Abstract
The combination of QTL mapping studies of synthetic lines and association mapping studies of natural diversity represents an opportunity to throw light on the genetically based variation of quantitative traits. With the positional information provided through quantitative trait locus (QTL) mapping, which often leads to wide intervals encompassing numerous genes, it is now feasible to directly target candidate genes that are likely to be responsible for the observed variation in completely sequenced genomes and to test their effects through association genetics. This approach was performed in grape, a newly sequenced genome, to decipher the genetic architecture of anthocyanin content. Grapes may be either white or colored, ranging from the lightest pink to the darkest purple tones according to the amount of anthocyanin accumulated in the berry skin, which is a crucial trait for both wine quality and human nutrition. Although the determinism of the white phenotype has been fully identified, the genetic bases of the quantitative variation of anthocyanin content in berry skin remain unclear. A single QTL responsible for up to 62% of the variation in the anthocyanin content was mapped on a Syrah × Grenache F1 pseudo-testcross. Among the 68 unigenes identified in the grape genome within the QTL interval, a cluster of four Myb-type genes was selected on the basis of physiological evidence (VvMybA1, VvMybA2, VvMybA3, and VvMybA4). From a core collection of natural resources (141 individuals), 32 polymorphisms revealed significant association, and extended linkage disequilibrium was observed. Using a multivariate regression method, we demonstrated that five polymorphisms in VvMybA genes except VvMybA4 (one retrotransposon, three single nucleotide polymorphisms and one 2-bp insertion/deletion) accounted for 84% of the observed variation. All these polymorphisms led to either structural changes in the MYB proteins or differences in the VvMybAs promoters. We concluded that the continuous variation in anthocyanin content in grape was explained mainly by a single gene cluster of three VvMybA genes. The use of natural diversity helped to reduce one QTL to a set of five quantitative trait nucleotides and gave a clear picture of how isogenes combined their effects to shape grape color. Such analysis also illustrates how isogenes combine their effect to shape a complex quantitative trait and enables the definition of markers directly targeted for upcoming breeding programs.
IN the past decade, research on plant quantitative trait loci (QTL) has successfully identified numerous loci that control the genetic variation of complex traits in plants. Price (2006) reported no fewer than 30 studies of successful QTL cloning concerning nine plant species, but these researches have been conducted only in biparental populations, revealing only a slice of the genetic architecture for the trait (Holland 2007). Now that we are entering the post-genomic era, the challenge for geneticists is to fully decipher the molecular bases of quantitative genetic variation in highly diversified resources and to integrate the existing phenotypic variation extensively (Nordborg and Weigel 2008). The widespread availability of plant genomic and genetic resources has triggered the need for more integrated research (Flint-Garcia et al. 2003) that is likely to combine the findings of different approaches from various experimental designs, as has already been done for humans (Hirschhorn and Daly 2005), animals (Ron and Weller 2007), and plants (Osterberg et al. 2002; Aranzana et al. 2005). The combination of linkage and association genetics for this purpose constitutes a powerful tool (Yu and Buckler 2006), and newly considered model crops such as grape will greatly benefit from these advances.
The number of grape genomic resources has increased considerably over the past few years and the sequencing of the grape genome has recently been completed (Jaillon et al. 2007; Velasco et al. 2007). Taking advantage of these new resources will enable considerable progress in complex trait dissection, given that the access to candidate genes is straightforward, and will allow the cloning of QTL. Grape shows extended genetic variation with a high level of linkage disequilibrium (LD) (Barnaud et al. 2006) that makes an association genetics strategy feasible, as has already been performed in model (Flint-Garcia et al. 2005) or other perennial plants (Gonzalez-Martinez et al. 2007).
The variation in anthocyanin content is responsible for the continuous reddish color tones of most plant species (grape, apple, petunia, sweet potato, snapdragon, and Arabidopsis). In plant secondary metabolism, the anthocyanin pathway is one of the best described; the genes coding structural enzymes have been cloned in different plant systems (Sparvoli et al. 1994; Holton and Cornish 1995; Boss et al. 1996). Various mutated genes affecting both enzymes and transcription factors have been reported in maize, snapdragon, strawberry, and petunia (Mol et al. 1998; Aharoni et al. 2001; Winkley-Shirley 2001; Quattrocchio et al. 2006). Grape, Vitis vinifera L., is an important edible source of concentrated anthocyanins; their importance for final wine quality (Flanzy 1998) and their antioxidant benefits for human health (Joseph et al. 2005) call for the need to increase the accumulation of anthocyanin in edible plants.
In the case of grape, extensive molecular physiology studies provide evidence that two adjacent transcription factors, VvMybA1 and VvMybA2, are able to induce the VvUFGT transcription needed for berry pigmentation (Ageorges et al. 2006; Walker et al. 2007). Recently, a final transcription factor, VvMyb5b, was also shown to marginally induce the VvUFGT (Deluc et al. 2008). Furthermore, white/colored variation in grape cosegregates as a monogenic locus with the VvMybA1 locus (Doligez et al. 2006a,b; Lijavetzky et al. 2006). The white grape phenotype has been linked to the homozygous presence of a transposable element, Gret1, in the promoter of the VvMybA1 locus (Kobayashi et al. 2002, 2005; Lijavetzky et al. 2006; This et al. 2007). The quantitative variation in anthocyanin content in the skin of the berry was also shown to display considerable fluctuation (Mazza 1995), but despite its economic importance, the determinism of anthocyanin accumulation in berry skin until now has not been elucidated in grape.
For the analysis of genetic variation of small effect, such as traditionally observed for QTL, traditional molecular physiology resources have reached their limit. First, this is true because transient assays in grape only validate the effect of a single mutation in a bimodal way (Torregrosa et al. 2002; Vidal et al. 2006), and stable transformation is both labor- and time-consuming in the case of a long-cycle crop such as grape (Bouquet et al. 2008). Second, if the overall genetic effect is due to many combined mutations, it may affect either the level of expression of a gene when located in the promoter or the functionality of the protein when located in the gene coding sequence. Both modifications can hardly be analyzed in single transformation experiments. The overall resulting genotypic effect can be determined only through linkage studies (Neale and Savolainen 2004). A QTL mapping approach in which no assumptions are made about the architecture of the trait and that provides accurate positional information is a powerful tool for identifying loci that control phenotypic variability (Salvi and Tuberosa 2005). Previous studies have illustrated the feasibility of cloning genes that underlie QTL, but starting with the initial QTL detection on a recombinant inbred line population, these researches often took 5–7 years, as in the case of the Tb1 locus in maize (Doebley and Stec 1993; Doebley and Wang 1997), the COL1 gene in Brassica (Lagercrantz et al. 1996; Osterberg et al. 2002), the fw2.2 gene in tomato (Alpert et al. 1995; Frary et al. 2000), or the ERECTA gene in Arabidopsis (Alonso-Blanco et al. 1998; Masle et al. 2005). In upcoming years, the process of QTL detection to quantitative trait nucleotide (QTN) identification and, soon, the functional validation of the effect of the identified polymorphisms will become immediate even in the case of perennials such as grape, thanks to a more simplified molecular data analysis. Finally, the quality of quantitative genetics studies will depend more on the careful definition of the experimental design and the acquisition of accurate phenotypes for the sample in question.
Relevant work concerning the genetic bases of flavonoid and phenylpropanoid biosynthesis have involved maize, for which abundant genetic resources are available. Great efforts have been made to fully dissect the loci underlying the genetic variation for maysin accumulation (Lee et al. 1998; McMullen et al. 1998). The QTL identified gave rise to a very extensive survey of loci controlling maysin accumulation. This involved the use of both traditional molecular genetics tools, as for the identification of the orange1 locus with epigenetic effect (Chopra et al. 2003), and association genetics techniques for the p and c2 loci (Szalma et al. 2005). This revealed a complex regulatory mechanism involving both transcription factors and enzymes of the flavonoid pathway. Anthocyanin, as one of the main flavonoids in grape, displays a continuous variation among cultivars, and its genetic bases thus necessarily are affected by many genes and/or many mutations of small effect. The purpose of this study was to describe the genetic bases of the quantitative variation of anthocyanin in grape on the basis of the fewest possible assumptions about the genetic architecture of the trait. The most straightforward method of dealing with the genetic architecture of a trait is to carry out QTL mapping as successfully done in grape (Doligez et al. 2002; Mejia et al. 2007). Furthermore, thanks to the extensive information obtained with the sequencing of the grape genome, we aimed to refine the findings of our QTL mapping strategy with a candidate gene approach through the use of an association study on a collection of genetic resources of great diversity, as has already been done for plant color with other models (Szalma et al. 2005; Chagné et al. 2007). This will allow the definition of genic markers believed to be the cause of the variation instead of targeting linked neutral markers in breeding perspectives.
Nonetheless, these methods often lead to a huge number of positive associations, especially in the presence of LD, which makes the genetic effect of each polymorphism confusing. We established an original procedure for selecting the most stringent nonredundant associations. Moreover, as the associated polymorphisms are randomly combined in planta to shape a trait, we aimed to establish a single multivariate model that would provide an optimal fit of the anthocyanin berry content variation at the genotype level. Subsequently, a set of five putative QTNs, corresponding to one retrotransposon insertion, one 2-bp insertion/deletion (indel), and three single nucleotide polymorphisms (SNPs), was selected and integrated into a multivariate model that accounts for 84% of the anthocyanin content of a highly diversified collection of cultivars. This study enabled us to explain most of the variation in grape color only on the basis of the information of five polymorphisms on three distinct genes within a single gene cluster.
MATERIALS AND METHODS
Plant materials:
The plant material consisted of two populations: one cross-derived mapping population for QTL mapping and one natural population for the association study. The mapping population Syrah × Grenache (S×G) was a F1 progeny of 191 individuals from a reciprocal cross between clone 73 of Syrah and clone 516 of Grenache. Each offspring genotype was randomly displayed on two blocks (A and B). Both Syrah and Grenache are typical cultivars in the south of France. The natural population sample is a core collection (CC) of 141 individuals, which maximizes the agromorphological diversity for 50 qualitative and quantitative traits (Barnaud et al. 2006).
Phenotyping:
Grapes were harvested at maturity (20° Brix) at different times: (1) in 2005 in the A block of S×G and in the CC and (2) in 2006 in both A and B blocks of S×G and in the CC. Eight representative clusters were harvested among the clones. Twenty-five berries with a density of between 130 and 160 g/liter were randomly selected for further analysis. Their skins were powdered under liquid nitrogen, and anthocyanins were analyzed by high performance liquid chromatography according to Fournand et al. (2006).
Total anthocyanin content was then log-transformed with a ln(1 + x) function to unskew their distribution while conserving the power of the 0 class. The normality of the distribution was checked using the Shapiro–Wilks test for individuals carrying anthocyanins (nonwhite cultivars). To improve the extraction of the genetic variance components, the data from 2005 and 2006 were treated together using the mixed procedure of SAS software (SAS Institute, Cary, NC; http://www.sas.com) to extract the best linear unbiased predictor (BLUP) for each genotype.
DNA extraction, genotyping, and sequencing:
One square inch (80–100 mg) of fresh young leaf was harvested for each genotype. DNA was extracted using a Qiagen DNA Plant Mini kit (QIAGEN S.A., Courtaboeuf, France; http://www.qiagen.com) with slight modifications as described in Adam-Blondon et al. (2004).
The 191 S×G offspring were genotyped for 97 short sequence repeat (SSR) markers. Marker selection was carried out using both (1) the position of SSR markers on reference maps (Adam-Blondon et al. 2004; Doligez et al. 2006a,b) to cover the genome with minimal intermarker space (maximum of 10 cM) and (2) the most informative polymorphisms (priority given to 1:1:1:1 segregating markers).
SSR markers were genotyped as described by Doligez et al. (2006a,b). Amplified fragments were analyzed with an ABI PRISM 3100 Genetic Analyzer (APPLERA, Norwalk, CT; http://www3.appliedbiosystems.com/index.htm). An additional indel corresponding to a 10-kbp Gret1 retroelement in the MybA1 locus promoter was genotyped as previously described (Kobayashi et al. 2002; This et al. 2007). PCR conditions were identical to those described by Kobayashi et al. (2002). Amplified fragments of the two pairs described above were then bulked and run together on a 1% agarose gel, stained with ethidium bromide, and photographed under UV light.
Amplification primers were designed using the Primer 3 software and are listed in the supporting information, Table S1. PCR fragments were amplified, sequenced, and analyzed as described by Le Cunff et al. (2008).
Framework genetic maps:
Segregation distortions for parental and consensus data were assessed with χ2 tests. The parental maps were constructed according to the pseudo-testcross strategy (Grattapaglia and Sederoff 1996) using Carthagene 0.999R software (de Givry et al. 2005). The best marker order was determined starting with a first order and then optimizing this order. Finally, a sliding-window likelihood calculation was used to detect local changes, and markers with uncertain order at LOD 2 were discarded. A consensus map was built using the same procedure. Kosambi's mapping function was applied to all maps for the computation of genetic distance.
QTL detection:
QTL detection was performed on both parental and consensus maps. Composite interval mapping (CIM) on total anthocyanin content was performed separately on the 2005 A, 2006 A, and 2006 B block data with MapQTL 4.0 (Van Ooijen et al. 2002) for the consensus map and with QTL Cartographer (Basten et al. 2001) for the parental maps. For CIM on the parental maps, we first used the forward-and-backward regression method for cofactor selection with 0.1 as the in-and-out threshold for the P-value of the partial F-test. Then, a genome scan was performed with a maximum of five cofactors within a 10-cM window. LOD thresholds corresponding to an experimentwise error rate of 5% were then determined through 1000 permutations. For CIM on the consensus map, we determined the simple interval mapping LOD thresholds through 1000 permutations with a genomewide error rate of 5%. Confidence intervals of the first and second rank of the QTL's position were determined as one- and two-LOD support intervals.
Local blast:
The intervals defined by QTL mapping were blasted on the grape's genome browser (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis), and supercontigs were isolated. A local blast on the NCBI UniGene set was performed on the same supercontigs to determine the total number of genes present in the QTL intervals (ftp://ftp.ncbi.nlm.nih.gov/repository/UniGene/Vitis_vinifera).
Haplotype reconstruction:
Haplotypes of the VvMybA genes have been reconstructed using a partition-ligation expectation maximization algorithm described in Qin et al. (2002) and implemented in PHASE v2.1 (Stephens et al. 2001), using a 200 burn-in with 200 iterations in total and a thinning interval of 1; this was repeated 10 times until convergence was validated. The algorithm was run again on the most highly associated polymorphisms to reconstruct an entire macrohaplotype combining the linkage signal of all three genes.
Association tests:
Association tests were carried out using SAS and TASSEL software packages (http://www.maizegenetics.net/index.php?option=com_content&task=view&id=89&Itemid=). To test for associations, we used the structured association (SA) method (Thornsberry et al. 2001). First, population structure was calculated via a Bayesian approach implemented in the STRUCTURE software (Pritchard et al. 2000) that used 20 SSR markers well scattered throughout the 19 grape linkage groups (LGs) (Lacombe et al. 2007). A total of 500,000 iterations were performed for each population number between 1 and 10 with a burn-in period of 500,000. The optimal subpopulation model was selected using Evanno's correction (Evanno et al. 2005). The best population subdivision was obtained for K = 2 subpopulations, and the corresponding Q matrix was used for association analyses. Second, kinship was calculated in two ways, as implemented in TASSEL according to Ritland's (1996) calculation: first by using the 20 SSRs described above and then by using 129 SNPs from 10 unigenes of the anthocyanin pathway (VvAM1, VvAM3, VvMYB4, VvCHI1, VvCHI2, VvF3′5'H2.1, VvF3′5'H2.2, VvCHS3, VvLDOX2, and VvDFR, respectively).
A naive general linear model (GLM) test, an SA test, and two mixed linear model (MLM) tests using the two different kinship matrices were performed with TASSEL on each gene to identify the highly associated polymorphisms; this was based on P-values for all models and on the adjusted R2 of the GLMs (TASSEL). The LD calculations were performed using the LD option implemented in TASSEL, and the effect of the gene on the LD level was tested using the ANOVA procedure in SAS. Only a reduced set of closely associated and nonredundant markers was selected on the basis of a stepwise cofactor selection of the GLM procedure with partial risk set to α = 0.01.
Regression model selection:
The genotypes of the most closely associated polymorphisms selected above were broken down into additive (allele doses) and dominance (heterozygote vs. homozygote contrast) effects. The set of polymorphisms was then entered in a multivariate regression model using the REG procedure of SAS software. First, the population structure effect was included as an initial effect. Second, for model ranks ranging from 1 to n covariates, n being the number of covariates selected in the previous step, we calculated all alternative models and selected the best model of each rank on the basis of the adjusted R2. Finally, the model with the optimal number of parameters was selected according to both Akaike and Bayesian (Schwarz) information criteria.
RESULTS
QTL mapping:
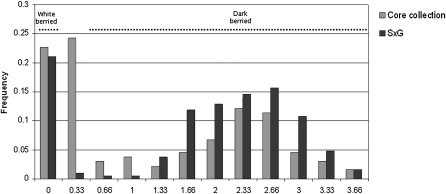
The S×G sample showed extensive variation ranging from 0 mg of anthocyanin for white cultivars to a maximum content of 31.6 mg of anthocyanin per gram of fresh skin with a variance of σS×G = 0.40 (data for 2005 are shown in Figure 1). In the S×G progeny, 30% of the individuals were white/green and 70% were dark-berried cultivars showing all 15 anthocyanin compounds, suggesting the 1:3 segregation of a major locus. QTL mapping was performed on the consensus map and on both parental maps—separately on each block for each year and collectively using BLUPs. In all detections, the BLUPs tended to improve both the score intensity and the position accuracy of the QTL (Table 1).
Figure 1.—
Distribution of the anthocyanin content in berry skin for the year 2005, expressed in logarithm of milligrams of anthocyanin/gram of fresh berry skin. The quantity of anthocyanin displays a continuous variation from 0 to 33.2 mg of anthocyanin per gram of fresh berry skin, with an overrepresented sample of white-berried cultivars displaying no anthocyanin.
TABLE 1.
Properties of the QTL detected on LD2 between markers VVMC5G7 and VVMybA1 in CIM for anthocyanin content of berry skin detected on the S×G map
| QTL confidence interval size in cM |
|||||
|---|---|---|---|---|---|
| Map | Year/block | LOD | LOD-1 | LOD-2 | R2 in MLM |
| Consensus | 2005/A | 18.37 | 9.3 | 12.9 | 0.479 |
| 2006/A | 18.37 | 7.6 | 11.5 | 0.486 | |
| 2006/B | 17.22 | 7.2 | 8.2 | 0.506 | |
| BLUP | 29.78 | 5.9 | 10.6 | 0.623 | |
| Syrah | 2005/A | 11.07 | 10.8 | 17.3 | 0.445 |
| 2006/A | 16.22 | 12 | 21.8 | 0.560 | |
| 2006/B | 18.36 | 14.5 | 22.8 | 0.533 | |
| BLUP | 30.72 | 9.2 | 10.6 | 0.543 | |
| Grenache | 2005/A | 7.91 | 2.2 | 4.2 | 0.320 |
| 2006/A | 10.86 | 1.8 | 3.6 | 0.390 | |
| 2006/B | 11.75 | 3.5 | 5.8 | 0.422 | |
| BLUP |
15.67 |
1.7 |
3.6 |
0.473 |
|
Four detections were carried out separately on three different maps. BLUPs of the genotypic effect extracted from mixed models were used to synthesize the information in the two blocks and 2 years and performed systematically better in QTL detection. R2 = adjusted percentage of variance explained by the model.
A single QTL located on LG2 between the VMC6B11 and VVIU20 markers was identified in all analyses with a 5.9-cM confidence interval at LOD 1 defined on the consensus map (Table 1). This locus accounted for 48–62% of the total variation in anthocyanin content in the berry and was repeated across all blocks and years. It was restricted to a 1.7-cM interval on the Grenache map, centered on the VvMybA1 Gret1 and accounting for 47% of total variation, and to a wider interval of 9.2 cM on the Syrah map located between the VMC6B11 marker and the VvMybA1 locus, accounting for 54% of total variation.
Candidate genes and polymorphisms within the QTL interval:
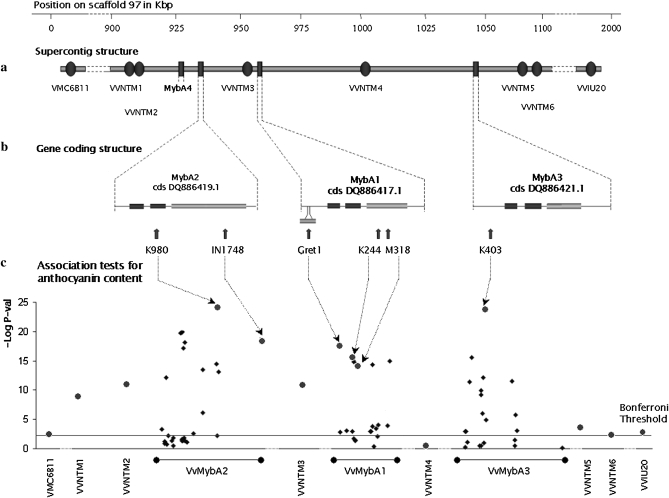
The 5.9-cM confidence interval at LOD 1 corresponded to a physical region of 2.85 Mbp. This segment was anchored on scaffold 97 of LG2 from the grape reference genome sequence (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis). The local blast on the grape nonredundant EST database enabled us to identify 68 unigenes with a perfect match.
Among these was a cluster of transcription factors belonging to the VvMybA family including four isogenes, of which VvMybA1 and VvMybA2 had already been described as being involved in the anthocyanin biosynthesis pathway (Kobayashi et al. 2002; Walker et al. 2007). The four VvMybA genes were retained as candidate genes (Table S1). The sequencing of a 6.2-kbp region in 141 genotypes (Figure 2 and Table S2) enabled the identification of a total of 75 SNPs, two indels, and one retrotransposon (Gret1): for VvMybA1, 20 polymorphisms on 1.5 kbp, including Gret1 in the promoter; for VvMybA2, 28 polymorphisms on 2.2 kbp, including an indel in the ORF; for VvMybA3, 21 polymorphisms on 1.75 kbp, including an indel in the ORF; for VvMybA4, 9 SNPs on 0.75 kbp.
Figure 2.—
(a) Presentation of VvMybA gene cluster and SSR markers, (b) position of the QTNs, and (c) level of association between markers and total anthocyanin content of berry skin along scaffold 97 of the grape genome browser. Along the x-axes, the dashed lines correspond to nonlinear scales. In the association tests, the microsatellite markers are presented in red, and the genic polymorphisms in blue; for the genic polymorphisms, dots correspond to QTNs and diamonds to the other polymorphisms. The Bonferonni threshold is equal to 6.25E-4.
Association genetics:
The CC sample showed greater variation than the S×G sample (σCC = 1.15), as was expected, with anthocyanin concentration ranging from 0 mg of anthocyanin to a maximum content of 33.2 mg of anthocyanin per gram of fresh grape skin. The association tests were performed following the SA (Thornsberry et al. 2001) and MLM (Yu and Pressoir et al. 2006) procedures to control for false positives, using the kinship calculated either on SSRs or on SNPs in independent genes. The GLM test of the population structure effect was significant (P-value = 0.0019) according to the hypothesis that the sample was divided into two admixed subpopulations. Thus, the population structure effect was included in all the association tests. Considering that we performed 78 tests, we applied a Bonferroni correction for multiple hypotheses testing, whereby the threshold of the tests was set to 6.25E-4. At this threshold, SA and MLM with kinship calculated on SSRs provided the same number of significant associations, while MLM with SNP kinship appeared slightly less conservative (Figure S1), detecting one additional significant association. We thus relied on the SA method.
Of the 78 tests performed on a BLUP predicting anthocyanin berry content across 2 years, 32 gave a significant result, with P-values ranging from 3.83E-04 to 5.72E-25 (Table 2), which explains 10–59% of the (anthocyanin) variation. Of these 32 associated polymorphisms, 10 were identified on VvMybA1, 12 on VvMybA2, 10 on VvMybA3, but none on VvMybA4. The location of the candidate polymorphisms on each gene, the effect on protein sequences, and the test statistics for association are presented in Table 2.
TABLE 2.
Markers and results of association tests for 32 associated polymorphisms, three haplotypes, and eight SSRs framing the candidate genes
| Gene | Marker | Type | Promoter/exon/intron/3′-UTR | Sy/NS | Frequency or no. of alleles | F-test | P-value | Marker R2 |
|---|---|---|---|---|---|---|---|---|
| VMC6B11 | SSR | 9 | 2.08 | 4.40E-03 | 0.36 | |||
| VVNTM1 | SSR | 6 | 9.63 | 1.41E-09 | 0.45 | |||
| VVNTM2 | SSR | 4 | 25.35 | 7.03E-12 | 0.45 | |||
| VvMybA2 | S84 | SNP | Promoter | 0.07 | 13.36 | 3.83E-04 | 0.10 | |
| R153 | SNP | Promoter | 0.23 | 33.69 | 2.45E-12 | 0.35 | ||
| W393 | SNP | Promoter | 0.23 | 65.25 | 6.63E-20 | 0.50 | ||
| W417 | SNP | Promoter | 0.24 | 65.31 | 5.80E-20 | 0.50 | ||
| R440 | SNP | Promoter | 0.21 | 53.29 | 2.53E-17 | 0.45 | ||
| S465 | SNP | Promoter | 0.24 | 58.18 | 1.98E-18 | 0.47 | ||
| Y754 | SNP | Promoter | 0.12 | 16.69 | 4.54E-07 | 0.22 | ||
| R762 | SNP | Exon | NS | 0.16 | 38.80 | 2.00E-13 | 0.40 | |
| K980 | SNP | Exon | NS | 0.27 | 87.08 | 3.19E-24 | 0.57 | |
| R984 | SNP | Exon | Sy | 0.23 | 42.98 | 7.43E-15 | 0.40 | |
| R1020 | SNP | Exon | Sy | 0.23 | 39.31 | 7.00E-14 | 0.38 | |
| Indel 1748 | Indel | Exon | NS | 0.21 | 65.01 | 2.31E-19 | 0.53 | |
| Haplotype | 24 | 10.69 | 2.66E-15 | 0.71 | ||||
| VVNTM3 | SSR | 10 | 7.45 | 8.59E-12 | 0.43 | |||
| VvMybA1 | Gret1 | Retro | Promoter | 0.39 | 67.30 | 3.03E-18 | 0.56 | |
| K244 | SNP | Exon | NS | 0.25 | 51.00 | 1.79E-16 | 0.46 | |
| K259 | SNP | Exon | NS | 0.3 | 47.96 | 7.12E-16 | 0.44 | |
| M318 | SNP | Exon | NS | 0.19 | 45.31 | 2.87E-15 | 0.43 | |
| Y566 | SNP | 3′-UTR | 0.18 | 46.07 | 2.17E-15 | 0.44 | ||
| R567 | SNP | 3′-UTR | 0.13 | 9.66 | 1.32E-04 | 0.14 | ||
| Y620 | SNP | 3′-UTR | 0.04 | 13.69 | 3.31E-04 | 0.10 | ||
| D660 | SNP | 3′-UTR | 0.04 | 6.74 | 7.42E-05 | 0.21 | ||
| W811 | SNP | 3′-UTR | 0.13 | 9.90 | 1.10E-04 | 0.15 | ||
| Y847 | SNP | 3′-UTR | 0.4 | 52.10 | 4.77E-16 | 0.52 | ||
| Haplotype | 26 | 13.30 | 1.90E-19 | 0.81 | ||||
| VVNTM4 | SSR | 4 | 0.82 | 4.43E-01 | 0.02 | |||
| VvMybA3 | Y166 | SNP | Promoter | 0.39 | 33.29 | 2.47E-12 | 0.33 | |
| M209 | SNP | Promoter | 0.1 | 50.34 | 1.24E-16 | 0.44 | ||
| M357 | SNP | Promoter | 0.05 | 14.61 | 5.19E-11 | 0.38 | ||
| W362 | SNP | Promoter | 0.06 | 25.93 | 4.26E-10 | 0.29 | ||
| R379 | SNP | Promoter | 0.42 | 14.20 | 3.36E-06 | 0.20 | ||
| K392 | SNP | Promoter | 0.11 | 36.42 | 3.79E-13 | 0.36 | ||
| K403 | SNP | Promoter | 0.39 | 92.54 | 5.72E-25 | 0.58 | ||
| R440 | SNP | Promoter | 0.48 | 12.92 | 8.45E-06 | 0.17 | ||
| R869 | SNP | Promoter | 0.03 | 34.52 | 1.50E-12 | 0.35 | ||
| R929 | SNP | Exon | Sy | 0.47 | 20.30 | 1.61E-05 | 0.15 | |
| Haplotype | 42 | 68.74 | 2.88E-10 | 0.7365 | ||||
| VVNTM5 | SSR | 4 | 5.79 | 2.74E-04 | 0.34 | |||
| VVNTM6 | SSR | 5 | 2.86 | 4.40E-03 | 0.18 | |||
|
VVIU20 |
SSR |
2 |
6.86 |
1.80E-03 |
0.15 |
P-values <6.25E-4 were considered to be significant. R2 mk is the percentage of anthocyanin variation explained by the polymorphism alone when integrating the structure. 3′-UTR, 3′ untranslated region; Sy, synonymous change; NS, nonsynonymous change. Frequencies are given for the SNP minority allele and the number of alleles are given for SSRs and haplotypes.
To determine whether the positive association was due to LD throughout the zone or to a particular effect of each polymophism, we genotyped the core collection with polymorphic SSR loci from scaffold 97. Two SSRs at both ends of scaffold 97 were already available (VMC6B11 and VVIU20), and six more were developed within scaffold 97 (VVNTM1–6), scattered between the VvMybA genes (Figure 2 and Table S1). We performed the same GLM tests as in the association genetics analysis, including eight SSR markers surrounding the three loci of interest. Each of the neutral markers located between each VvMybA isogene showed a high level of association. Only one test performed on SSR VVNTM4 was nonsignificant, revealing the presence of LD along the supercontig (Table 2 and Figure 2c). Nonetheless, 21 of the 32 polymorphisms linked to the phenotype had a smaller P-value than the most closely associated flanking SSR marker (VVNTM2, Table 2). Ten of the 32 associated polymorphisms and 5 of 6 polymorphisms selected using the stepwise cofactor selection method showed an adjusted R2 higher than that of VVNTM2, the most closely associated SSR. With R2 and P-values of the SSRs bordering each side of the supercontig that decrease with increasing distance from the VvMybA genes, we successfully restricted the associated interval to the VvMybA gene cluster only.
Linkage disequilibrium and haplotype structure among associated polymorphisms:
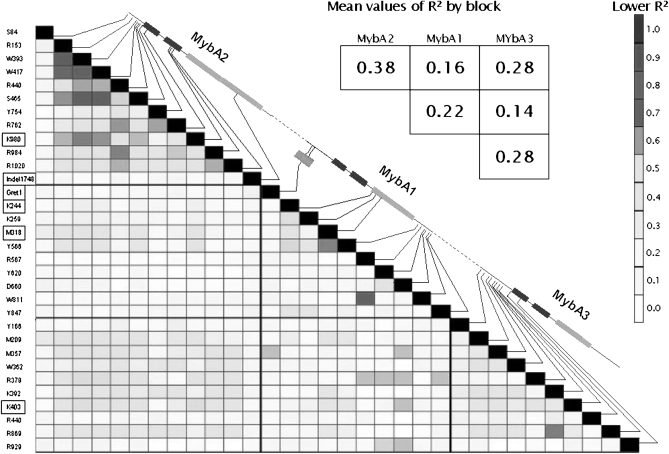
On the overall associated polymorphisms, intragenic LD was observed to be higher than intergenic LD in this region (Figure 3), resulting in significantly higher intragenic r2 values than in the overall SNP markers. The overall level of LD differed among genes (ANOVA P-value < 0.0001): tight LD on VvMybA2, moderate on VvMybA3, and moderate to low on VvMybA1 (rA22 = 0.38, rA32 = 0.28, and rA12 = 0.22, respectively). VvMybA2 and VvMybA3 appeared to be in strong intergenic LD (rA2A32 = 0.28) and significantly different from other intergenic patterns of LD (VvMybA1 vs. VvMybA2 and VvMybA1 vs. VvMybA3; P-value < 0.0001).
Figure 3.—
LD plot based on R2 values for the SNPs and indels associated with the total anthocyanin content of berry skin on VvMybA1, -A2, and -A3 genes in the lower diagonal and overall level of LD for the full genes in the upper diagonal. QTNs selected through stepwise cofactor selection (far left column) are framed in black. Presented R2 values are estimated according to Remington et al. (2001).
Phased haplotypes of each gene were then reconstructed using associated polymorphisms. On all three genes, the reconstruction enabled us to identify 96 haplotypes with a single haplotype representing a frequency of 45%. We identified 26 haplotypes on VvMybA1, 24 on VvMybA2, and 42 on VvMybA3 separately on each gene. In the haplotype group carrying the A1 Gret1 insertion, only two polymorphisms segregated with a frequency >5% (Y847 on VvMybA1 and R1020 on VvMybA2), while their frequencies ranged from 11 to 50% in the haplotype group carrying no A1 Gret1. This reveals that most of the associated polymorphisms were variable only in the absence of A1 Gret1; hence, their effect may be distinguished from that of A1 Gret1. Although unbalanced haplotype classes were involved, the tests performed on single-gene haplotypes led to reinforced association and LD levels within and among genes (Table 2). Due to weak statistical power, we did not carry out the tests on the haplotype set combining all 32 polymorphisms.
Stepwise cofactor selection for QTN identification:
To define a set of highly associated and independent polymorphisms within a strong LD context, we applied a stepwise cofactor selection method for the associated polymorphisms separately on each gene. A set of six putative QTNs was retained (Figure 2c). On VvMybA1, three polymorphisms were retained: the retrotransposon A1 Gret1 (promoter), SNP A1 K244 (change from R to S in position 188 of the amino acid sequence), and A1 M318 (change from Q to P in position 213). On VvMybA2, two polymorphisms were integrated: SNP A2 K980 (change from R to L in position 44) and A2 indel 1748 (frameshift leading to a truncated 265-amino-acid protein instead of 344); on VvMybA3, only one, SNP A3 K403 (promoter), was integrated; and none were integrated on VvMybA4.
The PHASE reconstruction with the six retained QTNs led to the identification of 17 distinct macrohaplotypes combining the linkage information of the three genes for the 141 individuals. A single macrohaplotype represented an overall frequency of 0.58 and 9 haplotypes were present in fewer than four individuals (frequency of 0.01). The macrohaplotype was highly associated with anthocyanin berry content in a structured association model with a P-value of 1.22E-30 and a marker r2 of 0.82. Among the six putative QTNs, there were only two pairs of loci showing substantial LD: A1 Gret1 and A2 indel 1748 that were moderately linked with r2 = 0.25 and D′ = 0.73 and A2 K980 and A3 K403 that were more closely linked with r2 = 0.51 and D′ = 0.90. All other pairs showed low LD, considering their physical linkage with an r2 ranging from 0.17 to 0.25 and a D′ ranging from 0.52 and 0.61.
Multivariate regression model and haplotype association:
To further dissect the combinatory effect of each of the six selected polymorphisms in terms of additive and dominance effects at the genotypic level (Table 3), we applied a regressive procedure for multivariate model testing. Both Akaike and Bayesian information criteria led to the selection of a seven-cofactor model explaining 84% of the variation in anthocyanin content (including population structure, additivity and dominance of A1 Gret1, additivity of A1 K244, dominance of A2 K980, additivity of A2 indel 1748, and additivity of K403). We showed that A1 Gret1 was the polymorphism that explained most of the variation (the partial additive effect of Gret1 in an SA model accounted for 59% of the variation in anthocyanin content of the berry) and was the only one included in the final model with its two genetic components. Including four other polymorphisms enabled us to account for an additional 23% of the variance. SNP A1 M318, which was initially selected as a putative QTN, appeared to be useless in the full-rank model, limiting the final set of putative QTNs to five.
TABLE 3.
Structured multivariate regression models
| No. cofactors in model | R2 | BIC | Cofactors | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 61 | −42.2 | Gret1 a | |||||||||||
| 2 | 77 | −78.8 | K244 a | In1748a | ||||||||||
| 3 | 81 | −91.1 | K244 a | K980 d | In1748a | |||||||||
| 4 | 82 | −94.9 | K244 a | K980 d | In1748a | K403 a | ||||||||
| 5 | 83 | −96.4 | Gret1 d | K244 a | K980 d | In1748a | K403 a | |||||||
| 6 | 84 | −97.4 | Gret1 a | Gret1 d | K244 a | K980 d | In1748a | K403 a | ||||||
| Partial R2 | 58.6 | 3.6 | 2.8 | 1.2 | 3.2 | 11.3 | ||||||||
| 7 | 84 | −96.6 | Gret1 a | Gret1 d | K244 a | K980 a | K980 d | In1748a | K403 a | |||||
| 8 | 84 | −95.5 | Gret1 a | Gret1 d | K244 a | M318 a | K980 a | K980 d | In1748a | K403 a | ||||
| 9 | 84 | −94.3 | Gret1 a | Gret1 d | K244 a | K244 d | M318 a | K980 a | K980 d | In1748a | K403 a | |||
| 10 | 84 | −92.1 | Gret1 a | Gret1 d | K244 a | K244 d | M318 a | K980 a | K980 d | In1748a | K403 a | K403 d | ||
| 11 | 84 | −89.6 | Gret1 a | Gret1 d | K244 a | K244 d | M318 a | M318 d | K980 a | K980 d | In1748a | K403 a | K403 d | |
| 12 |
83 |
−87.2 |
Gret1 a |
Gret1 d |
K244 a |
K244 d |
M318 a |
M318 d |
K980 a |
K980 d |
In1748a |
In1748d |
K403 a |
K403 d |
The models presented correspond to the best models for each cofactor number; after the Gret1 model, all higher models are nested, allowing the classification of the identified QTNs from the least to the most important. Models in boldface correspond to the optimal model, i.e., minimizing AIC and BIC criteria. R2, percentage of the variance explained by the model; partial R2, percentage of the variance explained by individual polymorphism in the model; AIC, Akaike information criteria; BIC, Bayesian (Schwarz) information criteria (the smaller the better); a is the additive component of the marker genetic effect, and d is the dominant component of the marker genetic effect.
DISCUSSION
In the previous section, we presented the results of a detailed study of QTL to QTNs for a quantitative trait in grape. These results elucidate the primordial role played by the cluster of the MybA genes in the quantitative determinism of grape berry color, since only five polymorphisms in the three genes are enough to explain 84% of the total phenotypic variation of the CC with reasonable statistical power (d.f.model = 10 and d.f.error = 67). While great emphasis was previously placed on the role played by allelic variants of the VvMybA1 promoter in explaining the qualitative distinction between white and dark-colored grapes (Kobayashi et al. 2002; Lijavetzky et al. 2006; This et al. 2007), here, we have instead developed an accurate model for a quantitative understanding of variation in grape color.
Increasing the accuracy of the analysis of grape color—molecular vs. empirical classification:
One way of increasing the accuracy of the analysis is to improve the estimation of the genotypic value. In this work, the data repetition across years and blocks was valorized through the definition of BLUP calculated from mixed models already used for the extraction of genetic variance components (Borevitz et al. 2002). Consequently, the overall annual effect for 2 years was approximately taken into account and was shown to be helpful in explaining the phenotype. Other environmental effects such as saccharose concentration or light exposure proved to have an influence on Myb genes related to grape anthocyanin biosynthesis (Matus et al. 2009). Full control of environmental effects can be achieved by repeated phenotyping across environments, years, and their covariance, as successfully done in apple by Segura et al. (2008).
A complex trait with a simple genetic architecture:
Previous work in quantitative genetics emphasizes that the Gret1 homozygous presence is the determinant for white color (This et al. 2007); our study confirms this finding because a single QTL was found and no other polymorphism was able to explain the white-to-colored grape bimodal variation.
Genetic variation for anthocyanin berry content was convincingly (LOD score up to 29.78, and R2 up to 0.62) and consistently (detected across years and blocks) explained by a single QTL. Nonetheless, the anthocyanin content of the berry shows a high level of heritability (Barritt and Einset 1969) and a continuous variation; thus, the anthocyanin content of the berry is necessarily polygenically determined. Knowledge of the grape genome sequence enabled the reduction of an interval of 5.9 cM into a set of 68 genes. Given previous evidence (Kobayashi et al. 2005; Lijavetzky et al. 2006; This et al. 2007; Walker et al. 2007), we reduced a priori this number to a single gene cluster. One of the main limitations of this study is the fact that we did not sequence all remaining 65 unigenes present in the QTL confidence interval, and thus there may be other loci that are also involved in anthocyanin biosynthesis in the same region; clearly, we have made a candidate gene assumption. Furthermore, the testing of new putative variation factors as candidate genes has recently been suggested (Deluc et al. 2008). Nonetheless, both the high scores of the QTL and the association tests and the overall goodness-of-fit of the final model are sufficient to thereby confirm a posteriori that most of the genetic effect has been included and unambiguously analyzed in different genetic backgrounds.
The LD pattern led us to consider a gene haplotype structure strongly associated with anthocyanin content. Nontheless, with unbalanced frequency in the haplotypes set, the test results have to be interpreted with caution because exaggerated numbers of classes lead to overparametered nonconservative tests. To avoid redundancy in the presence of LD and to identify the most informative sites, we used a regression technique with stepwise cofactor selection, which is more convenient in the case of a huge number of linked markers. Starting with 32 associated polymorphisms, we reduced the data set to five putative QTNs with a nonredundant genetic effect. In the future, an alternative method would be to adopt a composite association mapping strategy similar to the CIM in QTL detection but applied to natural populations (Boitard et al. 2006).
The very high percentage of explained variance leads us to consider that nearly all the variation is determined by the VvMybA gene cluster. A huge number of genes underlying QTL are described as transcription factors (Salvi and Tuberosa 2005; Price 2006). Previous studies in maize emphasized the c1 locus, encoding a transcription factor, as the major control for anthocyanin pigmentation of the aleurone layer of the kernel (Cone et al. 1986). Furthermore, this locus appeared to be duplicated on chromosome 6 (PL; Cone et al. 1993). Because PL is involved in anthocyanin regulation in the floral and vegetative tissues, both paralogs are involved in a tissue-specific control of the anthocyanin metabolism. Nonetheless, only a few studies identified a transcription factor gene cluster as shaping a quantitative trait (Francia et al. 2007). Such a cluster structure with multiple small effects on regulation is thus an original finding. An interesting feature is that each gene of the cluster appears to have a dose effect. Nearly all the SNPs except one (A2 K980) essentially display additive behavior (Table 3), which is coherent with the fact that we were investigating a transcription factor family in which such a dose effect is to be expected: more functional MYBA protein synthesis leads to more VvUFGT transcript abundance and thus to more anthocyanin synthesis. The retroelement of VvMybA1, Gret1, is the main component in generating color variation; nevertheless, the four other polymorphisms explaining an additional 23% of variance represent a very significant contribution to the phenotype (Table 3). For example, in the case of the S×G population, Syrah and Grenache had the same genotype at the Gret1 locus but differed in their A2 K980 genotype, Syrah being heterozygous while Grenache was homozygous for the weak allele. This explained both the lack of color observed in Grenache and the smaller size of the QTL compared to Syrah on the Grenache parental map.
These findings are also relevant in terms of breeding due to the close physical linkage between the VvMybA genes. Any new breeding strategy for anthocyanin potential in grape should first ensure the presence of the appropriate combination of VvMybA alleles in the parents at the haplotypic level. Otherwise, the low recombination rate will be very restrictive.
Validity of the detected associations:
The starting point in dissecting a quantitative phenotype is to obtain accurate positional information about the loci responsible for the variation. Both QTL studies and genome scans are very straightforward approaches as they do not rely on a priori selection of the polymorphisms to be tested. The use of collections of natural diversity enables very accurate resolution in quantitative genetics studies, leading to putatively causal polymorphisms (Yu and Buckler 2006). Furthermore, the results are easily transposed as the tests are performed in different genetic backgrounds, contrary to a cross-derived population where the results are cross-dependent (Flint-Garcia et al. 2005).
A general concern in association genetics on natural samples is the control of false positives due to genetic neutral covariance, generally referred to as “population structure” (Pritchard et al. 2000), and distinct levels of kinship. Methods of dealing with such problems are widely available but depend on the technical possibilities of the crop being studied (Thornsberry et al. 2001; Yu et al. 2006; Zhao et al. 2007). Convenient structure matrices to be used as cofactors in association studies rely on inferences about the past history of the crop concerned. The hypothesis relating to two major populations corresponding to Western and Eastern genetic pools was emphasized by Arroyo-García et al. (2006) and Le Cunff et al. (2008) and was integrated as such into our model. Recent studies have shown that the relative relatedness between individuals (kinship) performed better than overall population covariance (structure) in controlling false positives (Zhao et al. 2007). In the case of grape, we tested both structured association and mixed models using either the 20 SSRs or a set of 129 SNPs located in 10 genes of the anthocyanin biosynthesis pathway to calculate kinship. All alternative models appeared in our case to perform equally well (Figure S1), probably owing to the fact that either structure or kinship effects are weak compared to the very strong detected associations. Therefore, we simply relied on the structured association procedure, a purely deterministic model. Multiple testing is also challenging in association studies where the number of markers is often huge. We chose to apply a Bonferroni correction systematically and set an extreme threshold level for each test to ensure a very high level of stringency.
The use of a stepwise cofactor selection method and the definition of a multivariate model is the most precise way to interpret the genetic determinism of the in vivo accumulation of anthocyanin. The stepwise selection method ensures that all polymorphisms selected within a gene are independent; the linkage information is therefore synthesized in the best possible way. The haplotypic treatment of the QTNs led to a higher level of association than by taking each polymorphism independently, but the R2 is 2% lower than the selected multivariate model. These results must be interpreted with caution due to the non-homoscedasticity of the haplotype classes with the overrepresentation of a single haplotype and numerous low-frequency ones. In terms of the information provided, the multiple regression method allowed the ranking of the various QTNs in order of association magnitude and guaranteed the independence of the various effects tested, so as to ultimately establish the best nonredundant model. This approach represents a step forward in the field of complex trait dissection, where the combined effects of different genes are believed to exist but have yet to be demonstrated. In a system where all three genes may interact and have many different alleles, a statistical approach is very helpful in defining new hypotheses and in creating a hierarchy of the many polymorphisms that have been shown to shape the complex phenotype. Nonetheless, due to the extent of LD between VvMybA2 and VvMybA3 in the sample, the additive effect of A2 K980 cannot be differentiated from the effect of A3 K403. Replacing the additive effect of A3 K403 with A2 K980 decreased the R2 by only 0.2%.
Functional effect of the polymorphisms:
Polymorphisms on VvMybA2 and VvMybA3 were essentially located in the promoter and the first exon, while the VvMybA1 promoter and the first exon appeared to be less variable, showing more variability in the 3′-UTR instead. On these three genes, the third exon was rather monomorphic, showing only three SNPs on MybA1 that were all associated, one associated indel on VvMybA2, and one unassociated indel on VvMybA3. This exon corresponds to a cystein-rich (CR) domain putatively involved in DNA-binding activity. Previous work led by Walker et al. (2007) showed the capacity of both VvMybA1 and VvMybA2 to activate anthocyanin synthesis, and Kobayashi et al. (2002) showed that the CR domain in VvMybA3 was truncated, leading to a nonfunctional protein. One possible explanation for the involvement of nonfunctional VvMybA3 in berry pigmentation is the steric competition between VvMybA3 and the other two functional isogenes. This hypothesis is supported by the fact that, on VvMybA3, only one polymorphism within the promoter is linked, while, on VvMybA1 and VvMybA2, exonic polymorphisms also appear to be linked with anthocyanin content. Nonetheless, given the strong LD level between A2 K980 and A3 K403, we may also consider that the association of VvMybA3 is purely due to linkage. For locus VvMybA4, reported as being unexpressed in berries (Walker et al. 2007), we were unable to detect any effect on grape color due to the limited variability of the gene.
Of the six polymorphisms, three had already been identified in previous works as being associated with qualitative color variation (Kobayashi et al. 2005; This et al. 2007) because of gene silencing or because of a change in the structure of the white allele (Kobayashi et al. 2005; Walker et al. 2007). Nevertheless, for the first time, we have detected an association between A1 K244 and A1 M318 and the anthocyanin content of the berry, with both mutations leading to a change in the amino acid in the VvMybA1 protein structure: from R to S in position 188 and from Q to P in position 213, respectively. For the association characterized on VvMybA3 with A3 K403, only a statistics-based quantitative approach could enable the identification of putative effects on transcript regulation.
A final validation of all the effects described here could be performed through transient assays. In the case of a quantitative trait, due to very high technical variance in the phenotype of the transformed plants, this would require a huge number of replicates of independent transformations, which is not technically feasible at the present time. This study directly investigated the polymorphism that may cause the variation at the phenotypic level. As the revealed associations are very strong and are supported by additional physiological information (data not shown here), we believe that association genetics per se is sufficient validation to draw final conclusions about the genetic determinants of naturally occurring variation. The color quality aspect, a highly relevant trait for viticulturists, may now be easily targeted in any grape-cross offspring, thereby providing the opportunity to identify genotypes of interest very early, from light red for rosé wines to the most intense purple.
Acknowledgments
We thank V. Segura for many fruitful and inspiring discussions and two anonymous reviewers for helpful comments. We also thank the technical staff for the phenotypic analysis, in particular, M. Farnos, P. Ortigosa, C. Morel, and the staff of Domaine du Chapitre and Domaine de Vassal for grape cultivation. This work was funded in part by the European project, FLAVO (no. 513960); a French Genoplante project, COREGRAPGEN (Trilat 017); and a Ph.D. grant from the Languedoc-Rousillon region for A.F-L.
This article is dedicated to the memory of A. Bouquet and D. Fournand.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103929/DC1.
References
- Adam-Blondon, A. F., C. Roux, D. Claux, G. Butterlin, D. Merdinoglu et al., 2004. Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor. Appl. Genet. 109 1017–1027. [DOI] [PubMed] [Google Scholar]
- Ageorges, A., L. Fernandez, S. Vialet, D. Merdinoglu, N. Terrier et al., 2006. Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 170 372–383. [Google Scholar]
- Aharoni, A., C. H. R. De Vos, M. Wein, Z. K. Sun, R. Greco et al., 2001. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28 319–332. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., S. E. D. El-Assal, G. Coupland and M. Koornneef, 1998. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde islands ecotypes of Arabidopsis thaliana. Genetics 149 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert, K. B., S. Grandillo and S. D. Tanksley, 1995. Fw-2.2: a major QTL controlling fruit weight is common to both red-fruited and green-fruited tomato species. Theor. Appl. Genet. 91 994–1000. [DOI] [PubMed] [Google Scholar]
- Aranzana, M. J., S. Kim, K. Y. Zhao, E. Bakker, M. Horton et al., 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo-García, R., L. Ruiz-Garcia, L. Bolling, R. Ocete, M. A. Lopez et al., 2006. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 15 3707–3714. [DOI] [PubMed] [Google Scholar]
- Barnaud, A., T. Lacombe and A. Doligez, 2006. Linkage disequilibrium in cultivated grapevine, Vitis vinifera L. Theor. Appl. Genet. 112 708–716. [DOI] [PubMed] [Google Scholar]
- Barritt, B. H., and J. Einset, 1969. The inheritance of three major fruit colors in grapes. J. Am. Soc. Hortic. Sci. 94 87–89. [Google Scholar]
- Basten, C. J., B. S. Weir and Z. B. Zeng, 2001. QTL Cartographer. Department of Statistics, North Carolina State University, Raleigh, NC.
- Boitard, S., J. Abdallah, H. de Rochambeau, C. Cierco-Ayrolles and B. Mangin, 2006. Linkage disequilibrium interval mapping of quantitative trait loci. BMC Genomics 7 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J. O., J. N. Maloof, J. Lutes, T. Dabi, J. L. Redfern et al., 2002. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss, P. K., C. Davies and S. P. Robinson, 1996. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquet, A., L. Torregrossa, P. Iocco and M. R. Thomas, 2008. Grapes, pp. 189–232 in Compendium of Transgenic Crop Plants: Transgenic Temperate Fruits and Nuts, edited by C. Kole and T. Hall. Blackwell Publishing, Oxford.
- Chagne, D., C. M. Carlisle, C. Blond, R. K. Volz, C. J. Whitworth et al., 2007. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, S., S. M. Cocciolone, S. Bushman, V. Sangar, M. D. McMullen et al., 2003. The maize unstable factor for orange1 is a dominant epigenetic modifier of a tissue specifically silent allele of pericarp color1. Genetics 163 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., F. A. Burr and B. Burr, 1986. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., S. M. Cocciolone, F. A. Burr and B. Burr, 1993. Maize anthocyanin regulatory gene Pl is a duplicate of C1 that functions in the plant. Plant Cell 5 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Givry, S., M. Bouchez, P. Chabrier, D. Milan and T. Schiex, 2005. CAR(H)(T)AGene: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21 1703–1704. [DOI] [PubMed] [Google Scholar]
- Deluc, L., J. Bogs, A. R. Walker, T. Ferrier, A. Decendit et al., 2008. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and A. Stec, 1993. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics 134 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and R. L. Wang, 1997. Genetics and the evolution of plant form: an example from maize. Cold Spring Harbor Symp. Quant. Biol. 62 361–367. [PubMed] [Google Scholar]
- Doligez, A., A. Bouquet, Y. Danglot, F. Lahogue, S. Riaz et al., 2002. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor. Appl. Genet. 105 780–795. [DOI] [PubMed] [Google Scholar]
- Doligez, A., A. F. Adam-Blondon, G. Cipriani, V. Laucou, D. Merdinoglu et al., 2006. a An integrated SSR map of grapevine based on five mapping populations. Theor. Appl. Genet. 113 369–382. [DOI] [PubMed] [Google Scholar]
- Doligez, A., E. Audiot, R. Baumes and P. This, 2006. b QTLs for muscat flavor and monoterpenic odorant content in grapevine (Vitis vinifera L.). Mol. Breed. 18 109–125. [Google Scholar]
- Evanno, G., S. Regnaut and J. Goudet, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 2611–2620. [DOI] [PubMed] [Google Scholar]
- Flanzy, C., 1998 Les composés phénoliques in Oenologie: Fondements Scientifiques et Technologiques, Tec & Doc, Paris.
- Flint-Garcia, S. A., J. M. Thornsberry and E. S. Buckler, 2003. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54 357–374. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia, S. A., A. C. Thuillet, J. M. Yu, G. Pressoir, S. M. Romero et al., 2005. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 44 1054–1064. [DOI] [PubMed] [Google Scholar]
- Fournand, D., A. Vicens, L. Sidhoum, J. M. Souquet, M. Moutounet et al., 2006. Accumulation and extractability of grape skin tannins and anthocyanins at different advanced physiological stages. J. Agric. Food Chem. 54 7331–7338. [DOI] [PubMed] [Google Scholar]
- Francia, E., D. Barabaschi, A. Tondelli, G. Laido, F. Rizza et al., 2007. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor. Appl. Genet. 115 1083–1091. [DOI] [PubMed] [Google Scholar]
- Frary, A., T. C. Nesbitt, A. Frary, S. Grandillo, E. van der Knaap et al., 2000. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289 85–88. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez, S. C., N. C. Wheeler, E. Ersoz, C. D. Nelson and D. B. Neale, 2007. Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia, D., F. L. G. Bertolucci, R. Penchel and R. R. Sederoff, 1996. Genetic mapping of quantitative trait loci controlling growth and wood quality traits in Eucalyptus grandis using a maternal half-sib family and RAPD markers. Genetics 144 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn, J. N., and M. J. Daly, 2005. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6 95–108. [DOI] [PubMed] [Google Scholar]
- Holland, J. B., 2007. Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 10 156–161. [DOI] [PubMed] [Google Scholar]
- Holton, T. A., and E. C. Cornish, 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon, O., J. M. Aury, B. Noel, A. Policriti, C. Clepet et al., 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–465. [DOI] [PubMed] [Google Scholar]
- Joseph, J. A., B. Shukitt-Hale and G. Casadesus, 2005. Reversing the deleterious effects of aging on neuronal communication and behavior: beneficial properties of fruit polyphenolic compounds. Am. J. Clin. Nutr. 81 313S–316S. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., M. Ishimaru, K. Hiraoka and C. Honda, 2002. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215 924–933. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., N. Goto-Yamamoto and H. Hirochika, 2005. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin-color mutants. J. Jpn. Soc. Hortic. Sci. 74 196–203. [Google Scholar]
- Lacombe, T., J. M. Boursiquot, V. Laucou, F. Dechesne, D. Vares et al., 2007. Relationships and genetic diversity within the accessions related to malvasia held in the Domaine de Vassal grape germplasm repository. Am. J. Enol. Vitic. 58 124–131. [Google Scholar]
- Lagercrantz, U., J. Putterill, G. Coupland and D. Lydiate, 1996. Comparative mapping in Arabidopsis and Brassica, fine scale genome collinearity and congruence of genes controlling flowering time. Plant J. 9 13–20. [DOI] [PubMed] [Google Scholar]
- Le Cunff, L., A. Fournier-Level, V. Laucou, S. Vezzulli, T. Lacombe et al., 2008. Construction of nested genetic core collections to optimize the exploitation of natural diversity in Vitis vinifera L. subsp sativa. BMC Plant Biol. 8 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. A., P. F. Byrne, M. D. McMullen, M. E. Snook, B. R. Wiseman et al., 1998. Genetic mechanisms underlying apimaysin and maysin synthesis and corn earworm antibiosis in maize (Zea mays L.). Genetics 149 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky, D., L. Ruiz-Garcia, J. A. Cabezas, M. T. De Andres, G. Bravo et al., 2006. Molecular genetics of berry colour variation in table grape. Mol. Genet. Genomics 276 427–435. [DOI] [PubMed] [Google Scholar]
- Masle, J., S. R. Gilmore and G. D. Farquhar, 2005. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870. [DOI] [PubMed] [Google Scholar]
- Matus, J. T., R. Loyola, A. Vega, A. Pena-Neira, E. Bordeu et al., 2009. Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J. Exp. Bot. 60 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza, D., 1995. Anthocyanins in grapes and grapes products. Crit. Rev. Food Sci. Nutri. 35 341–371. [DOI] [PubMed] [Google Scholar]
- McMullen, M. D., P. F. Byrne, M. E. Snook, B. R. Wiseman, E. A. Lee et al., 1998. Quantitative trait loci and metabolic pathways. Proc. Natl. Acad. Sci. USA 95 1996–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia, N., M. Gebauer, L. Munoz, N. Hewstone, C. Munoz et al., 2007. Identification of QTLs for seedlessness, berry size, and ripening date in a \seedless × seedless table grape progeny. Am. J. Enol. Vitic. 58 499–507. [Google Scholar]
- Mol, J., E. Grotewold and R. Koes, 1998. How genes paint flowers and seeds. Trends Plant Sci. 3 212–217. [Google Scholar]
- Neale, D. B., and O. Savolainen, 2004. Association genetics of complex traits in conifers. Trends Plant Sci. 9 325–330. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., and D. Weigel, 2008. Next-generation genetics in plants. Nature 456 720–723. [DOI] [PubMed] [Google Scholar]
- Osterberg, M. K., O. Shavorskaya, M. Lascoux and U. Lagercrantz, 2002. Naturally occurring indel variation in the Brassica nigra COL1 gene is associated with variation in flowering time. Genetics 161 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. H., 2006. Believe it or not, QTLs are accurate! Trends Plant Sci. 11 213–216. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Z. H. S., T. H. Niu and J. S. Liu, 2002. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am. J. Hum. Genet. 71 1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., W. Verweij, A. Kroon, C. Spelt, J. Mol et al., 2006. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland, K., 1996. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res. 67 175–185 [Google Scholar]
- Ron, M., and J. I. Weller, 2007. From QTL to QTN identification in livestock—winning by points rather than knock-out: a review. Anim. Genet. 38 429–439. [DOI] [PubMed] [Google Scholar]
- Salvi, S., and R. Tuberosa, 2005. To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci. 10 297–304. [DOI] [PubMed] [Google Scholar]
- Segura, V., C. Cilas and E. Costes, 2008. Dissecting apple tree architecture into genetic, ontogenetic and environmental effects: mixed linear modelling of repeated spatial and temporal measures. New Phytol. 178 302–314. [DOI] [PubMed] [Google Scholar]
- Sparvoli, F., C. Martin, A. Scienza, G. Gavazzi and C. Tonelli, 1994. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis-Vinifera L). Plant Mol. Biol. 24 743–755. [DOI] [PubMed] [Google Scholar]
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalma, S. J., E. S. Buckler, M. E. Snook and M. D. McMullen, 2005. Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor. Appl. Genet. 110 1324–1333. [DOI] [PubMed] [Google Scholar]
- This, P., T. Lacombe, M. Cadle-Davidson and C. L. Owens, 2007. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor. Appl. Genet. 114 723–730. [DOI] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Torregrosa, L., C. Verries and C. Tesniere, 2002. Grapevine (Vitis vinifera L.) promoter analysis by biolistic-mediated transient transformation of cell suspensions. Vitis 41 27–32. [Google Scholar]
- Van Ooijen, J. W., M. P. Boer, R. C. Jansen and C. Maliepaard, 2002. MapQTL 4.0, Software for the Calculation of QTL Positions on Genetic Maps. Plant Research International, Wageningen, Netherlands.
- Velasco, R., A. Zharkikh, M. Troggio, D. A. Cartwright, A. Cestaro et al., 2007. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2 e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, J., J. Kikkert, B. Donzelli, P. Wallace and B. Reisch, 2006. Biolistic transformation of grapevine using minimal gene cassette technology. Plant Cell Rep. 25 807–814. [DOI] [PubMed] [Google Scholar]
- Walker, A. R., E. Lee, J. Bogs, D. A. J. Mcdavid, M. R. Thomas et al., 2007. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49 772–785. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley, B., 2001. Flavonoid biosynthesis. A Colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. M., and E. S. Buckler, 2006. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 17 155–160. [DOI] [PubMed] [Google Scholar]
- Yu, J. M., G. Pressoir, W. H. Briggs, I. V. Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38 203–208. [DOI] [PubMed] [Google Scholar]
- Zhao, K. Y., M. J. Aranzana, S. Kim, C. Lister, C. Shindo et al., 2007. An Arabidopsis example of association mapping in structured samples. PloS Genet. 3 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]