves of diversification (Oates et al., 2009).
Intrinsic heterogeneity (see Glossary, Box 1), by contrast, cannot be explained in an obvious, straightforward way by an external cause. Instead, it denotes the spontaneous diversification of a clonal population into continuous or discretely distinct variants. Intrinsic diversification has a profound meaning in complex systems sciences, for it is essential for the spontaneous generation of complex patterns - the paradigm of an emergent property that is characteristic of a living system (Goodwin, 1993; Kauffman, 1993). Intrinsic diversification of an initially unbiased, symmetric state requires two processes: first, a symmetry-breaking event (as exemplified by the bifurcation in Fig. 1A) that creates discrete, alternative options; and second, a stochastic process (such as gene expression noise, as discussed below) that drives the choice of these options. In complexity theory, such intrinsic diversification has been linked to instabilities and irreversibility in non-equilibrium systems (Nicolis and Prigogine, 1989; Prigogine, 1997).
Measuring heterogeneity
The introductory example (Fig. 1A) alerts us to the fact that population-averaging techniques that involve lysing entire populations of cells of a culture or tissue specimen mask substantial information on population distribution. However, even among single-cell analysis techniques, there is a broad spectrum of methods that differ fundamentally in the information they provide on the nature of heterogeneity. Rather than giving an overview of methods, we discuss three approaches that illustrate key distinctive principles and span a range of techniques.
Flow cytometry
Flow cytometry is, strictly speaking, not a single-cell analysis technique, but measures the property X of a cell, typically the cellular abundance of a specific protein, in an entire population at the resolution of individual cells. It offers the most lucid illustration of population heterogeneity. The histogram displays a roughly bell-shaped frequency distribution [most typically, a log-normal distribution for the frequency of X in the population, so that a Gaussian-like curve appears in the log-scale presentation (Kaneko, 2006; Mantzaris, 2006)]. The spread of cellular levels of a single protein in a clonal cell population typically covers a range of 10- to 1000-fold, substantially more than measurement error, which, as determined using standardized fluorescence beads, accounts for a less than 10-fold range of variability (Chang et al., 2008). Unlike single-cell analysis techniques, flow cytometry provides the statistical distribution of a trait X in a population - a layer of information that is crucial for studying gene expression noise and population dynamics, but is often ignored by biologists. Specifically, analyzing the distribution of X helps to detect the presence of multiple quasi-discrete subpopulations, as opposed to a continuous (smooth) distribution (Figs 1, 2) (Chang et al., 2006; Huang et al., 2009; Krutzik et al., 2004), and thus helps to distinguish between macro- and micro-heterogeneity (Fig. 2).
Single-cell real-time digital imaging
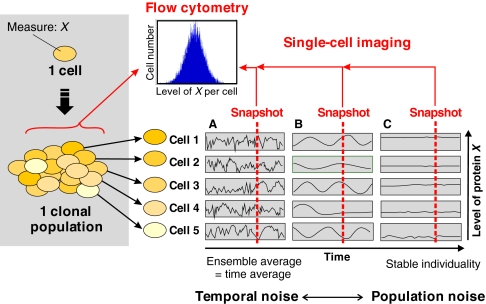
Flow cytometry provides population snapshots and cannot monitor temporal changes of a trait X within individual cells. At the other end of the spectrum of methods, single-cell monitoring of property X in real time using live video-microscopy tracks individual cells over time (Fig. 3). This permits the longitudinal monitoring of X; for example, the time course of the change in the cellular levels of a fluorescent protein. Such measurements deliver kinetic information on the temporal structure of the fluctuations of gene expression that evades flow cytometry (Austin et al., 2006). They also help to distinguish between ‘population noise’ and ‘temporal noise’, as discussed below. However, single-cell imaging captures only a handful to hundreds of cells at a time and hence barely produces sufficient data on the population as a statistical ensemble. Of biological interest is that single-cell monitoring allows the tracking of cell fate history and the construction of entire cell-lineage pedigrees (Kaufmann et al., 2007; Ravin et al., 2008; Schroeder, 2008).
Fig. 3.
Trait heterogeneity in cell populations and at the individual cell level. (A-C) Two approaches can in principle be applied to the analysis at the individual cell level of a cell population heterogeneous with respect to a trait X (e.g. the cellular abundance of a particular protein, as represented by shades of yellow): population distribution versus single-cell real-time tracking. Note that the ensemble snapshot provided by flow cytometry does not distinguish between the possible scenarios in individual cells. (A) Fast stochastic fluctuations attributable to gene expression noise. (B) Asynchronous, possibly deterministic, oscillations. (C) Fixed cell individuality. The two extremes of this spectrum represent (A) temporal noise and (B) population noise.
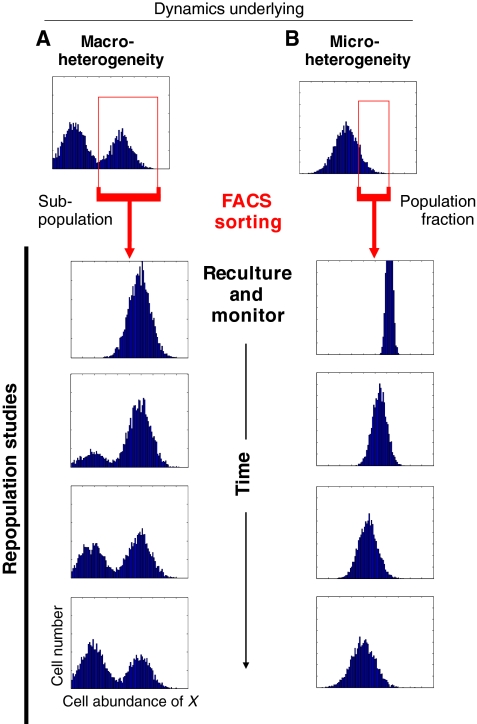
FACS and the dynamics of population distribution
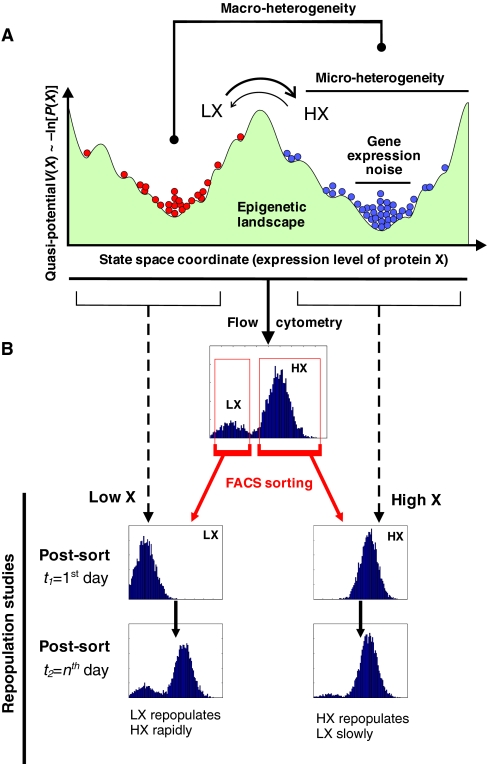
The sorting of cell populations using FACS (fluorescence-activated cell sorting) allows for the physical isolation of any segment of the total cell population with respect to the distribution of property X. This can provide insight into the dynamics that underlie heterogeneity at the population level and thus complements single-cell monitoring. Two scenarios can be distinguished (Fig. 4A,B): the sorting and reculturing of either a distinct subpopulation (Fig. 4A) or of a population fraction (Fig. 4B). In both cases, flow cytometry can be used subsequently to monitor how the sorted cells behave over time, i.e. whether and at what rate they restore the original distribution of X. This is most readily achieved when X is the expression of a gene of interest and is reported by a fluorescent protein (Kalmar et al., 2009; Kashiwagi et al., 2006), but it can also be achieved through the direct immunolabeling of cells, provided that the antibody is removed before the sorted cells are recultured (Chang et al., 2008).
Fig. 4.
Analyzing the dynamics underpinning non-genetic heterogeneity. (A,B) Schematic representation of fluorescence-activated cell sorting (FACS), based on simulations. The sorting and reculturing of a subpopulation in the case of (A) macro-heterogeneity, or of a tail fraction (‘outliers’) in the case of (B) micro-heterogeneity, can provide information on the nature of the processes that generate heterogeneity among clonal cells. (A) The presence of multiple separate peaks (representing subpopulations) with respect to a single trait X does not imply the presence of inconvertible, irreversibly committed cell types. Often, transitions occur. (B) Typically, outliers in mammalian cell populations slowly (within time frames of up to several days) repopulate the naïve distribution, which indicates the presence of an attractor state in a rugged epigenetic landscape (see Fig. 5). The rate at which a subpopulation repopulates the entire distribution (including subpopulations) provides information about the transition rate between distinct subpopulations.
Sorting for a discrete subpopulation (Fig. 4A) and monitoring whether it repopulates the other subpopulations readily establishes whether a subpopulation is a separate, stable and irreversible cell (sub)type - the default assumption in cell biology - or is in a dynamic, reversible state (Fig. 5, see next section) (Chang et al., 2006; Kalmar et al., 2009). In the latter case, the bimodal macro-heterogeneity most likely represents a dynamic equilibrium, with cells transitioning back and forth between the two subpopulations.
Fig. 5.
The epigenetic landscape and practical implications for network dynamics. (A) A projected epigenetic landscape with two attractors [low X (LX), high X (HX)] and their sub-attractors, which contribute to heterogeneity (see Box 2). Each circle represents a network state (i.e. a cellular phenotype) determined by the level of X as indicated by the position on the horizontal axis (i.e. one state space dimension, trait X, of the high-dimensional state space). The vertical axis displays the ‘potential’ V (X), as explained in Boxes 1 and 2. The height of the accumulation of circles reflects the density distribution as a function of X. (B) Associated flow cytometry histograms of cell population distributions with respect to X. Subpopulation sorting (see Fig. 4) can reveal the reversibility and the transition rates between the subpopulations.
By contrast, the utility of sorting out a fraction from a single-peaked distribution (Fig. 4B) is particularly counterintuitive because the dispersion is usually considered ‘noise’. However, recent studies demonstrate that the tail fractions exhibit distinct transcriptome-wide gene expression patterns of biological significance (Chang et al., 2008; Kobayashi et al., 2009).
A limitation of the physical sorting of populations that should be kept in mind is errors in sorting that can create ‘false-positive heterogeneity’, which needs to be separated from intrinsic population heterogeneity (Enver et al., 2009). Mis-sorted cells that have an inappropriate property X can contaminate a subpopulation and affect subsequent population behavior. This is particularly relevant if the sorted property X correlates with the rate of cell proliferation, such that a contaminating cell might proliferate much faster and ‘take over’ the sorted subpopulation with its progeny [discussed by Chang et al. (Chang et al., 2008)].
Source and dynamics of heterogeneity
Among the various facets and forms of heterogeneity, non-genetic, intrinsic micro-heterogeneity emerges as the most intriguing in biological terms because there is no straightforward explanation of its source and because its biological implications are only beginning to be explicitly studied. This form of heterogeneity is the focus of the next sections.
Temporal versus population noise
Given the recent explosion in the study of ‘gene expression noise’ (Kaern et al., 2005; Raj and van Oudenaarden, 2008) we need, for a more rigorous discussion of the source of heterogeneity, to introduce another apparent dichotomy: temporal versus population noise (Fig. 3).
Temporal noise (see Glossary, Box 1) refers to fluctuations in a property X (e.g. the cellular content of a particular protein) in an individual cell over time. This is what is commonly referred to by the term ‘gene expression noise’ (Kaern et al., 2005; Raj and van Oudenaarden, 2008). Because cellular fluctuations of X are generally not synchronized between cells, they collectively give rise to a heterogeneous population at any time point.
The study of gene expression noise has been facilitated by the availability of fluorescent protein reporters that enable the monitoring of gene expression in individual living cells. Briefly, the temporal ‘noisy’ fluctuations in gene expression (Fig. 3A) are thought to stem from the random (i.e. stochastic), synthesis and breakdown of individual molecules, such as mRNA and proteins. They are ultimately a manifestation of the thermal nature of chemical reactions, which constitute probabilistic molecular events. As these biomolecules are present at relatively small copy numbers in the cell, their stochastic fluctuations are, unlike in classical test-tube chemistry, not averaged out. This is known as the ‘finite number effect’ (Kaern et al., 2005). Temporal noise is accentuated when the assembly and dissociation of transcriptional complexes follow slow kinetics and when the subsequent uneven transcript production is amplified by a high translation rate (Kaern et al., 2005; Kaufmann and van Oudenaarden, 2007). Most studies of gene expression rely on bacteria and yeast as model systems, and on the use of flow cytometry to measure the population distribution of the abundance of a trait X (generally, a fluorescent protein) in individual cells as a snapshot. The (normalized) standard deviation of the observed, typically bell-shaped histogram of X is then taken as a measure of temporal noise.
However, as mentioned above, a population of cells can, in principle, also exhibit a broad distribution of a trait X even if X does not fluctuate in time (Fig. 3A), i.e. if X is a time-invariant, stable trait of an individual cell (Fig. 3C). Such cell-to-cell variation is referred to here as population noise (see Glossary, Box 1) and is due to stable differences between individual members of a population, much in the same way as the measurement of shoe size in the adult human population will give rise to a bell-shaped histogram. In the context of the heterogeneity discussed here, however, the absence of genetic differences must be kept in mind.
Temporal and population noise are scenarios at opposing ends of a spectrum and are introduced here as general principles to allow one to place the real case somewhere within the spectrum that is flanked by these two extremes (Fig. 3). Importantly, this general framework does not exclude heterogeneity due to fluctuations at intermediate time scales. These can stem from a variety of complicated, possibly deterministic (i.e. not noise-generated), but regulated changes in the abundance of a molecule. An example of this would be gene circuit-generated oscillations of X in individual cells (Hirata et al., 2002; Lahav et al., 2004; Kobayashi et al., 2009) that, if randomly out of phase between the cells, would produce the cell-to-cell variability seen in snapshot population measurements (Fig. 3B). Clearly, in reality, the ‘net’ fluctuation will result from the combination of several sources of fluctuations that occur at various time scales, as at least the stochastic component is inevitable.
Relationship between temporal and population noise
It is not possible to tell from a single population distribution snapshot (histogram) whether heterogeneity is mainly due to temporal or to population noise. However, the broad distribution is often taken by default as reflecting solely temporal fluctuations in individual cells. This inference makes the implicit assumption of ergodicity (see Glossary, Box 1) (Patrascioiu, 1987). The ergodic hypothesis states that one can determine the time average of the fluctuating X (Fig. 3A) of an individual cell by simply measuring N independent elements (i.e. cells) of an ensemble (i.e. cell population) just once (a snapshot histogram), instead of taking a number N repeated measurements of the same cell, provided that the fluctuations of an individual cell are so fast that the individual measurements at some typical time interval of the same cell give uncorrelated results (lack of autocorrelation) - the condition of ergodicity.
In brief, ergodicity implies that the time average equals the ensemble average. Ergodicity breaks down if the fluctuations are so slow that the length of time intervals needed to make the repeated measurements (avoiding autocorrelation) for determining a representative time average becomes impracticably long (approximating the case depicted in Fig. 3C).
Temporal noise, but not population noise, can satisfy the ergodic condition. Many of the early experimental studies on gene expression noise have implicitly assumed ergodicity and did not consider the time scale of fluctuations, which would entail the careful analysis of fluctuations of individual cells in real time. However, at least in mammalian cell populations, slow fluctuations of X (see below) that can span up to several cell division cycles appear to contribute substantially to population heterogeneity. This component of fluctuations is too slow for considering the distribution of flow cytometry histograms as a pure manifestation of noisy (thermal) fluctuations.
Although this Primer is not about gene expression noise, it is worth mentioning that in studies of gene expression noise the distinction between intrinsic and extrinsic noise has been prevalent (Elowitz et al., 2002; Volfson et al., 2006). Originally, these terms were defined operationally in conjunction with the experimental set-up used to distinguish between: (1) gene-intrinsic noise attributable to the probabilistic nature of the reactions mentioned above; and (2) cell environment-derived contributions (such as fluctuating upstream influences common to two genes) to the noisy expression of a particular protein. However, the semantics of the term ‘extrinsic noise’ have not been uniform and must be dealt with carefully, given that the definition of the system considered (a chemical reaction, a cellular process, a compartment or the entire cell) determines what is to be regarded as ‘intrinsic’ or ‘extrinsic’. Furthermore, the time scale of fluctuations, as discussed above, must also be considered. Thus, extrinsic noise has also been interpreted as being due to stochastic fluctuations that occur over a longer time scale than intrinsic noise and has been modeled as fluctuations in the ‘control parameters’ of gene circuit models (Lei, 2009). In this view, extrinsic noise might correspond to slowly changing cell-individual traits (e.g. nucleus size, ribosome content) and approaches population noise.
Finally, the drastic (but necessary) simplification used in mathematical models of noise that shrink the processes of gene expression from hundreds of chemical reactions that participate in transcription, splicing and protein synthesis and maturation, to the two-step ‘central dogma’ (transcription and translation), further blurs the formal interpretation of noise measurements. As such, consistency has thus far been achieved only for the basic principles of gene expression noise, not for the details (Kaufmann and van Oudenaarden, 2007; Paulsson, 2004). Real-time measurements of trait fluctuations in multiple individual cells in a population, as discussed above (see ‘Measuring heterogeneity’), will be crucial to address these problems and to determine the relative contributions of temporal versus population noise.
Slow fluctuations in mammalian cells
Recent kinetic studies of mammalian cells suggest that the wide dispersion of the expression level X of a given protein is due to fluctuations of X that have a significant component that occurs over several days, in addition to the fast temporal component caused by gene expression noise. Thus, heterogeneity is perhaps more aptly viewed as largely due to population noise when considering functional studies of mammalian developmental processes that typically extend over days to weeks. Nevertheless, brief bursts of protein expression due to temporal (gene expression) noise may also affect cell fate choice (Enver et al., 2009; Losick and Desplan, 2008; Raj and van Oudenaarden, 2008).
Slow fluctuations have been demonstrated by the real-time monitoring of protein expression in individual mammalian cells, using genomically integrated fluorescent protein reporters (Sigal et al., 2006), or by flow cytometry analysis of the time needed for FACS-sorted ‘outlier’ cells with ‘extreme’ levels of X to relax back and re-establish the native distribution of X (Fig. 4B) (Chang et al., 2008). The finding that heterogeneity is associated with the lasting, cell-specific expression of fate-determining proteins only in some, but not all, cells in early embryos (‘salt-and-pepper’ appearance) and the correlation of a cell's gene expression profile with its prospective fate (Graf and Stadtfeld, 2008; Hayashi et al., 2008; Kalmar et al., 2009; Silva and Smith, 2008; Zernicka-Goetz et al., 2009) support the view that heterogeneity is predominantly caused by slow changes that are substantially influenced by deterministic (non-stochastic) extrinsic factors. Thus, heterogeneity appears to have a developmental role, rather than simply reflecting epiphenomenal statistical blips due to temporal noise.
The epigenetic landscape
The source of these slow fluctuations is currently poorly understood (see Table 1 for specific explanations that have been proposed). Here, we summarize in qualitative terms a general conceptual framework from the field of non-equilibrium systems dynamics (Nicolis, 1986; Nicolis and Prigogine, 1989) that provides a different perspective for understanding non-genetic heterogeneity (see Box 2).
Table 1.
Possible mechanisms for slow fluctuations in a trait X as a cause of intrinsic non-genetic heterogeneity
| Mechanism | Example reference(s) |
| Gene expression noise in trait X bounded by an attractor (Ornstein-Uhlenbeck process), but with very weak mean-resetting force, further complicated by a rugged potential landscape and high dimensionality of the system | Chang et al., 2008 |
| Action-potential-like excitability, causing noise-induced wide excursions of X | Kalmar et al., 2009 |
| Deterministic oscillations in X with long periods, possibly due to ‘state cycles’ in genome-wide dynamics (this causes heterogeneity when not synchronized between cells) | Hirata et al., 2002; Kauffman, 1969a; Klevecz et al., 2004; Lahav et al., 2004 |
| Deterministic and stochastic oscillations of X due to non-cell-autonomous effects (cell-cell communication), such as array-enhanced coherence resonance | Skupin et al., 2008 |
| Deterministic chaotic dynamics (strange attractor) | Furusawa and Kaneko, 2001 |
| Random, unequal partitioning of transcript or protein X during cell division | Mantzaris, 2007 |
| Varying chromatin state at locus X between individual cells (not an independent cause in itself, but might be part of any of the above mechanisms) | Hayashi et al., 2008; Zernicka-Goetz et al., 2009 |
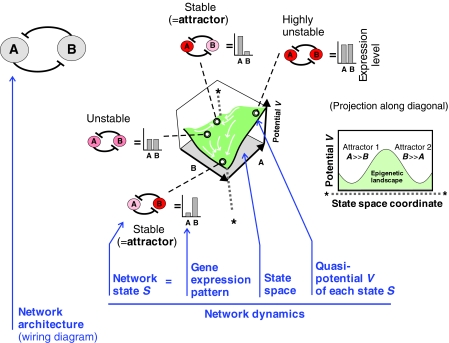
Box 2. Basic concepts in network dynamics
We consider a two-gene circuit as a minimal gene regulatory network consisting of the mutually inhibiting genes A and B, as indicated by the network architecture depicted below. The term ‘network dynamics’ denotes the concerted change in the expression of A and B, the expression levels of which collectively define a network state S=S(A, B), measurable as a gene expression pattern. The network state S is a point in the state space, the two axes of which represent the expression levels of A and B, so that each point in the state space is a combinatorially possible network state S(A, B). However, not all states S(A, B) are equally likely to exist because the network interactions constrain the way in which A and B change. In this example, as gene A suppresses gene B (and vice versa), a network state S with high expression levels of both A and B would be unstable and hence unlikely to exist. The network will settle down in a stable (likely) state instead, e.g. with A high and B low or vice versa. Such a stable equilibrium state that complies with the network interactions is called an attractor. In the network depicted, there are two attractors. The coexistence of multiple attractors in one network is referred to as ‘multi-stability’ [for a more formal description, see Huang et al. (Huang et al., 2007) and Huang and Kauffman (Huang and Kauffman, 2009)].
The expression level of a protein X is determined by a vastly complicated gene regulatory network (GRN) that encompasses virtually the entire genome. Thus, heterogeneity is actually multidimensional, not just with respect to a gene X as discussed so far. The integrated high-dimensional dynamics of a network can be formalized as a quasi-potential landscape (see Glossary, Box 1) (Bar-Yam et al., 2009; Huang and Kauffman, 2009; Wang et al., 2008), in which each point represents a network state (see Glossary, Box 1). This is, by definition, a gene expression pattern and approximately represents a cell phenotype (see Box 2 for an introduction to network dynamics). Thus, a movement in this landscape represents a change of the gene expression profile and, hence, of the cell phenotype. Cellular development, then, is represented by a flowing movement on the landscape, wherein the cell's state seeks the lower regions. This quasi-potential landscape is the formal equivalent of Waddington's metaphoric ‘epigenetic landscape’ (Waddington, 1957) and captures in an intuitive manner the global behavior of complex GRNs. It is in this sense that the term ‘epigenetic’ was proposed by Waddington (Waddington, 1942) and is still used by physicists, as discussed above. Importantly, the specific landscape topography carries key information on the potential behaviors of a cell, as it is a direct visualization of how the GRN dictates the manner in which a gene expression pattern as a whole can change under the regulatory constraints (Box 2).
In the epigenetic landscape, the lowest point in a ‘valley’ (i.e. a ‘potential well’ in terms of a generalized quasi-potential) represents an attractor state (see Glossary, Box 1) of the network. An attractor is a stationary self-stabilizing network state, the associated gene expression pattern of which is interpreted as defining a cell type (Huang et al., 2005; Kauffman, 1993). Cells (or, more precisely, GRNs) tend towards the attractor states and stay there, resisting minor perturbations. However, external regulatory signals and large perturbations (e.g. strong gene expression noise) that alter the expression of typically multiple genes can induce transitions of network states across the ‘hills’ that separate the attractor states. Such quasi-discontinuous state transitions appear as the all-or-none cell-type switching in individual cells (Fig. 1B) (Huang and Kauffman, 2009).
Distribution on the epigenetic landscape as a source of heterogeneity
The precise topography of the landscape, which is mathematically determined by the gene network architecture [for a more formal, accessible discussion, see Huang and Kauffman (Huang and Kauffman, 2009)], is not known in detail. Nevertheless, the general concept of an epigenetic landscape could explain non-genetic macro- and micro-heterogeneity, as well as temporal and population noise, within one formal framework. Because of cell-cell variation, a population of cells is represented not by a point, but by a ‘cloud’, i.e. by an ensemble of points distributed across a certain region in the landscape. With the incorporation of heterogeneous cell populations and of instabilities (‘hills’) that repel and partition the clouds, multicellular development can be thought of as the movement of a cloud in the landscape along the valleys, seeking their lowest points, akin to water flowing down the mountains and collecting into lakes. Thus, multicellular development could be formalized as the time-irreversible change of a complicated distribution (Petrosky and Prigogine, 1993; Prigogine, 1997).
The landscape of a complex network [at least, near the order-chaos regime boundary (Kauffman, 1993)] is generally ‘rugged’, that is, contains multiple, possibly nested valleys (attractors with ‘sub-attractors’) rather than a single dominating valley surrounded by smooth slopes. Thus, much as with real valleys, one can find a hierarchy of main valleys, sub-valleys and so forth, which would explain the distinct scales of heterogeneity encoded by the network. Macro-heterogeneity is then due to the simultaneous occupation by the cloud of multiple major valleys (attractors), each of which roughly corresponds to a distinct cell type (Huang et al., 2005; Kauffman, 1969b). They are separated by major ‘hills’ that are high enough to permit only rare noise-induced attractor transitions. Conversely, micro-heterogeneity is due to the dispersion of cells within a cell-type-specifying attractor into multiple sub-attractors that are separated by low energy barriers. The slow kinetics of the heterogeneity-producing fluctuations would be a manifestation of noise-induced transitions between these sub-attractors, which occur on a much slower time scale than thermal fluctuations between continuous expression levels (Fig. 5). Recent kinetic measurements of the non-exponential relaxation of outlier cells to the center of a main attractor indicate the presence of distinct intermediate states between the outlier and the ‘median’ states, which suggests the presence of sub-attractors (Chang et al., 2008).
Interestingly, this picture is reminiscent of an established concept employed to understand the folding of proteins, in which the coexistence of multiple energetic substates allows for many intermediate subconformations in addition to the main conformation of a protein. The dynamics of transitions between these (higher-energy) subconformations can be represented by a multi-well ‘washboard potential’ (akin to Fig. 5A) that traps rare outlier subconformations away from the central equilibrium state, so that molecules in the higher substates only slowly (possibly non-exponentially) relax back to the native state (Frauenfelder et al., 1991). The transient, but continual occupation of the substates corresponds to the presence of outlier cells at equilibrium and contributes to the spreading of the population histogram.
The general complex systems dynamics framework set out above can accommodate modifications to include alternative sources of slow dynamics and heterogeneity (see Table 1), such as strange attractors due to chaotic dynamics (Furusawa and Kaneko, 2001) or noise-induced action-potential-like excursions around an attractor state (Kalmar et al., 2009).
Conclusions
The epigenetic landscape offers a new vista on the dynamics of non-genetic heterogeneity and their biological consequences. In the case of micro-heterogeneity, the extreme expression values of protein X in outliers at the tail of a distribution could have biological consequences if X is a fate-determining protein, as the relatively long persistence of an outlier state is in the time scale (days) of gene regulation. In other words, non-ergodicity means that outliers, not just the mean, matter. Noise is more than just statistics that need to be averaged out. In the case of macro-heterogeneity, it should be kept in mind that two subpopulations (separate ‘peaks’ in the histogram) are not necessarily inconvertible cell types, but might transition into each other, depending on the relative height of the potential barrier (hill) that separates them. This is contrary to many biologists‘ deeply rooted intuition that discretely distinct subpopulations represent irreversibly committed cell types, and instead provides a conceptual framework for cell type plasticity (Graf, 2002; Joshi and Enver, 2002).
Evidence for the biological relevance of non-genetic diversity and slow dynamics is accumulating. The associated spontaneous and lasting phenotypic individuality of cells appears to play a central role in driving the diversification of cell types in metazoans, and hence could be a crucial phenomenon in multipotency.
However, the biological significance of non-genetic heterogeneity extends beyond multipotency and development. It also might be important for cell population survival by producing the phenotypic diversity for non-genetic selection (’mutationless evolution’), notably in tumors responding to chemotherapy (Brock et al., 2009; Cohen et al., 2008; Spencer et al., 2009). Almost no drug designed to alter a cell phenotype (e.g. by inducing apoptosis or differentiation) does so (at reasonable doses) in all of the cells of a population, perhaps because of the vast intrinsic non-genetic heterogeneity with regard to the responsiveness of the cells. This drastically limits the efficacy of even modern target-selective drugs or of reprogramming efforts (Brock et al., 2009; Huang, 2009).
Much as biologists have learned to appreciate random, inheritable genetic variability as a source of species diversification in evolution, they might need to embrace random non-genetic heterogeneity of inheritable traits in cell populations as a source of cell type diversification, instead of regarding it as a nuisance, in order to understand the essence of multicellular life.
Acknowledgements
We acknowledge the U.S. Air Force Office of Scientific Research and the National Institutes of Health for supporting this work and thank Alfonso Martinez Arias and Stuart Kauffman for valuable discussions. Deposited in PMC for release after 12 months.
References
- Aird W. C. (2004). Endothelial cell heterogeneity: a case for nature and nurture. Blood 103, 3994 [Google Scholar]
- Aurell E., Sneppen K. (2002). Epigenetics as a first exit problem. Phys. Rev. Lett. 88, 048101 [DOI] [PubMed] [Google Scholar]
- Austin D. W., Allen M. S., McCollum J. M., Dar R. D., Wilgus J. R., Sayler G. S., Samatova N. F., Cox C. D., Simpson M. L. (2006). Gene network shaping of inherent noise spectra. Nature 439, 608-611 [DOI] [PubMed] [Google Scholar]
- Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622-1625 [DOI] [PubMed] [Google Scholar]
- Baldinger K. (1980). Semantic Theory Oxford: Blackwell; [Google Scholar]
- Bar-Yam Y., Harmon D., de Bivort B. (2009). Systems biology. Attractors and democratic dynamics. Science 323, 1016-1017 [DOI] [PubMed] [Google Scholar]
- Bonifer C., Hoogenkamp M., Krysinska H., Tagoh H. (2008). How transcription factors program chromatin-lessons from studies of the regulation of myeloid-specific genes. Semin. Immunol. 20, 257-263 [DOI] [PubMed] [Google Scholar]
- Brock A., Chang H., Huang S. (2009). Non-genetic heterogeneity-a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 10, 336-342 [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230-1234 [DOI] [PubMed] [Google Scholar]
- Chang H. H., Oh P. Y., Ingber D. E., Huang S. (2006). Multistable and multistep dynamics in neutrophil differentiation. BMC Cell Biol. 7, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. H., Hemberg M., Barahona M., Ingber D. E., Huang S. (2008). Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544-547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. A., Geva-Zatorsky N., Eden E., Frenkel-Morgenstern M., Issaeva I., Sigal A., Milo R., Cohen-Saidon C., Liron Y., Kam Z., et al. (2008). Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511-1516 [DOI] [PubMed] [Google Scholar]
- Diercks A., Kostner H., Ozinsky A. (2009). Resolving cell population heterogeneity: real-time PCR for simultaneous multiplexed gene detection in multiple single-cell samples. PLoS One 27, e6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J. E., Hiiragi T. (2007). Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219-4231 [DOI] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183-1186 [DOI] [PubMed] [Google Scholar]
- Enver T., Pera M., Peterson C., Andrews P. W. (2009). Stem cell states, fates, and the rules of attraction. Cell Stem Cell 4, 387-397 [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Sligar S. G., Wolynes P. G. (1991). The energy landscapes and motions of proteins. Science 254, 1598-1603 [DOI] [PubMed] [Google Scholar]
- Furusawa C., Kaneko K. (2001). Theory of robustness of irreversible differentiation in a stem cell system: chaos hypothesis. J. Theor. Biol. 209, 395-416 [DOI] [PubMed] [Google Scholar]
- Goldberg A. D., Allis C. D., Bernstein E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635-638 [DOI] [PubMed] [Google Scholar]
- Goodwin B. (1993). How the Leopard Changed Its Spots: The Evolution of Complexity Princeton: Princeton University Press; [Google Scholar]
- Graf T. (2002). Differentiation plasticity of hematopoietic cells. Blood 99, 3089-3101 [DOI] [PubMed] [Google Scholar]
- Graf T., Stadtfeld M. (2008). Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 3, 480-483 [DOI] [PubMed] [Google Scholar]
- Grundel R., Rubin H. (1988). Maintenance of multiplication rate stability by cell populations in the face of heterogeneity among individual cells. J. Cell Sci. 91, 571-576 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S. M., Tang F., Surani M. A. (2008). Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K., Kageyama R. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840-843 [DOI] [PubMed] [Google Scholar]
- Huang A. C., Hu L., Kauffman S. A., Zhang W., Shmulevich I. (2009). Using cell fate attractors to uncover transcriptional regulation of HL60 neutrophil differentiation. BMC Syst. Biol. 3, 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. (2009). Reprogramming cell fates: reconciling rarity with robustness. BioEssays 31, 546-560 [DOI] [PubMed] [Google Scholar]
- Huang S., Kauffman S. (2009). Complex gene regulatory networks-from structure to biological observables: cell fate determination. In Encyclopedia of Complexity and Systems Science (ed. Meyers R. A.). Berlin: Springer; [Google Scholar]
- Huang S., Eichler G., Bar-Yam Y., Ingber D. E. (2005). Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 94, 128701 [DOI] [PubMed] [Google Scholar]
- Huang S., Guo Y. P., May G., Enver T. (2007). Bifurcation dynamics of cell fate decision in bipotent progenitor cells. Dev. Biol. 305, 695-713 [DOI] [PubMed] [Google Scholar]
- Jablonka E. (1994). Inheritance systems and the evolution of new levels of individuality. J. Theor. Biol. 170, 301-309 [DOI] [PubMed] [Google Scholar]
- Jablonka E., Lamb M. J. (2002). The changing concept of epigenetics. Ann. New York Acad. Sci. 981, 82-96 [DOI] [PubMed] [Google Scholar]
- Joshi C. V., Enver T. (2002). Plasticity revisited. Curr. Opin. Cell Biol. 14, 749-755 [DOI] [PubMed] [Google Scholar]
- Kaern M., Elston T. C., Blake W. J., Collins J. J. (2005). Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6, 451-464 [DOI] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Munoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. (2009). Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K. (2006). Life: An Introduction to Complex Systems Biology Berlin: Springer; [Google Scholar]
- Kaneko K. (2007). Evolution of robustness to noise and mutation in gene expression dynamics. PLoS One 2, e434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi A., Urabe I., Kaneko K., Yomo T. (2006). Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. PLoS ONE 1, e49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B. B., van Oudenaarden A. (2007). Stochastic gene expression: from single molecules to the proteome. Curr. Opin. Genet. Dev. 17, 107-112 [DOI] [PubMed] [Google Scholar]
- Kaufmann B. B., Yang Q., Mettetal J. T., van Oudenaarden A. (2007). Heritable stochastic switching revealed by single-cell genealogy. PLoS Biol. 5, e239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S. (1969a). Homeostasis and differentiation in random genetic control networks. Nature 224, 177-178 [DOI] [PubMed] [Google Scholar]
- Kauffman S. A. (1969b). Metabolic stability and epigenesis in randomly constructed genetic nets. J. Theor. Biol. 22, 437-467 [DOI] [PubMed] [Google Scholar]
- Kauffman S. A. (1993). The Origins of Order New York: Oxford University Press; [Google Scholar]
- Klevecz R. R., Bolen J., Forrest G., Murray D. B. (2004). A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc. Natl. Acad. Sci. USA 101, 1200-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. (2009). The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870-1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693-705 [DOI] [PubMed] [Google Scholar]
- Krutzik P. O., Irish J. M., Nolan G. P., Perez O. D. (2004). Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin. Immunol. 110, 206-221 [DOI] [PubMed] [Google Scholar]
- Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A. J., Elowitz M. B., Alon U. (2004). Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36, 147-150 [DOI] [PubMed] [Google Scholar]
- Lei J. (2009). Stochasticity in single gene expression with both intrinsic noise and fluctuation in kinetic parameters. J. Theor. Biol. 256, 485-492 [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K. W., Vogelstein B. (1998). Genetic instabilities in human cancers. Nature 396, 643-649 [DOI] [PubMed] [Google Scholar]
- Levsky J. M., Shenoy S. M., Pezo R. C., Singer R. H. (2002). Single-cell gene expression profiling. Science 297, 836-840 [DOI] [PubMed] [Google Scholar]
- Lewontin R. (2000). The Triple Helix: Gene, Organism, and Environment Cambridge: Harvard University Press; [Google Scholar]
- Loeb L. A. (2001). A mutator phenotype in cancer. Cancer Res. 61, 3230-3239 [PubMed] [Google Scholar]
- Losick R., Desplan C. (2008). Stochasticity and cell fate. Science 320, 65-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantzaris N. V. (2006). Stochastic and deterministic simulations of heterogeneous cell population dynamics. J. Theor. Biol. 241, 690-706 [DOI] [PubMed] [Google Scholar]
- Mantzaris N. V. (2007). From single-cell genetic architecture to cell population dynamics: quantitatively decomposing the effects of different population heterogeneity sources for a genetic network with positive feedback architecture. Biophys. J. 92, 4271-4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morange M. (2001). The Misunderstood Gene Cambridge: Harvard University Press; [Google Scholar]
- Nicolis G. (1986). Dissipative systems. Rep. Prog. Phys. 49, 873-949 [Google Scholar]
- Nicolis G., Prigogine I. (1989). Exploring Complexity: An Introduction New York: W.H. Freeman & Company; [Google Scholar]
- Novick A., Weiner M. (1957). Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 43, 553-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates A. C., Gorfinkiel N., Gonzalez-Gaitan M., Heisenberg C. P. (2009). Quantitative approaches in developmental biology. Nat. Rev. Genet. 10, 517-530 [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Zon L. I. (2002). Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat. Immunol. 3, 323-328 [DOI] [PubMed] [Google Scholar]
- Patrascioiu A. (1987). The Ergodic-hypothesis, a complicated problem in mathematics and physics. Los Alamos Science 15, 263-279 [Google Scholar]
- Paulsson J. (2004). Summing up the noise in gene networks. Nature 427, 415-418 [DOI] [PubMed] [Google Scholar]
- Petrosky T., Prigogine I. (1993). Poincare resonances and the limits of trajectory dynamics. Proc. Natl. Acad. Sci. USA 90, 9393-9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigogine I. (1997). The End of Certainty New York: Free Press; [Google Scholar]
- Ptashne M. (2007). On the use of the word ‘epigenetic’. Curr. Biol. 17, R233-R236 [DOI] [PubMed] [Google Scholar]
- Raj A., van Oudenaarden A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin R., Hoeppner D. J., Munno D. M., Carmel L., Sullivan J., Levitt D. L., Miller J. L., Athaide C., Panchision D. M., McKay R. D. (2008). Potency and fate specification in CNS stem cell populations in vitro. Cell Stem Cell 3, 670-680 [DOI] [PubMed] [Google Scholar]
- Rubin H. (1992). Mechanisms for enduring biological change. Am. J. Physiol. 262, L111-L113 [DOI] [PubMed] [Google Scholar]
- Schroeder T. (2008). Imaging stem-cell-driven regeneration in mammals. Nature 453, 345-351 [DOI] [PubMed] [Google Scholar]
- Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Liron Y., Rosenfeld N., Danon T., Perzov N., Alon U. (2006). Variability and memory of protein levels in human cells. Nature 444, 643-646 [DOI] [PubMed] [Google Scholar]
- Silva J., Smith A. (2008). Capturing pluripotency. Cell 132, 532-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. M., Hamazaki T., Hankowski K. E., Terada N. (2007). A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 25, 2534-2542 [DOI] [PubMed] [Google Scholar]
- Skupin A., Kettenmann H., Winkler U., Wartenberg M., Sauer H., Tovey S. C., Taylor C. W., Falcke M. (2008). How does intracellular Ca2+ oscillate: by chance or by the clock? Biophys. J. 94, 2404-2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack J. M. W. (2002). Timeline-Conrad Hal Waddington: the last renaissance biologist? Nat. Rev. Genet. 3, 889-895 [DOI] [PubMed] [Google Scholar]
- Spencer S. L., Gaudet S., Albeck J. G., Burke J. M., Sorger P. K. (2009). Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Koshland D. E., Jr (1976). Non-genetic individuality: chance in the single cell. Nature 262, 467-471 [DOI] [PubMed] [Google Scholar]
- Stockholm D., Benchaouir R., Picot J., Rameau P., Neildez T. M., Landini G., Laplace-Builhe C., Paldi A. (2007). The origin of phenotypic heterogeneity in a clonal cell population in vitro. PLoS ONE 2, e394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohman R. (1994). Epigenesis: the missing beat in biotechnology? Biotechnology (NY) 12, 156-164 [DOI] [PubMed] [Google Scholar]
- Strohman R. C. (1997). The coming Kuhnian revolution in biology. Nat. Biotechnol. 15, 194-200 [DOI] [PubMed] [Google Scholar]
- Takasuka N., White M. R., Wood C. D., Robertson W. R., Davis J. R. (1998). Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology 139, 1361-1368 [DOI] [PubMed] [Google Scholar]
- Tomlinson I., Sasieni P., Bodmer W. (2002). How many mutations in a cancer? Am. J. Pathol. 160, 755-758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfson D., Marciniak J., Blake W. J., Ostroff N., Tsimring L. S., Hasty J. (2006). Origins of extrinsic variability in eukaryotic gene expression. Nature 439, 861-864 [DOI] [PubMed] [Google Scholar]
- Waddington C. H. (1942). The epigenotype. Endeavour 1, 18-20 [Google Scholar]
- Waddington C. H. (1957). The Strategy of the Genes London: Allen and Unwin; [Google Scholar]
- Walczak A. M., Onuchic J. N., Wolynes P. G. (2005). Absolute rate theories of epigenetic stability. Proc. Natl. Acad. Sci. USA 102, 18926-18931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xu L., Wang E. (2008). Potential landscape and flux framework of nonequilibrium networks: robustness, dissipation, and coherence of biochemical oscillations. Proc. Natl. Acad. Sci. USA 105, 12271-12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Bryder D., Weissman I. L., Quake S. R. (2006). Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl. Acad. Sci. USA 103, 17807-17812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Morris S. A., Bruce A. W. (2009). Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 10, 467-477 [DOI] [PubMed] [Google Scholar]