Abstract
Flavocytochrome P450BM-3, a bacterial monooxygenase, contains a flavin mononucleotide (FMN) binding domain bearing a strong structural homology to the bacterial flavodoxin. The FMN serves as the one-electron donor to the heme iron but, in contrast to the electron transfer mechanism of mammalian cytochrome P450 reductase, the FMN semiquinone state is not thermodynamically stable and appears transiently as the anionic rather than the neutral form. A unique loop region comprised of residues -536Y-N-G-H-P-P541-, which forms a Type I′ reverse turn, provides several interactions with the FMN isoalloxazine ring, was targeted in this study. Nuclear magnetic resonance studies support the presence of a strong hydrogen bond between the backbone amide of Asn537 and FMN N5, the anionic ionization state of the hydroquinone, and for a change in the hybridization state of the N5 upon reduction. Replacement of Tyr536, which flanks the flavin ring, by the basic residues histidine or arginine did not significantly influence the redox properties of the FMN or the accumulation of the anionic semiquinone. The central residues of the Type I′ turn (-Asn-Gly-) were replaced with various combinations of glycine and alanine as a means to alter the turn and its interactions. Gly538 was found to be crucial in maintaining the type I′ turn conformation of the loop and the strong H-bonding interaction at N5. The functional role of the tandem –Pro-Pro- sequence which anchors and possible “rigidifies” the loop was investigated through alanine replacements. Despite changes in stabilities of the oxidized and hydroquinone redox states of the FMN, none of the replacements studied significantly altered the two-electron midpoint potentials. Pro541 does contribute to some degree to the strength of the N5 interaction, the formation of the anionic semiquinone. Unlike the flavodoxin, it would appear that the conformation of the FMN rather than the loop changes in response to reduction in this flavoprotein.
Flavocytochrome P450BM-3 (BM3)1, a fatty acid monooxygenase from Bacillus megaterium (ATCC 14581), is the first known bacterial P450 enzyme that belongs to the microsomal P450 class, resembling the eukaryotic P450 systems in both structure and function. However, unlike the others the BM3 holoenzyme consists of single 119-kDa polypeptide chain containing all the functionally independent domains, i.e. the heme or “oxygenase” domain and the diflavin or “reductase” portion (BMR) comprised of the NADPH/FAD- and FMN-binding subdomains. BMR displays about 35% identity and 56% similarity with mammalian NADPH-cytochrome P450 reductase (CPR) (1, 2). The NADPH/FAD- and FMN-binding domains are structurally homologous to ferredoxin NADP+ reductase and the bacterial flavodoxins, respectively (3).
Despite the structural and functional similarities between the CPR and BMR, each protein differs markedly in the extent of reduction during steady-state turnover and in the couple utilized by the FMN. Electron transfer to the heme-iron is generally considered to be mediated by the anionic FMN semiquinone (SQ) in BM3 (4–6). The FMN hydroquinone (HQ) is thought to be the primary donor in CPR (7) although the neutral SQ redox species has been proposed to be the only kinetically competent species under steady-state turnover conditions, but these studies were conducted using cytochrome c as the acceptor (8). Nonetheless, each reductase manages the FMN radical differently. CPR thermodynamically stabilizes the neutral form of the FMNSQ, while BM3 appears only to transiently utilize the thermodynamically unstable anionic state of the FMN radical as the catalytically relevant species (4, 6, 9). The “over reduction” of BM3, such that the FMN achieves the HQ state, results in a significantly lower overall monooxygenase activity, however (4, 6).
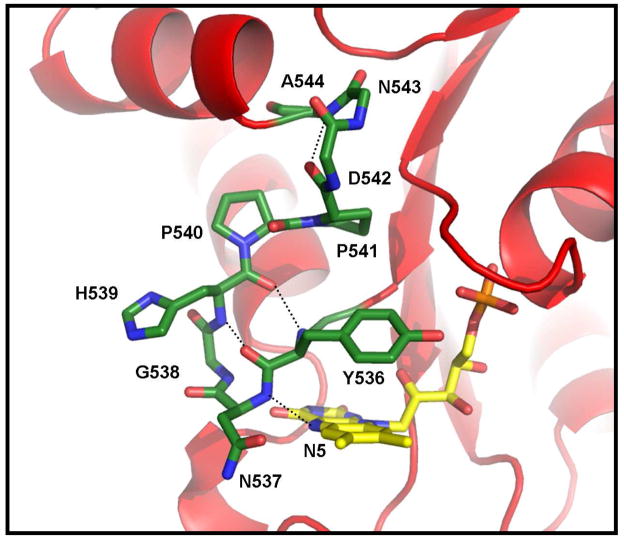
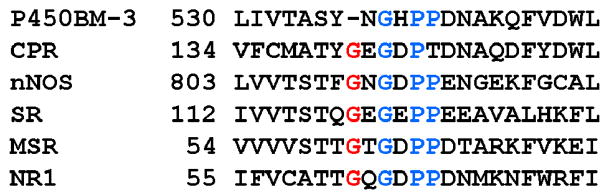
The structural basis for the regulation of the redox potentials of the flavin cofactor in flavoproteins has been a primary focus of our research. One of the intriguing and unique features that is evident in the x-ray crystal structure of the FMN domain of the BM3 is the polypeptide loop, -536Y-N-G-H-P-P541-, that provides several important contacts with the N5/C4O edge and the re- (inner) face (with the side chain of Tyr536) of the FMN cofactor (3). This loop displays several potentially important differences in both the structure and the manner in which it interacts with the cofactor relative to other homologous FMN-binding proteins/domains including CPR (10–11). The interactions provided by a corresponding loop in the flavodoxin have been shown to play important roles in establishing the redox properties of the FMN cofactor, particularly in the stabilization of the neutral semiquinone state (12–17). Sequence and structural alignments disclose that this loop in BM3 is the shortest; adopting a nearly classical four-residue type I′ reverse turn structure (Figure 1). The highly-conserved glycine residue found in the flavodoxin (Gly57 and Gly61 in Clostridium beijerinckii and Desulfovibrio vulgaris, respectively), CPR (Gly141), and several other diflavin reductase enzymes is absent in BM3 (Figure 2). This is of interest because a redox-linked conformational change involving this glycine residue plays a crucial role in the thermodynamic stabilization of the neutral flavin SQ and modulation of the flavin potentials in the flavodoxin (12, 17) and perhaps in CPR and nitric oxide synthase (18, 19). The shorter loop in BM3 also places the backbone amide NH of Asn537 in a position to serve as a hydrogen (H)-bond donor to the N5 of the FMN in the OX state (Figure 1) (3), an interaction that is not apparent in the other proteins. This interaction may also be facilitated by further “rigidifying” the loop by the tandem Pro-Pro sequence that supports the loop in this protein. These features may preclude the type of conformational changes observed in the flavodoxin that are responsible for the highly stabilized neutral SQ species. The significance of the loop size on the redox properties of the FMN was demonstrated recently through the insertion of a glycine residue after Asn537 (20). As a result, a dramatic increase in the stability of the neutral SQ species and altered redox and enzymatic activities was observed. Interestingly, the equivalent glycine residue in the larger loop within the FMN-binding domain of neuronal nitric oxide synthase was deleted in a recent study (21). The results mirrored those within the BM3 system in that the FMN now proceeds through the anionic SQ state with the reversal of the one-electron reduction potentials. Encouraged by these results and the crucial role of this loop in these proteins, a series of systematic amino acid replacements were introduced within this loop in this study with the intention of further probing in greater detail the role of the unique structural features of this loop in establishing the unique properties of BM3.
Figure 1.
View of the 536Y-N-G-H539 Type I′ turn that flanks the N5/C4O edge of the flavin cofactor in the FMN-binding domain of BM3. The putative hydrogen bonds between the backbone amide groups of Asn537 and Gly538 and the N5 and C4O atoms of the FMN, respectively, are indicated along with those that stabilize the turn. Included also is the adjacent anchoring Type I turn comprised of residues 541–544 and the tandem Pro-Pro sequence. Hydrogen atoms overall and the side chains for residues 542–544 have been omitted for clarity.
Figure 2.
Amino acid sequence alignment for the binding region flanking the re-face of the FMN cofactor for six members of the diflavin reductase family including flavocytochrome P450BM-3, human cytochrome P450 reductase (CPR), rat neuronal nitric oxide synthase (nNOS), E. coli sulfite reductase (SR), human methionine synthase reductase (MSR), and human novel reductase 1 (NR1). The conserved glycine residue highlighted in red is absent in BM3. The conserved glycine and tandem proline residues are highlighted in blue.
EXPERIMENTAL METHODS
Materials
Sodium dithionite and benzyl viologen was from Aldrich Chemical Company. Indigodisulfonate, anthraquinone-2-sulfonate and anthraquinone-2, 6-disulfonate were purchased from Fluka Chemicals. Safranin T was also from Fluka Chemicals but was recrystallized from ethanol before use. Phenosafranin was obtained from Allied Chemicals. Flavin mononucleotide was extracted from recombinant wild-type Clostridium beijerinckii flavodoxin and purified by anion-exchange chromatography. Sodium 2, 2-dimethyl-2-silapentane-5-sulfonate and 15NH4Cl (99%) was from Cambridge Isotope Laboratories. All other chemicals were of analytical reagent grade.
Mutagenesis, expression and purification of the FMN binding domain of flavocytochrome BM3
The amino acid substitutions were introduced into the coding region for the FMN-binding domain (FMN domain) (residues 471–649) of the flavocytochrome P450 from Bacillus megaterium (ATCC 14581) (BM3) which had been cloned into the pT7-7 expression vector (originally obtained from Worthington Biochemical Corp) (22, 23). Site-directed variants were prepared using either the two-step “megaprimer” PCR approach (24) or the QuikChange™ (Strategene) method using mutagenic primers containing the appropriate base changes to introduce the stated amino acid replacements. Automated DNA sequencing was used to confirm the mutations as well as to verify the sequence integrity of the entire coding region in all the constructs. The various recombinant variants were expressed as soluble proteins in high yield in the BL21 (DE3) strain of Escherichia coli (Novagen) after induction by IPTG and purified from the soluble cell extracts as described previously (20, 23). The purity of all protein preparations was confirmed by SDS-polyacrylamide gel electrophoresis.
Reductive Titrations, Kinetics, and Determination of Two-Electron Oxidation-Reduction Potentials
The midpoint potentials were determined spectrophotometrically during the anaerobic titration of the proteins with sodium dithionite2 in the presence of indicator dyes by methods previously described (14). All UV-visible spectra were recorded on Hewlett-Packard/Agilent photodiode array spectrophotometers at 25 °C in 50 mM sodium phosphate buffer, pH 7.0. The two-electron midpoint potentials were established using the Nernst equation with the system potentials established using the indicator dye anthraquinone-2, 6-disulfonate (Em,7: −184 mV at 25 °C relative to the standard hydrogen electrode) (26). Where indicated, reductive titrations were also conducted at pH 6.0 either in 50 mM sodium acetate or 50 mM sodium phosphate at a constant ionic strength. The pH of the solution was determined at both the beginning and the end of the titration. Midpoint potential determinations were conducted at least in triplicate with the error in the reported average midpoint values estimated to be within ± 5 mV.
Transient reductive kinetic analyses were performed on a Hi-Tech Scientific SF-61 stopped-flow spectrophotometer in 100 mM Tris-HCl buffer, pH 7.4. The various FMN-binding domains and freshly made sodium dithionite solutions (0.4 – 2.0 mM) were prepared in buffer that had been thoroughly flushed with pre-purified argon gas. Kinetic data for all protein variants were collected under pseudo-first order conditions with sodium dithionite in excess. The final concentration of the FMN-binding domains after mixing was 20 μM. Absorbance changes at 388 and 468 nm were recorded as a function of time. Apparent pseudo-first order rate constants for each kinetic trace were established using the Marquardt-Levenberg iterative non-linear regression algorithm and are reported as the average of at least three separate measurements. All protein variants were mixed with the same dithionite solutions in the stopped-flow instrument to aid in the direct comparison of the reductions kinetics.
Fluorescence Spectroscopy and Determination of FMN Dissociation Constants in the Oxidized State
The dissociation constant (Kd) for oxidized FMN was determined by non-linear regression analyses of either the changes in absorbance or fluorescence (excitation, 445 nm. emission, 522 nm) associated with FMN binding as a function of added apoprotein under the same conditions as those used for establishing the midpoint potentials. Apoprotein was prepared by the trichloroacetic acid precipitation technique (27, 28) or by dialysis against a 2M potassium bromide (29). Extinction coefficients used were 12,500 M−1cm−1 for FMN (30) and 28,700 M−1cm−1 for the apoprotein as determined by the method of Pace et al. (31). The dissociation constants for the HQ state, which cannot be determined directly, were calculated using a thermodynamic cycle linking the Kd for oxidized FMN complex and the two-electron midpoint potentials for both the bound and free FMN (32).
Reconstitution of Apoflavodoxin with of 15N-Enriched FMN and NMR Spectroscopy
FMN uniformly enriched at all four nitrogen atoms with >95% 15N was prepared and used to reconstitute the apoprotein according to the procedure detailed by Chang et al. (13). 1D 15N-NMR spectra were recorded for approximately 1–2 mM solutions of the oxidized and reduced protein samples in 50–100 mM phosphate buffer, pH 7, containing 10% D2O as described previously (13). Identical conditions were maintained for the 1D-15N-NMR spectra recorded under non-decoupled conditions and applying the “distortionless enhancement by polarization transfer” (DEPT) pulse sequence (33). Where applicable, these samples were also used for the 1H-15N HSQC NMR experiments. Reduction was achieved by the addition of freshly prepared sodium dithionite under anaerobic conditions. Proton chemical shifts were referenced to an internal standard of sodium 2, 2-dimethyl-2-silapentane-5-sulfonate set at 0.0 ppm and the nitrogen chemical shifts were referenced to an external standard of 15N-urea set at 76.0 ppm.
RESULTS
This study was initiated to evaluate a functional role of the short loop spanning residues Tyr536 to Pro541 in establishing the H-bonding interaction between the backbone amide group of Asn537 and the N5 of the FMN cofactor that is apparent in the crystal structure of the flavocytochrome P450BM-3 from Bacillus megaterium (ATCC 14581) (3). The FMN-binding domain spanning Arg471 to Ser649 was used for most of these studies to facilitate the analysis of changes to the properties of the FMN cofactor without the interference of the FAD and to allow for NMR data acquisition. The isolated domain displays physical and redox properties that are similar to those of the intact holoprotein. The midpoint potential at pH 7 was determined to be −194 ± 5 mV, a value very similar to that determined by electrochemical methods (−192 ± 13 mV) (5, 22, 23).
Evidence for strong hydrogen bond at the FMN N5 in the oxidized state and a change in the hybridization of the N5 upon reduction
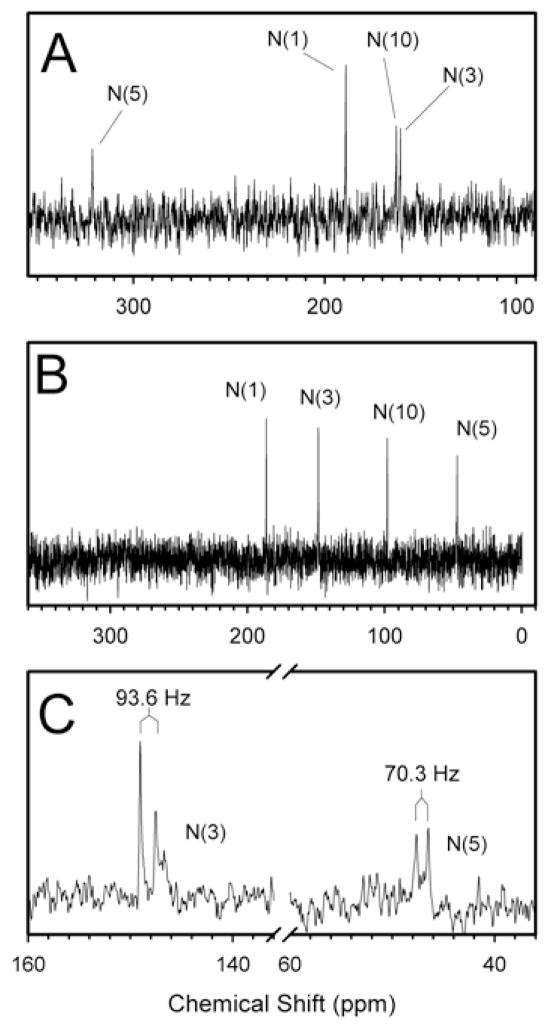
The N5 of the FMN cofactor in BM3 appears to be strongly H-bonded to the backbone amide NH of Asn537, an interaction that is not apparent in other FMN-binding proteins/domains. The importance of this interaction became quite apparent in a recent study in which this interaction was disrupted by the enlargement of this loop by one residue (through the insertion of a glycine) (20). To gain greater insight into changes in H-bonding and the hybridization state during reduction of the bound FMN, 15N-NMR spectroscopic analysis of FMN domain reconstituted with 15N-enriched FMN was performed in both OX and HQ states (Figures 3A & 3B) of the wild-type isolated domain and several of the loop variants. The 15N chemical shift values for the N1 and N5 atoms for oxidized FMN bound to the FMN domain were shifted upfield by >10 ppm relative to TARF in chloroform and from FMN in aqueous solution (34) (Table 1). Such shifts indicate the presence of strong H-bonds at the N1 and N5 in the oxidized state. The chemical shift value of 160.5 ppm was unambiguously assigned to the N3 by applying the 1H-15N DEPT (distortionless enhancement of polarization transfer) pulse sequence which distinguishes nitrogen atoms with and without hydrogen atoms, i.e. N3 versus N10, respectively (33). Based on comparisons to the values obtained for the flavin ring in aqueous or apolar solutions, the N3H appears to be H-bonded with the protein and remains so in the reduced state. The resonance peak at 162.6 ppm could then be assigned to the N10 atom. The downfield shift for this atom on going from an apolar to a polar solution has been attributed to an increase in sp2 hybridization that occurs when the polarization of the flavin ring is stabilized by H-bonding at C2O and C4O (34). The more upfield chemical shift value for this atom in the FMN domain relative to FMN in aqueous solution suggests that this atom is not fully sp2 hybridized and that the central pyrazine ring has a slightly bent conformation. Because the hybridization state at N10 also depends on the polarization state of the ring, an alternative interpretation is that H-bonding interaction(s) at C2O and/or C4O in the protein is (are) weaker then in aqueous solvent. Both C4O and C2O are within H-bonding distance with the side chain of Thr577 and backbone amide protons, respectively.
Figure 3.
1D 15N-NMR spectra of the wild-type FMN domain in the oxidized state (Panel A) and the fully reduced state (Panel B). Panel C: 1D 15N-NMR spectrum of the wild-type FMN domain in the fully reduced state using the DEPT pulse sequence without broad-band decoupling. The coupling constants for the 15N3 and 15N5 atoms were observed to be 93.6 and 70.3 MHz, respectively, as indicated.
Table 1.
15N Chemical Shift Values (ppm) for Free and Bound FMN in the Oxidized and Reduced States at pH 7.0, 300 °K
| FMN in the Oxidized State | FMN in the Fully Reduced State | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Atom | FMNa | TARFa,b | WT FMN domain |
-537Ala-Ala- Variant |
P540A Variant |
P541A Variant |
FMNH2a | FMNH−a | TARFH2a,b | WT FMN domain |
P540A Variant |
P541A Variant |
| N1 | 190.8 | 199.9 | 189.0 | 188.8 | 189.5 | 189.0 | 128.0 | 182.6 | 116.7 | 186.4 | 186.8 | 187.1 |
| N3 | 160.5 | 159.8 | 160.5 | 158.6 | 160.8 | 160.5 | 149.7 | 149.3 | 145.8 | 148.4 | 148.6 | 149.1 |
| N5 | 334.7 | 344.3 | 321.5 | 332.8 | 322.1 | 320.0 | 58.0 | 57.7 | 60.4 | 47.1 | 47.4 | 45.5 |
| N10 | 164.6 | 150.2 | 162.6 | 162.3 | 162.2 | 162.8 | 87.3 | 97.2 | 72.2 | 98.1 | 98.4 | 98.9 |
From Vervoort et al. (39).
Tetraacetylriboflavin in chloroform.
The chemical shift values in the fully reduced state were particularly revealing (Figure 3B). The chemical shift values for both N1 and N10 conclusively prove that the HQ is anionic in this protein. Of perhaps even greater significance is the chemical shift value of N5, which is far upfield of that of FMNH2 and FMNH−. This suggests that the N5 atom is more sp3-hybridized in the anionic HQ with the flavin ring bent out of plane at this position. This conclusion was unexpected. Therefore, the change in hybridization state was verified by measuring the coupling constants by one-dimensional 15N-NMR spectroscopy under proton non-decoupling conditions applying the DEPT pulse sequence (Figure 3C). Semi-empirical relationships between the experimentally observed 15N-1H coupling constants and the hybridization state of the nucleus under investigation have yielded values of about 72 Hz for sp3 hybridized nitrogen atoms and values of about 93 Hz for sp2 hybridized nitrogen atoms (35, 36). For the FMN domain in the oxidized state, N3 exhibits a coupling constant of 90.2 Hz3, indicative of its sp2 hybridization state and the planarity of the ring. These conclusions are consistent with the crystal structure (3). When the FMN cofactor is reduced, the N3 retains a coupling constant of 93.6 Hz and, thus, the sp2 hybridization state. However, a coupling constant of 70.3 Hz was observed for N5, which provides more persuasive evidence for its almost complete sp3 character. This is a significant observation because it suggests that in the reduced state, the N5 atom is lifted out of the plane of the ring. This result is most likely correlated to the nearly complete absence of an absorption band in the visible region of the reduced protein as it has been suggested that the absorption coefficient at 450 nm of a reduced flavin molecule is probably entirely related to the hybridization state of the N5 atom (37).
Quantitative estimates of the H-bonding strengths of the nitrogen atoms that are involved in a H-bond, specifically, N3, and N5 in the reduced state cannot be obtained from one-dimensional 15N-NMR experiments alone. However, we have made use of the temperature coefficient for hydrogen atoms involved in H-bonding (representing the systematic upfield shift in its resonance with increasing temperature) as an indicator of the strength of H-bonding (13). The 1H-15N HSQC spectra of oxidized and fully reduced FMN domain at pH 7.0 revealed the presence of the HSQC signal for the N3H and, when appropriate, the N5H at all temperatures tested. This implies that these protons are exchanging slowly with solvent due to their relative inaccessibility and strong H-bonding interactions with the protein. The relatively low values obtained for the temperature coefficients of −2.96 and −1.65 ppb/°K for the N3H in the oxidized and reduced states, respectively, and −0.647 ppb/°K for the N5H in the reduced flavin are consistent with strong H-bonding at these positions (13).
Y536H and Y536R Variants
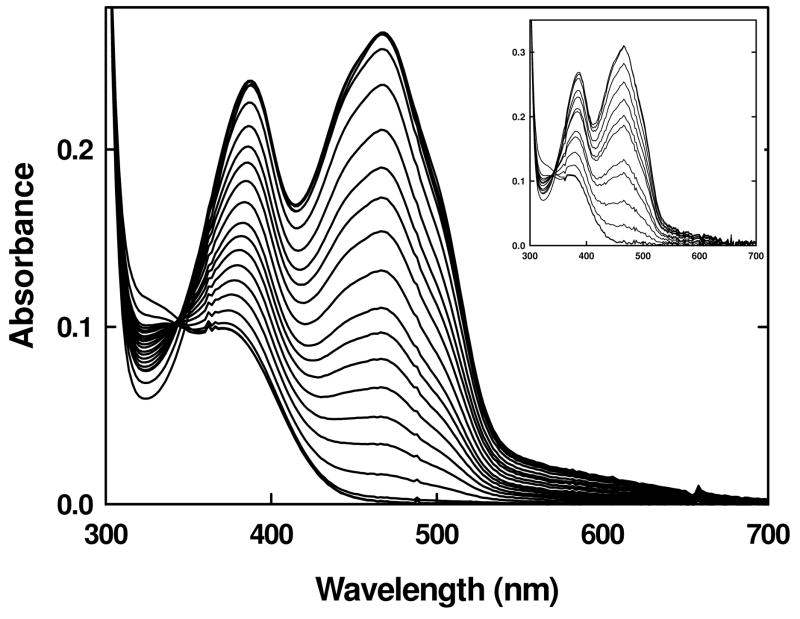
A positively charged amino acid positioned near the flavin ring might be expected to be influential in stabilizing the anionic reduced states of the cofactor through favorable electrostatic interactions. To test this hypothesis, a basic amino acid side chain could be introduced by replacing either the re-face Tyr536 or si-face Trp574 which flank the flavin ring (3). Because the replacement of Trp574 by non-polar residues has been shown to abolish FMN binding (38), Tyr536 was targeted for substitution by histidine and arginine residues. The Y536H variant exhibited spectral characteristics similar to wild type, including the characteristic weak charge-transfer band above 550 nm. FMN binding was affected, with the variant exhibiting a dissociation constant of 350 ± 72 nM or about 9-fold higher then wild type. Much of this loss of ~1.3 kcal/mol in binding free energy could be ascribed to the elimination of the H-bond between the phenolic –OH group of Tyr536 and the phosphate moiety on the FMN observed in the crystal structure of the wild-type protein (3). Reduction by sodium dithionite under anaerobic conditions followed a single two-electron process with no evidence for the accumulation of the semiquinone species (Figure 4). The two-electron midpoint potential at pH 7.0 was determined to be −188 mV, which is similar to wild type.
Figure 4.
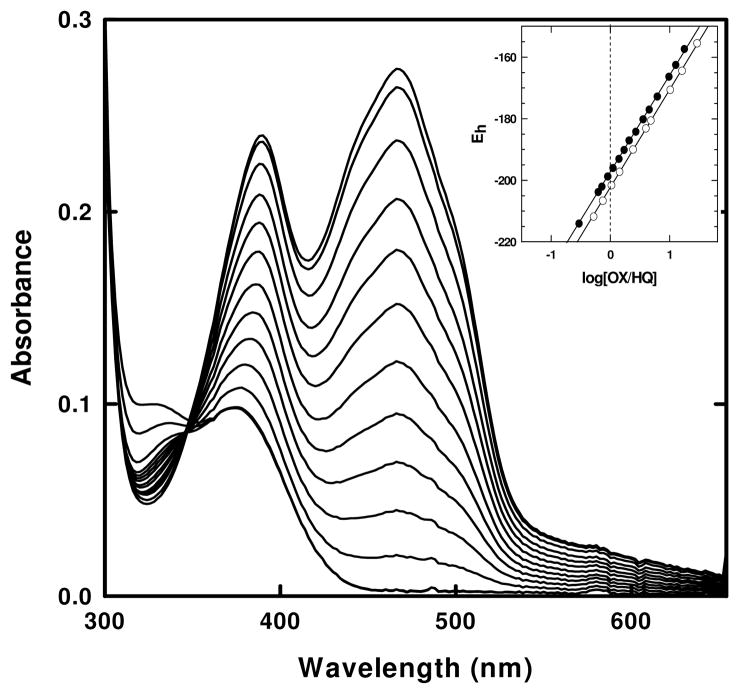
UV-visible absorbance spectra of the Y536H variant during a reductive titration with sodium dithionite under anaerobic conditions in 50 mM sodium phosphate buffer, pH 7.0 (main panel) and 50 mM sodium acetate or 50 mM sodium phosphate, pH 6.0, at a constant ionic strength (inset) at 25 °C.
Because the pKa of a histidine residue is in the range of 6.0 to 6.8, it is likely not to be completely ionized at pH 7.0. Thus, the anaerobic reductive dithionite experiments were performed at lower pH in order to observe what effect having a fully charged residue at that position would have. However, the protein was relatively unstable at lower pH, resulting in aggregate formation with the release of flavin. As glycerol is known to have a stabilizing effect on proteins, the experiment was repeated at different concentrations of glycerol, ranging from 5 – 15% and it was observed that at 15% glycerol, the protein was relatively stable. Under these conditions, a reductive dithionite experiment was performed and that also followed a single two-electron process similar to that at pH 7.0 (Figure 4 inset). The formation of the semiquinone species was not observed.
Unlike the Y536H mutant, the Y536R variant lost most of the FMN during the purification process. Reconstitution of the protein with flavin was feasible; however, the dissociation constant was substantially higher, established to be 2.1 ± 0.6 μM. Again, reductive dithionite titrations proceeded as a two-electron process with no discernible evidence for the formation of the anionic semiquinone species, adding to the conclusion that electrostatics do not seem to be a major determinant of the reduction potential. Given the instability of this variant, its midpoint potential could not be established reliably. The evidence obtained from both these variants strongly suggests that factors other than favorable electrostatic interactions are influencing the ability of the FMN-binding domain in BM3 to stabilize and accumulate the SQ species in either its anionic or neutral forms. Recent results obtained from the glycine insertion variant provide evidence that H-bonding interactions provided by the re-face loop are crucial so the following substitutions were introduced to further probe this hypothesis (20).
N537P, N537A, and N537G Variants
The structural features of the re-face loop that may be responsible for establishing and/or maintaining the H-bonding between the backbone amide NH of Asn537 and the FMN N5 were systematically evaluated (Figure 1). Because of this direct interaction with the cofactor, Asn537 was the first target for study. As an initial strategy, Asn537 was replaced by a proline residue in an effort to directly abolish this interaction through the elimination of the backbone NH group. Although the N537P variant could be expressed as a stable and soluble protein with a circular dichroism spectrum similar to wild type (data not shown), it was severely impaired in its ability to bind FMN. This outcome was not surprising in that proline residues cannot assume a type I′ conformation that is adopted by this turn (see below) due to its restricted φ angle of −60 ± 25°. Instead, a greater preference is observed for this residue to occupy the second position within type I and II turns (39). Indeed, molecular modeling coupled with geometry optimization using the AMBER molecular mechanics force field resulted in the complete distortion of the type I′ turn in favor of the optimal φ angles for proline, with the loop adopting more of a type II turn structure favored by X-Pro-Gly-X four-residue turns. As a result, residues 535–536 moved into the FMN binding site cavity and substantially altered the position of the Tyr536 side chain, which in the wild-type structure appears to H-bond with the 5′-phosphate group of the FMN. Though disappointing in its inability to bind FMN precluding further study, the results support an absolute requirement for the type I′ turn conformation in maintaining the stability and FMN binding characteristics in this domain.
The next set of variants, N537G and N537A (also designated -537Gly-Gly- and -537Ala-Gly-, respectively, to emphasize the critical two central residues in the turn), were designed to evaluate any contributions made by the side chain at position 537 and, as will be discussed later, for the evaluation of the role of turn stability. Both variants were expressed as holoproteins exhibiting UV-visible absorbance spectra in the oxidized state similar to wild type. Both variants displayed similar Kd values of 30 ± 5 nM (an average of at least 4 independent titrations) (Table 2). These values are comparable to wild type (41 ± 18 nM, 31 ± 8 nM) (this study, 27, respectively). Just as for wild type, anaerobic reductions by sodium dithionite followed a single two-electron process with a single isosbestic point maintained at 348 nm throughout the titration (Figure 5). There was no evidence for the accumulation of the SQ species. Two-electron midpoint potential values of –196mV and –202mV for N537A and N537G, respectively, were very similar to the wild-type value (Figure 5, inset and Table 2).
Table 2.
Summary of the re-face loop mutations, rationale, and effects on FMN binding and midpoint potentials
| Variants | Loop sequence | Rationale for substitution | Kd (nM) | EOX/HQb (mV) |
|---|---|---|---|---|
| Wild type | 536Y-N-G-H-P-P541 | -- | 41 ± 18 | −192c |
| Y536H | 536H-N-G-H-P-P541 | introduce positive charge near flavin | 350 ± 72 | −188 |
| N537A | 536Y-A-G-H-P-P541 | eliminate Asn sidechain (interactions) | 30 ± 5 | −196 |
| N537G | 536Y-G-G-H-P-P541 | alter turn stability | 30 ± 5 | −202 |
| N537G/G538A | 536Y-G-A-H-P-P541 | alter turn structure/stability | 400 ± 100 | −225 |
| N537A/G538A | 536Y-A-A-H-P-P541 | alter turn structure/stability | 1000 ± 200 | −204 |
| N537P | 536Y-P-G-H-P-P541 | eliminate N5 H-bond | N.D. a | N.D. a |
| P540A | 536Y-N-G-H-A-P541 | alter conformation/flexibility | 43 ± 7 | −194 |
| P541A | 536Y-N-G-H-P-A541 | alter conformation/flexibility | 140 ± 33 | −180 |
| P540A/P541A | 536Y-N-G-H-A-A541 | alter conformation/flexibility | N.D. a | N.D. a |
Not determined due to lack of cofactor binding.
Midpoint potential values are reported as average of at least two determinations with an estimated error of ± 5 mV.
Potentiometric value from Ref 27 which is also similar to the value of −194 ± 5 mV determined in this study using an indicator dye.
Figure 5.
UV-visible absorbance spectra of the N537A (-537Ala-Gly-) variant during a reductive titration with sodium dithionite in 50 mM sodium phosphate buffer, pH 7.0 at 25 °C under anaerobic conditions. The spectral changes shown were also qualitatively similar to those obtained for the N537G (-537Gly-Gly-) variant. Inset: Representative Nernst plots for the determinations of two-electron midpoint potentials for the N537A (-537Ala-Gly-) (closed circles) and the N537G (-537Gly-Gly-) (open circles) variants.
N537A/G538A and N537G/G538A Double Variants
The role of the stability and conformation of the type I′ turn adopted by this loop was probed by targeting the two central residues. Type I′ turns are highly dependent on the amino acid residue present at primarily the third positions of the turn (39, 40) A glycine is by far the most frequently observed residue at this position because of the absence of interference of a side chain with the protein backbone (41–43). This is the situation in the FMN domain, with Gly538 targeted in this study by its substitution with alanine, which is very infrequently found here. The introduction of a minimal side chain was expected to altering the loop stability and, perhaps, its interactions with the cofactor. In addition, Asn537 was again replaced by alanine as above, but also by glycine both to conform to its frequency at this position and unique functional role in other FMN-binding proteins/domains. The intention here was to alter or disrupt the H-bond made with N5 while minimally altering the loop structure by changing the positional preferences within the Type I′ turn. This approach has been used successfully to reveal aspects of backbone-flavin interactions in the flavodoxin (14). Thus, the double variants N537A/G538A and N537G/G538A (also designated -537Ala-Ala- and -537Gly-Ala-, respectively) were prepared.
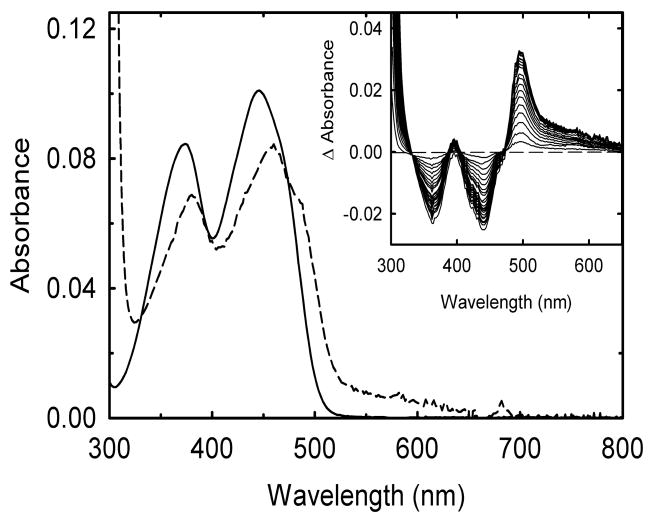
Both proteins lost much of their flavin cofactor during purification but were capable of reconstitution with FMN as evidenced by the distinct spectral changes associated with the titration of the apoprotein with the cofactor (Figure 6). The wavelength maxima for two major flavin visible absorbance bands were 380 ± 4 and 460 ± 2 nm for these variants, which are red shifted relative to FMN in solution just as for wild type, although not quite to the same extent (Figure 6). Extinction coefficients are also similar to wild type. The appearance of the broad absorbance band between 550 and 700 nm in both variants indicates that the FMN is binding in a manner similar to wild type in forming a charge transfer complex with Trp574. Binding was weaker than for wild type, however. Dissociation constants of 0.4 ± 0.1 and 1.0 ± 0.2 μM were obtained for the -537Gly-Ala- and -537Ala-Ala- variants, respectively (Table 2). Both are at least 10-fold higher than for wild type.
Figure 6.
Changes in the absorbance spectrum of FMN when bound to the apoprotein form of the -537Gly-Ala- variant. An FMN solution (~5 μM) in 50 mM sodium phosphate buffer pH 7.0 at 25 °C was titrated with sub-stoichiometric amounts of freshly prepared apoprotein. The main panel shows the absorbance of FMN before (solid line) and after (dashed line) the addition of saturating amounts of the apoprotein. Inset: Difference spectra obtained during the titration. All spectra are corrected for dilution and, if necessary, for some light scattering.
As with the wild-type domain, reductive titrations with sodium dithionite under anaerobic conditions proceeded by a two-electron reduction process with no spectral evidence for the formation of the semiquinone species. Midpoint potential values of −225 ± 9 mV and −204 ± 5 mV were obtained for -537Gly-Ala- and -537Ala-Ala- variants, respectively (Table 2). The values are slightly more negative than for the wild type. In addition to significantly weakening the binding of the OX form of the FMN (by up to 2 kcal/mol relative to wild type), based on the linked thermodynamic analyses (44), the potential changes also suggest that these substitutions induce an additional loss in binding energy for the HQ state of from 0.5 to 1.5 kcal/mol. It should be noted that the difference in binding energy between the OX and HQ states for the wild type and the -537Ala-Gly- and 537Gly-Gly- variants are also relatively small (<0.9 kcal/mol).
The effects of the Gly538 to alanine replacement in the context of -537Ala-Ala- variant on the interaction(s) between this loop and the flavin were evaluated by 15N NMR spectroscopy on apoprotein samples reconstituted with 15N-enriched FMN. The value for the N3 atom was shifted upfield by about 2 ppm (Table 1) which could indicate some weakening of the H-bond at this position. In the crystal structure, the backbone carbonyl group of Thr577 provides the H-bond donor to N3 (3). In contrast, the chemical shift value for N5, which is quite high in the wild-type domain, was shifted down field by over 10 ppm, placing it closer to that of free flavin in solution4. These results seem to suggest that the introduction of the methyl side chain at the third position in this Type I′ turn has primarily and significantly disrupted the strong H-bond at N5 by altering the structure of this loop. This disruption may have in turn affected to a lesser extent some of the other interactions made with the FMN, particularly that with N3. This type of synergy has been noted for the glutamate-59 variants in the C. beijerinckii flavodoxin (45). It was of interest to evaluate any changes in the N5 environment upon reduction of the FMN. Unfortunately, repeated efforts to obtain NMR spectra of the -537Ala-Ala- variant in its reduced state were unsuccessful due to sample instability over the prolonged data acquisition time required.
P540A and P541A Variants
The C-terminal end of the loop is anchored by a tandem –Pro-Pro- sequence. Because proline residues have greater stereochemical constraints than other amino acid, it is possible that these residues might contribute significantly to structure, placement, and “rigidity” of the re-face loop and thereby contribute to the strength of the H-bond with the FMN N5 and other interactions. The tandem proline sequence is situated at the end of a short helix (spanning residues 544–551) causing the polypeptide chain to form a “kink” prior to forming the Type I′ turn near the FMN. The phi dihedral angle of proline residues are restricted to values close to −60° (± 25°) as observed for Pro540 and Pro541 (−74° and −61°, respectively). These angles seem to be crucial for the placement of the turn relative to the flavin ring. To test the structural significance of the tandem proline sequence, alanine residues were independently substituted for each of the proline residue. It was reasoned that alanine, which can adopt similar phi dihedral angles as proline, can also assume a variety of other phi (and psi) angle relationships and introduce greater conformational flexibility in this loop region. The double variant, with both proline residues replaced by alanine, was also generated. However, this variant was purified as the apoprotein and because it could not be reconstituted with FMN cofactor in vitro, no further characterizations were conducted.
Both the P540A and P541A variants were expressed and purified as the holoprotein with stoichiometric levels of bound FMN. Each displayed absorbance spectra that were similar to wild type (data not shown). The dissociation constant for the FMN bound to the P540A variant, 43 ± 7 nM, was very similar to wild type (41 ± 18 nM). The P541A variant displayed a slightly weaker binding (140 ± 33 nM) (Table 2). Neither of the proline mutations resulted in substantial changes in the two-electron reduction potential values. The value for the P540A variant was −194 mV, which is nearly identical to the wild type while P541A variant has a slightly higher value of –180 mV. Using a thermodynamic cycle involving linked equilibria (44), the Kd values for the FMNHQ in each of the proline variants were calculated to be 18 nM and 20 nM for P540A and P541A, respectively, both being similar to that of the wild type (15 nM) (Table 2). The loss in binding of the OX form is somewhat greater than for the HQ form for the P541A variant (by 0.7 and 0.2 kcal/mol, respectively).
NMR analyses were again conducted on the P540A and P541A variants after reconstitution with 15N-enriched FMN. Both proline replacements generated NMR spectra very similar to that of the wild type (as in Figure 3) for the N1, N3, and N10 atoms. The N5 resonances were again upfield relative to FMN in aqueous solution, suggesting that H-bonding is retained at this position. However, small but significant upfield chemical shifts (1.5–1.6 ppm) relative to wild type were noted for the N5 atom in the P541A variant in both the oxidized and reduced states which indicates a somewhat altered N5 environment in this variant. The changes for the oxidized state may reflect alterations in H-bonding at this position while those in the reduced state could be reflecting an increase in sp3 character for this atom (Table 1). The chemical shift values for the P540A, exhibiting only minor changes in the N5 chemical shifts (Table 1) suggesting that strong H-bonding interactions are maintained in these variants.
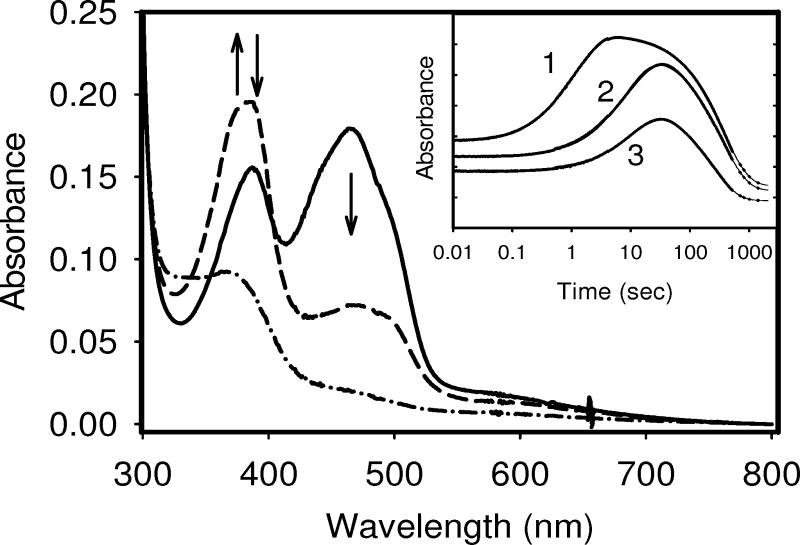
The relative stability of the anionic semiquinone is more difficult to establish as, like wild type, neither variant thermodynamically stabilizes this redox state. Just as for the wild type, the anionic SQ was formed transiently in both variants during reduction by excess sodium dithionite in the stopped-flow spectrophotometer (Figure 7). The biphasic absorbance changes at 388 nm were consistent with the formation and decay of the anionic SQ species. No spectral changes were observed around 580 nm indicating that the neutral flavin radical was not formed during the reduction process. While the absolute rates can not be related to physiological reduction rates, the relative rates obtained under identical conditions offer useful comparisons. The formation of the anionic SQ was found to be about 10-fold faster for P541A (kobs = 0.91 sec−1) than that for P540A (0.09 sec−1) and the wild type (0.11 sec−1), each obtained under identical conditions. The rate of the formation of HQ (or decay of SQ) was quite comparable (wild type, 0.006 sec−1; P540A, 0.007 sec−1; and P541A, 0.004 sec−1). The data suggest that the proline residues do not greatly influence the conversion of the SQ to the HQ. However, the higher rate of SQ formation observed in P541A suggests the OX species seems to become less stable and quickly reduced to SQ species in this variant.
Figure 7.
Time courses for the reduction by excess sodium dithionite of the FMN-binding domains for the wild type, P540A, and P541A variants in a stopped-flow spectrophotometer in 100mM Tris HCl, pH 7.4 at 25 °C under anaerobic conditions. The main panel shows spectra recorded at the start (solid line: fully oxidized), during the reduction (at approximately 15 sec) (dashed line: anionic SQ formation), and the end of the experiment (dot-dashed line: fully reduced). Arrows indicate the direction of the absorbance changes. Inset: Time course (shown in log scale) for the reduction of each protein as recorded at 388 nm, which represents the wavelength maximum for the anionic form of the SQ. Solid lines are non-linear regression fits to two exponential functions. Absorbance scale for each time traces have been arbitrarily offset for clarity. Trace 1: P541A variant; Trace 2: wild type; Trace 3: P540A variant.
DISCUSSION
Flavin cofactor-protein interactions play a major role in the “tuning” of the redox potentials and activities of flavoproteins through the differential stabilization of each redox state (12–17). Within flavodoxin-like FMN-binding proteins/domains, several functionally-important interactions are provided by a loop that flanks the re-face of the flavin ring. This loop adopts a variety of structures within this family in part because of their differing lengths. BM3, having the shortest loop comprised exactly of the four residues required for a beta-turn, i.e. 536Y-N-G-H539, adopts a classic Type I′ turn configuration with phi/psi parameters very close to consensus values. The analogous loop in the flavodoxin from Clostridium beijerinckii adopts a type II-like beta-turn while those in CPR, rat neuronal nitric oxide synthase (nNOS), and the Desulfovibrio vulgaris flavodoxin are described more as being part of “hair pin loops” adopting structural parameters not associated with various beta-turns.
In the flavodoxin and quite possibly in the FMN domain of CPR and nNOS, a redox-dependent conformational change has been linked to the modulation of the midpoint potentials through the thermodynamic stabilization of the neutral FMN SQ (12). This conformational change has been described as the “flipping” of the backbone carbonyl group of the glycine at the second position in the turn from an orientation pointing away from the flavin ring to one in which it can form a H-bond with the N5H of the FMN SQ (12), an interaction that is not apparent in the oxidized state. However, in the crystal structure of BM3 the turn is positioned much differently relative to the flavin ring such that the hydrogen atom of the backbone amide group of Asn537 is within H-bonding distance of the N5 of the FMN in the oxidized state (see Figure 1) (3). A strong H-bond is confirmed by the large up-field shift of the N5 resonance that was observed in the 15N NMR spectra of the oxidized FMN domain in this study (Table 1). As will be addressed further later, this interaction and its fate during the reduction of the flavin are expected to differentially influence the stabilities of each redox species and, therefore, affect the reduction potentials.
Unlike the flavodoxin, our evidence suggests that the type I′ turn in BM3 does not undergo a redox-dependent conformational change affecting the N5 interactions. But, what then becomes of the H-bonding interaction between the backbone amide NH of Asn537 and the N5 of the FMN upon reduction? Because of the high pKa for the N5 in the fully reduced state, protonation of this atom is required (46). However, protonation would likely establish unfavorable H-bond donor/acceptor and dipolar mismatches between the hydrogen atom on N5 and that on the backbone amide group of Asn537. Also, steric overlap may also arise between the hydrogen atoms on each group. These issues could be avoided in the one-electron reduced state through the formation of the anionic form of the SQ but less so for the HQ species. NMR evidence suggests that the flavin isoalloxazine ring undergoes a change in configuration upon reduction to the HQ state. Based on the 15N5-1H coupling constant, the N5 atom is more sp3 hybridized in the reduced state. This result is interpreted to suggest that the N5H moiety moves out of the plane of the ring to overcome any steric hindrance that might occur between the hydrogen atom and the loop. The two nitrogen atoms in the flavin ring can take on configurations that are independent of each other (37, 47). The presence of an aromatic stacking interaction would tend to inhibit bending of the flavin ring along the N5-N10 axis (48). In FMN domain, the coplanar Trp574 that is stacked tightly against the isoalloxazine ring could play this very role, resulting in only the N5 atom being distorted out of the plane of the molecule. This would effectively reduce the unfavorable steric and dipolar interactions with the protein without the need for structural changes in the FMN-binding loop that would be associated with a greater energetic cost due to the rigidity of the loop. Furthermore, sp3 hybridization of N5 prevents electron delocalization into the isoalloxazine ring, which increases the electron density at N5 and results in the large upfield shift for that atom. The enhanced basicity of the nitrogen atom also increases its proton affinity and therefore, a lack of a H-bond partner is tolerated. BM3 is not unique in this behavior. The N5 atom of the FMN in Old Yellow Enzyme is also H-bonded with the backbone amide hydrogen (from Thr37) and is also fully sp3 hybridized in the fully reduced state (47). However, unlike for BM3, Old Yellow Enzyme thermodynamically stabilizes the anionic semiquinone state to the level of 15–20% under equilibrium conditions (49).
Our ability to experimentally modulate the interactions between the flavin and main chain atoms is limited and more challenging than for those involving side chains for obvious reasons. However, addressing the positional frequencies (preferences) of amino acids at each of the four positions of beta-turns provides an alternative approach, one that has been very effectively used in the flavodoxin. In those studies, all four combinations of glycine and alanine residues were introduced at the two central positions of the reverse turn which alters the turn stability and conformational switching during flavin reduction (14). For type I′ turns, the backbone torsion angles (phi, psi) adopted by the central residues have idealized values of (60°, 30°) and (90°, 0°). Note that the values for Asn537 and Gly538 in BM3 of (48°, 38°) and (92 °, 2°), respectively, which fall within the range observed for the left-handed helical region of the Ramachandran plot. In this region, ~60% of the residues are populated by glycine (41). The high proportion of glycine residues is the consequence of unfavorable local steric interactions between the backbone and the Cβ atom of amino acids with side chains in this conformation (39, 40). Thus, the substitution of residues with side chains for the glycine in such turns should destabilize the turn depending on its particular conformation.
Data for the Gly/Ala substitutions introduced in this study at the central position of the turn correlate well with the predicted turn stability and the role of loop rigidity in BM3. The variants N537A (-537Ala-Gly-) and N537G (-537Gly-Gly-), which both retain a glycine at the third position favored by Type I′ turns, resembled the wild-type protein in their ability to bind FMN with high affinity and in their redox properties. The similarity of the two-electron midpoint potential values for both the N537A and N537G variants to wild type indicates that the side chain at this position is not having a significant influence. This was not surprising in that previous results on the flavodoxin clearly indicate that the influence of reverse turn stability on midpoint potentials is dictated by the presence or absence of side chains at the two central positions and not by the physico-chemical nature of the particular side chain (14). On the other hand, both the -537Gly-Ala- and -537Ala-Ala- variants, where Gly538 is replaced with alanine, were significantly impaired in their ability to bind FMN as tightly as in wild type. Why is this?
The Type I′ turn that is found in the wild-type structure becomes strongly disfavored when Gly538 is replaced with an alanine due to steric interference between the β-methyl group and the backbone atoms. Thus, the turn in these variants must adopt an alternate structure. Structural alternatives were explored based on the positional preferences of the various relevant turn types and by computational molecular modeling. The reconfiguration of the turn to a type I configuration would alleviate the steric issues imposed by the alanine replacement (42). However, to accommodate the backbone phi/psi parameters of this type of turn the side chain of Tyr536 would have to be repositioned dramatically. Molecular modeling indicates that the phenol side group would have to swing out of the FMN binding pocket into a more solvent exposed orientation leaving a large gap in the isoalloxazine-binding site. The H-bond between the phenolic –OH and the 5′-phosphate of the FMN is eliminated and the methyl group of the Ala537 appears to clash with the C4O/N5 edge of the ring unless the position of the flavin ring is altered. This configuration is expected to preclude FMN binding and is not favored.
A simpler alterative would be for the turn to adjust to a Type II′ configuration through the rotation of the central peptide bond by ~180°. Indeed, such turns show a good tolerance for amino acids with side chains at this position. However, as a result the favorable H-bond between the amide hydrogen of the central peptide bond with the C4O must be disrupted and a potential steric problem would arise between the C4O and the backbone carbonyl. Such changes would likely alter (weaken) the interaction at N5. The type II′ turn configuration would be particularly favored in the -537Gly-Ala- variant because of the glycine residue at the second position in the turn in this case. We favor this outcome based on the loss of up to 2 kcal/mol of binding free energy for the oxidized FMN in the -537Gly-Ala- and -537Ala-Ala- variants and the downfield shift in the chemical shift of the N5 and N10 atoms for -537Ala-Ala- variant which is consistent with the loss or significant disruption of the H-bonding at the C4O and N5 positions in these variants. The binding energy for the HQ state was also increased by up to an additional 0.5 kcal/mol resulting in the two-electron reduction potentials for this variant to be slightly higher than wild type. Unfortunately, we were unable after many attempts to obtain satisfactory NMR data for the -537Ala-Ala- variant in the reduced state to aid in our analysis of the changes occurring in this state. However, if one concludes that these amino acid replacements primarily alter the N5 interaction [and perhaps additionally that with the C4O], the disruptions do not significantly additionally affect the stability of the reduced states, certainly not to the same extent as in the flavodoxin (14).
Taken together, this evidence supports the general conclusion that the unique N5 interaction in this FMN domain (relative to the flavodoxin) is not a major factor in establishing the relative stabilities of the redox states of the FMN and, thus, its midpoint potentials. It is likely that this loop does not undergo any substantial redox-dependent conformation changes as in the flavodoxin. The NMR evidence suggests that the flavin isoalloxazine ring undergoes a change in configuration upon reduction to the HQ state.
Role of the tandem Pro-Pro sequence and extended turn
Just as in CPR and the D. vulgaris flavodoxin, the C-terminus of the re-face FMN binding loop in BM3 is supported by another beta-turn with the sequence, -541P-D-N-A- in BM3 (Figure 1). This turn has torsion parameters (phi, psi) for the two central residues (−54, −32 and −94, 20) that are close to a consensus Type I turn configuration (−60, −30 and −90, 0). A similar sequence is found in nNOS, E. coli sulfite reductase, human methionine synthase reductase, and human novel reductase 1 (Figure 2) suggesting a homologous structure in these diflavin reductases as well. Unlike for Type I′ turns, a proline residue is frequently found at the first position of Type I turns as found in all of these proteins except CPR. In BM3 this residue is Pro541. Interestingly, the two turns are linked (or bridged) by a proline residue in all of these proteins, i.e. Pro540 in BM3, generating a tandem proline sequence, again frequently observed in this class of flavoprotein.
The tandem proline sequence that anchors this loop could contribute to the strength of the interactions made with the FMN in BM3 through the positioning of the re-face loop (see Figure 1). It has been stated that this feature may serve to “rigidify” the loop (3). Thus, to assess a possible functional significance, these residues were targeted as part of this study. Somewhat surprising to us, the replacement of the bridging proline, Pro540, with an alanine residue had no discernible impact on the binding of the FMN in either the oxidized or reduced states and, the two-electron midpoint potentials (Table 2). However, the Pro541 to alanine substitution resulted in a loss of about 0.7 kcal/mol in binding energy for the oxidized FMN. It is possible that the greater flexibility or slight conformation changes introduced by this replacement may weaken the H-bonds made with the N5 and the C4O of the FMN. It has been noted that alanine is rather infrequently found at the first position of Type I turns (42, 43). The binding energy for the reduced FMN was not significantly affected by the alanine replacement as might be expected since the N5 interaction is likely to be abolished when the FMN is fully reduced. It was noted, however, that the NMR chemical shift of the N5 in the oxidized and reduced states are not substantially different from the wild type or the P540A variant. The steady-state kinetic studies conducted in BMR and BM3 proteins mimicked the physical studies in that the P540 to alanine replacement did not significantly alter the reductase and hydroxylase specific activities while those of the P541A variant were slightly decreased. The P541A variant also displayed a 10-fold higher rate for the formation of the anionic SQ during transient reductions by sodium dithionite while that of the P540A variant remained unchanged from wild type. As mentioned, the simultaneous replacement of both proline residues by alanine disrupted FMN binding altogether. Thus, these results firmly establish the functional importance of the tandem proline sequence and the somewhat greater influence of Pro541 on the FMN cofactor in BM3.
Lack of effects of electrostatics
The inability to favor an appreciable thermodynamic stabilization of the anionic SQ in BM3 by altering the interactions with the re-face loop prompted an investigation as to whether favorable electrostatic interactions may play a role in stabilizing the reduced forms of the flavin as has been observed in other proteins. With this aim in mind, the Y536H and Y536R variants were generated. While the histidine mutant would have an ionizable side chain, the effective charge of which could be regulated with pH (50), the arginine mutant would retain the positive charge in the pH range testable. There was no evidence for the substantial stabilization of the anionic SQ species. In fact and somewhat surprisingly, no significant change was observed in the two-electron reduction potential. There exists contradictory data on electrostatic control of the isoalloxazine environment. Just as was done in this study, the introduction of histidine and arginine residues in the FMN-binding site of the D. vulgaris flavodoxin afforded nearly 4.0 kcal/mol stabilization to the anionic HQ state with the associated increase in the midpoint potential. (51). The replacement of Arg237, again adjacent to the flavin ring, by alanine in the electron-transferring flavoprotein (ETF) from Methylophilus methylotrophus (sp W3A1), results in a decrease of ~200 mV in the midpoint potential of OX/SQ couple due to the destabilization of the anionic SQ (52). The anionic SQ is quite substantially stabilized by the wild-type protein, however. In contrast, several variants involving Arg238 in morphinone reductase, which again flanks the flavin cofactor near N1/C2O, suggests that the presence of this positively charged side chain is not a requirement for stabilization of the reduced flavin (53). Other protein-specific factors may also influence the role of electrostatic interactions in the modulation of the redox properties of the bound flavin cofactor. For example, modification of the H-bonding interaction to the N5 of the FAD by alphaSer254 in the W3A1 ETF protein also results in a substantial destabilization of the anionic SQ as well as the elimination of a kinetic barrier to its reduction (54). Other interactions must also play a more predominate role in destabilizing this redox species in the FMN-binding domain of BM3 as well. The demonstration that the enlargement of the re-face loop along with concomitant changes in the FMN interactions can achieve the stabilization of the neutral form of the SQ (20) but is resistant to the more moderate alterations described in this study illustrate the importance of the unique characteristics of this loop in BM3 and related proteins.
Footnotes
This study was supported in part by Grant GM36490 from the National Institutes of Health.
The abbreviations used are: BM3, flavocytochrome P450BM-3; BMR, diflavin reductase portion of BM3; CPR, microsomal cytochrome P450 reductase; FMN, flavin mononucleotide; OX, oxidized; SQ, semiquinone; HQ, hydroquinone; HSQC, heteronuclear single quantum correlation NMR spectroscopy.
The reactivity and avidity of FMN domain with sulfite ions was tested by titration with freshly prepared sodium sulfite. Spectral changes indicative of a flavin-sulfite adduct formation, which are similar to that of fully reduced flavin (25), were not observed. Thus, the use of sodium dithionite as a reductant is feasible (sulfite being a product of its anaerobic oxidation).
All coupling constants were determined with an accuracy of ± 0.8 Hz.
Given the similarity of the chemical shift values to unbound flavin and the weaker binding of FMN to the -537Ala-Ala- variant, a concern is that the 15N-FMN did not remain bound throughout the entire experiment. However, the Kd for the FMN complex of this variant is approximately 2 μM. Because the concentration of the holoprotein was significantly higher than this in the NMR experiment (~ 1 mM), it seems reasonable to conclude that the 15N-FMN should remain bound throughout unless the protein denatures during the extended experiment (overnight at room temperature). Visible absorbance and fluorescence spectroscopy and gel filtration of the NMR sample were also used to confirm that the 15N-FMN remained bound at the end of the experiment.
References
- 1.Miles CS, Ost TW, Noble MA, Munro AW, Chapman SK. Protein engineering of cytochromes P-450. Biochim Biophys Acta. 2000;1543:383–407. doi: 10.1016/s0167-4838(00)00236-3. [DOI] [PubMed] [Google Scholar]
- 2.Ruettinger RT, Wen LP, Fulco AJ. Coding nucleotide, 5′ regulatory, and deduced amino acid sequences of P-450BM-3, a single peptide cytochrome P-450:NADPH-P-450 reductase from Bacillus megaterium. J Biol Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- 3.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci U S A. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevrioukova I, Shaffer C, Ballou DP, Peterson JA. Equilibrium and transient state spectrophotometric studies of the mechanism of reduction of the flavoprotein domain of P450BM-3. Biochemistry. 1996;35:7058–7068. doi: 10.1021/bi960060a. [DOI] [PubMed] [Google Scholar]
- 5.Daff SN, Chapman SK, Turner KL, Holt RA, Govindaraj S, Poulos TL, Munro AW. Redox control of the catalytic cycle of flavocytochrome P-450 BM3. Biochemistry. 1997;36:13816–13823. doi: 10.1021/bi971085s. [DOI] [PubMed] [Google Scholar]
- 6.Sevrioukova IF, Peterson JA. NADPH-P-450 reductase: structural and functional comparisons of the eukaryotic and prokaryotic isoforms. Biochimie. 1995;77:562–572. doi: 10.1016/0300-9084(96)88172-7. [DOI] [PubMed] [Google Scholar]
- 7.Murataliev MB, Feyereisen R, Walker FA. Electron transfer by diflavin reductases. Biochim Biophys Acta. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez A, Lian LY, Wolf CR, Scrutton NS, Roberts GC. Stopped-Flow Kinetic Studies of Flavin Reduction in Human Cytochrome P450 Reductase and Its Component Domains. Biochemistry. 2001;40:1964–1975. doi: 10.1021/bi001719m. [DOI] [PubMed] [Google Scholar]
- 9.Hazzard JT, Govindaraj S, Poulos TL, Tollin G. Electron transfer between the FMN and heme domains of cytochrome P450BM-3. Effects of substrate and CO. J Biol Chem. 1997;272:7922–7926. doi: 10.1074/jbc.272.12.7922. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Q, Modi S, Smith G, Paine M, McDonagh PD, Wolf CR, Tew D, Lian LY, Roberts GC, Driessen HP. Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution. Prot Sci. 1999;8:298–306. doi: 10.1110/ps.8.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludwig ML, Pattridge KA, Metzger AL, Dixon MM, Eren M, Feng Y, Swenson RP. Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes. Biochemistry. 1997;36:1259–1280. doi: 10.1021/bi962180o. [DOI] [PubMed] [Google Scholar]
- 13.Chang FC, Swenson RP. The midpoint potentials for the oxidized-semiquinone couple for Gly57 mutants of the Clostridium beijerinckii flavodoxin correlate with changes in the hydrogen-bonding interaction with the proton on N(5) of the reduced flavin mononucleotide cofactor as measured by NMR chemical shift temperature dependencies. Biochemistry. 1999;38:7168–7176. doi: 10.1021/bi982203u. [DOI] [PubMed] [Google Scholar]
- 14.Kasim M, Swenson RP. Conformational energetics of a reverse turn in the Clostridium beijerinckii flavodoxin is directly coupled to the modulation of its oxidation-reduction potentials. Biochemistry. 2000;39:15322–15332. doi: 10.1021/bi001519a. [DOI] [PubMed] [Google Scholar]
- 15.Drennan CL, Pattridge KA, Weber CH, Metzger AL, Hoover DM, Ludwig ML. Refined structures of oxidized flavodoxin from Anacystis nidulans. J Mol Biol. 1999;294:711–724. doi: 10.1006/jmbi.1999.3151. [DOI] [PubMed] [Google Scholar]
- 16.Hoover DM, Drennan CL, Metzger AL, Osborne C, Weber CH, Pattridge KA, Ludwig ML. Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials. J Mol Biol. 1999;294:725–743. doi: 10.1006/jmbi.1999.3152. [DOI] [PubMed] [Google Scholar]
- 17.O’Farrell PA, Walsh MA, McCarthy AA, Higgins TM, Voordouw G, Mayhew SG. Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants. Biochemistry. 1998;37:8405–8416. doi: 10.1021/bi973193k. [DOI] [PubMed] [Google Scholar]
- 18.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural Basis for Isozyme-specific Regulation of Electron Transfer in Nitric-oxide Synthase. J Biol Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 19.Barsukov I, Modi S, Lian LY, Sze KH, Paine MJ, Wolf CR, Roberts GC. 1H, 15N and 13C NMR resonance assignment, secondary structure and global fold of the FMN-binding domain of human cytochrome P450. J Biomol NMR. 1997;10:63–75. doi: 10.1023/a:1018313830207. [DOI] [PubMed] [Google Scholar]
- 20.Chen H-C, Swenson RP. Effect of the insertion of a glycine residue into the loop spanning residues 536 – 541 on the semiquinone state and redox properties of the flavin mononucleotide-binding domain of flavocytochrome P450BM-3 from Bacillus megaterium. Biochemistry. 2008;47:13788–13799. doi: 10.1021/bi800954h. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Das A, Sibhatu H, Jamal J, Sligar SG, Poulos TL. Exploring the electron transfer properties of neuronal nitric-oxide synthase by reversal of the FMN redox potential. J Biol Chem. 2008;283:34762–34772. doi: 10.1074/jbc.M806949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sevrioukova I, Truan G, Peterson JA. The flavoprotein domain of P450BM-3: expression, purification, and properties of the flavin adenine dinucleotide- and flavin mononucleotide-binding subdomains. Biochemistry. 1996;35:7528–7535. doi: 10.1021/bi960330p. [DOI] [PubMed] [Google Scholar]
- 23.Kasim M. PhD Thesis. The Ohio State University; Columbus, OH: 2002. [Google Scholar]
- 24.Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 25.Müller F, Massey V. Flavin-sulfite complexes and their structures. J Biol Chem. 1969;244:4007–4016. [PubMed] [Google Scholar]
- 26.Clark WM. Oxidation-reduction potentials of organic systems. Robert E. Krieger Publishing Company; Huntington, N.Y.: 1972. [Google Scholar]
- 27.Haines DC, Sevrioukova IF, Peterson JA. The FMN-binding domain of cytochrome P450BM-3: resolution, reconstitution, and flavin analogue substitution. Biochemistry. 2000;39:9419–9429. doi: 10.1021/bi000255p. [DOI] [PubMed] [Google Scholar]
- 28.Wassink JH, Mayhew SG. Fluorescence titration with apoflavodoxin: a sensitive assay for riboflavin 5′-phosphate and flavin adenine dinucleotide in mixtures. Anal Biochem. 1975;68:609–616. doi: 10.1016/0003-2697(75)90656-9. [DOI] [PubMed] [Google Scholar]
- 29.Mayhew SG. Studies on flavin binding in flavodoxins. Biochim Biophys Acta. 1971;235:289–302. doi: 10.1016/0005-2744(71)90207-5. [DOI] [PubMed] [Google Scholar]
- 30.Whitby LG. A new method for preparing flavin-adenine dinucleotide. Biochem J. 1953;54:437–442. doi: 10.1042/bj0540437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Prot Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson RF. Energetics of the one-electron reduction steps of riboflavin, FMN and FAD to their fully reduced forms. Biochim Biophys Acta. 1983;722:158–162. doi: 10.1016/0005-2728(83)90169-x. [DOI] [PubMed] [Google Scholar]
- 33.Doddrell DM, Pegg DT, Bendall MR. Distortionless enhancement of NMR signals by polarization transfer. J Mag Resonance. 1982;48:323–327. [Google Scholar]
- 34.Vervoort J, Müller F, Mayhew SG, van den Berg WA, Moonen CT, Bacher A. A comparative carbon-13, nitrogen-15, and phosphorus-31 nuclear magnetic resonance study on the flavodoxins from Clostridium MP, Megasphaera elsdenii, and Azotobacter vinelandii. Biochemistry. 1986;25:6789–6799. doi: 10.1021/bi00370a010. [DOI] [PubMed] [Google Scholar]
- 35.Bourn AJR, Randall EW. Proton-proton double resonance studies of formamide-15N and N-methylformamide-15N. Mol Phys. 1964;8:567–571. [Google Scholar]
- 36.Binsch G, Lambert JB, Roberts BW, Roberts JD. Nitrogen-15 Magnetic Resonance Spectroscopy. II Coupling Constants. J Am Chem Soc. 1964;86:5564–5570. [Google Scholar]
- 37.Moonen CT, Vervoort J, Müller F. Reinvestigation of the structure of oxidized and reduced flavin: carbon-13 and nitrogen-15 nuclear magnetic resonance study. Biochemistry. 1984;23:4859–4867. doi: 10.1021/bi00316a007. [DOI] [PubMed] [Google Scholar]
- 38.Klein ML, Fulco AJ. Critical residues involved in FMN binding and catalytic activity in cytochrome P450BM-3. J Biol Chem. 1993;268:7553–7561. [PubMed] [Google Scholar]
- 39.Yang AS, Hitz B, Honig B. Free energy determinants of secondary structure formation: III. beta-turns and their role in protein folding. J Mol Biol. 1996;259:873–882. doi: 10.1006/jmbi.1996.0364. [DOI] [PubMed] [Google Scholar]
- 40.Yan Y, Erickson BE, Tropsha A. Free Energies for Folding and Refolding of Four Types of. beta Turns: Simulation of the Role of D/L Chirality. J Am Chem Soc. 1995;117:7592–7599. [Google Scholar]
- 41.Takano K, Yamagata Y, Yutani K. Role of non-glycine residues in left-handed helical conformation for the conformational stability of human lysozyme. Proteins. 2001;44:233–243. doi: 10.1002/prot.1088. [DOI] [PubMed] [Google Scholar]
- 42.Hutchinson EG, Thornton JM. A revised set of potentials for beta-turn formation in proteins. Prot Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guruprasad K, Rajkumar S. Beta- and gamma-turns revisited: A new set of amino acid turn-type dependent positional preferences and potentials. J Biosci. 2000;25:143–156. [PubMed] [Google Scholar]
- 44.Dubourdieu M, le Gall J, Favaudon V. Physicochemical properties of flavodoxin from Desulfovibrio vulgaris. Biochim Biophys Acta. 1975;376:519–532. doi: 10.1016/0005-2728(75)90172-3. [DOI] [PubMed] [Google Scholar]
- 45.Bradley LH, Swenson RP. Role of glutamate-59 hydrogen bonded to N(3)H of the flavin mononucleotide cofactor in the modulation of the redox potentials of the Clostridium beijerinckii flavodoxin. Glutamate-59 is not responsible for the pH dependency but contributes to the stabilization of the flavin semiquinone. Biochemistry. 1999;38:12377–12386. doi: 10.1021/bi991172f. [DOI] [PubMed] [Google Scholar]
- 46.Dudley KH, Ehrenberg A, Hemmerich P, Müller F. Spektren und Strukturen der am Flavin-Redoxsystem beteiligten Partikeln. Studien in der Flavinreihe IX. Helv Chim Acta. 1964;47:1354–1382. [Google Scholar]
- 47.Beinert WD, Ruterjans H, Müller F. Nuclear magnetic resonance studies of the Old Yellow Enzyme. 1 15N NMR of the enzyme recombined with 15N-labeled flavin mononucleotides. Eur J Biochem. 1985;152:573–579. doi: 10.1111/j.1432-1033.1985.tb09234.x. [DOI] [PubMed] [Google Scholar]
- 48.Haynes CA, Koder RL, Miller AF, Rodgers DW. Structures of nitroreductase in three states: effects of inhibitor binding and reduction. J Biol Chem. 2002;277:11513–11520. doi: 10.1074/jbc.M111334200. [DOI] [PubMed] [Google Scholar]
- 49.Stewart RC, Massey V. Potentiometric studies of native and flavin-substituted Old Yellow Enzyme. J Biol Chem. 1985;260:13639–13647. [PubMed] [Google Scholar]
- 50.Chang FC, Swenson RP. Regulation of oxidation-reduction potentials through redox-linked ionization in the Y98H mutant of the Desulfovibrio vulgaris [Hildenborough] flavodoxin: Direct proton nuclear magnetic resonance spectroscopic evidence for the redox-dependent shift in the pKa of Histidine-98. Biochemistry. 1997;36:9013–9021. doi: 10.1021/bi970783+. [DOI] [PubMed] [Google Scholar]
- 51.Swenson RP, Krey GD. Site-directed mutagenesis of tyrosine-98 in the flavodoxin from Desulfovibrio vulgaris (Hildenborough): regulation of oxidation- reduction properties of the bound FMN cofactor by aromatic, solvent, and electrostatic interactions. Biochemistry. 1994;33:8505–8414. doi: 10.1021/bi00194a015. [DOI] [PubMed] [Google Scholar]
- 52.Talfournier F, Munro AW, Basran J, Sutcliffe MJ, Daff S, Chapman SK, Scrutton NS. Alpha-Arg-237 in Methylophilus methylotrophus (sp. W3A1) electron-transferring flavoprotein affords approximately 200-millivolt stabilization of the FAD anionic semiquinone and a kinetic block on full reduction to the dihydroquinone. J Biol Chem. 2001;276:20190–20196. doi: 10.1074/jbc.M010853200. [DOI] [PubMed] [Google Scholar]
- 53.Craig DH, Barna T, Moody PC, Bruce NC, Chapman SK, Munro AW, Scrutton NS. Effects of environment on flavin reactivity in morphinone reductase: analysis of enzymes displaying differential charge near the N-1 atom and C-2 carbonyl region of the active-site flavin. Biochem J. 2001;359(Pt 2):315–323. doi: 10.1042/0264-6021:3590315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang KY, Swenson RP. Modulation of the redox properties of the flavin cofactor through hydrogen-bonding interactions with the N(5) atom: role of alphaSer254 in the electron-transfer flavoprotein from the methylotrophic bacterium W3A1. Biochemistry. 2007;46:2289–2297. doi: 10.1021/bi0616293. [DOI] [PubMed] [Google Scholar]