Abstract
We have evaluated the extent to which SNPs identified by genomewide surveys as showing unusually high levels of population differentiation in humans have experienced recent positive selection, starting from a set of 32 nonsynonymous SNPs in 27 genes highlighted by the HapMap1 project. These SNPs were genotyped again in the HapMap samples and in the Human Genome Diversity Project–Centre d'Etude du Polymorphisme Humain (HGDP–CEPH) panel of 52 populations representing worldwide diversity; extended haplotype homozygosity was investigated around all of them, and full resequence data were examined for 9 genes (5 from public sources and 4 from new data sets). For 7 of the genes, genotyping errors were responsible for an artifactual signal of high population differentiation and for 2, the population differentiation did not exceed our significance threshold. For the 18 genes with confirmed high population differentiation, 3 showed evidence of positive selection as measured by unusually extended haplotypes within a population, and 7 more did in between-population analyses. The 9 genes with resequence data included 7 with high population differentiation, and 5 showed evidence of positive selection on the haplotype carrying the nonsynonymous SNP from skewed allele frequency spectra; in addition, 2 showed evidence of positive selection on unrelated haplotypes. Thus, in humans, high population differentiation is (apart from technical artifacts) an effective way of enriching for recently selected genes, but is not an infallible pointer to recent positive selection supported by other lines of evidence.
IN the last 50,000–100,000 years (KY), humans have expanded from being a rare species confined to parts of Africa and the Levant to their current numbers of >6 billion with a worldwide distribution (Jobling et al. 2004). Paleontological and archaeological evidence suggests that key aspects of modern human behavior developed ∼100–50 KYA in Africa (Henshilwood et al. 2002) and behaviorally modern humans then expanded out of Africa ∼60–40 KYA (Mellars 2006). The physical and biological environments encountered outside Africa would have been very different from those inside and included climatic deterioration reaching a glacial maximum ∼20 KYA and subsequent amelioration that permitted the development of agricultural and pastoral lifestyles in multiple independent centers after ∼10 KYA. Neolithic lifestyles would have led to further changes including higher population densities, close contact with animals, and novel foods, in turn leading to new diseases (Jobling et al. 2004). It is likely that genetic adaptations accompanied many of these events.
Adaptation, or positive natural selection, leaves an imprint on the pattern of genetic variation found in a population near the site of selection. This pattern can be identified by comparing the DNA variants in multiple individuals from the same and different populations and searching for signals such as unusually extended haplotypes (extended haplotype homozygosity, EHH) (Voight et al. 2006; Sabeti et al. 2007; Tang et al. 2007), high levels of population differentiation (International Hapmap Consortium 2005; Barreiro et al. 2008; Myles et al. 2008), or skewed allele frequency spectra (Carlson et al. 2005). These signals become detectable at different times after the start of selection and are all transient, being gradually eroded by both molecular processes such as mutation, recombination, or further selection and population processes such as migration or demographic fluctuations, with the survival order extended haplotypes < population differentiation < allele frequency spectra (Sabeti et al. 2006). The absolute timescales of survival are not well understood, but extended haplotype tests typically detect selection within the last 10 KY (Sabeti et al. 2006) while unusual allele frequency spectra may detect much older selection. For example, it has been suggested that the signal associated with the FOXP2 gene (Enard et al. 2002) may predate the modern human–Neanderthal split ∼300–400 KYA (Krause et al. 2007), although such an interpretation has been questioned (Coop et al. 2008). However, despite significant uncertainties and limitations, population-genetic analyses are well placed to provide insights into many of the important events within the timescale of recent human evolution.
In principle, it should be possible to survey the genome for sites of selection and then interpret this catalog in the light of archaeological, climatic, and other records. Progress toward such a goal has, however, been limited: many factors can confound the detection of selection and only genotype data from previously ascertained SNPs, rather than full resequence data, have thus far been available throughout the whole genome. In practice, the strategy used has therefore been to search the genome for signals that can be detected in available genotype data, such as extended haplotypes or population differentiation, and evaluate the significance of the regions identified by comparing them with empirical distributions of the same statistic, models that incorporate information about the demography, or biological expectations (McVean and Spencer 2006). However, it remains unclear how effective this strategy is: What false positive and false negative rates are associated with its applications? Further evaluation is desirable.
The International HapMap Project has carried out the highest-resolution study so far of genetic variation in a set of human populations. In an article published in 2005, genotypes of >1 million SNPs were reported from 270 individuals with ancestry from Africa (Yoruba in Ibadan, Nigeria: YRI), Europe (Utah residents with ancestry from northern and western Europe: CEU), China (Han Chinese in Beijing, China: CHB), and Japan (Japanese in Tokyo, Japan: JPT) (International HapMap Consortium 2005). This article highlighted 32 SNPs from 27 genes that showed particular evolutionary interest because of a combination of two factors: they were nonsynonymous, that is, they changed an amino acid within a protein-coding gene and thus were likely to alter biological function, and they also exhibited a high level of population differentiation equal to or exceeding that of rs2814778, a SNP that is associated with strong biological evidence for population-specific selection. This SNP underlies the FY*0 (Duffy blood group negative) phenotype; FY*0 homozygotes do not express the Duffy blood group antigen on red blood cells and are consequently highly resistant to infection by the malarial parasite, Plasmodium vivax. The *0 allele is nearly fixed in Africa and rare outside, and it is widely believed that this is due to selection for resistance to vivax malaria.
However, a number of studies have emphasized that large differences in allele frequency between populations can arise without positive selection: for example, a highly differentiated SNP in the Neuregulin I gene was not accompanied by unusual patterns in adjacent SNPs (Gardner et al. 2007), and large frequency differences can be quite common in empirical data sets, particularly in comparisons between Africa or America and the rest of the world, where population bottlenecks and “allele surfing” may have occurred during the exit from and entrance to these continents, respectively (Hofer et al. 2009). We wished to measure the extent to which the high population differentiation observed at the 27 HapMap genes might have resulted from positive selection and the extent to which it reflected other origins such as demographic factors, chance, or errors. We therefore retyped the same SNPs in the HapMap samples and in a large additional set of human populations and applied alternative tests for selection, either based on long-range haplotypes or based on full resequence data. For the latter, sequence data for 5 of the genes were available from public sources, and four new data sets were generated for this project. We found that, while genotyping errors led to some artifactual high differentiation signals, population differentiation was a useful but by no means infallible guide to recent selection detected by other methods.
MATERIALS AND METHODS
DNA samples and genotyping:
HapMap samples (International HapMap Consortium 2005) and extended HapMap samples were purchased from the Coriell Institute for Medical Research (Camden, NJ), the Brahui (BRU, Pakistan) samples were from the collection of S. Q. Mehdi, the Human Genome Diversity Project–Centre d'Etude du Polymorphisme Humain (HGDP–CEPH) collection (Cann et al. 2002) was kindly provided by Howard Cann (CEPH, Paris, France), and one chimpanzee (Pan troglodytes) sample was purchased from the European Collection of Cell Cultures (Salisbury, Wiltshire, UK). The HGDP–CEPH samples were whole-genome amplified before use (GenomiPhi HY DNA amplification kit; Amersham Biosciences, Piscataway, NJ).
The 32 SNPs were assembled for SNaPshot genotyping into eight small multiplexes (multiplexes 1–8) containing 4 SNPs each. Primers to amplify a different-sized fragment for each SNP within a multiplex were designed using Primer3 (http://frodo.wi.mit.edu/) and extension primers again differing in length within a multiplex were picked from the sequence immediately up- or downstream of the SNP (supporting information, Table S1). Primer interactions within the multiplex were evaluated and minimized using the AutoDimer program (http://www.cstl.nist.gov/div831/strbase/AutoDimerHomepage/AutoDimerProgramHomepage.htm). PCRs contained 10–50 ng DNA, 1× Invitrogen (Carlsbad, CA) Platinum Taq buffer, 4 mm MgCl2, 200 μm of each dNTP, 0.04 μm of each primer, and 1 unit of Invitrogen Platinum Taq DNA polymerase in a 10-μl reaction volume. A touchdown PCR program was used: denaturation at 94° for 15 min and then 20 cycles of 94° for 30 sec, annealing at 70° for 30 sec, and extension at 72° for 45 sec, decreasing the annealing temperature by 1° per cycle. This was followed by 15 cycles of denaturation at 94° for 30 sec, annealing at 50° for 30 sec, and extension at 72° for 45 sec and a final extension at 72° for 7 min. The PCR products were purified by treatment with Exonuclease I (USB Corporation, Cleveland; 1.5 units) and Shrimp Alkaline Phosphatase (USB Corporation, 2.0 units) at 37° for 1 hr followed by 85° for 15 min. The extension reaction contained 1× ABI Prism SNaPshot Multiplex ready reaction mix (Applied Biosystems, Foster City, CA), 0.5 μm of each primer, and 1 μl of each PCR product and was carried out as recommended (Applied Biosystems). The extension PCR products were purified using 1 unit Shrimp Alkaline Phosphatase and then run on an ABI 3100 Genetic Analyzer. SNP calling was carried out using GeneMapper software v. 3.0 (Applied Biosystems). Since peak heights in heterozygotes are not always equal, we needed to choose a threshold to distinguish between heterozygotes and homozygotes. For this, we calculated (allele 1 peak height – allele 2 peak height)/(allele 1 peak height + allele 2 peak height) using an in-house Perl script to give a value between +1 and −1 for each sample. We then inspected the distribution of values from each SNP for all samples and set an appropriate threshold.
For quality control, we included 13 pairs of blind duplicates and observed seven discrepancies in 294 comparisons (1.2% error), mostly reflecting ambiguities in calling heterozygotes in samples that lay near the threshold (Table S2). Hardy–Weinberg equilibrium was also calculated, and no significant departures were observed within individual populations. Comparisons with HapMap data were not used for genotyping quality control (although they were used for sequencing quality control) and are discussed in the results section.
Resequencing:
Two overlapping ∼6-kb fragments covering ∼12 kb spanning each of the high-differentiation SNPs in the F2, HERC1, ZNF646, and GNB1L genes (primers in Table S3) were amplified by long PCR with Platinum High Fidelity Taq polymerase (Invitrogen). The PCR contained 1× Platinum High Fidelity Taq polymerase reaction buffer, 2 mm MgSO4, 200 μm of each dNTP, 1 unit Taq polymerase, and 10 μm of each primer in a 25-μl volume. The reaction was carried out by touchdown PCR starting with denaturation at 94° for 15 min and then 94° for 30 sec, annealing at 71° for 30 sec, and extension at 68° 10 min for 20 cycles, with the annealing temperature decreasing by 0.5° per cycle. This was followed by 94° for 30 sec, 63° for 30 sec, and extension at 68° for 10 min for another 15 cycles and a final extension at 68° for 10 min.
The long PCR products were then used as templates for a second round of nested PCR to produce fragments for sequencing. Sequenced fragments were designed to be ∼500 bp ± 15% long and overlap by 240 bp ± 30%, so that >95% of the regions were covered ≥4× on both strands (Table S3). The nested PCR contained a 200-fold dilution of the long PCR products, 1× Invitrogen Platinum Taq PCR reaction buffer, 1.6 mm MgCl2, 200 μm of each dNTP, 0.5 unit Taq polymerase, and 10 μm of each primer in a 15-μl reaction volume. Reactions were set up by a Beckman Coulter Biomek FX robot. The PCRs were carried out for 30 cycles of 94° for 30 sec, 60° for 30 sec, and extension at 68° for 45 sec, followed by a final extension at 68° for 3 min. Standard capillary sequencing reactions were performed by the Sanger large-scale sequencing service.
Potential SNPs were flagged by Mutation Surveyor v. 2.0 software (SoftGenetics) and then all were checked manually. SNP calling quality was assessed by including four duplicate samples in the resequencing experiment and comparing the SNP calls with HapMap calls. No discrepancies were found between our duplicates; comparisons with HapMap data showed 0 of 816 (HERC1), 6 of 635 (ZNF646), 17 of 1492 (F2), and 34 of 1088 (GNB1L) discrepancies. Reexamination of the discrepant positions suggested that the ratios of our miscalls:HapMap miscalls were 2:4, 4:13, and 18:16. The higher error rate in our data for GNB1L reflected lower sequence quality from this gene. Significant proportions of the HapMap errors might be accounted for by two immediately adjacent SNPs that could affect genotyping but not resequencing (F2) and a misassigned individual contributing 11 of the 16 miscalls (GNB1L). Overall, after distinguishing between likely sequencing and HapMap errors, the mean reliability of our variant position calls was estimated at 99.4%. Data are reported in Table S4, Table S5, Table S6, and Table S7.
Statistical analysis:
The frequency of each SNP allele in each sample was determined by counting. FST was used to measure population differentiation and was calculated using the Hierfstat R package (Goudet 2005). To evaluate the significance of the observed FST values, we compared them with the empirical distribution of genomewide SNPs in the HapMap and HGDP (J. Z. Li et al. 2008) panels. Genomewide SNPs were divided into 20 mean frequency classes (0–5%, 5–10%, etc.) and the 95th and 99th percentiles were calculated for each class. The mean frequency in the test SNP in HapMap samples was compared to the 95th and 99th percentiles of its frequency class. Linkage disequilibrium was visualized using Haploview (Barrett et al. 2005). Haplotypes were reconstructed by PHASE 2.1 (Stephens and Donnelly 2003) from both our own resequence data and data obtained from the literature (Hamblin et al. 2002) or downloaded from the SeattleSNPs (http://pga.mbt.washington.edu/) and National Institute of Environmental Health Sciences (NIEHS) SNPs (http://egp.gs.washington.edu/) websites. Median-joining networks of inferred haplotypes from regions of high linkage disequilibrium surrounding the SNP of interest (“Network region size” in Table 3) were constructed using Network 4.50 (Bandelt et al. 1999).
TABLE 3.
Summary statistics calculated from resequence data
| Gene | Population | Sample size | Segregating sites | Resequenced region size (kb) | Network region size (kb) | Nucleotide diversity (× 104) | P-value for ML-HKA test | Tajima's D | P-value (best-fit model) | P-value (best-fit model unascertained) | P-value (SeattleSNP database empirical) | Fu and Li's D | P-value (best-fit model) | P-value (best-fit model unascertained) | Fu and Li's F | P-value (best-fit model) | P-value (best-fit model unascertained) | Fay and Wu's H | P-value (best-fit model) | P-value (best-fit model unascertained) | Fu's Fs | P-value (best-fit model) | P-value (best-fit model unascertained) | P-value (SeattleSNP database empirical) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FY | Italian | 32 | 30 | 8.6 | 8.6 | 8.50 | −0.02 | 0.426 | 0.441 | 0.038 | 0.05 | 0.218 | 0.213 | 0.00 | 0.447 | 0.439 | −7.72 | 0.061 | 0.061 | −6.65 | 0.009 | 0.009 | 0.094 | |
| FY | Chinese | 32 | 13 | 8.6 | 8.6 | 5.45 | 1.53 | 0.083 | 0.079 | 0.087 | 1.05 | 0.075 | 0.084 | 1.38 | 0.164 | 0.156 | 0.41 | 0.445 | 0.493 | 1.14 | 0.447 | 0.447 | 0.289 | |
| FY | Hausan | 32 | 18 | 8.6 | 8.6 | 4.84 | −0.20 | 0.228 | 0.284 | 0.285 | 0.50 | 0.090 | 0.090 | 0.38 | 0.132 | 0.135 | −5.64 | 0.056 | 0.053 | −1.44 | 0.334 | 0.346 | 0.191 | |
| FY | Pakistani | 28 | 27 | 8.6 | 8.6 | 8.87 | 0.41 | 0.388 | 0.371 | 0.466 | 0.76 | 0.155 | 0.150 | 0.75 | 0.314 | 0.294 | 2.29 | 0.156 | 0.133 | −4.24 | 0.036 | 0.036 | 0.154 | |
| FY | Worldwide | 124 | 43 | 8.6 | 8.6 | 9.13 | −0.01 | 0.083 | 0.086 | 0.190 | −0.06 | 0.022 | 0.023 | −0.09 | 0.023 | 0.024 | −7.70 | 0.045 | 0.043 | −27.97 | 0.013 | 0.011 | 0.065 | |
| EDAR | EurAm | 48 | 85 | 22.3 | 5.7 | 3.55 | 0.201 | −2.09 | 0.007 | 0.008 | 0.032 | −2.26 | 0.041 | 0.043 | −2.67 | 0.088 | 0.091 | −31.37 | 0.000 | 0.001 | −6.09 | 0.012 | 0.012 | 0.094 |
| EDAR | AsAm | 46 | 27 | 22.3 | 5.7 | 0.74 | 0.340 | −2.42 | 0.002 | 0.002 | 0.003 | −2.23 | 0.040 | 0.044 | −2.78 | 0.069 | 0.074 | −21.44 | 0.003 | 0.003 | −7.58 | 0.004 | 0.004 | 0.079 |
| EDAR | AfAm | 48 | 121 | 22.3 | 5.7 | 10.37 | 0.150 | −0.55 | 0.446 | 0.456 | 0.468 | −1.30 | 0.251 | 0.245 | −1.21 | 0.487 | 0.484 | −3.73 | 0.087 | 0.085 | −6.34 | 0.152 | 0.147 | 0.383 |
| EDAR | HisAm | 48 | 68 | 22.3 | 5.7 | 4.47 | −1.29 | 0.079 | 0.085 | 0.089 | −1.03 | 0.216 | 0.220 | −1.41 | 0.242 | 0.250 | −24.63 | 0.001 | 0.001 | −1.87 | 0.167 | 0.164 | 0.305 | |
| EDAR | Worldwide | 188 | 166 | 22.3 | 5.7 | 6.48 | −1.58 | 0.168 | 0.150 | 0.083 | −3.81 | 0.061 | 0.057 | −3.25 | 0.476 | 0.459 | −31.28 | 0.001 | 0.001 | −39.55 | 0.001 | 0.001 | 0.036 | |
| ADH1B | EurAm | 44 | 33 | 15.7 | 14.7 | 6.46 | 0.652 | 1.14 | 0.081 | 0.114 | 0.119 | 0.69 | 0.169 | 0.167 | 1.04 | 0.202 | 0.192 | −0.57 | 0.306 | 0.332 | −0.63 | 0.328 | 0.328 | 0.471 |
| ADH1B | AsAm | 48 | 26 | 15.7 | 14.7 | 4.04 | 0.715 | 0.26 | 0.440 | 0.421 | 0.361 | 1.28 | 0.035 | 0.034 | 1.09 | 0.224 | 0.217 | −13.52 | 0.017 | 0.021 | 4.99 | 0.087 | 0.085 | 0.047 |
| ADH1B | AfAm | 54 | 62 | 15.7 | 14.7 | 6.80 | — | −0.75 | 0.439 | 0.428 | 0.338 | −1.66 | 0.154 | 0.150 | −1.56 | 0.391 | 0.390 | −0.55 | 0.193 | 0.191 | −8.73 | 0.060 | 0.058 | 0.246 |
| ADH1B | HisAm | 44 | 37 | 15.7 | 14.7 | 7.08 | 1.05 | 0.133 | 0.129 | 0.154 | 1.55 | 0.011 | 0.011 | 1.74 | 0.087 | 0.085 | 1.08 | 0.533 | 0.559* | 0.91 | 0.418 | 0.419 | 0.326 | |
| ADH1B | Worldwide | 190 | 76 | 15.7 | 14.7 | 7.65 | −0.25 | 0.144 | 0.152 | 0.720 | −2.80 | 0.243 | 0.232 | −1.90 | 0.207 | 0.213 | −0.72 | 0.228 | 0.225 | −16.60 | 0.138 | 0.131 | 0.244 | |
| ERCC6 | PDR | 180 | 50 | 8.1 | 8.1 | 5.96 | −1.34 | 0.312 | 0.272 | 0.155 | −3.44 | 0.107 | 0.100 | −3.02 | 0.462 | 0.478 | 3.90 | 0.890 | 0.883 | −13.13 | 0.243 | 0.229 | 0.298 | |
| F2 | CEU | 44 | 29 | 10.3 | 10.3 | 3.60 | 0.355 | −1.49 | 0.048 | 0.053 | 0.070 | 0.79 | 0.141 | 0.139 | −0.06 | 0.398 | 0.400 | −4.55 | 0.111 | 0.126 | −0.39 | 0.366 | 0.367 | 0.497 |
| F2 | CHB | 46 | 23 | 10.3 | 10.3 | 5.98 | 0.391 | 0.60 | 0.251 | 0.307 | 0.362 | 0.08 | 0.419 | 0.133 | 0.33 | 0.594 | 0.392 | −2.52 | 0.185 | 0.213 | −2.39 | 0.105 | 0.107 | 0.282 |
| F2 | YRI | 44 | 27 | 10.3 | 10.3 | 2.58 | 0.835 | −1.91 | 0.011 | 0.010 | 0.014 | −3.39 | 0.004 | 0.004 | −3.45 | 0.067 | 0.063 | −5.78 | 0.055 | 0.052 | −2.55 | 0.488 | 0.497 | 0.324 |
| F2 | BRU | 44 | 35 | 10.3 | 10.3 | 4.11 | −1.62 | 0.038 | 0.040 | 0.054 | −0.76 | 0.272 | 0.282 | −1.31 | 0.231 | 0.243 | −3.74 | 0.137 | 0.157 | −2.51 | 0.096 | 0.098 | 0.268 | |
| F2 | Worldwide | 178 | 57 | 10.3 | 10.3 | 5.01 | −1.45 | 0.245 | 0.221 | 0.119 | −3.56 | 0.090 | 0.083 | −3.14 | 0.506 | 0.490 | −4.02 | 0.100 | 0.097 | −17.40 | 0.121 | 0.115 | 0.220 | |
| HERC1 | CEU | 44 | 27 | 9.9 | 9.9 | 4.93 | 0.833 | −0.90 | 0.180 | 0.190 | 0.144 | 0.20 | 0.358 | 0.350 | −0.25 | 0.490 | 0.500 | 0.86 | 0.489 | 0.517 | −1.78 | 0.176 | 0.173 | 0.322 |
| HERC1 | CHB | 46 | 17 | 9.9 | 9.9 | 1.56 | 0.799 | −1.90 | 0.014 | 0.018 | 0.040 | −1.40 | 0.122 | 0.141 | −1.90 | 0.147 | 0.158 | −9.21 | 0.038 | 0.044 | −2.32 | 0.109 | 0.110 | 0.289 |
| HERC1 | YRI | 44 | 38 | 9.9 | 9.9 | 6.79 | — | −0.79 | 0.423 | 0.409 | 0.317 | −1.02 | 0.352 | 0.349 | −1.13 | 0.507 | 0.504 | −0.10 | 0.222 | 0.219 | −0.87 | 0.253 | 0.257 | 0.160 |
| HERC1 | BRU | 44 | 29 | 9.9 | 9.9 | 5.47 | −0.63 | 0.240 | 0.258 | 0.191 | −0.40 | 0.398 | 0.409 | −0.58 | 0.372 | 0.383 | 1.88 | 0.760 | 0.802 | −5.18 | 0.019 | 0.020 | 0.092 | |
| HERC1 | Worldwide | 178 | 68 | 9.9 | 9.9 | 6.63 | −1.36 | 0.290 | 0.276 | 0.155 | −3.80 | 0.061 | 0.057 | −3.22 | 0.485 | 0.469 | −1.17 | 0.203 | 0.200 | −21.16 | 0.060 | 0.056 | 0.125 | |
| ZNF646 | CEU | 44 | 26 | 11.3 | 11.3 | 6.22 | 0.129 | 0.57 | 0.263 | 0.254 | 0.372 | −1.95 | 0.068 | 0.069 | −1.24 | 0.274 | 0.281 | 1.82 | 0.701 | 0.725 | 1.06 | 0.395 | 0.397 | 0.302 |
| ZNF646 | CHB | 46 | 22 | 11.3 | 11.3 | 1.54 | 0.098 | −2.14 | 0.006 | 0.008 | 0.030 | −1.11 | 0.185 | 0.193 | −1.79 | 0.161 | 0.171 | −9.46 | 0.037 | 0.042 | −3.72 | 0.067 | 0.050 | 0.181 |
| ZNF646 | YRI | 44 | 35 | 11.3 | 11.3 | 6.16 | 0.204 | −0.47 | 0.407 | 0.418 | 0.477 | −0.24 | 0.327 | 0.331 | −0.40 | 0.303 | 0.302 | −0.88 | 0.177 | 0.174 | −1.33 | 0.318 | 0.329 | 0.185 |
| ZNF646 | BRU | 44 | 18 | 11.3 | 11.3 | 5.73 | 1.79 | 0.047 | 0.051 | 0.047 | 0.06 | 0.212 | 0.209 | 0.81 | 0.289 | 0.280 | 1.41 | 0.669 | 0.711 | 3.70 | 0.156 | 0.155 | 0.094 | |
| ZNF646 | Worldwide | 178 | 64 | 11.3 | 11.3 | 9.87 | −1.15 | 0.432 | 0.410 | 0.250 | −4.15 | 0.032 | 0.030 | −3.34 | 0.454 | 0.437 | −0.26 | 0.258 | 0.254 | −12.09 | 0.285 | 0.268 | 0.327 | |
| CEACAM1 | AfAm | 48 | 57 | 19.7 | 19.7 | 7.06 | 0.403 | 0.73 | 0.051 | 0.052 | 0.056 | 0.84 | 0.035 | 0.037 | 1.00 | 0.058 | 0.056 | 4.10 | 0.881 | 0.877 | −0.69 | 0.229 | 0.235 | 0.841 |
| CEACAM1 | EurAm | 46 | 34 | 19.7 | 19.7 | 2.08 | 0.279 | −1.60 | 0.036 | 0.040 | 0.050 | −1.59 | 0.114 | 0.117 | −1.95 | 0.163 | 0.170 | −18.74 | 0.006 | 0.007 | −0.53 | 0.342 | 0.343 | 0.477 |
| CEACAM1 | Worldwide | 94 | 57 | 19.7 | 19.7 | 5.72 | 0.20 | 0.055 | 0.055 | 0.845 | 0.17 | 0.011 | 0.013 | 0.17 | 0.015 | 0.016 | −2.49 | 0.145 | 0.141 | −3.15 | 0.192 | 0.198 | 0.762 | |
| GNB1L | CEU | 44 | 27 | 9.3 | 7.9 | 7.45 | 0.186 | 0.38 | 0.327 | 0.317 | 0.477 | 0.69 | 0.169 | 0.167 | 0.70 | 0.276 | 0.269 | 0.95 | 0.507 | 0.534 | −0.76 | 0.306 | 0.305 | 0.460 |
| GNB1L | CHB | 46 | 32 | 9.3 | 7.9 | 9.73 | 0.022 | 0.81 | 0.250 | 0.238 | 0.262 | −0.20 | 0.459 | 0.467 | 0.22 | 0.434 | 0.421 | 2.34 | 0.852 | 0.860 | −0.99 | 0.240 | 0.240 | 0.436 |
| GNB1L | YRI | 46 | 45 | 9.3 | 7.9 | 6.18 | — | −1.53 | 0.074 | 0.068 | 0.045 | −2.28 | 0.055 | 0.053 | −2.42 | 0.206 | 0.199 | −0.59 | 0.191 | 0.189 | −4.47 | 0.292 | 0.286 | 0.477 |
| GNB1L | LWK | 46 | 50 | 9.3 | 7.9 | 6.38 | −1.68 | 0.042 | 0.038 | 0.024 | −2.64 | 0.026 | 0.024 | −2.76 | 0.146 | 0.141 | −5.75 | 0.055 | 0.052 | −9.69 | 0.040 | 0.037 | 0.223 | |
|
GNB1L |
Worldwide |
182 |
93 |
9.3 |
7.9 |
8.83 |
−1.53 |
0.199 |
0.178 |
0.101 |
−4.41 |
0.018 |
0.016 |
−3.65 |
0.377 |
0.360 |
−4.03 |
0.100 |
0.097 |
−40.48 |
0.001 |
0.001 |
0.030 |
EurAm, European-American; AsAm, Asian-American; AfAm, African-American; HisAm, Hispanic-American.
Extended haplotypes from the three populations YRI, CHB + JPT, and CEU were examined in three ways. EHH (Sabeti et al. 2002) was analyzed in HapMap2 200-kb phased haplotypes (http://www.hapmap.org/) surrounding each SNP, using the program Sweep (http://www.broad.mit.edu/mpg/sweep/) after transforming build 36 genomic coordinates to the build 35 coordinates required by Sweep. Control regions to evaluate the 95th and 99th percentiles were the ENCODE neutral regions (Birney et al. 2007). For integrated Haplotype Score (iHS) (Voight et al. 2006) and cross population (XP)-EHH (Sabeti et al. 2007), genotypes for all unrelated HapMap samples were downloaded from the HapMap project website; haplotypes were inferred using BEAGLE (Browning and Browning 2009); and ancestral states from the chimpanzee, orangutan, and macaque sequence at the corresponding position were provided by dbSNP. For cases where the ancestral state could not be inferred the major allele in YRI was assumed to be ancestral. iHS and XP-EHH were calculated using custom C scripts provided by J. Pickrell (Pickrell et al. 2009). For iHS, SNPs with a minor allele frequency <0.05 were discarded. Both scores were standardized as described by Sabeti et al. (2007) to give a mean score of 0 and standard deviation of 1. For iHS this standardization was performed separately for scores within 20 equally spaced bins of derived allele frequency before combining the scores together from all bins, following the protocol of Voight et al. (2006).
Nei's diversity (π) and the summary statistics Tajima's D (Tajima 1989), Fu and Li's D and F (Fu and Li 1993), Fu's Fs (Fu 1997), and Fay and Wu's H (Fay and Wu 2000) were calculated using an in-house program written in C. The significance of each diversity value was assessed using a maximum-likelihood HKA test, where each gene was compared with the random ENCODE region ENr321 using a model with free mutation and no selection vs. free mutation and selection (Wright and Charlesworth 2004). The significance of the other test results was evaluated by comparison with values from coalescent simulations (Hudson 2002) carried out in two ways. First, we controlled for demographic effects using the best-fit demographic model for each population, using the cosi-package (Schaffner et al. 2005). Second, we also controlled for the ascertainment of the SNPs via high population differentiation by retaining only the subset of simulations from the best-fit demographic model that each yielded at least one SNP with an FST value as high as those in the relevant HapMap SNPs. Our data consisted of regions with high LD and so we set recombination to zero in these simulations. For non-HapMap samples, we chose the demographic model from the closest HapMap population; for the two genes sequenced in African-Americans, this meant excluding admixture. Third, we calculated empirical P-values from the SeattleSNPs data for Tajima's D and Fu's Fs.
RESULTS
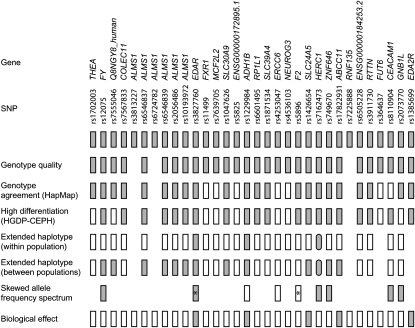
To determine how many of the 32 high-differentiation nonsynonymous SNPs from 27 genes were likely to have resulted from positive selection and how many could be accounted for in other ways, we applied a series of filters to the SNPs, regenotyping them in the HapMap samples and the HGDP–CEPH panel, examining their extended haplotype patterns, and finally testing allele frequency spectra from full resequence data for a subset of the genes. A summary of these stages and their outcomes is shown in Figure 1.
Figure 1.—
Summary of analyses of the 32 high-differentiation SNPs from 27 genes. Genotype quality: shaded rectangle, passed in our assays; space, failed. Genotype agreement (HapMap): shaded rectangle, good agreement between our data and HapMap1; space, no data. High differentiation (HGDP–CEPH): shaded rectangle, FST value >95th percentile of relevant frequency class; open rectangle, <95th percentile. Extended haplotype (within population): shaded rectangle, iHS value of derived SNP >95th percentile; open rectangle, <95th percentile. Extended haplotype (between populations): shaded rectangle, XP-EHH value of derived SNP >95th percentile in one or more pairwise comparisons; open rectangle, <95th percentile. Rounded rectangles: rs7162473 not tested, but signals in neighboring SNPs as described in the text. Skewed allele frequency spectrum: shaded rectangle, significant evidence for positive selection of derived high-differentiation allele; open rectangle, no significant departure from neutrality; space, no resequence data; *, significant evidence for positive selection on another allele. Biological effect: shaded rectangle, effect reported for derived allele; open rectangle, not reported.
As a first step, we retyped the SNPs in the DNA samples used by the HapMap project. Three SNPs failed in our assay, but 2 of these were from ALMS1, for which 4 additional SNPs were successfully typed, so most subsequent conclusions are based on 29 SNPs from 26 genes. In contrast to the HapMap findings, 4 of the SNPs were monomorphic in our assay, and 3 showed polymorphic, but significantly different, patterns. Several independent lines of evidence support our genotyping results (Table 1) and our conclusions were passed on to the HapMap Consortium with the result that most of the data were corrected in the HapMap2 release. Thus, after genotyping in the HapMap samples, 7 SNPs from 7 genes were found to have experienced genotyping artifacts, in five cases sufficient to neutralize evidence for positive selection, and 22–24 SNPs from 19–21 genes remained candidates for selection.
TABLE 1.
SNPs discrepant between HapMap1 and this study
| Derived allele frequency |
|||||
|---|---|---|---|---|---|
| Gene | Population | HapMap1 | This study | Other evidence | Corrected |
| FXR1 | YRI | 0 | 0 | Yes | |
| rs11499 | CEU | 0.992 | 0 | ||
| CHB | 0 | 0 | |||
| JPT | 0 | 0 | |||
| MCF2L2 | YRI | 0 | 0.574 | Yes | |
| rs7639705 | CEU | 0 | 0.788 | ||
| CHB | 0.733 | 0.733 | |||
| JPT | 0.705 | 0.645 | |||
| ENSG00000172895.1 | YRI | 0 | 0 | Yes | |
| rs5825 | CEU | 0.992 | 0 | ||
| CHB | 0 | 0 | |||
| JPT | 0 | 0 | |||
| ERCC6 | YRI | 1 | 0 | NIEHS SNPs: 0.010 in SNP discovery panel including African samples | Yes |
| rs4253047 | CEU | 0.034 | 0.034 | ||
| CHB | 0 | 0 | |||
| JPT | 0 | 0 | |||
| NEUROG3 | YRI | 0.983 | 0.733 | Changed but still incorrect | |
| rs4536103 | CEU | 0 | 0.309 | ||
| CHB | 0.978 | 0.744 | |||
| JPT | 1 | 0.8 | |||
| FUT6 | YRI | 0 | 0 | TSC-CSHL: 0 in CEPH sample | No current data |
| rs364637 | CEU | 1 | 0 | ||
| CHB | 0 | 0 | |||
| JPT | 0 | 0 | |||
| CEACAM1 | YRI | 0.567 | 0.567 | SeattleSNPs: 0.978 in European Americans | No |
| rs8110904 | CEU | 0.008 | 1 | ||
| CHB | 1 | 1 | |||
| JPT |
1 |
1 |
|||
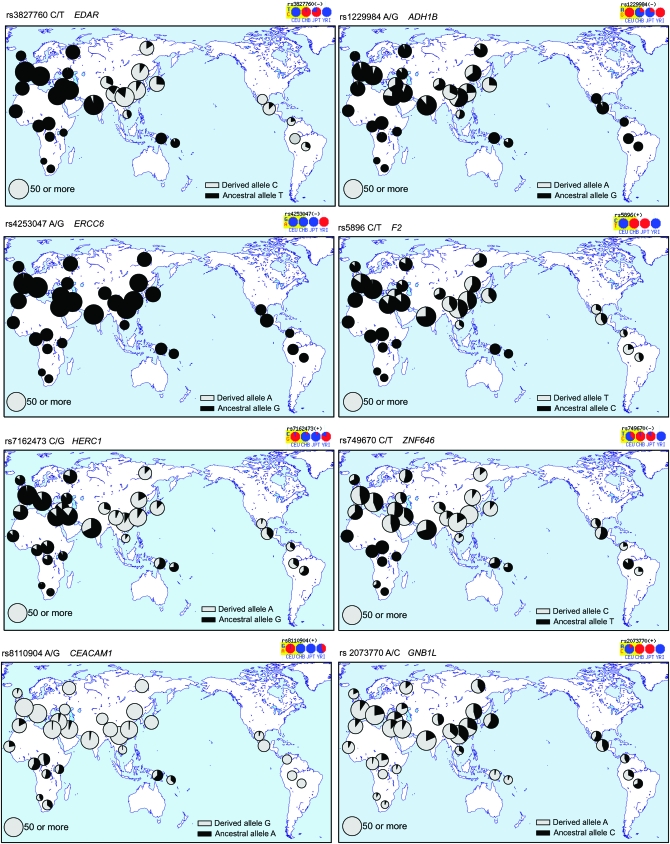
We next examined all 29 SNPs yielding high-quality genotypes in the HGDP–CEPH panel. This panel includes populations with similar geographic origins to the HapMap samples: Yoruba, French, Han Chinese, and Japanese, respectively, as well as populations from additional geographic regions. For all of the SNPs, the allele frequencies were similar between the (corrected) HapMap samples and their geographic equivalents. r2 values were as follows: YRI–Yoruba, 0.71; CHB–Han Chinese, 0.98; JPT–Japanese, 0.97; and CEU–French, 0.98. Worldwide frequencies for 8 genes illustrating a range of patterns are shown in Figure 2 and those for the remainder in Figure S1, Figure S2, and Figure S3. Population differentiation was evaluated using the statistic FST, and results are presented for the HapMap populations alone and the HGDP–CEPH populations considered by country (32 populations as in Figure 2; HGDP-32) or grouped according to the five genetic clusters identified by genomewide analyses (Rosenberg et al. 2002) (HGDP-5) (Table 2). These values were highly correlated (r2 = 0.75 HapMap vs. HGDP-32; r2 = 0.74 HapMap vs. HGDP-5). The values for 6 of the 7 SNPs identified above as representing artifacts were not unusually high. The seventh of these SNPs (rs8110904, CEACAM1) still showed high FST in both HapMap and HGDP data. The values for the remaining 22 SNPs were all relatively high: 17 lay outside the 99th percentile in all analyses and another 2 in the HapMap but not in the HGDP panel, a difference that might sometimes be expected because the SNPs were ascertained in the HapMap samples. Of the remaining 3 SNPs, 1 (rs5896 in F2) showed significant differentiation in the HGDP alone, and 2 (rs1702003 in THEA and rs2073770 in GNB1L) in none of the analyses. Thus analysis of a larger and more diverse set of populations confirms that 21/29 SNPs and 18/26 genes show unusually high levels of population differentiation (Figure 1).
Figure 2.—
Distributions of the ancestral (black) and derived (gray) alleles of the nonsynonymous high-differentiation SNPs (data for remaining SNPs are in Figure S1, Figure S2, and Figure S3).
TABLE 2.
FST values
|
FST |
||||
|---|---|---|---|---|
| SNP | Gene | HapMap | HGDP-32 | HGDP-5 |
| rs1702003 | THEA | 0.360 | 0.153 | 0.167 |
| rs12075 | FY | 0.632** | 0.380** | 0.429** |
| rs7555046 | Q8NGY8_human | 0.747** | 0.194 | 0.224 |
| rs7567833 | COLEC11 | 0.796** | 0.399** | 0.455** |
| rs6546837 | ALMS1 | 0.742** | 0.345** | 0.428** |
| rs6546839 | ALMS1 | 0.787** | 0.429** | 0.501** |
| rs2056486 | ALMS1 | 0.737** | 0.309** | 0.394** |
| rs10193972 | ALMS1 | 0.718** | 0.321** | 0.405** |
| rs3827760 | EDAR | 0.857** | 0.739** | 0.795** |
| rs11499 | FXR1 | 0.000 | 0.000 | 0.000 |
| rs7639705 | MCF2L2 | 0.040 | 0.033 | 0.034 |
| rs1047626 | SLC30A9 | 0.692** | 0.336** | 0.419** |
| rs5825 | ENSG00000172895.1 | 0.000 | 0.000 | 0.000 |
| rs1229984 | ADH1B | 0.688** | 0.313* | 0.318* |
| rs6601495 | RP1L1 | 0.851** | 0.512** | 0.576** |
| rs1871534 | SLC39A4 | 1.000** | 0.720** | 0.751** |
| rs4253047 | ERCC6 | 0.023 | 0.000 | 0.000 |
| rs4536103 | NEUROG3 | 0.184 | 0.089 | 0.106 |
| rs5896 | F2 | 0.353 | 0.233* | 0.283* |
| rs1426654 | SLC24A5 | 0.957** | 0.817** | 0.853** |
| rs7162473 | HERC1 | 0.707** | 0.425** | 0.488** |
| rs749670 | ZNF646 | 0.672** | 0.326** | 0.372** |
| rs17822931 | ABCC11 | 0.808** | 0.494** | 0.569** |
| rs6505228 | ENSG00000184253.2 | 0.799** | 0.501** | 0.574** |
| rs3911730 | RTTN | 0.827** | 0.391** | 0.437** |
| rs364637 | FUT6 | 0.000 | 0.000 | 0.000 |
| rs8110904 | CEACAM1 | 0.406** | 0.384** | 0.446** |
| rs2073770 | GNB1L | 0.292 | 0.119 | 0.148 |
| rs1385699 |
EDA2R |
0.790** |
0.428** |
0.526** |
*P < 0.05; **P < 0.01.
We then investigated the long-range haplotype structure surrounding the derived allele of each of the 29 SNPs. The EDA2R SNP rs1385699 could not be evaluated by our approach because it lay on the X chromosome, while for 2 of the other SNPs (rs7162473 in HERC1 and rs364637 in FUT6), map positions were missing from the 2009 HapMap data set. Of the remaining 26 SNPs, 2 (rs3827760 from EDAR and rs1229984 from ADH1B) showed a significant iHS signal in the CHB + JPT sample and 12 (including the same EDAR and ADH1B SNPs and SNPs from the FY, Q8NGY8_human, ALMS1, SLC30A9, SLC24A5, ZNF646, and ABCC11 genes) showed a significant XP-EHH signal at the 5% level in one or more populations or comparisons (Figure 1, Table S8). Overall, 5.9% of the SNPs tested for iHS showed a signal within the top 5%, not significantly different from the proportion expected by chance and probably reflecting the low power of iHS to detect selection around alleles at high or low frequency, combined with the ascertainment for SNPs with extreme frequencies in individual populations. In contrast, there was striking evidence for an overrepresentation of XP-EHH signals in these SNPs: 11.6% of the SNPs are in the 2% most extreme results (1% of each tail) and 30.4% are in the top 10% of results. The signals detected by simple population differentiation are often still reflected in the surrounding haplotype structures.
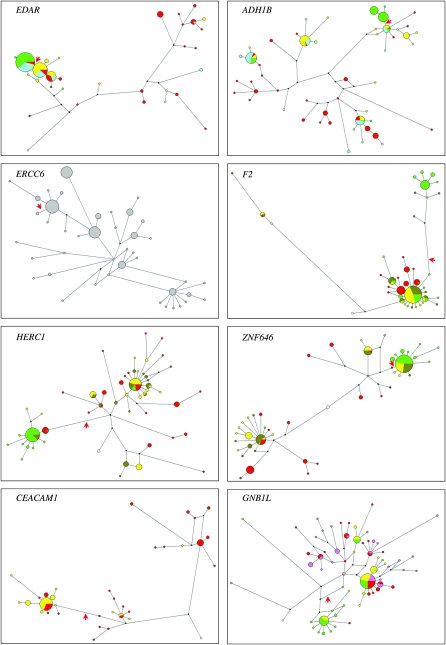
We finally examined full resequence data from a subset of the genes. Five genes in the set (four with high population differentiation) had already been resequenced by others: FY by Hamblin and colleagues (Hamblin et al. 2002); EDAR, ADH1B, and CEACAM1 by the SeattleSNPs project; and ERCC6 by the NIEHS SNPs project, and we resequenced an additional three high-differentiation genes, F2, HERC1, and ZNF646, and a fourth, GNB1L, because of the unusual geographic distribution of its derived allele. Nucleotide diversity and five statistics that summarize different aspects of the allele frequency spectrum were calculated; their significance was evaluated by comparison with (1) a multilocus HKA test to investigate whether the number of SNPs was lower than expected (Wright and Charlesworth 2004), (2) the best-fit demographic model for each population to test all statistics (Schaffner et al. 2005), (3) this model incorporating a modification for the ascertainment of the SNPs, and, for some, (4) the empirical data generated by the SeattleSNPs project (Table 3). The relationships between the inferred haplotypes were visualized using median-joining networks (Figure 3). Recent positive selection in a particular population is expected to lead to a number of characteristics in that population: low diversity, negative values of the summary statistics, and high-frequency haplotypes showing as large circles or clusters of circles in the median-joining networks.
Figure 3.—
Median-joining networks of resequenced genes. The red arrow indicates the nonsynonymous SNP. Color represents population of origin: red, Africa; yellow, Europe; dark green, Pakistan; bright green, East Asia; blue, Hispanic; gray, unknown; white, ancestral sequence deduced from chimpanzee.
Results for the nine genes were as follows:
(1) FY provides a paradigm of positive selection in humans with the FY*O allele as described earlier, but also carries a nonsynonymous SNP at high frequency in East Asia (Figure S1) defining the FY*A allele. Although an HKA test provided support for reduced diversity of the FY*O allele in Africa (Hamblin et al. 2002), summary statistics did not provide evidence for positive selection in the Chinese sample or indeed in the Hausan (African) sample where the *O allele predominates. In the combined worldwide sample, all statistics except Tajima's D showed evidence for a departure from neutral evolution (Table 3), and a complex pattern of haplotypes containing several local clusters was observed in the network (Figure S4). XP-EHH showed a strong signal indicative of positive selection in the YRI (Figure 1; Table S8) and so appears to be detecting selection on the *O allele rather than the *A allele.
(2) EDAR shows a very high frequency of the derived allele in East Asia and the Americas and a low frequency elsewhere (Figure 2A); the haplotype carrying this allele has correspondingly high frequency (45/46 chromosomes, 98%) and low diversity (44/45 C-allele chromosomes share a single haplotype and the 45th differs by a single SNP; Figure 3A). This pattern leads to a diversity value of 0.74 × 10−4 in the Asian-American sample, an order of magnitude lower than the average for chromosome 2 (Sachidanandam et al. 2001) and the lowest value in Table 3. The HKA test did not show significantly reduced diversity for this or any other gene (results included in Table 3), perhaps because its power to detect incomplete sweeps is low. Summary statistics are almost all significantly skewed, also showing many of the lowest P-values in Table 3. The same haplotype was present at moderate frequency in the Hispanic-American sample, where Fay and Wu's H, but not the other statistics, was significant. Somewhat surprisingly, the European-American sample showed significantly negative values for Tajima's D, Fu and Li's D, Fay and Wu's H, and Fu's Fs (Table 3), indicating possible positive selection. Such selection could not be acting on rs3827760 because the derived allele of this SNP is present only in 2/48 chromosomes. The network pattern (Figure 3A) suggests that it could be acting on the adjacent cluster of haplotypes where a central haplotype making up 28/48 chromosomes is surrounded by four one-step neighbors together contributing 15/48 chromosomes and two two-step neighbors consisting of 1 chromosome each and thus together forming 94% of the sample. This cluster lies in a region of the network that carries no other nonsynonymous SNPs, so there is no obvious second target of selection. An iHS signal is seen within the combined CHB + JPT sample where the frequency is 87%, and XP-EHH signals are seen in the comparisons involving this sample as previously (Sabeti et al. 2007), but also in the CEU–YRI comparison, supporting the hypothesis of additional independent positive selection in Europeans.
(3) The derived A allele of rs1229984 in ADH1B is present at relatively high frequency in Asia, particularly East Asia (Figure 2B), and, in the resequenced samples, was found only in the Asian-Americans where it lay in three neighboring haplotypes forming a small cluster in the network (Figure 3B). The frequency of derived alleles in this population was higher than expected, as reflected in a significantly negative Fay and Wu's H value (Table 3), but diversity was not unusually low, and several other statistics such as Tajima's D were actually positive, so evidence for selection was unconvincing, consistent with the conclusions of an independent study of this SNP (H. Li et al. 2008), which suggested that, if selection were acting, it was more likely to be on a nearby regulatory region SNP than on rs1229984. iHS did show a moderately significant value in the combined CHB + JPT sample, as did XP-EHH in the comparison of this sample with the CEU (Figure 1, Table S8), but both of these signals are also compatible with selection on a nearby SNP.
(4) The ERCC6 SNP rs4253047 initially identified by the HapMap study was represented by the ancestral allele in almost all of our samples (Figure 2C) and thus its inclusion was the result of a genotyping artifact; but since sequence data were available from the NIEHS SNPs study, we performed the same analyses as a negative control. Reassuringly, no test suggested evidence for positive selection.
(5) F2 showed the highest frequency of the derived allele in East Asia (Figure 2D), but this was associated with both higher diversity in the CHB than in the other populations examined and positive, albeit nonsignificant, values for most of the summary statistics (Table 3). The network showed a distinct branch marked by the nonsynonymous SNP and largely specific to the CHB and carrying 26/46 (57%) of CHB chromosomes (Figure 3D); the presence of two distant haplotype clusters in the CHB accounts for the positive summary statistics. Interestingly, Tajima's D and Fu and Li's D were significantly negative in the YRI (Table 3) and the YRI haplotypes were clustered in the network (Figure 3D), findings potentially pointing to an earlier episode of positive selection at the same locus that was unlinked to the nonsynonymous SNP that led to the ascertainment of the gene.
(6) HERC1 showed the highest frequency of the derived allele in East Asia, which reached >90% in several samples (Figure 2E). It therefore showed significantly negative values of Tajima's D and Fay and Wu's H in the CHB but not in other populations (Table 3), standard strong indicators of positive selection approaching fixation, and is illustrated by a large cluster in the network dominated by the CHB haplotypes (Figure 3E). Complications with the mapping of rs7162473 prevented the direct application of the extended haplotype tests (see materials and methods), but examination of all the HapMap2 SNPs in a 100-kb window surrounding this SNP revealed that 20/41 (49%) fell within the top 5% of iHS signals in the CHB + JPT sample, while XP-EHH showed an even stronger signal. For the CHB + JPT–YRI comparison, 70/75 (93%) fell within the top 1% of SNPs for selection in CHB + JPT and 100% of SNPs fell within the top 5%, whereas for the CEU–CHB + JPT comparison the values were 23/75 (31%) and 100%, respectively (this time in the negative tail, i.e., again pointing to selection in East Asia). These conclusions are consistent with those of Sabeti et al. (2007) and are indicated by the distinct symbols in Figure 1.
(7) The ZNF646 nonsynonymous SNP rs749670 derived allele is common in most populations outside sub-Saharan Africa, reaching ∼50% in Europe/West Asia and fixation in one East Asian sample (Figure 2F). Consequently, it shows low diversity and significantly negative values of Tajima's D and Fay and Wu's H in the CHB (Table 3), but higher diversity and positive values of several summary statistics in the CEU and BRU, reaching significance for Tajima's D in the BRU. The network (Figure 3F) shows a large cluster carrying most (45/46) of the CHB haplotypes but also 18/44 (41%) of the BRU haplotypes, explaining the negative and positive statistics, respectively. XP-EHH values are significant at the 5% level and indicate selection in the CHB + JPT sample (Figure 1, Table S8). Evidence for positive selection in East Asia is thus convincing.
(8) CEACAM1 rs8110904 showed a high frequency of the derived allele in all populations outside sub-Saharan Africa, reaching fixation in many of the samples (Figure 2G). Resequence data were available only for African-American and European-American samples, but showed low diversity and significantly negative Tajima's D and Fay and Wu's H in the European-Americans (Table 3). The corresponding network (Figure 3) shows a cluster specific for the derived allele carrying most of the European-American haplotypes, consistent with positive selection on the derived allele outside Africa.
(9) GNB1L stood out in this study as the only gene of the 27 that had the highest frequency of the derived allele in Africa, reaching fixation in one sample (Figure 2H), although its population differentiation was not unusually high (Table 2). It showed its lowest diversity and uniformly negative summary statistics in the two African samples, which are significant for Tajima's D, Fu and Li's D, and Fu's Fs in the Luhya in Webuye, Kenya (LWK) (Table 3). The network pattern is relatively complex, but there is one predominant derived haplotype, well represented along with its neighbors in the African samples (Figure 3H). Despite the unexceptional FST values, these unusual findings are consistent with positive selection in Africa.
Thus the resequencing studies provide powerful insights into the evolutionary history of this set of genes. The ERCC6 gene, with no significant population differentiation, showed no evidence for positive selection. Of the other eight genes, five showed evidence for positive selection acting on the derived allele from multiple significant summary statistic values. Thus using this stringent criterion, ∼60% of the nonsynonymous SNPs showing exceptionally high population differentiation are likely to result from positive selection. Two of the examples where clear evidence for positive selection on the derived SNP was not obtained, ADH1B and F2, both show recent expansions of haplotypes carrying the derived allele in network analyses, but these haplotypes are present at too low a frequency to lead to significant summary statistic values. The third example, FY, might have experienced complex rounds of selection that are not readily detected by the methods used (Hamblin et al. 2002). And two of the genes, EDAR and F2, show evidence of positive selection unrelated to the highly differentiated SNP, suggesting that they may have been the targets of multiple episodes of selection, although perhaps less complex than those of FY.
DISCUSSION
Two general conclusions emerge from this work. First, technical artifacts have contributed significantly to the apparent discoveries of highly differentiated SNPs. Second, if these artifacts are excluded, the extremely highly differentiated SNPs identified by the HapMap1 empirical survey do appear to have arisen predominantly as a result of population-specific positive selection, rather than genetic drift, a conclusion reached despite the limited power of all available methods to detect selection. We consider each of these conclusions in more detail and some of the biological implications of our findings.
The 32 SNPs that provided the starting point for this study resulted from a stringent ascertainment process by the HapMap1 project: more than 1 million SNPs were genotyped, and then the most highly differentiated nonsynonymous ones were picked out (International HapMap Consortium 2005). In retrospect, it is clear that this strategy also enriched for a rare “bookkeeping” error: allele switching. If a SNP that actually has little or no variation is labeled as having the wrong allele in one population, it will appear to show a very high level of population differentiation. This error accounts for most (five of seven) of the major genotyping discrepancies and associated low population differentiation in the HGDP–CEPH panel. Conversely all of the low-variability nonsynonymous SNPs in the HapMap set subject to this error would have been among the 32, suggesting that it affected only a very small proportion of HapMap genotypes, a low level of error that is probably inescapable in a project of this size, although one that would have been readily detected by simple replication of the small number of results highlighted in the final publication. We searched the 2008 HapMap data for remaining patterns that might indicate allele switching (one population sample fixed for the opposite allele to all the others, allowing one individual to depart from this pattern) and found 2 of 10,928 nonsynonymous SNPs compared with 6 of 832,520 noncoding SNPs that met our criteria. These numbers indicate a nominally significant enrichment of possible allele switching errors in nonsynonymous SNPs (P = 0.004 by Fisher's exact test), but this significance would be lost if one fewer nonsynonymous SNP with this pattern had been discovered, so we do not attach importance to the finding.
After excluding 8 genes because of low population differentiation and 1 because of our genotyping failure, we were left with a set of 18 in which to investigate whether or not high differentiation was due to selection. For this purpose, we had a set of somewhat independent indicators of selection: long-range haplotype structure within or between populations, summary statistics based on resequence data, and networks. The last did not provide a formal test for selection but nevertheless offered considerable insight into the history of the region. REHH and iHS analyses picked out only a few of the genes as showing evidence of unusually long haplotypes when analyzed within a population, a result that reflects the limited power of the method when applied to SNPs ascertained by high differentiation. Such SNPs are usually present at very high frequency in the population of most interest, so the absence of a signal is not evidence for a lack of selection. In contrast, XP-EHH analyses showed a strong enrichment of signals: 10/18 genes with confirmed high population differentiation show signals if the indirect HERC1 result is included, consistent with the higher power of this test relative to iHS for selected variants at higher allele frequencies (Sabeti et al. 2007).
In considering the summary statistic results, we need to take into account the fact that multiple (five) tests were used, but also the partial correlations between the tests, which are sensitive to different but related aspects of the data. We did this by simulating neutral loci in each population, with or without biased ascertainment as described in materials and methods. There was an ∼6% (range 5.8–6.6%) chance of obtaining two or more significant values (compared with <3% or obtaining three or more) and we therefore set a threshold of requiring a significant P-value in at least two tests. With this criterion, we obtained evidence for a departure from neutrality in the population with the high-frequency derived allele, in the direction expected from positive selection, in >60% of the genes examined. We take this as robust evidence for positive selection and also for the power and utility of the resequencing approach. However, some features of the tests applied deserve further discussion. They are best suited to detect positive selection when a single selected haplotype and its derivatives have risen to very high frequency, but not fixation. For example, the lowest and most significant values of Tajima's D were observed for EDAR and ZNF646 (Table 3), where the population samples of interest contained just one and two haplotypes lacking the selected SNP, respectively. These patterns are visualized in the networks as a large cluster containing the selected SNP with one or two small distant haplotypes (Figure 3). When a similar network pattern, in HERC1, contained four haplotypes in the distant cluster (<9%), the value of Tajima's D was not so low, although still significant. But when the number of haplotypes outside the largest cluster in the population of interest was as large as 23% or 54% (ADH1B, F2), nonsignificant statistics were observed. Two conclusions emerge from this discussion: with the current small sample sizes, excessive weight should not be placed on the exact summary statistic values and significance levels, and the networks can provide useful indications, although not firm evidence, that selection may be favoring haplotypes that do not show up in the tests used.
For some of the genes analyzed, there is evidence for biological consequences of the amino acid difference investigated here (Figure 1): EDAR rs3827760 influences NF-κB activity (Bryk et al. 2008) and hair thickness (Fujimoto et al. 2008a,b), including in a mouse model (Mou et al. 2008), and thus might have been sexually selected; the alcohol dehydrogenases metabolize alcohol and rs1229984 in ADH1B is associated with protection against alcoholism (H. Li et al. 2008); SLC24A5 is one of the major loci contributing to light skin color in Europeans (Lamason et al. 2005); ABCC11 rs17822931 determines wet/dry earwax type (Yoshiura et al. 2006); and rs1385699 in EDA2R has been associated with male-pattern baldness (Prodi et al. 2008). For other genes, more general information is available about possible biological functions of the gene, although not the consequences of the specific amino acid change studied: for example, ALMS1 is implicated in carbohydrate metabolism (Scheinfeldt et al. 2009), RNF135 in growth regulation (Douglas et al. 2007), and CEACAM1 in a wide range of functions including infection (Kuespert et al. 2006). These links, particularly with the specific SNP, make the case for selection more compelling. It is notable that among this small set of selection events are three that involve changes in visible appearance, suggesting that mate choice may have been a powerful selective force.
It was striking that two of the eight genes examined in detail showed evidence of positive selection on haplotypes different from those leading to the original ascertainment, and a third gene, FY, might also be placed in this category. Departures from neutral evolution are most readily detected by intraspecific tests when a single round of selection acts on a new mutation or rare SNP and have lower power to detect selection in more complex circumstances, emphasizing the remarkable nature of this observation. Positive selection is rare, and it appears that selection is focused preferentially on a small number of target genes, many of which may experience multiple independent selective events.
Finally, what conclusions can be drawn about the prospects for cataloging the sites of positive selection in our genome and linking them to a broader understanding of human evolution? The strategy of picking out highly differentiated functional SNPs yields (after excluding artifacts) a large enrichment for selected genes, with >60% of candidates being true positives. Thus follow-up of all hits identified using this approach by full resequencing and functional studies would be worthwhile. However, it is clear that this strategy identifies only a very small proportion of the regions that have experienced positive selection—its false negative rate is likely to be high. Furthermore, empirical approaches are inevitably biased toward detecting certain forms of selection (Teshima et al. 2006), although searching for outliers in population differentiation is one of the few effective ways of detecting selection on standing, rather than new, variants (Innan and Kim 2008). There is an urgent need for more effective ways of detecting selection, and resequence data from large numbers of individuals as planned by the 1000 Genomes Project (http://www.1000genomes.org/page.php) will provide an excellent data set to test these.
Acknowledgments
We thank the original donors of the HapMap, Brahui, and HGDP–CEPH samples; Howard Cann for supplying the HGDP–CEPH DNAs; Joe Pickrell for C scripts and helpful discussions on extended haplotype tests; and the Sanger Large-Scale Sequencing Team for generating the sequence data. This work was supported by The Wellcome Trust.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107722/DC1.
References
- Bandelt, H. J., P. Forster and A. Röhl, 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16 37–48. [DOI] [PubMed] [Google Scholar]
- Barreiro, L. B., G. Laval, H. Quach, E. Patin and L. Quintana-Murci, 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40 340–345. [DOI] [PubMed] [Google Scholar]
- Barrett, J. C., B. Fry, J. Maller and M. J. Daly, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 263–265. [DOI] [PubMed] [Google Scholar]
- Birney, E., J. A. Stamatoyannopoulos, A. Dutta, R. Guigo, T. R. Gingeras et al., 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, B. L., and S. R. Browning, 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk, J., E. Hardouin, I. Pugach, D. Hughes, R. Strotmann et al., 2008. Positive selection in East Asians for an EDAR allele that enhances NF-kappaB activation. PLoS ONE 3 e2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann, H. M., C. de Toma, L. Cazes, M. F. Legrand, V. Morel et al., 2002. A human genome diversity cell line panel. Science 296 261–262. [DOI] [PubMed] [Google Scholar]
- Carlson, C. S., D. J. Thomas, M. A. Eberle, J. E. Swanson, R. J. Livingston et al., 2005. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 15 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop, G., K. Bullaughey, F. Luca and M. Przeworski, 2008. The timing of selection at the human FOXP2 gene. Mol. Biol. Evol. 25 1257–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, J., D. Cilliers, K. Coleman, K. Tatton-Brown, K. Barker et al., 2007. Mutations in RNF135, a gene within the NF1 microdeletion region, cause phenotypic abnormalities including overgrowth. Nat. Genet. 39 963–965. [DOI] [PubMed] [Google Scholar]
- Enard, W., M. Przeworski, S. E. Fisher, C. S. Lai, V. Wiebe et al., 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418 869–872. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.-X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.-X., and W.-H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, A., R. Kimura, J. Ohashi, K. Omi, R. Yuliwulandari et al., 2008. a A scan for genetic determinants of human hair morphology: EDAR is associated with Asian hair thickness. Hum. Mol. Genet. 17 835–843. [DOI] [PubMed] [Google Scholar]
- Fujimoto, A., J. Ohashi, N. Nishida, T. Miyagawa, Y. Morishita et al., 2008. b A replication study confirmed the EDAR gene to be a major contributor to population differentiation regarding head hair thickness in Asia. Hum. Genet. 124 179–185. [DOI] [PubMed] [Google Scholar]
- Gardner, M., S. Williamson, F. Casals, E. Bosch, A. Navarro et al., 2007. Extreme individual marker FST values do not imply population-specific selection in humans: the NRG1 example. Hum. Genet. 121 759–762. [DOI] [PubMed] [Google Scholar]
- Goudet, J., 2005. HIERFSTAT, a package for R to compute and test variance components and F-statistics. Mol. Ecol. Notes 5 184–186. [Google Scholar]
- Hamblin, M. T., E. E. Thompson and A. Di Rienzo, 2002. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 70 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henshilwood, C. S., F. d'Errico, R. Yates, Z. Jacobs, C. Tribolo et al., 2002. Emergence of modern human behavior: Middle Stone Age engravings from South Africa. Science 295 1278–1280. [DOI] [PubMed] [Google Scholar]
- Hofer, T., N. Ray, D. Wegmann and L. Excoffier, 2009. Largeallele frequency differences between human continental groups are more likely to have occurred by drift during range expansions than by selection. Ann. Hum. Genet. 73 95–108. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Innan, H., and Y. Kim, 2008. Detecting local adaptation using the joint sampling of polymorphism data in the parental and derived populations. Genetics 179 1713–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, 2005. A haplotype map of the human genome. Nature 437 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling, M. A., M. E. Hurles and C. Tyler-Smith, 2004. Human Evolutionary Genetics. Garland Science, New York/Abingdon, UK.
- Krause, J., C. Lalueza-Fox, L. Orlando, W. Enard, R. E. Green et al., 2007. The derived FOXP2 variant of modern humans was shared with Neanderthals. Curr. Biol. 17 1908–1912. [DOI] [PubMed] [Google Scholar]
- Kuespert, K., S. Pils and C. R. Hauck, 2006. CEACAMs: their role in physiology and pathophysiology. Curr. Opin. Cell. Biol. 18 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason, R. L., M. A. Mohideen, J. R. Mest, A. C. Wong, H. L. Norton et al., 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310 1782–1786. [DOI] [PubMed] [Google Scholar]
- Li, H., S. Gu, X. Cai, W. C. Speed, A. J. Pakstis et al., 2008. Ethnic related selection for an ADH class I variant within East Asia. PLoS ONE 3 e1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. Z., D. M. Absher, H. Tang, A. M. Southwick, A. M. Casto et al., 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319 1100–1104. [DOI] [PubMed] [Google Scholar]
- McVean, G., and C. C. Spencer, 2006. Scanning the human genome for signals of selection. Curr. Opin. Genet. Dev. 16 624–629. [DOI] [PubMed] [Google Scholar]
- Mellars, P., 2006. Going east: new genetic and archaeological perspectives on the modern human colonization of Eurasia. Science 313 796–800. [DOI] [PubMed] [Google Scholar]
- Mou, C., H. A. Thomason, P. M. Willan, C. Clowes, W. E. Harris et al., 2008. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum. Mutat. 29 1405–1411. [DOI] [PubMed] [Google Scholar]
- Myles, S., K. Tang, M. Somel, R. E. Green, J. Kelso et al., 2008. Identification and analysis of genomic regions with large between-population differentiation in humans. Ann. Hum. Genet. 72 99–110. [DOI] [PubMed] [Google Scholar]
- Pickrell, J. K., G. Coop, J. Novembre, S. Kudaravalli, J. Z. Li et al., 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodi, D. A., N. Pirastu, G. Maninchedda, A. Sassu, A. Picciau et al., 2008. EDA2R is associated with androgenetic alopecia. J. Invest. Dermatol. 128 2268–2270. [DOI] [PubMed] [Google Scholar]
- Rosenberg, N. A., J. K. Pritchard, J. L. Weber, H. M. Cann, K. K. Kidd et al., 2002. Genetic structure of human populations. Science 298 2381–2385. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419 832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., S. F. Schaffner, B. Fry, J. Lohmueller, P. Varilly et al., 2006. Positive natural selection in the human lineage. Science 312 1614–1620. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., P. Varilly, B. Fry, J. Lohmueller, E. Hostetter et al., 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam, R., D. Weissman, S. C. Schmidt, J. M. Kakol, L. D. Stein et al., 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409 928–933. [DOI] [PubMed] [Google Scholar]
- Schaffner, S. F., C. Foo, S. Gabriel, D. Reich, M. J. Daly et al., 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt, L. B., S. Biswas, J. Madeoy, C. F. Connelly, E. E. Schadt et al., 2009. Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history. Mol. Biol. Evol. 26 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., and P. Donnelly, 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, K., K. R. Thornton and M. Stoneking, 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5 e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima, K. M., G. Coop and M. Przeworski, 2006. How reliable are empirical genomic scans for selective sweeps? Genome Res. 16 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen and J. K. Pritchard, 2006. A map of recent positive selection in the human genome. PLoS Biol. 4 e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., and B. Charlesworth, 2004. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura, K., A. Kinoshita, T. Ishida, A. Ninokata, T. Ishikawa et al., 2006. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat. Genet. 38 324–330. [DOI] [PubMed] [Google Scholar]