Abstract
Protein scaffolds bring together multiple components of a signalling pathway, thereby promoting signal propagation along a common physical ‘backbone'. Scaffolds play a prominent role in natural signalling pathways and provide a promising platform for synthetic circuits. To better understand how scaffolding quantitatively affects signal transmission, we conducted an in vivo sensitivity analysis of the yeast mating pathway to a broad range of perturbations in the abundance of the scaffold Ste5. Our measurements show that signal throughput exhibits a biphasic dependence on scaffold concentration and that altering the amount of scaffold binding partners reshapes this biphasic dependence. Unexpectedly, the wild-type level of Ste5 is ∼10-fold below the optimum needed to maximize signal throughput. This sub-optimal configuration may be a tradeoff as increasing Ste5 expression promotes baseline activation of the mating pathway. Furthermore, operating at a sub-optimal level of Ste5 may provide regulatory flexibility as tuning Ste5 expression up or down directly modulates the downstream phenotypic response. Our quantitative analysis reveals performance tradeoffs in scaffold-based modules and defines engineering challenges for implementing molecular scaffolds in synthetic pathways.
Keywords: MAP kinase signalling, pheromone, Saccharomyces cerevisiae, signal throughput, Ste5 scaffold
Introduction
Protein scaffolds bind concomitantly to multiple components of a signalling pathway, thereby organizing signal transmission onto a common physical backbone. Scaffold-based modules are broadly used to propagate signals that regulate cell cycle, proliferation, differentiation and motility in species ranging from yeast to human (Pawson and Scott, 1997). Scaffolds are also emerging as a promising platform for engineering synthetic signalling modules. Molecular redesign of scaffolds has been used to alter the repertoire of scaffold binding partners, thereby redirecting signal flow (Park et al, 2003) and altering signal dynamics (Bashor et al, 2008).
In addition to the molecular design of the scaffold, the quantitative performance of scaffold-based modules will depend on the expression level of the scaffold and its binding partners. Computational models predict that scaffolds may not always promote signal propagation (Levchenko et al, 2000). When scaffold concentration exceeds an optimal level, enzymes and substrates are predicted to bind to distinct scaffolds rather than onto a single backbone, thereby weakening signal transmission via combinatorial inhibition.
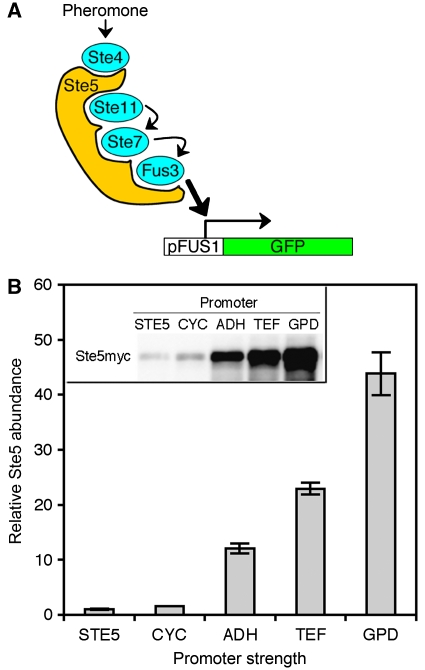
These model predictions, however, are based on idealized mathematical representations of scaffold-based signalling. In contrast, scaffold-mediated signalling in vivo is often far more intricate as exemplified for the prototypical scaffold Ste5 in yeast cells (Figure 1A). The Ste5 scaffold undergoes dimerization, which may contribute to apparent cooperativity (Ferrell, 2000), translocates between different subcellular compartments, which regulates its proteosomal degradation (Garrenton et al, 2009), is regulated by Fus3-mediated negative feedback (Bhattacharyya et al, 2006) and binds competitively to multiple proteins (Fus3 and Kss1) with different affinities (Kusari et al, 2004). This complex array of mechanisms conceals precisely how real scaffolds such as Ste5 quantitatively contribute to signal transmission in vivo.
Figure 1.
Modulating the expression level of the scaffold Ste5. (A) The Ste5 scaffold and the pheromone MAP kinase pathway in Sacchaormyces cerevisiae. Ste5 binds to Ste4, Ste11, Ste7 and the MAP kinase, Fus3. Another MAP kinase, Kss1 (not depicted for clarity), also binds Ste5 and is also activated by Ste7. Upon pheromone stimulation, Ste5 facilitates signal transmission from Ste4 to Fus3/Kss1. Active Fus3 and Kss1 trigger the transcription of FUS1, cell-cycle arrest and ultimately mating. (B) The relative expression levels of Ste5 in strains expressing Myc-tagged Ste5 behind a constitutive promoter (pCYC, pADH, pTEF or pGPD) or the wild-type STE5 promoter (pSTE5). Error bars denote s.e.m. (n=3). Source data is available for this figure at www.nature.com/msb
In fact, previous studies have shown that Ste5 overexpression augments signal throughput (Kranz et al, 1994; Garrenton et al, 2009), suggesting that it may not conform to the combinatorial inhibition model. In contrast, other scaffolds such as JIP and KSR behave more consistently with model predictions: their overexpression diminishes signal throughput (Dickens et al, 1997; Joneson et al, 1998). These apparent discrepancies are due, in part, to limitations of the classical binary approach of comparing wild-type cells to an overexpression mutant. Thus, in the case of Ste5, it is unclear whether the extent of overexpression was sufficient to enter the combinatorial inhibition regime. Meanwhile, even for scaffolds that seem to undergo combinatorial inhibition, the binary approach does not provide quantitative insight. It remains unclear what the optimum level of scaffold is and whether wild-type cells operate at this optimum. If the wild-type configuration is sub-optimal, how much signal throughput is forsaken and what might be the underlying reasons? Furthermore, the level of scaffold may influence other signalling properties, such as ultrasensitivity (Ferrell, 2000). To understand more comprehensively how scaffold expression level affects different aspects of pathway performance, we conducted an in vivo sensitivity analysis of the mating pathway to a broad range of perturbations in Ste5 abundance.
Results and discussion
Modulation of scaffold expression level
To better understand the quantitative effect of scaffold abundance on MAP kinase signalling, we engineered a panel of yeast strains that expresses Ste5 at different levels. Starting with a ste5 parent strain, we introduced a C-terminal, Myc-tagged version of STE5 under the regulation of various constitutive promoters and measured the relative expression level of Ste5 in the different strains by a quantitative immunoblot procedure (Materials and methods and Supplementary Figure S1). Ste5 expression in this panel of yeast strains spanned nearly two orders of magnitude (Figure 1B). The highest level of expression was 50-fold greater than that supported by the wild-type STE5 promoter.
Effect of scaffold on signal throughput and phenotypic response
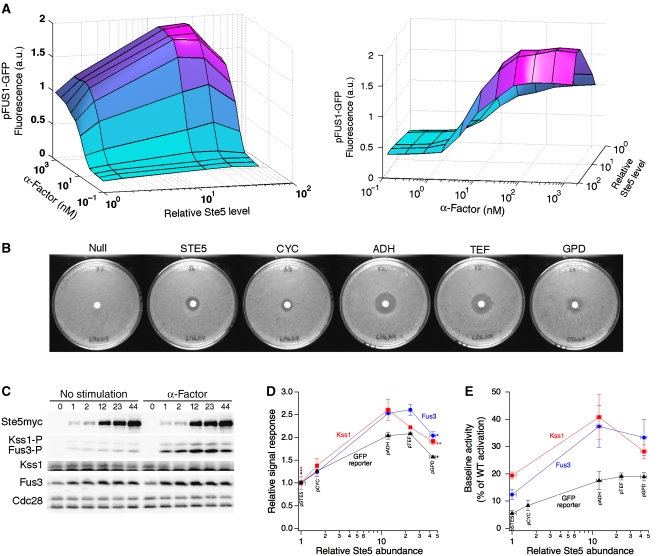
To quantify the sensitivity of the mating pathway to Ste5 abundance, we measured the mating transcriptional response over a broad range of α-factor concentrations in our panel of yeast strains. Variations in scaffold abundance had a significant effect on the transcriptional output of the mating pathway (Figure 2A). At every α-factor dose, the output was biphasic with respect to the level of Ste5, revealing that an optimum level of Ste5 scaffold is needed to maximize signal throughput. This biphasic relationship is consistent with model predictions (Levchenko et al, 2000) and with studies of scaffolds JIP and KSR (Dickens et al, 1997; Joneson et al, 1998). Previous studies involving Ste5 overexpression have reported only signal augmentation (Kranz et al, 1994; Garrenton et al, 2009). Our data shows however, that this may have been a limitation of binary comparisons between the wild-type and a single overexpression mutant. In fact, even in our study, comparing the wild-type to any individual strain overexpressing Ste5 would have led to the conclusion that increasing Ste5 expression enhances the mating pathway output (Figure 2D). By sampling multiple Ste5 expression levels, we identified the biphasic relationship where Ste5 promotes and ultimately thwarts pathway performance.
Figure 2.
Sensitivity analysis of mating pathway to perturbations in scaffold abundance. (A) The relative mean pFUS1-GFP reporter response for the strains expressing different levels of Ste5 and treated with different doses of α-factor for 2.5 h. Two different views of the surface plot are shown. (B) The mating halo assay for cells expressing different levels of Ste5. Results from a representative trial are shown (n=3). (C) Yeast cells were induced with α-factor or left unstimulated. The indicated proteins were analysed by immunoblotting. Relative Ste5 expression is indicated above the gel lanes. (D) Quantitative measurements of phospho-MAP kinase and pFUS1-GFP responses. Phospho-Fus3 was normalized by its respective total protein expression. Phospho-Kss1 was normalized by its total protein expression as measured in uninduced lysates. Error bars denote s.e.m. (n=3). The asterisks indicate the P-value between the marked data point and the maximum data point in the same curve: *P<0.01 and **P<0.05 (Student's t-test). (E) Baseline activation of phospho-MAP kinase and pFUS1-GFP. Phospho-Fus3 was normalized by its respective total protein expression. Phospho-Kss1 was normalized by an equal loading control. Error bars denote s.e.m. (n⩾4). Source data is available for this figure at www.nature.com/msb
This quantitative data set also enabled an assessment of other aspects of pathway performance beyond signal output. We characterized the effect of Ste5 abundance on the responsiveness of the mating pathway by fitting the Hill equation to the dose–response curves. This analysis revealed that both the Hill coefficient (nH) and the α-factor dose at which half-maximal response is achieved (EC50), are unaffected by Ste5 expression level (see Supplementary information, Supplementary Figure S2 and Supplementary Table SIII). Thus, the expression level of Ste5 affects signal transmission solely by modulating signal throughput of the mating pathway, without affecting the apparent cooperativity or stimulus potency.
To test whether changes in the transcriptional response translate to the ultimate biological response, we assessed the mating response of yeast cells using the halo assay. In this assay, pheromone diffuses from a central source and induces cell-cycle arrest up to a certain radius. Beyond this radius, the local pheromone concentration no longer induces the threshold level of signalling needed to illicit cell-cycle arrest. We hypothesized that cells expressing the optimum level of Ste5 may achieve this threshold level of signalling at lower doses of pheromone than cells operating with sub-optimal doses of Ste5 (Supplementary Figure S3A). If this hypothesis were accurate, then we would expect the halo size to be sensitive to changes in Ste5 expression. Indeed, the halo size varied as a function of Ste5 expression, reaching a maximum at an optimal dose of Ste5 (Figure 2B and Supplementary Figure S3B). Further increasing Ste5 expression level beyond this optimum reduced the size of the halo. The optimum level of Ste5 that maximized the halo size precisely correlates with the optimum Ste5 level for transcriptional response.
Closer examination of the Ste5 module
Transcriptional response and cell-cycle arrest are several steps downstream of the direct MAP kinase outputs of the Ste5 scaffold. To confirm that the effect of Ste5 perturbations on the downstream elements of the mating pathway truly emanates from the immediate outputs of the Ste5 module, we measured the phosphorylation of the mating MAP kinases, Fus3 and Kss1, by quantitative immunoblot. Following stimulation with 1.2 μM α-factor for 15 min, the levels of both phospho-Fus3 and phospho-Kss1 exhibit a biphasic dependence on Ste5 abundance, closely resembling the trend in transcriptional output and phenotypic response (Figure 2C and D). Meanwhile, the total expression levels of Fus3 and Kss1 vary negligibly, indicating that the biphasic phosphorylation of Fus3 and Kss1 are not due to Ste5-induced changes in the expression level of these MAP kinases (Supplementary Figure S4). Furthermore, detailed analysis of the phospho-MAP kinase dose–response curves showed no significant correlation between Ste5 expression level and the nH and EC50 parameters (Supplementary Figure S5 and Supplementary Tables SIV and SV), consistent with the FUS1 reporter response.
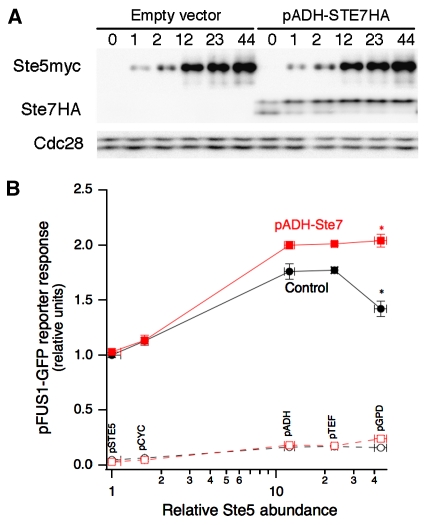
Interestingly, both phospho-Fus3 and phospho-Kss1 exhibit similar biphasic dependence on Ste5 abundance, suggesting that a common upstream factor, such as Ste7, may be the limiting component. To test this hypothesis, we overexpressed HA-tagged Ste7 in our panel of yeast strains that express Ste5 at different levels (Figure 3A). At low scaffold abundance, overexpression of Ste7 did not alter the mating reporter response relative to control cells carrying the empty vector (Figure 3B). However, at higher scaffold concentrations, overexpression of Ste7 did increase the reporter response and, importantly, eliminated the downturn in signal throughput.
Figure 3.
Scaffold-limited and Ste7-limited regimes of signalling. (A) Yeast strains expressing different levels of Ste5 were transformed with either an empty vector control or a vector encoding HA-tagged Ste7 downstream of an ADH promoter. The expression of Ste5myc and Ste7HA were confirmed by immunoblotting. Relative Ste5 expression is indicated above the gel lanes. (B) Yeast overexpressing Ste7 or not were stimulated with a saturating dose of α-factor (150 nM) for 2.5 h and the pFUS1-GFP reporter response was quantified by flow cytometry. Error bars denote s.e.m. (n=5). The asterisks indicate the P-value between the marked data points: *P<0.01 (Student's t-test). Source data is available for this figure at www.nature.com/msb
These results demonstrate a scaffold-limited and Ste7-limited regime of signalling. When the scaffold is the limiting factor to signal throughput (for scaffold doses below the optimum), increasing the expression of Ste7 had no effect on signal throughput. However, past the optimum dose of scaffold, signal throughput was limited by Ste7. Overexpression of Ste7 eliminated the biphasic dependence of signal throughput on scaffold amount, at least within the range of Ste5 expression explored. We reason that the optimum Ste5 dose has shifted to a level higher than that captured by our panel of yeast strains. These results demonstrate that the abundance of scaffold and its binding partners together shape the biphasic dependence of signal throughput and determine the optimum dose of scaffold.
Sensitivity of signal quality to scaffold abundance
Our data demonstrate that the optimum dose of Ste5 maximizes signal transmission and raises the question of whether there are tradeoffs in other metrics of pathway performance. Scaffolds play an important role in maintaining the fidelity of stimulus–response relationships. We tested whether changes in Ste5 expression level affect the activation of two closely related pathways, the pheromone and the high-osmolarity MAP kinase pathways (see Supplementary information and Supplementary Figure S6). Pheromone stimulation activated only the mating MAP kinases and did not stimulate phosphorylation of Hog1, the high-osmolarity MAP kinase. Meanwhile, stimulation with sorbitol appropriately activated Hog1 with no cross-activation of Fus3 in the pheromone pathway. Thus, across nearly 50-fold change in Ste5 expression level, signal fidelity is maintained.
Another important metric of the performance of signalling modules is the signal-to-noise ratio. High-quality signal transmission involves maintaining a low baseline signal in the absence of stimulation, while responding with a strong signal when the stimulus is present. To investigate the effect of increased scaffold abundance on baseline signalling, we examined the phosphorylation of Fus3 and Kss1 in the absence of pheromone. Our measurements show that increasing Ste5 expression elevates the basal activities of Fus3 and Kss1 (Figure 2C). To gauge more quantitatively the significance of this baseline activation, we compared the baseline signal to the signal generated in wild-type cells treated with a saturating dose (60 nM) of pheromone for 5 min. This analysis revealed that baseline activation of Fus3 and Kss1 increases up to two- to three-fold upon increasing Ste5 expression, reaching nearly 40% of the signal generated upon pheromone stimulation of wild-type cells (Figure 2E). This baseline drift is propagated to the transcriptional response: baseline activation of FUS1 reporter also increased up to three-fold following Ste5 overexpression, reaching approximately 20% of the signal generated in pheromone-stimulated wild-type cells. The consequence of this baseline drift is that the dynamic range of phospho-Fus3 and pFUS1-GFP signalling is reduced (Supplementary Tables SIII–V), suggesting that the ability of the signalling pathway to discriminate between the absence and presence of the stimulus is diminished.
Potential implications for natural and synthetic scaffold-based modules
Our results reveal that the wild-type expression of Ste5 is not set for maximum signal throughput and suggest at least three potential reasons for this sub-optimal configuration. The most straightforward explanation is that operating at half-maximal throughput permits regulatory flexibility to tune up or down module performance. Consistent with this possibility, it has recently shown been that Ste5 stability and expression level are regulated in vivo through mechanisms involving Ste5 subcellular compartmentalization (Garrenton et al, 2009). Our data suggest that such modulation of Ste5 expression level would have quantitative effects on the ultimate biological response. Second, operating with a sub-optimal dose of Ste5 places the pathway in the Ste5-limited regime. In this regime, pathway performance may be tuned solely by altering Ste5 expression level and experience less sensitivity to perturbations in other module components, such as Ste7. Finally, our analysis suggests that there may be a penalty for operating at the optimum level of Ste5: baseline activation of the mating pathway increases, thereby reducing the dynamic range of signalling.
Molecular scaffolds offer a promising platform for engineering synthetic regulatory circuits. While baseline activation may be a potential design constraint for scaffold-based synthetic pathways, our results show that shifting the scaffold expression level to an optimum provides a two to threefold improvement in signal throughput. This improvement has direct quantitative implications for the downstream response as we have demonstrated for pheromone-mediated cell-cycle arrest. In addition, changes of similar magnitude in the strength of Fus3 signalling can have significant qualitative effects on the phenotypic response to pheromone stimulation (Hao et al, 2008). Even in other biological contexts, small differences in signals, particularly MAP kinases, lead to drastic switch-like changes in cell decisions (Ferrell, 1996). Thus, evaluating and appropriately modulating scaffold-mediated contributions to signal flux could be a significant consideration in designing scaffolded synthetic circuits. Our results suggest both opportunities and potential challenges for the utilization of scaffolds in regulatory networks. By quantitatively delineating these tradeoffs, our results help to define the engineering challenges that must be addressed to effectively implement scaffolds in synthetic circuits.
Materials and methods
Strains
The strains used in this study are listed in Supplementary Table SI. Strain CB011 was kindly gifted by Wendell Lim of UCSF, and was used for all reporter and immunoblot experiments. Strain EY1775 was generously provided by Elaine Elion of Harvard University, and was used for the halo assay.
Plasmid constructs
The plasmids used in this study are listed in Supplementary Table SII. Vectors containing the STE5 allele, the STE7 allele, the 13Myc and 3HA epitope tags, and the ADH/CYC1/GPD/TEF promoters were kindly provided by Elaine Elion (Harvard University), Christina Smolke (Caltech), Ray Deshaies (Caltech) and David Chan (Caltech), respectively. The STE5 allele was sub-cloned by PCR from plasmid pSKM12 and was ligated into the base shuttle vector pRS416 (low-copy CEN/ARS, URA3). The STE7 allele was sub-cloned from plasmid pVS10 and was ligated into the base shuttle vector pRS415 (low-copy CEN/ARS, LEU2). The 13Myc and 3HA epitope tags were sub-cloned from plasmids pFA6a-13Myc-His3MX6 and pFA6a-3HA-His3MX6, respectively, and were fused to the C-terminus of the gene of interest in the base shuttle vectors. The various constitutive promoters were sub-cloned from the following vectors: p416ADH, p416CYC1, p416GPD and p416TEF. The native STE5 promoter was cloned from W303 genomic DNA by PCR, encompassing a sequence of 800 bp upstream to the start codon. All promoters were inserted into the base shuttle vector immediately upstream of the start codon of the gene of interest.
Flow cytometry
Yeast cells grown in selective media to mid-log phase growth (OD ∼0.1–1.0) were induced with the indicated amount of α-factor or sorbitol and shaken for 2.5 h at 30°C. One millilitre of ice-cold TE buffer was added to 0.5 ml cells. Cells were briefly vortexed to break up cell clumps and GFP fluorescence was detected using the Cell Lab Quanta SC flow cytometer from Beckman Coulter. To avoid complications that arise from pheromone-induced cell-cycle arrest, fluorescent measurements were recorded for strain CB011, which contains a far1Δ mutation that uncouples the cell-cycle response from the transcriptional mating response (Bhattacharyya et al, 2006). Data were analysed as described previously (Bhattacharyya et al, 2006), with some modifications. Electronic volume, an approximate measurement of cell size, was used instead of forward scatter. Cells were first gated on a side scatter versus electronic volume plot, and then cells were gated on a GFP versus side scatter plot to quantify fluorescence. Background fluorescence was subtracted from all fluorescent measurements.
Western blot
Cell growth and lysis
Yeast cells (CB011) grown on selective media at mid-log phase growth (OD ∼1.0, 1.3e7 cells/ml) were induced with the indicated amount of α-factor or sorbitol and shaken for 5 or 15 min at 30°C. TCA was added to 5–8 ml cells at a final concentration of 20%, and incubated on ice for 5 min. Cells were then collected and washed 3 × with 1 ml Tris–HCl (pH=8.0) by centrifugation to ensure good solubility of protein. SDS–urea buffer (50 μl water and 100 μl of 125 mM Tris–HCl (pH=7.5), 8 M urea, 4% (wt (g)/vol (ml)) SDS, 2% (vol/vol) β-mercaptoethanol, 0.02% (wt g/vol ml) bromophenol blue) was added with ∼50 μl acid-washed glass beads (425–600 μm). Cells were homogenized using Fast Prep (Bio101 Savant) at speed 6.5 for 45 s, and then the whole-cell lysate was incubated at 42°C for 15 min to promote protein solubilization. After centrifugation for 15 min at maximum speed in a tabletop centrifuge, 50 μl lysate was recovered and diluted by SDS–loading buffer (300 μl of 50 mM Tris–HCl (pH=6.8), 12% (vol/vol) glycerol, 2% (wt g/vol ml) SDS, 1% DTT, 0.01% (wt g/vol ml) bromophenol blue).
SDS–PAGE—Quantitative western blots only
To obtain quantitative data, many modifications to the standard western blot protocol were made. To validate the linear comparison of samples within a gel, a standard curve consisting of ∼7 data points was included with each gel as an internal control. To minimize variability of quantification, samples to be compared in a given gel were loaded in quadruplicate. Supplementary Figure S1A displays a typical quantitative western blot for Ste5myc measurement. This approach requires concomitant analysis of multiple samples on a single gel; thus, all quantitative gels were run using a wide-gel apparatus (TV-200YK from Topac) that accommodated 30 lanes in a single gel.
The dynamic range of the western blot protocol is limited. To successfully detect all samples within a common dynamic range (as defined by the standard curve), samples were diluted as required in the whole-cell lysate of equivalent protein concentration, but lacking the antigenic protein of interest. (Finding the proper dilutions for each blot was accomplished through an iterative procedure.) We loaded the lanes, whenever possible, with an equivalent lysate volume and protein concentration. This was done to mitigate pipetting error during gel loading, and to prevent horizontal band dispersion during electrophoresis (this effect complicates the box-drawing step of quantitation).
Immunoblotting
Blots were transferred to nitrocellulose (BioRad) and were blocked for 1 h in 3% milk TBST solution. Primary antibody incubation was conducted in blocking buffer overnight at 4°C. Primary antibodies and dilutions used in this study were as follows: anti-Myc for detection of Ste5myc, 1:10 000 (9e10; Covance); anti-Cdc28 for equal loading control, 1:1000 (sc-53; Santa Cruz Biotechnologies); anti-phospho-p44/42 MAPK for activity of both Fus3 and Kss1, 1:1000 (9101; Cell Signaling Technology); anti-Fus3 for total Fus3, 1:1000 (sc-6773; Santa Cruz Biotechnologies); anti-Kss1 for total Kss1, 1:500 (sc-28547; Santa Cruz Biotechnologies); anti-HA for detection of Ste7HA, 1:10 000 (MMS-101R; Covance) and anti-phospho-p38 MAPK for phospho-Hog1, 1:1000 (9211; Cell Signaling Technology). HRP-conjugated secondary antibodies (BioRad) were used at dilution 1:10 000. Blots were treated with Supersignal West Pico or Femto substrate (Pierce) and images were recorded using the Versa-Doc 3000 imager (BioRad).
Analysis—Quantitative western blots only
Signal intensities were quantified using the Volume tool in Quantity1 software. For each blot, equivalently sized, rectangular boxes were drawn around each band. A global background measurement was taken and was subtracted from all band intensities.
For each blot, a standard curve was constructed by linear regression. The signal intensities for experimental samples were averaged and then interpolated using the standard curve (Supplementary Figure S1B). The interpolated values were then adjusted for the differential volumes used during loading by dividing by the respective volume loaded. The output of this calculation yields the final data from a single quantitative western blot.
Data from anti-Myc Ste5, anti-phospho-Fus3 and anti-phospho-Kss1 blots were subsequently normalized by the equal loading controls total Cdc28, total Fus3 and total Kss1, respectively, unless otherwise indicated. Signal intensities for the equal loading controls were determined by the same quantitative procedure described above.
Halo assays for α-factor sensitivity
Halo assays were performed for strain EY1775 (which is FAR1+) as previously described, except that assays were performed in normal selective media with neutral pH (Sprague, 1991).
Supplementary Material
Supplementary text, Supplementary tables SI–V, Supplementary figures S1–6
Supplementary Figure S2A
Supplementary Figure S4
Supplementary Figure S5
Complete Dataset
Acknowledgments
We thank the members of the Asthagiri and Deshaies laboratories for helpful discussions and guidance with experiments; D Chan, R Deshaies, C Smolke, E Elion and W Lim for reagents and E Davidson, R Deshaies and P Sternberg for comments on the manuscript. This work was supported by the Institute for Collaborative Biotechnologies through Grant DAAD19-03-D-0004 from the US Army Research Office. SAC was supported by the NIH Molecular Cell Biology Training Grant NIH/NRSA 5T32GM07616.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bashor CJ, Helman NC, Yan S, Lim WA (2008) Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319: 1539–1543 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA (2006) The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311: 822–826 [DOI] [PubMed] [Google Scholar]
- Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ (1997) A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277: 693–696 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (1996) Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci 21: 460–466 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (2000) What do scaffold proteins really do? Sci STKE 2000: PE1. [DOI] [PubMed] [Google Scholar]
- Garrenton LS, Braunwarth A, Irniger S, Hurt E, Kunzler M, Thorner J (2009) Nucleus-specific and cell cycle-regulated degradation of mitogen-activated protein kinase scaffold protein Ste5 contributes to the control of signaling competence. Mol Cell Biol 29: 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG (2008) Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell 30: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joneson T, Fulton JA, Volle DJ, Chaika OV, Bar-Sagi D, Lewis RE (1998) Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J Biol Chem 273: 7743–7748 [DOI] [PubMed] [Google Scholar]
- Kranz JE, Satterberg B, Elion EA (1994) The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev 8: 313–327 [DOI] [PubMed] [Google Scholar]
- Kusari AB, Molina DM, Sabbagh W Jr, Lau CS, Bardwell L (2004) A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J Cell Biol 164: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg PW (2000) Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA 97: 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Zarrinpar A, Lim WA (2003) Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299: 1061–1064 [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278: 2075–2080 [DOI] [PubMed] [Google Scholar]
- Sprague G (1991) Assay of yeast mating reaction. Methods Enzymol 194: 77–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text, Supplementary tables SI–V, Supplementary figures S1–6
Supplementary Figure S2A
Supplementary Figure S4
Supplementary Figure S5
Complete Dataset