Abstract
Telomere binding proteins protect chromosome ends from degradation and mask chromosome termini from checkpoint surveillance. In Saccharomyces cerevisiae, Cdc13 binds single-stranded G-rich telomere repeats, maintaining telomere integrity and length. Two additional proteins, Ten1 and Stn1, interact with Cdc13 but their contributions to telomere integrity are not well defined. Ten1 is known to prevent accumulation of aberrant single-stranded telomere DNA; whether this results from defective end protection or defective telomere replication is unclear. Here we report our analysis of a new group of ten1 temperature-sensitive (ts) mutants. At permissive temperatures, ten1-ts strains display greatly elongated telomeres. After shift to nonpermissive conditions, however, ten1-ts mutants accumulate extensive telomeric single-stranded DNA. Cdk1 activity is required to generate these single-stranded regions, and deleting the EXO1 nuclease partially suppresses ten1-ts growth defects. This is similar to cdc13-1 mutants, suggesting ten1-ts strains are defective for end protection. Moreover, like Cdc13, our analysis reveals Ten1 promotes de novo telomere addition. Interestingly, in ten1-ts strains at high temperatures, telomeric single-stranded DNA and Rad52-YFP repair foci are strongly induced despite Cdc13 remaining associated with telomeres, revealing Cdc13 telomere binding is not sufficient for end protection. Finally, unlike cdc13-1 mutants, ten1-ts strains display strong synthetic interactions with mutations in the POLα complex. These results emphasize that Cdc13 relies on Ten1 to execute its essential function, but leave open the possibility that Ten1 has a Cdc13-independent role in DNA replication.
GENOME stability is critically dependent upon functional telomeres. DNA ends that lack telomeres, or that have dysfunctional telomeres, are metabolized by DNA repair processes; without an appropriate repair template, such chromosome ends can be resected or joined inappropriately with other chromosome ends. Thus, genomic integrity can be significantly compromised by telomere dysfunction, particularly in proliferating cells where cycles of instability may ensue due to creation of dicentric chromosomes (Bailey and Murnane 2006). Protein complexes that bind to the duplex and single-stranded telomere repeats are key for stabilizing the chromosome ends (de Lange 2005). In proliferating cells, this job is complicated not only because the terminal chromatin must be opened during the process of chromosome replication, but also because additional processes that metabolize DNA ends are active. For example, while nonhomologous end joining processes are preferentially used in repair of DNA double-strand breaks in G1, homologous recombination is preferentially used for this repair in S and G2 (Ferreira and Cooper 2004; Zierhut and Diffley 2008). Given these complexities, it is not surprising that our molecular understanding of how telomere proteins protect chromosomes ends is incomplete.
Budding yeast has been useful for dissecting how cells correctly metabolize their chromosome ends. In Saccharomyces cerevisiae, the terminal DNA comprises approximately 300 bp of TG1-3/C1-3A sequences, ending with a short single-stranded overhang of the G-rich repeats. This 3′ overhang is ∼12–14 nucleotides, although during the late S/G2 phase of the cell cycle, it becomes longer, >30 nucleotides in length (Wellinger et al. 1993b; Dionne and Wellinger 1996; Larrivee et al. 2004). Central among factors that prevent inappropriate telomere degradation in S. cerevisiae is Cdc13, a protein that binds to single-stranded telomere G-rich repeats (Garvik et al. 1995; Lin and Zakian 1996; Nugent et al. 1996). Reducing Cdc13 function through either the cdc13-1 temperature sensitive (ts) allele or the cdc13-td conditional null (degron) allele results in telomere C-strand loss, with degradation continuing into the subtelomeric chromosomal regions (Garvik et al. 1995; Vodenicharov and Wellinger 2006). Correspondingly, homologous recombination at chromosome termini increases in cdc13-1 strains (Carson and Hartwell 1985; Garvik et al. 1995). The loss of Cdc13 unmasks the telomeres, provoking activation of the DNA damage checkpoint (Weinert and Hartwell 1993; Garvik et al. 1995). This protective role of Cdc13 is most likely its essential function.
A thorough, mechanistic understanding of how Cdc13 mediates chromosome end protection is hampered in part because the activities responsible for the loss of the telomere C strand are not fully known. At normal telomeres, the Mre11-Rad50-Xrs2 complex has a role regulating resection required for telomere addition, whereas the Exo1 nuclease, Rad9 and Rad24 checkpoint proteins each influence the resection process at uncapped telomeres (Lydall and Weinert 1995; Maringele and Lydall 2002; Larrivee et al. 2004; Zubko et al. 2004). The 5′-to-3′ resection of both normal and uncapped telomeres is regulated by the activity of Cdk1, the yeast cyclin-dependent kinase (Frank et al. 2006; Vodenicharov and Wellinger 2006). Similar to the activities that promote 5′-to-3′ degradation of DNA ends at double-strand breaks (Aylon et al. 2004; Ira et al. 2004), the activities that lead to telomere resection are active in late S and G2 cell cycle phases (Wellinger et al. 1993a, 1996; Marcand et al. 2000; Vodenicharov and Wellinger 2006). Interestingly, Cdc13 is required to prevent degradation at telomeres only in proliferating cells and not when cells are blocked in stationary phase (Vodenicharov and Wellinger 2006). Additional factors, such as the S. cerevisiae Rap1 protein, prevent chromosome fusions by nonhomologous recombination during the G1 phase of the cell cycle (Pardo and Marcand 2005; Marcand et al. 2008).
At least two additional proteins, Stn1 and Ten1, aid the capping role of Cdc13. Like CDC13, both STN1 and TEN1 are essential, and loss of their function leads to excessive single-stranded telomeric DNA (Grandin et al. 1997, 2001; Petreaca et al. 2007). STN1 was originally identified as a high copy suppressor of cdc13-1 temperature sensitivity (Grandin et al. 1997), and TEN1 was similarly isolated as a dosage suppressor of stn1-13 (Grandin et al. 2001). Combining either the cdc13-1 allele with stn1 mutations or the ten1-31 allele with stn1-13 is lethal (Grandin et al. 2001; Petreaca et al. 2007). The essential nature of these genes makes it difficult to clearly differentiate whether these genes operate in the same, or in parallel pathways to protect telomeres. A compelling argument that Cdc13, Stn1, and Ten1 likely function in a common pathway is that, in addition to these genetic interactions, Stn1 and Ten1 proteins interact with one another both in vivo and in vitro (Grandin et al. 2001; Gao et al. 2007), and each associates with Cdc13 in the yeast two-hybrid assay (Grandin et al. 1997, 2001; Petreaca et al. 2007). From these data, Cdc13, Stn1, and Ten1 are suggested to function as a single complex that mediates chromosome end protection in S. cerevisiae. Such a complex would share some similarities with the single-stranded DNA binding complex RPA (Gao et al. 2007). Whether these proteins normally operate exclusively as a heterotrimeric complex is still not entirely clear. Stn1 and Ten1 can make contributions to capping that are independent of Cdc13, as shown in experiments where overproducing the Stn1 essential domain with Ten1 replaced the essential function of Cdc13 (Petreaca et al. 2006). In addition, while the Schizosaccharomyces pombe Stn1 and Ten1 homologs are critical for telomere protection, they do not interact with Pot1, the single-stranded telomere binding protein that is also critical for telomere capping (Martin et al. 2007).
The role of Ten1 in maintaining both telomere integrity and length homeostasis is not understood. It has been assumed that Stn1 and Ten1 play the same role as Cdc13 in maintaining telomere integrity, namely, preventing inappropriate terminal resection. However, whether this is in fact the case is not entirely clear. For one, disrupting the DNA replication machinery can give rise to an excess of terminal single-stranded DNA, although in this case, the ssDNA accumulation is attributed to a failure to synthesize the lagging DNA strand rather than removing a block to telomere resection (Diede and Gottschling 1999; Adams Martin et al. 2000). Although both Cdc13 and Stn1 are thought to act as capping proteins, each can interact with Polα subunits (Qi et al. 2003; Grossi et al. 2004; Petreaca et al. 2006), making it important to evaluate Ten1 function more carefully. Our goal here was to compare how Cdc13 and Ten1 promote chromosome end protection, first by testing whether Ten1 acts to prevent telomere resection from activities comparable to those that degrade telomeres in cdc13-1, and second by determining the impact of ten1 dysfunction upon Cdc13. The cdc13-1 allele has been extremely useful in analyzing the CDC13 essential function; TEN1 analysis has been hindered by a lack of equivalent genetic reagents. Here we have created a collection of ten1-ts alleles useful for probing the essential role of TEN1. Analysis of these alleles, which show constitutive telomere elongation, reveals that Ten1 promotes telomere capping with a similar cell cycle dependency as Cdc13, protecting ends during the period in which mitotic forms of Cdk1 are active. Critically, by showing that single-stranded DNA is generated in ten1-ts strains under conditions where semi-conservative replication is complete, we conclude that Ten1 truly can function as a capping protein. Moreover, the ten1-ts strains fail to restrain degradation of chromosome ends and induce formation of Rad52 repair foci, despite the association of wild-type Cdc13 with telomeres, indicating not only that Cdc13 binds telomeres independent of Ten1 function, but also that Cdc13 telomere localization is not sufficient for end protection. Finally, although the ten1-ts capping-deficient phenotypes parallel cdc13-1, only the ten1-ts strains are highly sensitive to impaired POL1 function, leaving open the possibility that TEN1 function additionally impacts terminal replication.
MATERIALS AND METHODS
Plasmids:
Plasmids used in this study are listed in Table 1. Plasmids pCN284 and pCN250 each contain a 2-kb fragment surrounding the TEN1 open reading frame, with 1 kb of upstream and 0.6 kb of downstream sequences. This insert was amplified from genomic DNA using primers CO13 and CO14. BamHI and HindIII restriction sites within the primers were used to subclone this fragment into YCplac22 and pRS416. Plasmids containing the mutant ten1-ts alleles were created by PCR mutagenesis (below). The TEN1 two-hybrid plasmids were PCR amplified (oligos CO15 and CO16) and subcloned into pAS1.
TABLE 1.
Plasmid list
| Plasmid | Description | Reference |
|---|---|---|
| pCN250 | CEN URA3 native romoter TEN1 | This study |
| pCN284 | CEN TRP1 native promoter TEN1 | This study |
| pCN309 | CEN TRP1 native promoter ten1-101 | This study |
| pCN311 | CEN TRP1 native promoter ten1-103 | This study |
| pCN358 | CEN TRP1 native promoter ten1-105 | This study |
| pCN359 | CEN TRP1 native promoter ten1-106 | This study |
| pCN124 | 2μ TRP1 ADH promoter GAL4 DBD TEN1 | Petreaca et al. (2006) |
| pCN445 | 2μ TRP1 ADH promoter GAL4 DBD ten1-101 | This study |
| pCN441 | 2μ TRP1 ADH promoter GAL4 DBD ten1-105 | This study |
| pCN447 | 2μ TRP1 ADH promoter GAL4 DBD ten1-106 | This study |
| pCN452 | 2μ TRP1 ADH promoter GAL4 DBD ten1-103 | This study |
| pCN181 | 2μ LEU2 ADH promoter GAL4 AD STN1 | Petreaca et al. (2006) |
| pVL835 |
2μ LEU2 ADH promoter GAL4 AD CDC13Δ585-677 |
This study |
Strains:
Yeast strains were propagated following standard procedures. Strains are listed in Table 2. To knock out TEN1, the KANMX2 cassette was PCR amplified from pFA6-KanMX2 (Wach et al. 1994) using oligos CO1 and CO2 and transformed into a diploid strain. Correct knockouts were verified by PCR. The ten1Δ∷kanMX2/pCN250 haploids (hc558, hc1832) were obtained by dissecting the ten1Δ∷kanMX2/TEN1 [pCN250] diploid. The ten1-ts strains were obtained by transforming ten1-ts plasmids into hc1832 and shuffling out the pCN250 (TEN1 URA3) plasmid by plating cells on SD −Trp plates containing 1 mg/ml 5-fluoro-orotic acid (5-FOA). Double mutant strains were created through hc1832 matings, followed by tetrad dissection and plasmid shuffling.
TABLE 2.
Strain list
| Strain | Description | Reference |
|---|---|---|
| hc160 | MATaura3-52 ade2-101 lys2-801 leu2-Δ1 his3-Δ200 | Petreaca et al. (2006) |
| hc558 | MATα ten1Δ∷kanMX2 [pCN250] | This study |
| hc1832 | MATa ten1Δ∷kanMX2 [pCN250] | This study |
| hc1862 | MATa ten1Δ∷kanMX2 [pCN309] | This study |
| hc1863 | MATa ten1Δ∷kanMX2 [pCN311] | This study |
| hc1864 | MATa ten1Δ∷kanMX2 [pCN358] | This study |
| hc1865 | MATa ten1Δ∷kanMX2 [pCN359] | This study |
| hc1212 | MATα ten1Δ∷kanMX2 [pCN358] | This study |
| hc1676 | MATa cdc13-1myc18x ten1Δ∷kanMX2 [pCN250] | This study |
| hc1722 | MATa rad52Δ∷LYS2 ten1Δ∷kanMX2 [pCN250] | This study |
| hc1723 | MATa est2Δ∷kanMX2 ten1Δ∷kanMX2 [pCN250] | This study |
| hc2025 | MATa ten1Δ∷kanMX2 [pCN250, pCN284] | This study |
| hc2026 | MATa ten1Δ∷kanMX2 [pCN250, pCN358] | This study |
| hc2027 | MATa rad52Δ∷LYS2 ten1Δ∷kanMX2 [pCN250, pCN284] | This study |
| hc2028 | MATa rad52Δ∷LYS2 ten1Δ∷kanMX2 [pCN250, pCN309] | This study |
| hc2029 | MATa cdc13-1myc18x ten1Δ∷kanMX2 [pCN250, pCN284] | This study |
| hc2030 | MATa cdc13-1myc18x ten1Δ∷kanMX2 [pCN250, pCN358] | This study |
| hc2031 | MATa est2Δ∷kanMX2 ten1Δ∷kanMX2 [pCN250, pCN284] | This study |
| hc2032 | MATa est2Δ∷kanMX2 ten1Δ∷kanMX2 [pCN250, pCN358] | This study |
| hc2009 | MATa rad52Δ∷LYS2 | This study |
| hc1841 | MATa rad52Δ∷LYS2 ten1Δ∷kanMX2 [pCN358] | This study |
| hc1848 | MATa rad52Δ∷LYS2 ten1Δ∷kanMX2 [pCN309] | This study |
| hc1892 | MATa rad52Δ∷LEU2 GALHO∷LEU2 TG1-3HOsite∷LYS2 ten1Δ∷kanMX2 [pCN250] | This study |
| hc1943 | MATa rad52Δ∷LEU2 GALHO∷LEU2 TG1-3HOsite∷LYS2 ten1Δ∷kanMX2 [pCN284] | This study |
| hc1946 | MATa rad52Δ∷LEU2 GALHO∷LEU2 TG1-3HOsite∷LYS2 ten1Δ∷kanMX2 [pCN358] | This study |
| hc1841 | MATa rad52Δ∷LYS2 ten1Δ∷kanMX2 [pCN250,pCN358] | This study |
| hc18 | MATa yku80Δ∷kanMX2 CDC13myc18x∷HIS3 | Petreaca et al. (2006) |
| hc1997 | MATa cdc13-1 | This study |
| hc1998 | MATa cdc13-1 cdc28-as1 | This study |
| hc2000 | MATa ten1Δ∷kanMX2 cdc28-as1 [pCN250] | This study |
| hc2005 | MATa ten1Δ∷kanMX2 cdc28-as1 [pCN358] | This study |
| hc2035 | MATa CDC13myc18x∷HIS3 ten1Δ∷kanMX2 [pCN309] | This study |
| hc1721 | Mat a RAD52-YFP ten1Δ∷kanMX2 [pCN284] | This study |
| hc1840 | Mat a RAD52-YFP ten1Δ∷kanMX2 [pCN358] | This study |
| hc 579 | Mat a exo1Δ∷kanMX2 | This study |
| hc1970 | Mat a exo1Δ∷kanMX2 ten1Δ∷kanMX2 [pCN309] | This study |
| hc1972 | Mat a exo1Δ∷kanMX2 ten1Δ∷kanMX2 [pCN358] | This study |
| Hc1989 |
Mat a exo1Δ∷kanMX2 cdc13-1 |
This study |
Screen for ten1ts alleles:
Random mutations in TEN1 were created by PCR amplifying genomic DNA with oligos CO13, CO14 under mutagenic conditions. Each 25-μl PCR reaction contained ∼5 ng of genomic DNA in 2.5 mm MgCl2, 0.05 mm MnCl2, with 0.02 mm each of dATP and dGTP, 1.0 mm each of dCTP and dTTP, 1-μM primers, and 10 units Taq DNA polymerase (New England Biolabs). The reaction had an initial denaturation step of 2 min at 94°, followed by 35 cycles of 1 min at 94°, 2 min at 45°, 1.5 min at 72°, and finished with a final 10 min extension at 72°. pCN284 was digested with PpuMI and BsmI, removing a 0.6-kb fragment that contains the TEN1 ORF. This “gapped” plasmid was purified and cotransformed with the PCR product into hc558, allowing recombination in vivo. Colonies on SD −Trp plates were streaked on SD −Trp 5-FOA, and tested for temperature sensitivity. pCN250 was retransformed into the ts candidates to confirm complementation by TEN1. ten1-ts plasmids were rescued and sequenced; each allele contains at least one mutation that alters the Ten1 protein sequence. Two alleles, ten1-101 and ten-105, were integrated into the genome. The integrated alleles show phenotypes similar to the plasmid-borne alleles (see supporting information, Figure S1 and Figure S2).
Serial dilutions:
Cultures inoculated from single colonies were incubated 4 days at 23°. For each strain, 10-fold serial dilutions from the same initial concentration of cells were done in a 96-well microtiter dish and stamped onto appropriate plates. Plates were incubated 4 days at 23°, 3 days at 30° or 36° before pictures were taken.
Southern blot:
Genomic DNA was prepared as described from cultures incubated at 23° for 4 days (Lundblad and Szostak 1989). For blots analyzing telomere restriction fragments, the genomic DNA was digested with XhoI, fragments separated on 0.8% agarose gel, then transferred to a Hybond-XL membrane (Amersham). Blots were probed with [32P]-dGT/CA and exposed on film.
Colony plating efficiency:
Cells were inoculated into 5-ml SD −Trp media and incubated for 3–4 days. Cultures were then diluted and sonicated, and the concentration determined using a hemocytometer. Three hundred cells were plated on SD −Trp plates and 104 cells were plated on SD −Trp 5-FOA plates; for wild-type strains, 103 cells were plated on SD −Trp 5-FOA. The efficiency of colony formation on SD −Trp 5-FOA plates was normalized to the colony efficiency on SD −Trp plates, using the following calculation: CE5FOA = number of colonies on SD −Trp 5-FOA/[104 × (number of colonies on SD−Trp/300)].
Telomere healing assay:
The telomere healing assay was performed as described (Diede and Gottschling 1999). Cells were incubated in SD −Lys media overnight and then arrested in YP-raffinose with 15 mg/liter nocodazole for 3 hr until >80% of cells were arrested as large-budded cells. Galactose was added into the media to 3%, and the cells were harvested at 0-, 1-, 3-, and 5-hr time points. The nocodazole arrest was maintained throughout the experiment. Genomic DNA was prepared from cells at each time point and was digested overnight with SpeI. Southern blots were done to visualize the DNA bands, probing with [32P]-ADE2 as in Diede and Gottschling (1999). The signal on the blots was quantified with National Institutes of Health (NIH) Image J. The signals from the uncut and cut ADE2 bands at each time point were added and normalized to the signal of the 0-hr time point. To determine the fraction of viable Ade+Lys− cells, aliquots from time points were spread on SD −Ade plates and grown at 23°. Colonies were replica-plated to SD −Ade and SD −Ade −Lys media.
Single-stranded TG repeat analysis:
Cells were incubated at 23° overnight until OD600 was ∼1.0. Equivalent amounts of cells were shifted to the indicated temperatures (30° or 36°) for 4 hr. For assays with synchronized cell cultures, cells were treated for 3 hours at 23° with α-factor to arrest in G1, with 200 mm hydroxyurea (HU) to arrest in S phase, and with 15 mg/liter nocodazole to arrest in G2/M phase. Only after cells had arrested were they were shifted to different temperatures. After the temperature shift, additional α-factor, HU, or nocodazole was added to the culture to keep the cells arrested. For cdc28-as1 strains, following the arrest in G2/M induced by nocodazole treatment, 5 μM 1-NMPP1 (Calbiochem) was added to the culture and incubated at 23° for 1 hr before the temperature was shifted. Genomic DNA was prepared as described (Dionne and Wellinger 1996). One hundred micrograms of the genomic DNA was incubated with ExoI (New England Biolabs and USB) or mock treated with buffer prior to XhoI digestion. Digested DNA was separated on a 0.75% agarose gel and the nondenatured gel was probed with a [32P]-dCA oligo (Dionne and Wellinger 1996). After exposure of the gel, it was denatured in a high pH buffer and reprobed with [32P]-dCA oligo to measure total amount of TG1-3 repeats. The gels were exposed on an Amersham phosphorimager screen.
Chromatin Immunoprecipitation:
Chromatin immunoprecipitations (ChIPs) were performed as previously described (Strahl-Bolsinger et al. 1997), with the following modifications. Two-hundred-milliliter cells were grown into OD600 ∼1, then cultures were split into four 50-ml cultures. Two cultures were treated with 15 mg/liter nocodazole for 4 hr, with one culture at 23° and the other at 36°, while the two other 50-ml cultures were diluted to an OD600 = 0.4 and grown at 23° or 36° for 4 hr (asynchronous cultures). Cells were lysed in ChIP lysis buffer (50 mm HEPES/KOH pH 7.5, 150 mm NaCl, 1mm EDTA, 1% Triton X-100, 0.1% Na-deoxycholate), supplemented with protease inhibitors (1 mm PMSF, 1 mm benzamidine, 1mg/ml bacitracin). Ten percent of the lysate was removed for input normalization and the rest was subjected to immunoprecipitation. Immunoprecipitations were performed in 1 ml volume in siliconized tubes. Lysates were incubated with 4 μl/ml antibody (mouse monoclonal anti-MYC 9E10) overnight, followed by a 2-hr incubation with protein A/G-Plus agarose beads (40-μl beads/sample). Beads were washed sequentially with ChIP lysis buffer, ChIP wash buffer (10 mm Tris–HCl pH 8, 0.25 m LiCl, 0.5% NP-40, 0.5% Na-deoxycholate) and 1× Tris–EDTA. DNA was isolated using a QIAGEN PCR purification kit. Tenfold serial dilutions of the isolated DNA were dot blotted onto nitrocellulose membrane and hybridized with [32P]dGT/CA, the same probe used in telomere Southern blots. The blot was exposed on a Phosphoimager screen (Amersham) and quantified using the Amersham Typhoon 9410.
Western blot:
Twenty-five-milliliter cell cultures were incubated at 23° overnight until the OD600 was ∼1.0. Cells were pelleted and resuspended in 20% TCA with proteinase inhibitors, and then lysed with glass beads in a bead beater. Fifty microliters of the cell lysate was loaded onto SDS-polyacrylamide gels. For the HA-Ten1, cells were lysed using a bead beater in Buffer A (25 mm HEPES, PH 7.5, 5 mm MgCl2, 150 mm KCl, 10% glycerol, and 0.5% Triton-X 100) with proteinase inhibitors; 100 μg of each cell lysate was loaded onto SDS-polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (Pierce) and probed with either an anti-myc primary antibody (9E10, Covance/Babco) or an anti-FLAG antibody (Sigma), followed by goat anti-mouse HRP conjugated secondary antibody. For loading controls, the blots were stripped and reprobed with an anti-Rad53 antibody (Santa Cruz). The Perkin Elmer Renaissance chemiluminescence reagent was used to detect the secondary antibody, and blots were exposed to Fuji film.
Yeast two hybrid:
TEN1 two-hybrid plasmids were cotransformed into pJB694a (LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ) (James et al. 1996) with pCN181 (pACT-STN1) or with pVL835 (pACT-CDC13Δ585-677). Colonies were inoculated in 1 ml of SD −Leu −Trp liquid media and incubated 1 day at 23°. Serial 10-fold dilutions of the cultures were then stamped onto SD −Leu −Trp, SD −Leu −Trp −Ade, and SD −Leu −Trp −His + [1mm] 3AT plates to examine ADE2 and HIS3 reporter gene expression. Plates were incubated 3–4 days at 23°, 30°, 32°, and 36°.
RESULTS
ten1-ts strains show conditional growth and constitutive interaction defects:
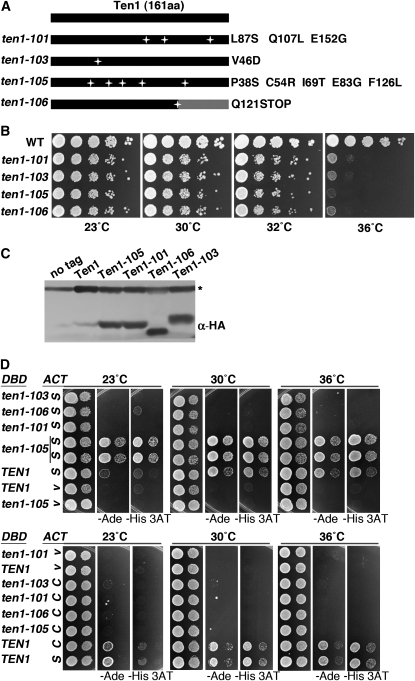
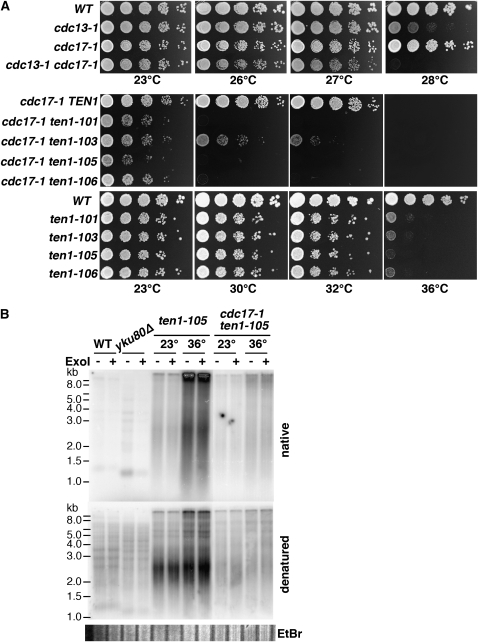
To develop TEN1 reagents useful for examining its role in telomere capping, a PCR mutagenesis strategy was used to generate temperature-sensitive ten1 alleles. Linear TEN1 PCR products generated under mutagenic conditions were cotransformed into cells with a linear vector containing the genomic regions flanking TEN1 at each end of the plasmid. DNA gap repair in vivo between the homologous sequences on the plasmid and the PCR fragments results in a circular plasmid encoding the PCR amplified TEN1. The four ten1-ts alleles that were chosen for further analysis show reproducible temperature-sensitive growth phenotypes at 36°, with maximum permissive temperatures of ∼32° (Figure 1, A and B). The growth defect of each allele is recessive, being fully complemented by one wild-type copy of TEN1 (data not shown). The ten1-105 allele has the most severe ts growth defect; five missense mutations are distributed throughout this protein. The ten1-106 allele has a mutation that introduces a stop codon at amino acid 121, effectively truncating 40 amino acids from the Ten1 carboxyl terminus. The only Ten1 residue altered in common with other published alleles is Q107; in ten1-3, Q107R was the sole lesion, and the strain showed telomere length alterations but no growth defects (Grandin et al. 2001). While conducting plasmid shuffle experiments, where cells retaining only the mutant ten1-ts plasmid are selected on plates containing 5-FOA, we noted that at 23° the plating efficiency of the ten1Δ/pten1-ts mutant strains was >10-fold lower than that of ten1Δ/pTEN1 (Table 3). This indicates that there must be some initial impediment to the viability of the ten1-ts strains that is overcome in the cells surviving on the 5-FOA plates.
Figure 1.—
ten1-ts alleles show progressive telomere elongation at permissive temperature. (A) Diagram of mutations in PCR-generated ten1 alleles. (B) Growth phenotype of ten1-ts alleles. Serial 10-fold dilutions were prepared from equivalent cell concentrations and stamped onto YPD plates. The plates were incubated at the indicated temperatures for 3–4 days. WT (wild-type, hc160), ten1Δ/pTEN1 (hc1832), ten1-101 (hc1862), ten1-103 (hc1863), ten1-105 (hc1864), and ten1-106 (hc1865). (C) Western blot showing expression of Ten1-ts proteins. The Ten1-ts two-hybrid plasmids were transformed into wild-type cells (hc160) and incubated at 30° overnight until the OD600 reached 0.8–1.0. One hundred micrograms of each cell lysate was run on a 10% SDS–PAGE gel, and each HA-Gal4DBD-Ten1-ts fusion protein was detected by anti-HA antibody (12CA5). A cross-reacting band present in the lysates is marked with an asterisk. Strains: No tag (hc160), Ten1 (pCN124), Ten1-101 (pCN445), Ten1-103 (pCN452), Ten1-105 (pCN441), and Ten1-106 (pCN447). (D) Yeast two-hybrid analysis of interaction of Ten1-ts with Stn1 and Cdc13. Ten1 and Ten1-ts proteins were expressed as a Gal4 DNA binding domain fusion (pCN124, pCN441, pCN445, pCN447, and pCN452). STN1 and CDC13 were expressed as Gal4 activation domain fusion proteins (S, pCN181; C, pVL835; V, vector, pACT2.2). An in-frame deletion removes a portion of the Cdc13 DNA binding domain in pVL835 (CDC13Δ585-677). Tenfold serial dilutions of cultures were stamped on SD −Leu −Trp, SD −Leu −Trp −Ade, and SD −Leu −Trp −His +1 mM 3AT plates to examine the expression of the ADE2 and HIS3 reporter genes.
TABLE 3.
Relative plating efficiencies of ten1-ts strains at 23° (percentage)
| Strain genotype |
TRP1 Plasmid |
||||
|---|---|---|---|---|---|
| TEN1 | ten1-101 | ten1-103 | ten1-105 | ten1-106 | |
| RAD52 ten1Δ | 47 ± 24 | 2.1 ± 0.8 | 1.0 ± 0.6 | 1.2 ± 1.0 | 0.6 ± 0.6 |
| rad52Δ ten1Δ | 40 ± 15 | 0.5 ± 0.5 | 0.2 ± 0.3 | 0.2 ± 0.1 | 0.4 ± 0.6 |
|
exo1Δ ten1Δ |
26 ± 6 |
5.9 ± 1.7 |
5.1 ± 1.9 |
5.9 ± 4.6 |
2.6 ± 1.6 |
Cultures were grown at 23° in −Trp liquid media for 3 days, and then the cell density was calculated by counting with a hemocytometer. Three hundred cells were plated on −Trp media and 10,000 cells plated on −Trp 5-FOA media. The number of colonies growing on the plates was determined after 7 days of growth at 23°. Multiple cultures were tested for each strain (n = 6). The plating efficiency was calculated for all strains, and the median fraction of cells that grow on −Trp 5-FOA relative to −Trp plates is expressed as a percentage. Thus, for ten1-101, only ∼2% of the colonies containing the pten1-101-TRP1 plasmid are viable on −Trp 5-FOA plates.
Since Ten1 is known to associate with both Cdc13 and Stn1, a disrupted interaction among these proteins could provide a molecular explanation for the mutant phenotype. To determine whether any of the mutations in TEN1 disrupt these potentially essential interactions, a two-hybrid assay was performed. First, all four Ten1-ts fusion proteins are expressed at similar levels, with the Ten1-106 protein migrating as expected for a truncation (Figure 1C). Wild-type Ten1 protein expression is not as robust; it is not clear if this is a factor in the apparently weaker two-hybrid interactions of wild-type Ten1 at 23° (Figure 1D). The Ten1-103 protein reproducibly shows altered mobility on SDS–PAGE; it was recently shown that mutating the adjacent residue (R47E) also impacts Ten1 gel migration (Qian et al. 2009). Next, Ten1 interaction was tested with Stn1 and Cdc13 across a range of temperatures. Among these four mutants, only Ten1-105 was capable of interacting with Stn1, activating each of the three reporter genes at all temperatures (Figure 1D, and data not shown). The other mutant Ten1-ts proteins failed to activate any reporter genes. None of the Ten1 mutants are competent for Cdc13 interaction (Figure 1D). Since these interactions fail even at temperatures permissive for growth, the ten1-ts conditional growth defects are not likely to be solely attributable to a specific interaction defect.
ten1-ts strains have extremely elongated telomeres:
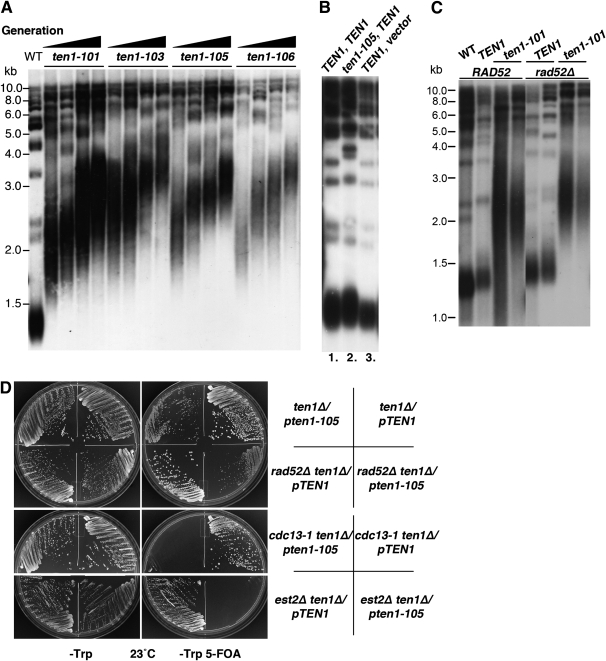
Each ten1-ts strain was found to exhibit extremely elongated and heterogeneous telomeres, even at temperatures permissive for growth (Figure 2A). Thus, the telomere length defect is not conditional. To determine whether the telomeres reach a new steady-state length or whether the mechanisms that negatively regulate telomere extension are deficient, leading to continual elongation as the cells are propagated, telomere lengths were examined in strains that were serially passaged for increasing numbers of generations (Figure 2A). These data show telomeres progressively elongate with successive generations, as examined here over ∼110 generations, and even continue lengthening beyond this point (data not shown). Thus, negative regulation of telomere length is severely crippled. This elongation phenotype is recessive, as shown in Figure 2B, where ten1-105 telomeres remain wild type in length if TEN1 is expressed from a CEN plasmid.
Figure 2.—
ten1-105 is synthetic lethal with cdc13-1 and est2-Δ, and progressively elongates telomere length independent of homologous recombination. (A) Telomere length increases as strains proliferate at 23°. The initial culture for each strain (1×) was inoculated from a colony arising on SD −Trp 5-FOA plates. To obtain successive generations of the ten1-ts strains, colonies from the SD −Trp 5-FOA plates were propagated by successive streak-outs on SD −Trp plates. To obtain cultures of later generations, cells were inoculated from colonies arising on these SD −Trp plates. Strains were cultured in liquid SD −Trp media at 23° for 3 days. Genomic DNA was digested with XhoI and the Southern blot was probed with 32P[dGT/CA]. WT (wild-type, hc160), ten1-101 (hc1862), ten1-103 (hc1863), ten1-105 (hc1864), and ten1-106 (hc1865). (B) Telomere length defect is recessive in ten1Δ/pTEN1, pten1-105 strains. Southern blot showing ten1Δ strains bearing the following plasmids: lane 1, pCN250 (TEN1) + pCN284 (TEN1); lane 2, pCN250 (TEN1) + pCN358 (ten1-105); lane 3, pCN284 (TEN1) + pCN416 (vector). Strains were grown at 23° for 4 days, and the Southern blot was performed as in A. (C) Southern blot comparing telomere length in ten1-101 and ten1-101 rad52Δ strains. Cell cultures were incubated at 23° for 4 days. Genomic DNA was digested with XhoI, and the blot was probed with 32P[dGT/CA]. Wild type (WT, hc160), ten1-101 (hc1862), rad52Δ TEN1 (hc1722), rad52Δ ten1-101 (hc1848). (D) Plates showing growth of ten1-105 double mutant strains that were streaked simultaneously on SD −Trp and SD −Trp 5-FOA plates. All strains initially contained the pTEN1-URA3 plasmid (pCN250); only cells losing this plasmid will grow on the SD −Trp 5-FOA plates. Plates were incubated at 23° for 5 days. Strains: hc2026, hc2025, hc2027, hc2028, hc2030, hc2029, hc2031, and hc2032.
Typically, telomerase activity is responsible for generating elongated telomeres in S. cerevisiae, although homologous recombination activities could also be involved. If telomerase is necessary for the ten1-ts telomere extension, then telomeres will remain short in telomerase-deficient strains. Conversely, if recombination is sufficient to generate and maintain the elongated ten1-ts telomeres, then the expectation is that loss of telomerase will not affect the length of telomeres. This was tested by creating a haploid strain, est2Δ ten1Δ/pTEN1-URA, pten1-ts-TRP1, which contains plasmids encoding both wild-type and mutant versions of the TEN1 gene. When the senescing strains were plated on media containing 5-FOA to select cells bearing only the mutant ten1-ts plasmid, no viable colonies were obtained at 23° (Figure 2D). Similar results were obtained for all four ten1-ts alleles (data not shown). These ten1-ts strains therefore show a synthetic phenotype with the est2Δ null allele, preventing further growth of the senescent strain. We infer from this data that these ten1-ts strains are not able to elongate their telomeres independently of telomerase activity. This conclusion would be consistent with a previous analysis of ten1 alleles, where loss of TLC1 in a GAL-ten1-31 strain led to telomere shortening (Grandin et al. 2001). The synthetic phenotype raises the possibility that the ten1-ts strains have a telomere capping deficiency at 23°, similar to cdc13-1 est2Δ strains (Nugent et al. 1996). In this case, the telomere shortening or lack of telomerase complex would be enhancing a ten1-ts protection defect present at permissive temperature.
To determine whether homologous recombination contributes to telomere length regulation, analogous plasmid shuffle experiments were conducted with rad52Δ ten1Δ strains. The ten1-ts rad52Δ strains are viable, with no change in their maximum permissive temperature (Figures 2D, 8A, Table 3). The telomeres in the ten1-ts strains become similarly elongated and heterogeneous in the absence of RAD52 (ten1-101 rad52Δ shown Figure 2C), agreeing with the conclusion that telomerase creates the elongated, heterogeneous telomeres in the ten1-ts strains.
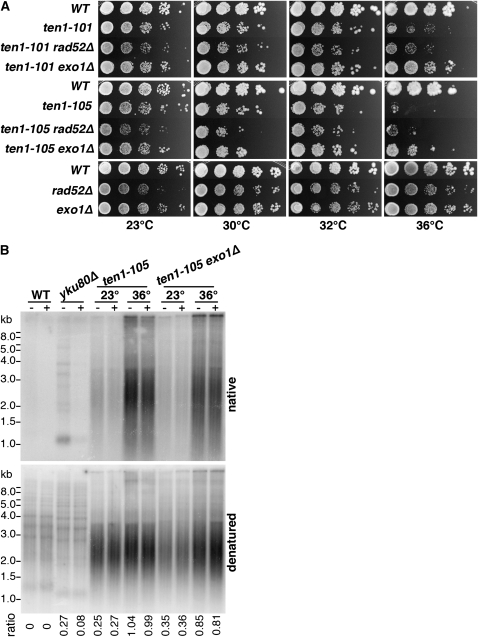
Figure 8.—
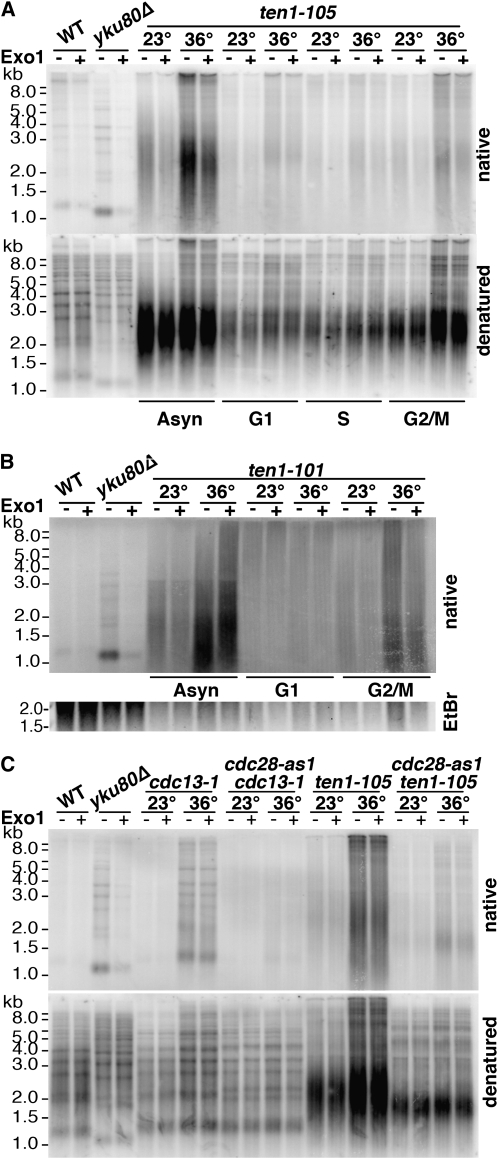
Suppression of ten1-ts temperature sensitivity and TG1-3 resection by exo1Δ. (A) Temperature sensitivity of ten1-101 and ten1-105 are suppressed by exo1Δ and not affected by rad52Δ. Serial 10-fold dilutions were plated on YPD and incubated at indicated temperatures. Wild-type (hc160), ten1-101 (hc1862), ten1-101 rad52Δ (hc1848), ten1-101 exo1Δ (hc1970), ten1-105 (hc1864), ten1-105 rad52Δ (hc1841), ten1-105 exo1Δ (hc1972), rad52Δ (hc2009), and exo1Δ (hc579). (B) exo1Δ reduces single-stranded TG1-3 generation in ten1-105 and cdc13-1 strains. Cultures were grown at 23°, then split, with half shifted to 36°. Incubation continued at 23° and 36° for 4 hr. Genomic DNA was then isolated and half was treated with ExoI prior to XhoI digestion. The upper panel shows the gel probed with a [32P]-CA oligo under native conditions, and the lower panel shows hybridization with the [32P]-CA oligo after gel denaturation. The number indicated below is the ratio of signal in the lane on the native gel/denatured gel, minus the ratio determined from the wild-type lanes. Strains: wild-type (hc160), yku80Δ (hc18), ten1-105 (hc1864), ten1-105 exo1Δ (hc1972), cdc13-1 (hc1997), and cdc13-1 exo1Δ (hc1989).
Cdc13 remains telomere associated in ten1-ts strains:
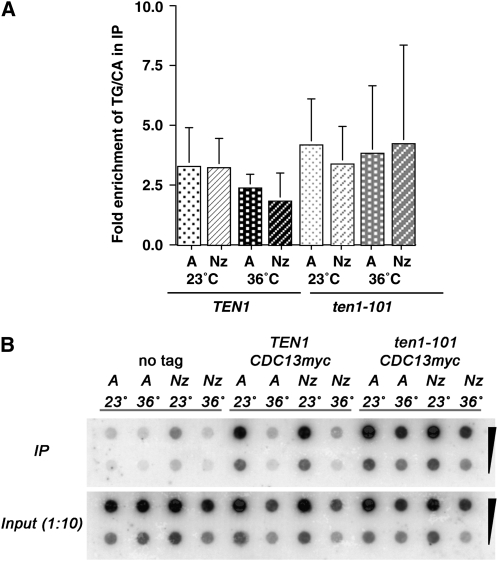
Consistent with the hypothesis that TEN1 functions in the same pathway as CDC13, we find that ten1-ts alleles have a synthetic lethal interaction with cdc13-1 (Figure 2D). It is possible that the compromised ten1 function directly affects Cdc13, leading to the similar capping-deficient phenotypes. Therefore, we next tested whether deficient ten1 function alters the extent of Cdc13 association with telomere chromatin. First, we determined that the total Cdc13 protein levels are similar in both wild-type and ten1-ts strains at permissive and restrictive temperatures (data not shown). Next, Cdc13 localization to telomere chromatin was tested by ChIP. Since the Cdc13-mediated protection from resection is critical in G2/M arrested cells, the immunoprecipitation of telomere sequences with Cdc13-myc18x was tested in both asynchronous and nocodazole arrested strains. The overall telomere length is quite different in TEN1 and ten1-ts strains; to correct for this, we quantified the amount of telomere repeat sequence present in the immunoprecipitate and the total lysate using dot blots hybridized with a telomere repeat probe. In addition, if Cdc13 localizes only to the ends of the telomeres, testing the association with the TG1-3 repeats rather than an adjacent subtelomeric locus removes caveats associated with the size of the sonication fragments generated in the ChIP procedure. Analysis of multiple experiments shows that Cdc13-myc18x remains associated with telomeres at 36° in ten1-101 cells at least as efficiently as in TEN1 strains (Figure 3, A and B). No enrichment of telomere sequences in either the Cdc13-myc18x TEN1 or Cdc13-myc18x ten1-101 strains is observed under our experimental conditions if formaldehyde is not added (data not shown). Since the ten1-ts telomere repeats are greatly elongated, these data indicate that, on average, each elongated telomere is associated with more Cdc13 than is found at normal, shorter telomeres. Consistent with the increased Cdc13 telomere binding observed in ykuΔ strains (Fisher et al. 2004) and in late S/G2 when telomeres transiently have longer single-stranded termini (Taggart et al. 2002), an increase in the single-stranded nature of the elongated ten1-ts telomeres would be expected to promote the overall extent of Cdc13 association in these strains. These data are consistent with the interpretation that Ten1 is not necessary for Cdc13 to bind telomeres in vivo, in agreement with Qian et al. (2009).
Figure 3.—
Cdc13-myc18x telomere association is not diminished in ten1-101. (A) Chromatin immunoprecipitation of Cdc13myc18x. Two hundred milliliters of wild-type untagged (hC160), CDC13myc(18) (hC1985) and ten1-101 CDC13myc18x (hC2035) strains were grown in YPD or SD −His media to an OD600 ∼1, and split into four 50-ml cultures. Two cultures were treated with nocodazole for 4 hr at 23° and 36° (Nz), and two cultures were diluted to an OD600 = 0.4 and incubated at 23° or 36° for 4 hr (asynchronous cultures, A). Immunoprecipitations from cell lysates were carried out with an anti-myc antibody (12CA5). The percentage of the total telomeric DNA present in each IP was determined by dot blot analysis, hybridizing with a [32P]d(GT/CA) probe, and exposing the blot on a Phosphorimager screen. To normalize the data, the percentage of telomere DNA recovered in each IP from Cdc13myc18x strains was divided by the percentage of telomere DNA recovered in the IP from the untagged Cdc13 strain. The averages of four independent experiments are graphed, and the error bars represent the standard deviation. (B) Dot blot of ChIP DNA samples from strains grown either asynchronously or arrested in G2 with nocodazole probed with [32P]d(GT/CA), as described in A. Shown are 10-fold dilutions of the immunoprecipitate and the input DNA for each strain.
Ten1 helps to promote de novo telomere addition:
Double-stranded breaks (DSB) in DNA activate a cellular checkpoint response, arresting the cell cycle and promoting activities that can lead to the repair of the broken DNA. DNA double-strand breaks are typically resected, initially being degraded from 5′ to 3′ to create single-stranded 3′ overhangs at the break (White and Haber 1990). However, it has been shown that when short tracts of telomere repeat sequences are placed adjacent to the DNA break site, the broken DNA end is protected from degradation (Diede and Gottschling 1999; Michelson et al. 2005; Hirano and Sugimoto 2007). The presence of this telomere “seed” sequence also greatly stimulates the addition of new telomere repeats to the DSB (Diede and Gottschling 1999). The ability of this de novo telomere to cap the new DNA end has been shown to be critically dependent upon Cdc13 (Diede and Gottschling 1999; Hirano and Sugimoto 2007). Since Cdc13 is proficient for telomere localization in ten1-ts strains and the mutant Ten1 proteins are deficient for Cdc13 interaction, we next tested whether Ten1 is required to assist Cdc13 in directing telomere addition at a DNA DSB that is flanked by a telomere repeat.
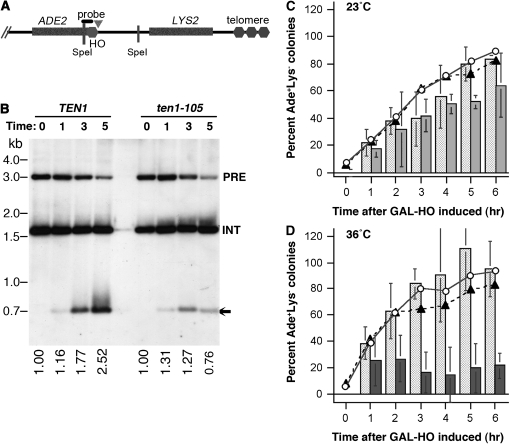
Strains with an 81-bp telomere seed sequence adjacent to an HO site were used to test whether Ten1 is similarly required to protect the HO endonuclease-generated DNA double-strand breaks from resection and to promote de novo telomere addition (diagrammed in Figure 4A). In this system, a cassette containing the ADE2 gene, 81 bp of TG1-3 repeats, and a 30-bp HO endonuclease recognition site was integrated at the ADH4 locus near the left end of chromosome VII (Diede and Gottschling 1999). The HO endonuclease gene is integrated into the genome, with its expression controlled by the GAL1 promoter, and the strain lacks the RAD52 gene. To test telomere addition and protection of the cut end in wild-type and ten1-105 strains, cells were arrested in G2/M using nocodazole, and the HO endonuclease was induced by switching to galactose media at the same time that the temperature was shifted to 36°. As the wild-type strain shows in Figure 4B, over time, the end that is released by the HO cut and has the adjacent telomere seed is “healed” by the addition of new telomere repeats. In contrast, telomere addition does not occur on the newly generated end in the ten1-105 strain (Figure 4B). Moreover, after 5 hr, less of the fragment generated by the HO cut remains, even though the extent of cutting by HO is comparable to the wild-type strain. In previous studies, such diminishment of this cut end corresponded with a failure to restrain resection; strains deficient only in telomere addition retain this cut end (Diede and Gottschling 1999). While it is possible that in the ten1-105 strain, extreme misregulation of telomerase creates a heterogeneous smear that is difficult to see, the simplest interpretation is that ten1-105 cells are deficient in de novo telomere addition and end protection while arrested in G2/M.
Figure 4.—
de novo telomere addition requires Ten1 in G2/M arrested cells. (A) Schematic diagram of the left arm of chromosome VII containing the HO endonuclease recognition site adjacent to 81 bp of telomeric repeats (Diede and Gottschling 1999). The ADE2 gene is located on the centromere proximal side of the inserted telomere repeat. The chromosome fragment from the HO site to the end of the chromosome does not contain any essential genes. A LYS2 marker is located in this region to retain this nonessential fragment. Note: this diagram is not to scale. (B) Telomere addition at 36°. Wild-type and ten1-105 strains containing the telomere healing cassette were grown in SD −Lys media at 23°, and then switched to YP-raffinose media containing nocodazole to arrest cells in G2/M. While maintaining the arrest, galactose was added to induce expression of the HO endonuclease, and the cultures were simultaneously shifted to 36°. Cells were collected at 0, 1, 3, and 5 hr following addition of galactose. Genomic DNA was isolated, digested with SpeI, and fragments were separated on a 1% agarose gel. The Southern blot was probed with a [32P]-labeled fragment that hybridizes to the ADE2 gene (Diede and Gottschling 1999). Both the native ADE2 locus (INT) and the HO-adjacent ADE2 gene are recognized by this probe. The gel was exposed on a Phosphorimager screen and the total amount of signal in the HO-adjacent ADE2 locus determined for each lane. The numbers below each lane represent the sum of the precut and cut fragments, normalized to the amount present at T = 0. PRE, fragment prior to HO digestion; INT, internal ADE2 control; arrow, fragment after HO digestion. WT (wild-type, hc1943), ten1-105 (hc1946). (C and D) Ade+Lys− colony formation as a means to assess the ability of ten1-105 strains to heal the DSB if released from the G2/M arrest and plated at 23°. In C, cells were incubated at 23° while HO was induced; in D, cells were shifted to 36° during the induction. Colonies on SD −Ade were replica-plated to SD −Ade −Lys (and SD −Ade) after 5 days at 23°. The lines on the graphs show the proportion of Ade+ colonies that are also Lys− at each time point [(Ade+ Lys−)/(Ade+)].  TEN1,
TEN1,  ten1-105. The bars on the graphs show the percentage of Ade+ Lys− colonies relative to the number of Ade+ colonies at T = 0; cells unable to heal the DSB are not expected to form colonies. Lighted shaded bars, TEN1; shaded bars, ten1-105 solid bars, ten1-105 .
ten1-105. The bars on the graphs show the percentage of Ade+ Lys− colonies relative to the number of Ade+ colonies at T = 0; cells unable to heal the DSB are not expected to form colonies. Lighted shaded bars, TEN1; shaded bars, ten1-105 solid bars, ten1-105 .
To address whether the mutant strains are capable of healing the induced break if they are released from the arrest and shifted to 23°, cells from each time point were plated on −Ade. The fraction of colonies that were Ade+ Lys−, indicating loss of the terminal fragment, was then determined by replica plating. When the HO site is induced in cells arrested at either 23° or 36°, the proportion of the viable Ade+ colonies that are “healed” at each time point is similar in wild-type and ten1-105 strains (lines in Figure 4, C and D). This indicates that the processes required to stabilize the broken end can function efficiently in ten1-105 strains, at least if shifted back to 23° and allowed to cycle. Nonetheless, relatively fewer ten1-105 cells are able to form colonies after 4–6 hr, even if the HO cut is created at 23° (bars in Figure 4, C and D). The drop in the relative number of ten1-105 cells capable of forming Ade+ Lys− colonies after the shift to 36° is striking. However, a similar drop in viability is also observed when no break is created while the cells are held at 36°. If ten1-105 cells are arrested in nocodazole and shifted to 36° for 4 hr, only ∼25% of the cells remain capable of forming colonies at 23° (data not shown). Thus, in the case where the DSB is induced, the small fraction of cells capable of forming a colony may reflect lethal events other than failure to stabilize the broken chromosome end.
Ten1-ts strains have a conditional defect in telomere capping:
To more directly determine whether the ten1-ts alleles have a defect capping chromosome ends, the integrity of the telomeres in the ten1-ts strains was analyzed at various temperatures to test for the presence of aberrant levels of single-stranded TG1-3 sequences. The G-rich strand was observed to be more single stranded in ten1-ts strains than in wild-type strains at 23° (Figure 5, A and B), consistent with the genetic data indicating a potential capping deficiency at permissive temperature. However, after incubation for 4 hr at 30° or 36°, the levels of single-stranded TG1-3 sequences dramatically increased, indicating significantly compromised chromosome end protection at these temperatures (Figure 5, A and B). Similar conditional capping deficiencies were observed for the ten1-101 and ten1-106 strains (Figure 5B, and data not shown). Thus, these ten1-ts alleles show a severe telomere integrity defect that is temperature dependent. Taking into account the ChIP data, telomere resection is occurring in the ten1-ts strains despite the presence of Cdc13 at telomeres. Interestingly, despite showing high levels of ssTG1-3 at 30°, the strains remain viable, without a significant drop in plating efficiency. The increase in temperature to 36° leads to a substantial drop in cell viability and to only a small further increase in ssTG1-3. The presence of single-stranded telomere repeat sequences in the ten1-ts strains is therefore not likely to be the only variable affecting cell growth at high temperature; it is possible that more internal sequences become single stranded at high temperature. At 23° and 30°, the extended telomere lengths may help mitigate an impact on cell viability.
Figure 5.—
ten1-103 and ten1-105 show conditional defects in telomere end protection. (A) Single-stranded telomeric DNA in ten1-105 at 23°, 30°, and 36°. Cultures were grown at 23° overnight and then shifted to the indicated temperature for 4 hr. Genomic DNA was isolated under nondenaturing condition and split, with half treated with 40 units ExoI (NEB) for 2.5 hr prior to digestion with XhoI. The gel was probed with a [32P]-labeled CA oligo that hybridizes to the G-rich telomere strand. The upper panel shows the gel under native conditions. After exposure on a Phosphorimager, the gel was denatured under alkaline conditions and hybridized with the [32P]-CA oligo to measure total TG1-3 levels. The lower panel shows the denatured gel. The number indicated below is the ratio of signal in the lane on the native gel/denatured gel, minus the ratio determined from the wild-type lanes. Strains: WT (wild-type, hc160), yku80Δ (hc18), and ten1-105 (hc1864). (B) Single-stranded telomeric DNA in ten1-103 strains at 23°, 30°, and 36°. The strains were grown, and DNA was isolated and processed as described in A. The native gel is shown in full; a representative slice from the denatured gel is shown in the lower panel. WT (wild-type, hc160), yku80Δ (hc18), and ten1-103 (hc1863).
To determine whether the single-stranded telomere repeats represent a contiguous loss of the C-rich strand from the end of the chromosome, we examined the single-stranded signal following digestion with the Escherichia coli ExoI enzyme. This exonuclease acts upon single-stranded 3′ DNA ends and does not degrade internal single-stranded DNA (Lehman and Nussbaum 1964). This experiment showed that the majority of the ssTG1-3 signal in the ten1-ts strains is resistant to digestion, particularly at high temperatures (Figure 5). A similar resistance to ExoI digestion was observed for cdc13-1 in these experiments (Figure 6). Neither overnight incubation of ten1-105 DNA with a large excess of ExoI nor using an ExoI enzyme from a different source yielded different results (data not shown). In contrast, the ssTG1-3 signal in the yku80Δ strains was sensitive to ExoI, as expected (Gravel et al. 1998; Polotnianka et al. 1998) (Figures 5 and 6). The partial resistance of the single-stranded telomere regions in the ten1-ts strains to digestion by ExoI may indicate that some portion of this signal is not terminal or reflect incomplete digestion of large amounts of single-stranded DNA. Alternatively, it is possible that in the strains with longer telomeres, some DNA secondary structure forms during isolation of the DNA that obstructs access by ExoI. At present, we cannot resolve these possibilities. Such ExoI-resistant, mung bean nuclease-sensitive TG1-3 signal has previously been observed in some stn1 strains (Petreaca et al. 2007).
Figure 6.—
ssTG1-3 generation in ten1-101 and ten1-105 is cell cycle and Cdk1 dependent. (A) In-gel hybridization showing cell cycle dependency of single-strand generation in ten1-105. Asynchronous cell cultures growing at 23° were divided, with equivalent portions arrested at 23° in G1 with α-factor, in S phase with hydroxyurea (HU), or in G2/M with nocodazole. Cultures were then split and shifted to the indicated temperature for 4 hr while maintaining the arrested state. One culture was maintained in logarithmic growth prior to the temperature shift (Asyn). Genomic DNA was then isolated and split, with half treated with ExoI prior to XhoI digestion. The upper panel shows the gel probed with a [32P]-CA oligo under native conditions, and the lower panel shows hybridization with the [32P]-CA oligo following denaturation. WT (wild-type, hc160), yku80Δ (hc18), and ten1-105 (hc1864). (B) In-gel hybridization showing cell cycle dependency of single-strand generation in ten1-101. Asynchronous cell cultures were divided and either allowed to continue growth at 23°, or arrested at 23° in G1 with α-factor or in G2/M with nocodozole. Cultures were then split and incubated at the indicated temperature for 4 hr while maintaining the arrested state. Genomic DNA was then isolated and split, with half treated with ExoI prior to XhoI digestion. The agarose gel was probed with a [32P]-CA oligo under native conditions. Only the native gel is shown; the total DNA loaded in each lane is shown in the ethidium bromide-stained gel slice. WT (wild-type, hc160), yku80Δ (hc18), and ten1-101 (hc1862). (C) In-gel hybridization showing the creation of ssTG1-3 is dependent on functional Cdk1. Cells were arrested at 23° in G2/M with nocodazole, and then split, with half shifted to 36° for 4 hr. To inhibit the Cdc28-as1 activity, 1-NMPP1 was added to the cultures 1 hr prior to the temperature shift. The cells were arrested in nocodazole throughout the experiment. Treatment of the genomic DNA and gel probing were as in A, with the upper panel showing the native gel and the lower panel showing the denatured gel. Wild-type (WT, hc160), yku80Δ (hc18), cdc13-1 (hc1997), cdc13-1 cdc28-as1 (hc1998), ten1-105 (hc1864), and ten1-105 cdc28-as1 (hc2005) strains are shown.
Ten1 is required to prevent telomere resection in G2/M arrested cells:
The conditional nature of the ten1-ts capping defects allowed us to probe when cells require the Ten1 end protection function. As cells progress through the cell cycle, the requirements for capping chromosomes vary. If the protection Ten1 provides to telomere termini is similar to that provided by Cdc13 (and Stn1), then its capping function should be critical in the G2/M phase of the cell cycle, but not in the G1 phase (Vodenicharov and Wellinger 2006). Using the conditional ten1-ts alleles, we tested at what cell cycle stage ten1-deficient cells acquire aberrant single-stranded TG1-3 DNA. Cells were first arrested at permissive temperature in G1 with α factor, in S phase with HU, or in G2/M with nocodazole treatment. After the cells had arrested, they were shifted to restrictive temperature while maintaining the cell cycle block. The analysis showed that the telomere C-strand loss in ten1-105 cells occurs most extensively when the cells are arrested in G2/M, with little loss occurring in G1 or S phase arrested cells (Figure 6A). Similar results were observed for the other ten1-ts alleles, such as ten1-101 (Figure 6B). It is possible that the low level of ssTG1-3 that is detected in the G1 arrested cells arises from the small fraction of cells that fail to arrest. In the α-factor-treated ten1-105 cultures, 85–90% of the cells arrested in G1; the small percentage of cells that did not arrest were large-budded cells (data not shown). The pattern of ssTG1-3 generation in these ten1-ts strains parallels that observed in cdc13-1 and cdc13-td strains (Vodenicharov and Wellinger 2006).
The cell cycle dependency of generating the single-stranded telomeres in ten1-ts may, like Cdc13, at least partially reflect the period of the cell cycle when the enzyme(s) that can extensively resect the telomere ends are active. In cdc13-deficient cells, it has been shown that telomere resection is regulated by the Cdk1 kinase (Vodenicharov and Wellinger 2006). This regulated resection activity is not active in G1, at least in response to endonuclease generated DSBs (Ira et al. 2004; Barlow et al. 2008). To determine whether the generation of single-stranded telomeres in ten1-105 cells is controlled by Cdk1, an analog-sensitive allele of CDC28, cdc28-as1, was used. The enlarged ATP binding site in Cdc28-as1 can accommodate 1-NM-PP1, a bulky ATP analog, allowing specific inhibition of the Cdk1 activity (Bishop et al. 2000). As previously reported (Vodenicharov and Wellinger 2006) and shown in Figure 6C, telomeres remain double stranded in cdc13-1 strains at 36° under conditions where Cdk1 activity is inhibited. The telomeres in the ten1-105 strain also remain largely intact at 36° following Cdc28 inhibition (Figure 6C). Thus, Ten1 and Cdc13 protect telomeres from similar activities, consistent with the interpretation that these proteins provide end protection through a shared mechanism. As will be discussed below, it is possible that the low levels of ssTG1-3 detected in the ten1-105 strain at 36° in the inhibited Cdc28-as1 cells reflect a failure to protect from additional activities.
End protection defects correlate with Rad52 foci formation and suppression by exo1Δ:
If Ten1 is in fact required to protect chromosome ends from resection, as indicated by the Cdk1-dependent resection of telomere DNA in ten1-ts mutants, it would be expected that Rad52 foci would be induced in ten1-ts mutants. Rad52 and other homologous recombination proteins form subnuclear repair foci in response to DNA damage such as that created by DNA double-strand breaks, single-stranded DNA, and eroded telomeres (Raderschall et al. 1999; Lisby et al. 2001; Khadaroo et al. 2009). To address whether ten1-ts inactivation can induce Rad52-YFP foci assembly after S phase, the ten1-ts strains were arrested in G2/M at permissive temperature, and then examined following a shift to restrictive temperature (36°). As compared to wild-type cells, the ten1-105 strain shows a significant increase in the number of cells with Rad52-YFP foci, even in the cells incubated at 23° (Figure 7A). Consistent with the conclusion that all the ten1-ts alleles show a capping defect, examination of asynchronous cultures reveals that each strongly induces repair foci in large-budded cells, as shown for cells grown overnight at 30°, a permissive temperature for growth (Figure 7, B and C). The finding of substantial induction of Rad52-YFP foci in the cells even at 30° is not surprising, given the large amount of telomere single-stranded DNA that is observed at 30° (Figure 5). Nonetheless, the absence of Rad52 does not alter the viability of these ten1-ts strains at any temperature (Figure 8A, data not shown).
Figure 7.—
Rad52-YFP foci form in large-budded ten1-ts cells at semi-permissive and restrictive temperatures. (A) The percentage of nocodazole-arrested cells with Rad52-YFP foci. Cells were grown at 23°, incubated in nocodazole until arrested in G2/M (3 hr), and then split, with half of the cells shifted to 36° for 4 hr while maintaining the arrest. One hundred large-budded cells per strain were scored, and the average and standard deviation from four replicates are shown. (B) Micrographs of TEN1 RAD52-YFP and ten1-105 RAD52-YFP cells were taken following overnight incubation of strains in liquid media at 30° and observed by fluorescence microscopy (Nikon Eclipse 800). (C) The percentage of large-budded cells with Rad52-YFP foci following overnight incubation in liquid media at 30°. One hundred or more cells with large buds were scored from each culture, with at least 3 cultures scored for each strain. The average number of cells with foci is graphed, with error bars showing the standard deviations. Strains: TEN1 RAD52-YFP (hC1720), ten1-101 RAD52-YFP (hc1847), ten1-103 RAD52-YFP (hc1857), ten1-105 RAD52-YFP (hc1840), and ten1-106 RAD52-YFP (hc1853).
EXO1 encodes a 5′-3′ nuclease that participates in metabolism of DNA in many contexts. Of particular relevance here, Exo1 is known to contribute to the generation of single-stranded DNA in cdc13-1, most notably in subtelomeric X and unique regions (Maringele and Lydall 2002). Deletion of EXO1 partially suppresses the cdc13-1 temperature sensitivity (Maringele and Lydall 2002), consistent with a reduction in lethal single-stranded lesions created by the loss of capping. In parallel with these results, we found that exo1Δ is also an extragenic suppressor of the ten1-101 and ten1-105 temperature sensitivity (Figure 8A), now allowing cell growth at 36°. The level of single-stranded TG DNA in the ten1-105 exo1Δ strain is modestly reduced relative to ten1-105 (Figure 8B). Similarly, as expected, comparatively less single-strand TG1-3 was also observed in cdc13-1 exo1Δ. The simplest interpretation is that the exo1Δ mutation improves ten1-ts viability because the extent of telomere resection is reduced below a critical threshold. The observation that exo1Δ also improves the initial plating efficiency of ten1-ts strains (Table 3) suggests that ten1-ts cells have insufficient end protection that the cells must overcome to proliferate. This reduced ability to form an initial colony is also observed by tetrad analysis of integrated ten1-105 alleles; ∼50% of ten1-105 cells form only microcolonies (data not shown). Further analysis of the initial events occurring in newly derived ten1-ts cells and the distribution and extent of single-stranded generation in the mutant strains could help resolve the mechanism of exo1Δ-mediated suppression.
Reduced Polα function significantly compromises ten1-ts growth:
Our data show that the ten1-ts alleles phenocopy aspects of both cdc13-1 and stn1 mutants. Interestingly, one characteristic of stn1 strains that appears distinct from cdc13 strains is a strong synergistic phenotype when both STN1 and Polα function (POL12 in particular) are compromised (Grossi et al. 2004). In particular, stn1-13 pol12-216 strains have a synergistic phenotype, with increased levels of ssTG1-3 generated. A similar synergism with cdc13 was not reported. To determine whether ten1-ts strains are similar to, or distinct from, cdc13 in this regard, we tested whether compromising Polα through the cdc17-1 mutation alters the viability of cdc13-1 or ten1-ts strains. As shown in Figure 9A and reported in (Garvik et al. 1995), cdc13-1 cdc17-1 strains are viable and show a slight reduction in maximum permissive temperature compared to cdc13-1. In contrast, the temperature sensitivity of each ten1-ts cdc17-1 strain is greatly enhanced, with a maximum permissive temperature of ∼28°, although growth is compromised even at 23°. Since cdc17-1 does alter terminal chromatin structure (Adams Martin et al. 2000), it is possible that resection of telomeres is enhanced in the ten1-ts cdc17-1 strain. To test this, the generation of ssTG1-3 was assessed in cells arrested in G2/M prior to a shift to restrictive temperature (Figure 9B). The double mutant ten1-105 cdc17-1 strain did not show enhanced ssTG1-3 generation when arrested at 23° or 36°, consistent with the interpretation that resection is not enhanced per se in this cell cycle window. The relative level of single-stranded telomeres was similar to that in ten1-105 (compare native to denatured signals). We note that the telomere signal consistently appears more heavily amplified in the ten1-ts single mutant strain.
Figure 9.—
cdc17-1 significantly enhances the growth defect in ten1-ts, but not cdc13-1 strains. (A) Growth of single and double mutants. Ten-fold serial dilutions of strains were incubated on YPD at the indicated temperatures. cdc13-1 cdc17-1 strains were obtained by tetrad dissection. cdc17-1 ten1-ts strains were obtained by plasmid shuffle from parental strain hC1678, using plasmids pCN284, pCN309, pCN311, pCN358, and pCN359. The ten1-ts single mutant plates were incubated at the same time as the double mutants. (WT is hC160). (B) In-gel hybridization analysis of ssTG1-3 in G2/M arrested cells. Strains were synchronized in nocodazole at 23°, and then split and incubated 3 more hr in nocodazole at 23° or 36°. DNA was isolated under native conditions; equivalent DNA concentrations were digested with XhoI and analyzed by in-gel hybridization. The total DNA loaded in each lane is similar, as visualized by ethidium bromide staining, shown below denatured gel.
DISCUSSION
Ten1 functions as a telomere capping protein:
Our data strongly support the hypothesis that Ten1, like Cdc13, protects telomeres from cell cycle-regulated activities that resect the telomere C-rich strand. Overall, the ten1-ts capping phenotypes parallel defects observed in cdc13-1 strains. First, similar to cdc13-1, cdc13-td, and stn1-td strains (Vodenicharov and Wellinger 2006), extensive resection occurs when ten1-ts strains are blocked in G2/M, but not when blocked in G1 or in early S phase. Second, using the ATP analog-sensitive allele of CDK1, cdc28-as1, it is clear that the resection for both cdc13-1 and ten1-105 is dependent upon Cdk1 activity. Third, deletion of the Exo1 nuclease partially restores ten1-ts viability and modestly reduces the level of single-stranded telomere DNA. Finally, the loss of ten1 function results in increased Rad53 phosphorylation (LX, data not shown), a strong induction of Rad52-YFP foci, and a RAD9-dependent cell cycle arrest (Grandin et al. 2001) (LX, data not shown), consistent with activation of the DNA damage checkpoint. Thus, like Cdc13, Ten1 is required to prevent resection of telomeres.
The comparable cdc13 and ten1 capping defects support the idea that the processes leading to creation of the single-stranded telomere repeats are similar in these strains. One difference from cdc13-1, however, was that despite Cdk1 inhibition in the cdc28-as1 ten1-105 strain, a small amount of single-stranded TG1-3 DNA was still generated at nonpermissive temperature. A trivial explanation for this would be that there was incomplete inhibition of Cdc28-as1, despite using a high concentration of 1-NM-PP1. A more speculative interpretation would be that telomere chromatin is disrupted in the ten1-105 strain such that some Cdk1-independent resection occurs, as has been shown at a DSB in strains that are deficient for dot1, a methyltransferase, or for rad9, a checkpoint mediator that binds to modified chromatin (Lazzaro et al. 2008).
Does Ten1 protect telomeres only by preventing resection?
While supporting a critical role for Ten1 in preventing telomere resection in G2/M, these data do not exclude the possibility that Ten1 affects telomere integrity during other cell cycle phases. We note that the extent to which telomeres become single stranded in ten1-ts strains is consistently higher in asynchronously dividing cells than in G2/M arrested cells. This difference could reflect the involvement of additional activities in generating the ssTG1-3 or that the processes that are acting in G2/M have higher activity in a different cell cycle window. There is some cell cycle specificity to the operation of DNA repair pathways that can influence how a DNA double-strand break is repaired (Kanaar et al. 2008). By extension, the outcome of losing telomere capping is likely to be influenced by the cell cycle phase when capping is lost. Since we did not observe significant single-stranded TG1-3 during either a G1 or early S phase arrest, we suggest that ten1-ts strains are likely to be highly susceptible to generating excessive single-stranded DNA during late S phase, in conjunction with the passage of replication forks through telomeres. The synthetic phenotype of ten1-ts cdc17-1 strains is consistent with this hypothesis. Whether such single-stranded DNA would arise because Ten1 has a role connected with the conventional replication machinery in duplicating telomeres or because the ten1-ts defect makes telomeres more prone to resection while they are undergoing replication remains to be explored. It will be important to more fully understand how chromosome capping is maintained during DNA replication. In this regard, it is particularly interesting that the likely Stn1 mammalian homolog, OBFC1/AAF-44, has been identified as part of a complex that not only can regulate Polα activity but also associate with telomeres (Casteel et al. 2008; Dejardin and Kingston 2009; Wan et al. 2009).
What is the molecular basis of the ten1-ts deficiency?
Since Cdc13, Stn1, and Ten1 are likely to function as a complex to promote capping (Grandin et al. 1997, 2001; Pennock et al. 2001), interactions among these proteins should be important for their function. However, three of the Ten1-ts proteins interact with neither Cdc13 nor Stn1, even at permissive temperatures. At a minimum, the Ten1–Stn1 interaction was expected to be criticial for the essential function of these proteins. The ten1-105 allele shows a more severe phenotype than ten1-101 or ten1-103, and yet Ten1-105 is competent for association with Stn1. Although the Ten1–Cdc13 and –Stn1 interactions must contribute to both telomere integrity and proper regulation of telomerase, these data indicate that the primary defect in these ten1-ts alleles that leads to impaired cell viability cannot simply be attributable to only a loss of interaction with either Cdc13 or Stn1.
Given the similarities between ten1-ts and cdc13-1 phenotypes, it was possible that the ten1-ts capping defect results from deficient Cdc13 localization to telomeres. However, we found that Cdc13 remains associated with telomeres in ten1-101 at high temperature, a condition where Ten1-101 no longer provides adequate protection. These results show that in vivo Cdc13 can bind to telomere repeats independent of Ten1, and demonstrate that the binding of Cdc13 to telomeres is not sufficient to provide a functional telomere cap. It was recently reported that Cdc13 association with a subtelomeric region is reduced in two ten1 strains (Qian et al. 2009). The lesions in these mutants perturb the Ten1–Cdc13 contact and the strains show significant telomere elongation, but no impairment of viability (Qian et al. 2009). Since these sets of ten1 alleles are all deficient for Cdc13 interaction, how Cdc13 telomere association is affected in ten1 strains may depend more on the overall structure of the chromosome ends than specifically upon the robustness of the interaction between Cdc13 with Ten1. It would not be surprising if the distribution of Cdc13 along telomere chromatin becomes altered in ten1-ts strains as a consequence of the increased single-stranded nature of their telomeres.
Cdc13-bound telomeres promote ten1-ts viability:
Ten1-ts strains rely on Cdc13 for viability even at permissive temperature. The presence of functional Cdc13 on telomeres in ten1-ts strains may not only lead to the extreme telomere lengthening that is observed, but also could help limit resection of subtelomeric regions. The binding of Cdc13 to the ssTG1-3, or possibly the elongated state of the telomeres, could be the reason ten1-ts cells can proliferate at 30° despite significant ssTG1-3 accumulation and strong damage foci induction. The growth defects of both cdc13-1 and ku deficient strains are also modulated by telomere length (Downey et al. 2006; Vega et al. 2007). A tolerance for disrupted telomere integrity is observed in yku70Δ strains, which exhibit single-stranded TG1-3 repeats at low temperatures (Gravel et al. 1998), and only lose viability when the temperature is raised to 37° (Feldmann and Winnacker 1993; Boulton and Jackson 1996). In these yku− strains, the loss of viability correlates with both the amount of single-stranded DNA in subtelomeric regions (Maringele and Lydall 2002) and the length of the duplexed telomere tract (Gravel and Wellinger 2002). Furthermore, short tracts of telomere repeats adjacent to a DSB require Cdc13 to prevent resection, whereas long tracts of telomere repeats have a reduced dependence upon Cdc13 for protection from resection (Negrini et al. 2007). Both the reduced initial plating efficiency of ten1-ts strains (when cells initially have wild-type telomeres) and the synthetic phenotype of ten1-ts est2Δ are consistent with the hypothesis that increased telomere length helps compensate for ten1 capping defects, although telomerase could also contribute to capping (Singh and Lue 2003; Vega et al. 2007). Thus, understanding the molecular basis for Ten1 telomere capping activity remains a significant issue for further exploration.
Acknowledgments
We are grateful to Dan Gottschling, David Morgan, and Rodney Rothstein for sharing strains. We thank lab members and Jeff Bachant for discussion and comments on this manuscript. This work was supported by a grant to C.I.N. from the National Institutes of Health (R01-CA96972).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.108894/DC1.
References
- Adams Martin, A., I. Dionne, R. J. Wellinger and C. Holm, 2000. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon, Y., B. Liefshitz and M. Kupiec, 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23 4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, S. M., and J. P. Murnane, 2006. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 34 2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, J. H., M. Lisby and R. Rothstein, 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell 30 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray et al., 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407 395–401. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, M. J., and L. Hartwell, 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42 249–257. [DOI] [PubMed] [Google Scholar]
- Casteel, D. E., S. Zhuang, Y. Zheng, F. W. Perrino, G. R. Boss et al., 2009. A DNA polymerase-alpha/primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 284 5807–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange, T., 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19 2100–2110. [DOI] [PubMed] [Google Scholar]
- Dejardin, J., and R. E. Kingston, 2009. Purification of proteins associated with specific genomic loci. Cell 136 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede, S. J., and D. E. Gottschling, 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99 723–733. [DOI] [PubMed] [Google Scholar]
- Dionne, I., and R. J. Wellinger, 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93 13902–13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey, M., R. Houlsworth, L. Maringele, A. Rollie, M. Brehme et al., 2006. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124 1155–1168. [DOI] [PubMed] [Google Scholar]
- Feldmann, H., and E. L. Winnacker, 1993. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 268 12895–12900. [PubMed] [Google Scholar]
- Ferreira, M. G., and J. P. Cooper, 2004. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, T. S., A. K. Taggart and V. A. Zakian, 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 11 1198–1205. [DOI] [PubMed] [Google Scholar]
- Frank, C. J., M. Hyde and C. W. Greider, 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell 24 423–432. [DOI] [PubMed] [Google Scholar]
- Gao, H., R. B. Cervantes, E. K. Mandell, J. H. Otero and V. Lundblad, 2007. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14 208–214. [DOI] [PubMed] [Google Scholar]
- Garvik, B., M. Carson and L. Hartwell, 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin, N., S. I. Reed and M. Charbonneau, 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11 512–527. [DOI] [PubMed] [Google Scholar]
- Grandin, N., C. Damon and M. Charbonneau, 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel, S., M. Larrivee, P. Labrecque and R. J. Wellinger, 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280 741–744. [DOI] [PubMed] [Google Scholar]
- Gravel, S., and R. J. Wellinger, 2002. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 22 2182–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi, S., A. Puglisi, P. V. Dmitriev, M. Lopes and D. Shore, 2004. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 18 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, Y., and K. Sugimoto, 2007. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol. Biol. Cell 18 2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani et al., 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar, R., C. Wyman and R. Rothstein, 2008. Quality control of DNA break metabolism: in the ‘end,’ it's a good thing. EMBO J. 27 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadaroo, B., M. T. Teixeira, P. Luciano, N. Eckert-Boulet, S. M. Germann et al., 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell. Biol. 11 980–987. [DOI] [PubMed] [Google Scholar]
- Larrivee, M., C. LeBel and R. J. Wellinger, 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro, F., V. Sapountzi, M. Granata, A. Pellicioli, M. Vaze et al., 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27 1502–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, I. R., and A. L. Nussbaum, 1964. The deoxyribonucleases Of Escherichia Coli. V. On the specificity of exonuclease I (phosphodiesterase). J. Biol. Chem. 239 2628–2636. [PubMed] [Google Scholar]
- Lin, J. J., and V. A. Zakian, 1996. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA 93 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby, M., R. Rothstein and U. H. Mortensen, 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V., and J. W. Szostak, 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57 633–643. [DOI] [PubMed] [Google Scholar]
- Lydall, D., and T. Weinert, 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270 1488–1491. [DOI] [PubMed] [Google Scholar]
- Marcand, S., V. Brevet, C. Mann and E. Gilson, 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10 487–490. [DOI] [PubMed] [Google Scholar]
- Marcand, S., B. Pardo, A. Gratias, S. Cahun and I. Callebaut, 2008. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 22 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele, L., and D. Lydall, 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, V., L. L. Du, S. Rozenzhak and P. Russell, 2007. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc. Natl. Acad. Sci. USA 104 14038–14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson, R. J., S. Rosenstein and T. Weinert, 2005. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 19 2546–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini, S., V. Ribaud, A. Bianchi and D. Shore, 2007. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 21 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, C. I., T. R. Hughes, N. F. Lue and V. Lundblad, 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274 249–252. [DOI] [PubMed] [Google Scholar]
- Pardo, B., and S. Marcand, 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock, E., K. Buckley and V. Lundblad, 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104 387–396. [DOI] [PubMed] [Google Scholar]
- Petreaca, R. C., H. C. Chiu, H. A. Eckelhoefer, C. Chuang, L. Xu et al., 2006. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell. Biol. 8 748–755. [DOI] [PubMed] [Google Scholar]
- Petreaca, R. C., H. C. Chiu and C. I. Nugent, 2007. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction With Cdc13p. Genetics 177 1459–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka, R. M., J. Li and A. J. Lustig, 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8 831–834. [DOI] [PubMed] [Google Scholar]
- Qi, H., T. K. Li, D. Kuo, E. K. A. Nur and L. F. Liu, 2003. Inactivation of Cdc13p triggers MEC1-dependent apoptotic signals in yeast. J. Biol. Chem. 278 15136–15141. [DOI] [PubMed] [Google Scholar]
- Qian, W., J. Wang, N. N. Jin, X. H. Fu, Y. C. Lin et al., 2009. Ten1p promotes the telomeric DNA-binding activity of Cdc13p: implication for its function in telomere length regulation. Cell Res. 19 849–863. [DOI] [PubMed] [Google Scholar]
- Raderschall, E., E. I. Golub and T. Haaf, 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. M., and N. F. Lue, 2003. Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl. Acad. Sci. USA 100 5718–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., A. Hecht, K. Luo and M. Grunstein, 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11 83–93. [DOI] [PubMed] [Google Scholar]
- Taggart, A. K., S. C. Teng and V. A. Zakian, 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297 1023–1026. [DOI] [PubMed] [Google Scholar]
- Vega, L. R., J. A. Phillips, B. R. Thornton, J. A. Benanti, M. T. Onigbanjo et al., 2007. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 3 e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenicharov, M. D., and R. J. Wellinger, 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell 24 127–137. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wan, M., J. Qin, Z. Songyang and D. Liu, 2009. OB-fold containing protein 1 (OBFC1), a human homologue of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 284 26725–26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert, T. A., and L. H. Hartwell, 1993. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger, R. J., A. J. Wolf and V. A. Zakian, 1993. a Origin activation and formation of single-strand TG1–3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol. Cell. Biol. 13 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger, R. J., A. J. Wolf and V. A. Zakian, 1993. b Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72 51–60. [DOI] [PubMed] [Google Scholar]
- Wellinger, R. J., K. Ethier, P. Labrecque and V. A. Zakian, 1996. Evidence for a new step in telomere maintenance. Cell 85 423–433. [DOI] [PubMed] [Google Scholar]
- White, C. I., and J. E. Haber, 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut, C., and J. F. Diffley, 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko, M. K., S. Guillard and D. Lydall, 2004. Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13–1 mutants. Genetics 168 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]