Abstract
The onset of prezygotic and postzygotic barriers to gene flow between populations is a hallmark of speciation. One of the earliest postzygotic isolating barriers to arise between incipient species is the sterility of the heterogametic sex in interspecies' hybrids. Four genes that underlie hybrid sterility have been identified in animals: Odysseus, JYalpha, and Overdrive in Drosophila and Prdm9 (Meisetz) in mice. Mouse Prdm9 encodes a protein with a KRAB motif, a histone methyltransferase domain and several zinc fingers. The difference of a single zinc finger distinguishes Prdm9 alleles that cause hybrid sterility from those that do not. We find that concerted evolution and positive selection have rapidly altered the number and sequence of Prdm9 zinc fingers across 13 rodent genomes. The patterns of positive selection in Prdm9 zinc fingers imply that rapid evolution has acted on the interface between the Prdm9 protein and the DNA sequences to which it binds. Similar patterns are apparent for Prdm9 zinc fingers for diverse metazoans, including primates. Indeed, allelic variation at the DNA–binding positions of human PRDM9 zinc fingers show significant association with decreased risk of infertility. Prdm9 thus plays a role in determining male sterility both between species (mouse) and within species (human). The recurrent episodes of positive selection acting on Prdm9 suggest that the DNA sequences to which it binds must also be evolving rapidly. Our findings do not identify the nature of the underlying DNA sequences, but argue against the proposed role of Prdm9 as an essential transcription factor in mouse meiosis. We propose a hypothetical model in which incompatibilities between Prdm9-binding specificity and satellite DNAs provide the molecular basis for Prdm9-mediated hybrid sterility. We suggest that Prdm9 should be investigated as a candidate gene in other instances of hybrid sterility in metazoans.
Author Summary
Speciation, the process by which one species splits into two, involves reproductive barriers between previously interbreeding populations. The question of how speciation occurs has rightly occupied the attention of biologists since before Darwin's “On the Origin of Species.” Studies of recently diverged species have revealed the presence of hybrid sterility genes (colloquially referred to as “speciation genes”), alleles of which are associated with sterility of interspecies hybrids. Mouse Prdm9 is the only known such gene in vertebrate animals. Here we report that the Prdm9 protein has evolved extremely rapidly in its DNA-binding domain, comprising an array of “zinc fingers.” This suggests that hybrid sterility may arise from a mismatch between the DNA-binding specificity of Prdm9 and rapidly evolving DNA. We propose that Prdm9 binds to satellite-DNA repeats evolving rapidly within and between different species. Prdm9 evolution is unusual because other hybrid sterility genes appear only to evolve rapidly in isolated bursts, whereas Prdm9 has evolved rapidly over 700 million years, in many rodent species, diverse primates and other metazoans. This leads to the tantalizing possibility that Prdm9 may have served as a “speciation gene” on other occasions in metazoan evolution, a possibility that will now need to be investigated.
Introduction
The question of how two species originate from one has fascinated biologists since before Darwin's iconic treatise on the subject [1]. Postzygotic reproductive barriers between species are thought to result from the acquisition of genetic incompatibilities as an incidental by-product of divergence between two populations. In its simplest form, this Dobzhansky-Muller model involves genetic interactions between two loci (e.g. a and b) [2]. In isolated populations, new alleles can arise and go to fixation in two isolated populations (A in one and B in the other) since they remain compatible with ancestral alleles. However, a negative epistatic interaction between the two new alleles (A with B) in hybrids might result in sterility or inviability, a hallmark of postzygotic isolation in hybrids between two species [3]. Theory predicts that additional incompatibilities will accumulate rapidly following an initial genetic incompatibility [4]. One of the earliest postzygotic isolating barriers in interspecies hybrids is the sterility of the heterogametic sex (XY males or ZW females), a pattern referred to as Haldane's rule that holds almost universally across animal taxa [3],[5]. Examination of early events in speciation that lead to hybrid sterility (for example [6],[7]) is thus vital to gain insight into this mysterious process.
The first hybrid sterility gene to be discovered was the Drosophila Odysseus-site homeobox (OdsH) gene. The D. mauritiana allele of OdsH causes hybrid male sterility when introgressed into D. simulans together with adjacent loci [8],[9]. OdsH encodes a presumptive DNA-binding protein which is exclusively expressed in male reproductive tissues [9]. OdsH function within Drosophila species remained unclear until recently (ablation of the gene in D. melanogaster has a very modest effect on male fertility [10]) as did the molecular basis for why it causes hybrid sterility. However, the manifestation of hybrid sterility appears to be correlated with rapid evolution of OdsH specifically in its DNA-binding homeobox domain, in the species clade that includes D. mauritiana and D. simulans [11].
A second hybrid sterility gene was discovered not as a Dobzhansky-Muller incompatibility but as a result of gene transposition. Hybrids between D. melanogaster and D. simulans, which carry two 4th chromosomes from D. simulans in an otherwise D. melanogaster genetic background, are sterile. This sterility is caused by the transposition of the JYAlpha gene away from the 4th chromosome in D. simulans [12]. Since JYAlpha is required for male fertility, D. melanogaster male flies that only possess D. simulans 4th chromosomes lack JYAlpha and are therefore sterile. In contrast to OdsH, the biological cause of hybrid sterility is well understood but involves no sequence divergence of the underlying sterility gene and only affects a fraction of F2 hybrids.
A third hybrid sterility gene was recently discovered in crosses between the Bogota and USA subpopulations of D. pseudoobscura. F1 males resulting from crosses between Bogota females and USA males are almost completely sterile when young. When aged, however, these F1 males recover partial fertility but produce all female progeny. Intriguingly, a single gene Overdrive (Ovd) was found to be causal for both the segregation distortion and hybrid male sterility [13]. Like OdsH, Ovd encodes a putative DNA-binding protein whose biological function is unclear. Like OdsH, rapid evolution of Ovd in the Bogota lineage appears to be associated with hybrid sterility. Genetic results with Ovd strongly suggest that hybrid sterility is a by-product of intraspecies genomic conflict, manifest as segregation distortion [13].
Prdm9 (Meisetz) is the fourth hybrid sterility gene, the first to be described in vertebrates. It was discovered in crosses between the mouse subspecies Mus musculus musculus and Mus musculus domesticus. Allelic differences at Prdm9 provide the genetic basis for the Hybrid sterility 1 (Hst1) locus, which together with other genetic loci [6],[7],[14], is responsible for spermatogenic failure in sterile hybrids between Mus m. musculus and Mus m. domesticus [15]. Polymorphism linked to Hst1 is associated with sterility traits not only for Mus m. domesticus strains but also, separately, for Mus m. musculus strains [16]. In natural Mus m. musculus populations these polymorphisms appear to have arisen very recently [16]. Prdm9 is a meiosis-specific gene that is only expressed in germ cells entering meiotic prophase in both female and male mice [17]. Loss of Prdm9 causes sterility in both sexes due to impaired meiotic progression at the pachytene stage. Furthermore, nonsynonymous SNPs in human PRDM9 are associated with infertility and azoospermia via meiotic arrest [18],[19]. Prdm9 encodes 3 protein isoforms, of which the largest isoform contains an N-terminal KRAB motif, a central histone H3 Lysine-4-methyltransferase (SET) domain, and several zinc fingers in its carboxy-terminal region (Figure 1). Similar zinc fingers in other proteins have been shown to mediate sequence-specific binding to DNA. The number of zinc fingers encoded in mouse Prdm9 appears to directly affect hybrid sterility. Whereas an allele of Prdm9 encoding 13 zinc fingers causes postzygotic hybrid sterility, an allele containing 14 zinc fingers does not (Figure 1) [15]. The finding that changes in a single DNA-binding determinant appears to be causal for hybrid sterility motivated our analysis to study the evolutionary constraints that shape the sequence and copy number of zinc finger motifs in Prdm9 across a broad taxonomic panel of metazoans, starting with rodents.
Figure 1. Schematic of Prdm9 protein encoded by M. musculus.
Schematic of the domain architecture for the long protein isoform encoded by the M. musculus Prdm9 gene. The Prdm9 protein contains KRAB, SSXRD, and SET domains and a single zinc finger in its N-terminal half, while the C-terminal half consists of an array of zinc finger domains [17]. The shorter Prdm9 protein isoforms lack the C-terminal zinc fingers and apparently do not localize to the nucleus. Sterility and fertility associated alleles of Prdm9 in M. m. musculus differ only in one extra zinc finger (red triangle) [15].
Results
Concerted evolution and positive selection of Prdm9-encoded zinc fingers in rodents
We sequenced the terminal zinc fingers from the final exons of Prdm9 from 11 rodent species to which we added the genomic sequences of mouse (C57BL/6J) and rat Prdm9 (Figure 2A), thereby sampling a ∼25 million year period of rodent phylogeny [20]. The C57BL/6J strain of mice is a mosaic of M.m. musculus, M.m. domesticus and M.m. castaneus [21]. The C57BL/6J mouse genome assembly harbours the M.m. domesticus Prdm9 allele [22]. We found that rodents vary greatly in their numbers of zinc fingers present in the C-terminal array: from 7 in Peromyscus polionotus to 12 in Mus musculus (Figure 2A). Even closely-related species pairs, such as field and water voles (Microtus agrestis and Arvicola terrestris), and M. macedonicus and M. spicilegus, differ in their numbers of zinc fingers (Figure 2A).
Figure 2. Concerted evolution among rodent Prdm9 genes.
(A) Prdm9 C-terminal zinc fingers for 13 rodent species are shown as pink rectangles. Zinc fingers whose nucleotide sequences are identical are joined by solid lines. Zinc fingers with identical sequences from the same species are consistent with gene conversion and/or intra-exon duplication. A phylogeny of these species is also shown with estimated divergence dates (indicated at nodes) given in millions of years (my) [20],[83]. Common names to species are listed in the legend to Figure 6. (B) The proportion of pairwise cDNA comparisons between aligned zinc fingers from the same gene (see Materials and Methods) which show greater than 90% identity. All mouse Prdm9 zinc fingers are more than 90% identical to all other C-terminal zinc fingers in the same protein (indicated in red), a much higher fraction than for any other zinc finger protein encoded by the mouse genome.
Rodent Prdm9 zinc finger sequences have been subject to concerted evolution. Many changes in numbers of zinc fingers have resulted from very recent lineage-specific duplications (Figure 2A). Twelve of the 13 rodent species we examined possess at least one pair of Prdm9 zinc fingers that were so recently duplicated that they have identical nucleotide sequences. In one case (Peromyscus leucopus, Figure 2A), Prdm9 encodes a cluster of five zinc fingers that are identical at the nucleotide level, together with another pair of identical zinc fingers. Consistent with concerted evolution, Prdm9 zinc fingers from the same species often form monophyletic clades, even in comparisons of closely related rodents (Figure S1). Such concerted sequence evolution may result from multiple rounds of zinc finger duplication and deletion (‘birth-and-death’ model [23]) to change zinc finger numbers. However, we favor non-allelic gene conversion as a dominant mechanism [24] since it more easily accounts for the many interdigitated and non-adjacent zinc finger duplications, as well as the complexity of the inferred zinc finger phylogeny. Although more occasional gain and loss of zinc finger sequences have been observed previously for other genes [25], the extreme degree of sequence similarity between different zinc finger pairs is far greater for Prdm9 than for any other zinc finger gene present in the C57BL/6J mouse genome sequence (Figure 2B).
In addition to concerted evolution, our analyses reveal evidence for positive selection at particular codons responsible for DNA binding specificities within Prdm9 zinc fingers in rodents. Due to the high degree of concerted evolution, it is not formally correct to carry out a pairwise analysis of the non-synonymous to synonymous rate ratio (dN/dS) when comparing Prdm9 sequences from two different species. Instead, by comparing all Prdm9 zinc fingers within a species, we find that all but one of these 13 rodent species have acquired more amino acid substitutions than would be expected under neutral evolution within their Prdm9 zinc fingers (Figure 3). For instance, in the Prdm9 encoded zinc fingers from Mus musculus strain C57BL/6J (Figure 3A), two codons are predicted to have evolved under positive selection (positions labelled −1 and 3 in Figure 3A). Intriguingly, positive selection is restricted to only a small number of positions within these zinc finger sequences. Sites labelled −1, 3, and 6 were identified as having evolved by positive selection in the majority of the 13 rodent species we examined when comparing all zinc fingers from a particular species (tabulated in Figure 3B). Codons at these sites are turned over rapidly. For instance, two recently diverged vole species, Microtus agrestis and Arvicola terrestris exhibit species-specific codons at positions −1, 3 and 6 (Figure S2) despite their independent evolution only over the last 0.5 million years [26]. In each case, we use the Sitewise Likelihood-ratio method (SLR) [27] with p-value thresholds of 0.05 after multiple testing correction. Since these methods can be strongly affected by tree topology, we tested both the most likely and other competing topologies to conservatively estimate non-synonymous substitutions; this will reduce the chance of false-positives in our analysis (see Materials and Methods). These unusually elevated values may reflect the sustained action of positive selection, consistent with the elevated rates observed for many rodent species (Figure 3). Rapid evolution and addition/deletion of zinc fingers (that provide the basis for hybrid sterility among M. musculus strains [15]) are thus recurrent across rodent evolution.
Figure 3. Positive selection of zinc fingers encoded by rodent Prdm9 genes.
(A) A multiple alignment of the zinc finger sequences from M. musculus C57BL/6J highlights the invariant Cys2His2 Zn2+-coordinating residues as well as positions −1, 3, and 6 that dictate the DNA-binding specificity of individual zinc fingers. Deviations from the consensus amino acid at each position are shown in boldface. In this species, positions −1 and 3 meet the criteria for positive selection [27] (highlighted in yellow and with red crosses). (B) Predicted positively selected sites in Prdm9 from diverse rodent lineages. Positive selection was inferred for each species [27] from intra-species Prdm9 zinc finger sequence alignments. Positively selected sites (P<0.05 after multiple testing correction) are shown mapped to the third mouse Prdm9 zinc finger sequence (MMM3). The majority of positively selected sites fall at positions −1, 3, and 6. (C) Estimated dN/dS values at four zinc finger positions (namely, −2, −1, 3, and 6) in a comparison of zinc fingers from all rodents (in contrast to the analyses of species-specific zinc fingers in (B)) for which there exists strong evidence of positive selection [27]. The P-values shown have been corrected for multiple testing. Common names to species are listed in the legend to Figure 6.
We also inferred evolutionary rates for each codon from an alignment of every Prdm9 zinc finger from all of these 13 rodent species. Rates for three sites (sites −1, 3 and 6), together with a fourth (site −2), greatly surpass the neutral rate with values of dN/dS up to 8 (Figure 3C). These ratios greatly exceed those found for corresponding positions in other mammalian zinc finger genes [28]–[30]. These three positions (namely −1, 3 and 6) correspond exactly to the positions known to be involved in sequence-specific DNA-binding [31],[32]; structural studies have shown that amino acids within the zinc finger α-helix at positions −1, 3 and 6 make contacts with bases 3, 2 and 1 in the primary DNA strand respectively, whilst the amino acid at position 2 interacts with the complement of base 4 [33]. Thus the finding that positive selection on residues −1, 3 and 6 indicates that it has specifically acted to alter DNA-binding preferences encoded by Prdm9.
Rapid evolution of PRDM9 in primates
Based on our findings in rodents, we next undertook a survey of PRDM9 divergence in the primate lineage to ask whether the extraordinary evolution of Prdm9 was limited to rodents alone. In humans, there appear to be two genes that are orthologous to a single mouse Prdm9, suggesting a recent gene duplication [34],[35]. These two genes, PRDM7 and PRDM9, are found at chromosomal locations 16q24.3 and 5p14, respectively. It is clear that since the gene duplication PRDM7 has acquired distinct tissue-specific patterns of expression and has undergone major structural rearrangements, dramatically altering the number of encoded zinc fingers (2 in macaques, 5 in orangutans) while diverging from ancestral patterns of transcript splicing [34]. Furthermore, there is evidence for a frame-disruption affecting PRDM7 in some humans. Consequently, we do not investigate PRDM7 further in this report.
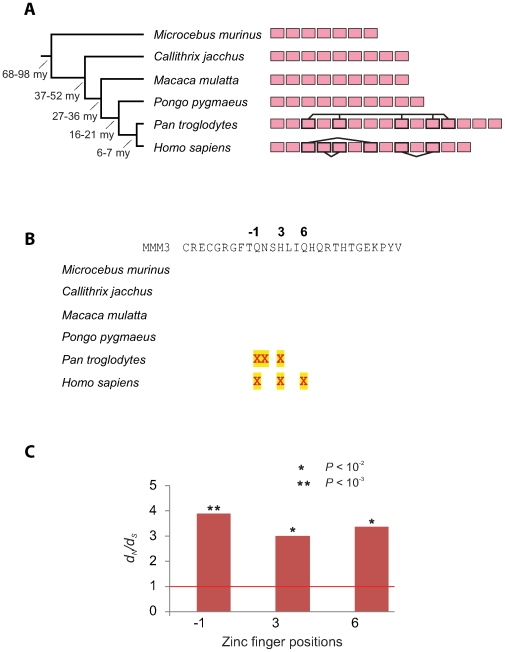
Primate PRDM9 appears to show a large variation in numbers of zinc fingers in its C-terminal array similar to what we found in rodents (Figure 2A). Chimpanzee, orangutan, rhesus macaque and marmoset PRDM9 genes encode 15, 10, 9, and 9 C-terminal zinc fingers as opposed to 13 in human PRDM9 (Figure 4A). As in rodents, primate zinc fingers also show evidence for concerted evolution. For example, there are three identical pairs out of the C-terminal array of 13 zinc fingers encoded by human PRDM9.
Figure 4. Concerted evolution and positive selection among primate PRDM9 genes.
(A) PRDM9 C-terminal zinc fingers for 6 primate species are shown as pink rectangles. Zinc fingers whose nucleotide sequences are identical are joined by solid lines. Zinc fingers with identical sequences from the same species are consistent with gene conversion and/or intra-exon duplication. (B) Predicted positively selected sites in Prdm9 from divergent primate lineages. Positive selection was inferred for each species [27] from intra-species Prdm9 zinc finger sequence alignments. Positively selected sites (P<0.05 after multiple testing correction) are shown mapped to the third mouse Prdm9 zinc finger sequence (MMM3) as shown in Figure 3A. (C) Estimated dN/dS values at three zinc finger positions (namely −1, 3, and 6) in a comparison of zinc fingers from all primates for which there exists strong evidence of positive selection [27]. The P-values shown have been corrected for multiple testing. Common names to species are listed in the legend to Figure 6.
When we compared the PRDM9 gene sequence between humans and chimpanzees, we found the nucleotide divergence to be 7.1%, over 5-fold higher than the divergence observed genome-wide (1.23% [36]) although the high degree of concerted evolution complicates this human-chimpanzee ortholog comparison. However, it does appear that much of the divergence has resulted from a combination of positive selection and concerted evolution. Estimated dN/dS values for positions −1, 3 and 6 of human PRDM9 zinc fingers are 12.6, 9.9 and 13.9 respectively, substantially greater than 1. Indeed, either by a species-specific zinc finger analysis (Figure 4B) or by a pooled analysis of all primate PRDM9 encoded zinc fingers (Figure 4C), we find strong evidence for positive selection at these positions.
Our findings suggest that positive selection and concerted evolution have directly and dramatically altered DNA-binding specificity of the encoded PRDM9 protein in primates as was observed in rodents. For instance, for 12 of the 15 C-terminal array of chimpanzee PRDM9 zinc fingers, codons at position −1 are not found in any human PRDM9 zinc finger at the same position; similarly, 6 human zinc fingers have codons at this position that are not present in the chimpanzee ortholog (Figure 5A and 5B). Like in rodents (Figure 2 and Figure 3), the PRDM9 genes of closely related primate species are differentiated not only by the numbers of zinc fingers they encode, but also by species-specific codons, particularly at key positions that dictate DNA-binding specificity (Figure 4 and Figure 5).
Figure 5. Sequence divergence and diversity among human and chimpanzee PRDM9 zinc finger sequences.
(A) Multiple sequence alignment of human (Homo sapiens) PRDM9 zinc finger sequences, with positively selected positions (P<0.05, after multiple testing correction) indicated by red asterisks interspersed among a consensus amino acid sequence. Positions −1, 3, and 6 (numbered relative to the start of the zinc finger α–helix) that represent sequence-variable positions frequently involved in DNA binding are also indicated. Codons highlighted in green are not found at the same position in any chimpanzee PRDM9 zinc finger. (B) Multiple sequence alignment of chimpanzee (Pan troglodytes) PRDM9 zinc finger sequences with a predicted positively selected site indicated as in panel (A). Note that several chimpanzee PRDM9 zinc finger codons (highlighted in green) at positions −1 and 3 are unique to this species, relative to humans (A). (C) Numbered and boxed codons in panel (A) contain human nonsynonymous SNPs. SNPs numbered 1–7 were identified in this study among 50 Chinese individuals whilst heterozygous SNPs numbers 3, 6, 8, and 9 are significantly enriched among fertile, as opposed to infertile, males in the study by Irie et al. [18].
PRDM9 evolution in humans
We next investigated whether positive selection on PRDM9 had left population genetic signatures of selection that still remained evident among modern humans. Each of the two methods we employed exploits SNP data and accounts for issues concerning population structure and growth (see Materials and Methods). Particularly recent selective sweeps are characterized by long extents of linkage disequilibrium (LD) that ensue when the haplotype carrying the advantageous allele rises in frequency more rapidly than a neutral allele. Conversely, tests based on this characteristic are particularly sensitive for detecting recent episodes of positive selection [37]. Looking at patterns of LD, we did not find evidence for very recent selective sweeps at PRDM9. In our test we computed the maximum correlation coefficient (r2) between SNP pairs spanning the PRDM9 locus, and compared these to the empirical distribution of this statistic across the genome. These maximum r2-statistics were not significantly different from the background (p values of 0.24, 0.23 and 0.24 for the African, European and Japanese/Chinese population panels).
Since tests based on long extents of LD or haplotypes are sensitive for very recent sweeps [37] only, while tests based on Tajima's D maintain power until some time after fixation of the advantageous allele [38], we also used a Tajima's D estimate to investigate whether polymorphisms linked to PRDM9 exhibit an unusual population frequency spectrum. When an advantageous allele has risen to fixation, the extended haplotype associated with it will, for a considerable time thereafter, carry young and low-frequency polymorphisms, which may be observed as a reduction of Tajima's D, defined as the scaled difference of two estimators of heterozygosity which are identical under the standard neutral model [39]. There are significant caveats to the calculation of Tajima's D from genotyping data which bias against the recovery of low frequency SNPs. The Perlegen genotyping data have been shown to provide useful Tajima's D statistics after empirically accounting for this ascertainment bias [40],[41]. Using these methods, we calculated Tajima's D at the PRDM9 locus [41] in African Americans (D = −0.130; p = 0.038), European Americans (D = −0.259, p = 0.068), and Asian Americans (D = 1.7). With the caveat that there might be uneven distribution of ascertainment biases across the genome, there appears to be weak evidence for a recent selective sweep in African Americans. In contrast to PRDM9, Tajima's D provides no evidence for recent sweeps in any of the three populations at the PRDM7 locus.
We were interested in using intraspecies human polymorphisms to gain further insight into the evolutionary forces that drive the concerted evolution of PRDM9. To this end, we sequenced the terminal PRDM9 zinc finger sequences from 50 Han Chinese individuals, seeking sequence polymorphisms that might have arisen by gene conversion. Under gene conversion, we would expect to observe a nucleotide polymorphism in one zinc finger that is identical to its fixed paralogous base in another. We observed 7 codons containing single nucleotide polymorphisms (SNPs; blue rectangles in Figure 5A and 5C). Of these, 4 (numbered 1, 2, 5 and 7 in Figure 5A and 5C) represent changes to codons that are not represented among any of the remaining zinc finger sequences and thus are unlikely to have arisen by gene conversion. The remaining 3 changes are to codons that are also present in at least one paralogous position within the other zinc fingers. A separate study identified 17 non-synonymous SNPs within human PRDM9 zinc fingers, of which 13 showed evidence for having arisen by gene conversion from paralogous sequences [18]. We infer, therefore, that non-allelic gene conversion has contributed to the rapid evolution of primate PRDM9, and this provides a likely mutational mechanism for many other PRDM9 orthologues.
What are the functional consequences of these non-synonymous SNPs in PRDM9? Two recent genetic association studies have investigated PRDM9 SNPs and their association with azoospermia. The first study [19] did not find correlated SNPs in the C-terminal zinc fingers. However, a second study found that individual nonsynonymous SNPs in the zinc finger domain are associated with a significantly decreased risk of infertility [18]. For instance, human non-synonymous SNPs (labelled 3, 6, 8 and 9 in Figure 5A and 5C) are associated with decreased risk of sterility in a cohort of Japanese men [18], of which two (numbers 3 and 6) were found among the 50 Han Chinese individuals we sequenced. In addition, 3 out of 4 non-synonymous SNPs associated with fertility are found at zinc finger position 6, a site predicted to determine DNA-binding specificity and which we show has evolved under positive selection in human PRDM9 (Figure 4 and Figure 5A). Surprisingly, in each instance, the ‘minor’ allele at each position is associated with protection against sterility in Japanese men [18]. Intriguingly, in both studies, the effect on ameliorating azoospermia or oligospermia was manifest even in the heterozygous condition [18],[19], suggesting that PRDM9's effect is semi-dominant (consistent with results of hybrid sterility seen in mouse Prdm9). In a situation where a minor allele provides a protective benefit against sterility, we might expect that high frequency retention of these alleles would be favored by balancing selection in this population. Consistent with this expectation, we point out that Asian American individuals had a striking Tajima's D of +1.7 in contrast to the negative Tajima's D in the other two populations in the Perlegen dataset, although this statistic by itself is not strong evidence of balancing selection given the ascertainment bias.
Rapid evolution of Prdm9 is an ancient feature in metazoans
The two evolutionary themes (concerted evolution and positive selection) that typify PRDM9 evolution in primates and in rodents also have occurred recurrently across metazoan evolution (summarized in Figure 6). For instance, we found evidence of concerted evolution among Prdm9-encoded zinc fingers in the sea anemone Nematostella vectensis, the gastropod snail Lottia gigantea, and the polychaete worm Capitella sp. I (Figure S3, S4, S5), organisms that last shared a common ancestor with mammals approximately 700 million years ago [42]. In addition, we find strong evidence of positive selection in zinc fingers of N. vectensis Prdm9 for the same 3 positions (namely, −1, 3 and 6) also identified from analyses of rodent and primate lineages (summarized in Figure 6). Estimated dN/dS values for these positions were exceptionally high, ranging between 25 and 32. A single codon of the Capitella worm Prdm9 zinc fingers also shows evidence of positive selection (Figure 6). Thus, even early branching metazoans show strong evidence of both concerted evolution and positive selection within Prdm9-encoded zinc fingers.
Figure 6. Predicted positively selected sites in Prdm9 from divergent metazoan lineages.
Results for previously presented rodent and primate lineages are also shown here for comparison (blue shading). Positive selection was inferred for each species [27] from intra-species Prdm9 zinc finger sequence alignments. Positively selected sites (P<0.05 after multiple testing correction) are shown mapped to the third mouse Prdm9 zinc finger sequence (MMM3). The majority of positively selected sites, across 700 million years of divergence from sea anemone to mammals, fall at positions −1, 3, and 6. The inferences of positive selection for Capitella were made on the basis of three sequences on separate unassembled genomic scaffolds. Despite their high sequence similarity, multiple uncorrelated point substitutions, especially among the zinc fingers, suggest that they may represent allelic copies or rapidly diverging paralogues.
Concerted evolution is also apparent in Prdm9 zinc fingers for many mammals including elephants (Loxodanta africana), cats (Felis catus), common shrews (Sorex araneus), cattle (Bos taurus), muntjak deer (Muntiacus reevesi and Muntiacus muntjak vaginalis), bats (Myotis lucifugus) and rabbits (Oryctolagus cuniculus) (data not shown). It is also evident among the zinc fingers of Prdm9 from the Atlantic salmon (Salmo salar) and the rainbow trout (Oncorhynchus mykiss). Of the four complete zinc fingers in rainbow trout Prdm9, two are identical in nucleotide sequence, and the remaining pair are more closely-related to each other than they are to those of Prdm9 for the Atlantic salmon (Figure S6), with which it last shared a common ancestor approximately 20 million years ago [43]. Evidence for positive selection is, however, less compelling outside of these fish, the sea anemone, rodents and primates. This is perhaps owing to the stringent multiple testing correction we employed, especially in cases where there are insufficient zinc fingers to obtain significant power for this kind of analysis (see Materials and Methods).
Despite strong evidence of concerted evolution and/or positive selection in many metazoan Prdm9 sequences, this pattern is not universal across all metazoans. In comparisons of Prdm9 in other ray-finned fishes (including Danio rerio) and in tunicates (including Ciona intestinalis), we found no evidence for either concerted evolution or positive selection within their zinc fingers. Among mammals, we found two homologs of Prdm9 in the platypus Ornithorhynchus anatinus, but evidence for neither concerted evolution nor positive selection. When we investigated the Prdm9 ortholog in the marsupial Monodelphis domestica and the nematode Caenorhabditis elegans, we were surprised to find a complete loss of all zinc fingers. Despite Prdm9 being essential for fertility in mice, Prdm9 appears lacking in chicken (Gallus gallus), frog (Xenopus tropicalis) and fly (Drosophila melanogaster) genomes, while the dog (Canis familiaris) genome has acquired multiple disruptive mutations (“pseudogenization”) within its Prdm9 ortholog [44]. This either implies that Prdm9 function in meiosis is carried out by another gene in these lineages, or that Prdm9's essential function in meiosis is itself lineage- or species-specific.
Discussion
Rapid evolution of DNA–binding specificity and insights into Prdm9 function
Our finding of recurrent and dramatic episodes of rapid evolution of Prdm9 in different lineages indicates that the protein-DNA interface at which Prdm9 acts, has frequently altered between, and within, species. These evolutionary observations allow us to revisit some key models of Prdm9 function and how its divergence might give rise to hybrid sterility. The currently prevailing model is that Prdm9 encodes a transcription factor for euchromatic genes during meiosis. Mouse Prdm9 (Meisetz) was first discovered for its essential role in meiotic prophase of both male and female meiosis [17]. Its SET domain was later found to catalyse the specific transition from di- to tri-methylation of the Lysine-4 residue on histone H3 (H3-K4), an activity that is characteristically associated with transcriptional activation [45]. Indeed, by tethering experiments, Prdm9 was shown to be able to activate transcription. Furthermore, in Meisetz−/− testes, the transcriptional regulation of close to 125 genes was disturbed. Thus, Prdm9 (Meisetz) was proposed be a master transcriptional regulator of entry into meiosis in mammals, and all data including the intriguing association with human azoospermia [18],[19] are consistent with this view [17].
However, the accelerated evolution of the Prdm9-DNA interface challenges whether Prdm9's only, or even primary, role is a transcription factor for euchromatic genes. Such a function would leave unexplained why cis-acting (promoter) sequences to which Prdm9 binds, would be subject to repeated positive selection over the long time course of metazoan evolution. Rapid evolution at the protein-DNA interface would be especially disfavoured if it was required for fertility. We cannot formally rule out the unprecedented possibility that a transcription factor may evolve rapidly in concert with all of its (at least 125 [17]) cis-acting binding sites if indeed Prdm9 directly mediates the transcription activation of meiotic promoters. However, in general, the larger the number of cis-acting sequences that Prdm9 has to bind, the more its DNA-binding would be expected to be evolutionarily constrained which, we suggest, argues against its primary role as a transcription factor.
An alternative model for Prdm9 function
We considered the possibility that the rapid evolution of Prdm9 was actually required for, rather than an impediment to, its function. One of the strongest observations in favor of the transcription model was the fact that the SET domain catalyzed transition from di- to tri-methyl H3-K4, a chromatin mark most often associated with transcriptional activation. And yet, this chromatin mark is not unique to transcriptional activation. Indeed, the same transition from di- to tri-methyl H3-K4, distinguishes canonical H3-nucleosomes at centromeric versus pericentric heterochromatic regions at mitotic centromeres of organisms as diverse as flies and humans [46]. Inactivation of a centromere on a human artificial chromosome directly results in loss of H3-K4 dimethylation and accumulation of H3-trimethylation [47].
We hypothesize that Prdm9's essential role in meiosis is directly related to its ability to bind rapidly-evolving DNA elements. While we do not know the identity of these DNA elements, we speculate that Prdm9 may function by binding directly to repetitive DNA sequences that are found at pericentric and centromeric regions (Figure 7A). Such repetitive DNA sequences (or ‘satellite repeats’) evolve exceedingly rapidly across multiple lineages [48]–[52]. It has been previously proposed that this rapid evolution results from centromere-drive [53],[54], a process in which meiotic products compete during female meiosis for retention in the egg versus exclusion as polar bodies. The genetic opportunity to ‘cheat’ during female meiosis is the evolutionary thread common among many repetitive DNA elements [55]–[58]. Further, DNA-binding proteins are thought to rapidly evolve their DNA-binding specificity to suppress this ‘meiotic drive’ [59]–[63]. Under this model, rapid changes in satellite-DNA sequences potentially ensuing from centromere-drive are followed by positive selection of non-synonymous substitutions within Prdm9 DNA-binding determinants to counter the deleterious effects of the meiotic (centromere) drive process. This would explain not only the rapid evolution and retention of Prdm9 in most metazoans but also the loss of Prdm9 genes in some lineages, when a second satellite-DNA binding protein may have taken over this suppressor function.
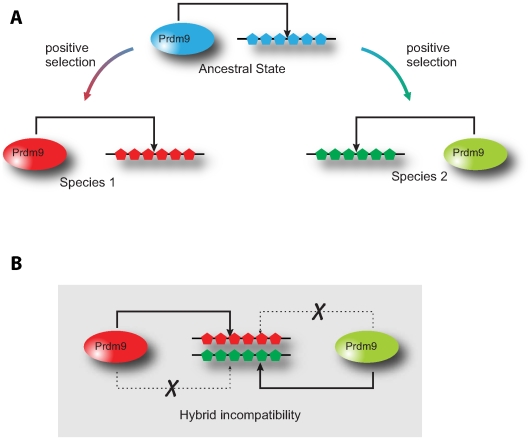
Figure 7. A novel satellite–DNA binding model for Prdm9 in hybrid sterility.
(A) Prdm9 could serve as a satellite-DNA binding protein that facilitates its heterochromatinization. Hybrid sterility ensues when the sterility-associated Prdm9 protein (blue) cannot bind to “newly expanded” satellite DNA repeats (red or green) potentially at pericentric regions of chromosomes that arose due to centromere-drive [63]. (B) Under this model, in isolated populations, satellite–DNAs diverge quickly by “centromere-drive” and Prdm9 DNA–binding specificity evolves rapidly to suppress this drive. However, in hybrid males, inappropriate localization of Prdm9 to diverged DNA–binding satellites from other species would result in either inappropriate chromosome condensation (as shown) or compromised centromere function (not shown), either of which would result in male sterility.
A recent study on the Drosophila OdsH hybrid sterility gene provides interesting parallels to the Prdm9 study [64]. Due to its evolutionary descent from the unc-4 transcription factor [11], OdsH was also believed to be a transcription factor. Since the DNA-binding homeobox domain had undergone rapid evolution, hybrid sterility was proposed to result from altered gene expression in Drosophila testis [65], much the same as it has been suggested for Prdm9. However, functional analyses of OdsH revealed it to function as a heterochromatin-binding protein, with altered DNA binding resulting in altered heterochromatic localization and chromosome decondensation [64]. A transcription factor function of Prdm9 (like in OdsH) may be directly tied to a chromosome decondensation function. Indeed, work from a number of model systems especially the fission yeast Schizosaccharomyces pombe has revealed that transcription of heterochromatic repeats is a prequel and often a pre-requisite for the deposition of heterochromatin-specific histone modifications and proteins required for transcriptional silencing and condensation [66]–[69]. Prdm9 binding to satellite-DNA may facilitate its heterochromatinization by virtue of its transcriptional activity (Figure 7A), and alterations of Prdm9's binding specificity could allow it to act on a wider array of satellite-DNAs, consistent with its semi-dominant effect in hybrid sterility and human azoospermia. The chromosome decondensation and synapsis defects in male meiosis observed in sterile hybrids between M. m. musculus and M. m. domesticus species [15] would be explained by an inability to correctly bind and package satellite DNA (Figure 7B). Indirect consequences of such decondensation could be the transcriptional misregulation of some genes, as observed in Prdm9−/− mice [17]. Alternatively, ‘mismatched’ binding of Prdm9 to centromeric satellite-DNA repeats would result in their inappropriate heterochromatinization, again leading to chromosome condensation defects and male sterility. Under this model, mismatched Prdm9-satellite DNA configurations would be predicted to result in sterility only in hybrid males, but not in hybrid females [53]. We would like to emphasize the current absence of functional data to support such a hypothesis. However, the precedence provided by the OdsH study [64] and the consistent rapid evolution seen at Prdm9's DNA-binding interface provides a simple, testable explanation for the onset of highly context-specific hybrid sterility. Variation in Hst1 (Prdm9) occasions a genetic incompatibility between the Prdm9 DNA-binding protein encoded by this locus and the satellite DNAs to which Prdm9 binds (or fails to bind). The finding that human azoospermia is rescued by heterozygous PRDM9 alleles [18],[19], including some that alter DNA-binding preferences, further suggests that a reduced repertoire of satellite-DNA binding ability may be responsible for the meiotic arrest at pachytene seen not only in the hybrid mice species [15], but also in the Prdm9 −/− mice [17], a possibility that directly lends itself to genetic and cytological scrutiny.
The proposal that episodes of meiotic drive and suppression drive hybrid incompatibilities is not new [70],[71]. Indeed, cryptic meiotic-drive suppressor systems have been uncovered by introgression analyses between different Drosophila species [72]. Moreover, recent studies of hybrid inviability amongst Drosophila species have revealed the very likely role that pericentric heterochromatin plays in the manifestation of genetic incompatibility [73],[74]. While the molecular function of Prdm9 remains to be fully elucidated, our findings directly implicate the Dobzhansky-Muller incompatibility underlying Prdm9-mediated sterility as residing at a rapidly evolving protein-DNA interface.
Recurrent rapid evolution of the Prdm9 hybrid sterility gene
The onset of interspecies hybrid incompatibilities is widely believed to ensue as the by-product of acquired genetic differences in geographically isolated populations. This process can be imagined to take place in the absence of any selective pressure, purely by genetic drift [75]. However, the accumulation of genetic incompatibilities is more likely with accelerated evolutionary change, especially if recurrent genetic conflicts were driving the divergence. Consistent with this, many hybrid incompatibility genes for both sterility and inviability are associated with dramatic episodes of positive selection [11],[73],[76].
Here, we have shown that the Prdm9 gene, which was identified as a hybrid sterility gene in mice [15], has evolved rapidly due to the dual forces of concerted evolution and positive selection. This rapid evolution is seen not just across the rodent lineage, but also in primates and especially humans, whereby some alleles at positively selected sites are associated with male sterility via azoospermia due to meiotic arrest. Strikingly, rapid evolution of Prdm9 is observed in some fish, in the sea anemone and a polychaete worm and thus, parsimoniously, is an ancestral feature of metazoan evolution, an evolutionary period spanning 700 million years. This recurrent evolution of Prdm9 is in stark contrast to both the Ovd and OdsH hybrid sterility gene in Drosophila, which appear to have evolved rapidly only in isolated lineages in which its role in hybrid sterility is manifest [11],[13] whereas the gene transposition of JYalpha is also highly lineage-specific [12]. From sequenced transcripts, Prdm9 is known to be expressed in male and female germ-line tissues across diverse metazoans such as trout, cattle, pig, sea urchin, and gastropod snail (accessions: CR372724, EF432552, EW634943, AM222434 and CAXX2975) in line with its previously described expression profile for mouse [17].
Hybrid sterility has been shown to arise from the simple deletion or insertion of a zinc finger domain in Prdm9 in mice [15]. The loss or gain of a single zinc finger is among the least perturbing of all changes in zinc finger number and sequence we have observed. For example, even closely related species, such as humans and chimpanzees, or bank and field voles, or rainbow trout and Atlantic salmon, differ much more dramatically at DNA-binding positions of their Prdm9 zinc fingers (Figure 3, Figure 4, Figure 5, Figure S2, Figure S6). Moreover, findings from human genetic association studies demonstrate that even individual amino acid changes in PRDM9 can affect male fertility even within species [18]. Finally, recent studies clearly demonstrate that Hst1 (Prdm9) associated genetic incompatibilities have evolved independently and are polymorphic in both M. m. musculus and M. m. domesticus mouse subspecies [16]. Our study has found even more radical alterations within Prdm9 zinc fingers than are observed in the M. m. musculus x M. m. domesticus cross. These changes, by themselves, may not be sufficient to result in reproductive isolation, as incompatibilities with a (as yet unknown) rapidly evolving DNA component would be required for hybrid sterility. In addition, hybrid sterility is clearly affected by multiple other loci [6],[7] whose discovery will lend further insight into the biological forces behind hybrid sterility. Nevertheless, our findings of recurrent rapid evolution of Prdm9 suggest its candidacy as a postzygotic hybrid sterility gene in other metazoan taxa.
Materials and Methods
Predicting Prdm9 genes
Prdm9 genes, and their 3′ (carboxy-terminal) arrays of zinc fingers (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6), were predicted from genome sequences available from UCSC, Ensembl and JGI genome browsers. Additional Prdm9 sequences were identified from the interrogation of nucleotide sequence databases using TBLASTn. Prediction of 3′ Prdm9 zinc finger sequences is greatly facilitated by their presence in the single 3′ terminal coding exon in all species. Orthology of Prdm9 sequences was confirmed using phylogenetic analysis [44], by consideration of the KRAB-SET-zinc finger domain architecture that is conserved among many but not all (including some fish, C. elegans and Monodelphis) Prdm9 proteins (see text), and by reciprocal best BLAST hits. Details of Prdm9 gene predictions from all species investigated are provided in Dataset S1.
Sequencing of Prdm9 zinc fingers in multiple rodent species
In addition to genomic data obtained for Mus musculus and Rattus norvegicus, sequencing of the final exon of Prdm9 was performed from genomic DNA purified from reproductive tract tissue from a total of 11 additional (sub-)species: Mus musculus castaneus, Mus macedonicus, Mus spicilegus, Coelomys pahari, Apodemus sylvaticus, Meriones unguiculateus, Peromyscus leucopus, Peromyscus maniculatus, Peromyscus polionotus, Microtus agrestis and Arvicola terrestris. PCR products were amplified using primers designed from the most highly conserved regions from mouse and rat genomic sequence flanking the last exon; either: Mus-Prdm9-F1 5′ CAAAGAACAAATGAGATCTGAG or Mus-Prdm9-F2 5′ AGAACAGGCCAGACAACAAAT with Mus-Prdm9-R1 5′ GTCTT(C/T)CTGTAATTGTTGAGATG or Mus-Prdm9-R2 5′ GCT(G/A)TTGGCTTTCTCATTC. Products were amplified using the proof-reading Pfx DNA polymerase (Invitrogen), purified from agarose gels using the Qiaquick gel purification kit (Qiagen) and sequenced in both directions from 2 or more independent amplification reactions. Sequence traces were initially curated and assembled using Chromas 2.0 (http://www.technelysium.com.au/chromas.html) and Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Genbank accessions are provided in Dataset S2.
Sequencing of PRDM9 zinc fingers from human and chimpanzee samples
Sequencing of the zinc finger repeat domain of PRDM9 was performed from the genomic DNA of 50 Chinese normal control samples. PCR amplification, purification and sequencing was carried out as above using the primers Hs-PRDM9-F 5′-GGCCAGAAAGTGAATCCAGG-3′ and Hs-PRDM9-R 5′-TGAAGCCACCTCACACAGCTG-3′. Products were gel purified and A-tailed prior to sub-cloning into the pCR4-TOPO vector (Invitrogen). T7 and T3 vector primers were used to sequence mini-prep DNA from positive clones. Genbank accessions are provided in Dataset S2.
Chimpanzee (Pan troglodytes) PRDM9 C-terminal zinc fingers were sequenced by PCR using the primers Pt-PRDM9-F 5′-GCCTGACCAAAACATCTACCCTGACC-3′ and Pt-PRDM9-R 5′-GTCATGAAAGTGGCGGATTTG-3′. PCR products were both directly sequenced as well as cloned into the pCR4-TOPO vector (Invitrogen) and six independent clones sequenced using vector-specific primers. The genomic DNA sample was obtained from Coriell (ID#NG03448). The Genbank accession can be found in Dataset S2.
Prediction of positively selected codons
For the prediction of positively selected sites, we included all zinc finger sequences from the 3′ terminal array only if they were complete (28-codon) and retained, at conserved positions, two cysteine and two histidine residues expected to coordinate a single Zn2+ ion. This excludes, for example, the first two zinc finger motifs in primates and rodents. Phylogenetic trees for each multiple alignment were constructed by applying the Fitch-Margoliash criterion to distance matrices of synonymous substitutions per synonymous site (dS) as calculated by the codeml programme [77],[78]. Tree topologies were accepted if they were corroborated by phyml [79] and treebest (http://treesoft.sourceforge.net/treebest.shtml) programs.
Amino acid sites under positive selection were inferred using “site likelihood method” (SLR) [27] with p-value thresholds of 0.05 after multiple testing correction. We observed that inferences of positive selection among sequence similar zinc fingers from the same species were sensitive to tree topology. Nevertheless, the use of alternative less-well supported topologies tended only to increase evidence for positive selection. As a result, we have, conservatively, used inferences from the most strongly supported tree.
SLR, and other maximum-likelihood approaches that take account of codon evolution, have proved reliable provided that assumptions in evolutionary models are not greatly violated. One such assumption is vertical inheritance without gene conversion, which is demonstrably violated for Prdm9. However, gene conversion is more likely to affect analyses of sequence-similar zinc fingers from the same species and is less of a factor in analyzing zinc fingers from all the rodent or primate clades due to the greater sequence divergences involved (for instance, all identical zinc fingers are essentially treated as one representative sequence in analyses). Our inferences of positive selection among all zinc fingers in rodent or primate clades (Figure 3C and Figure 4C) are accordingly the most robust to phylogeny variations and show high dN/dS values, and low and significant p-values.
Tests for positive selection in the human population
Rapid fixation of an advantageous allele changes the pattern of polymorphisms around the locus under selection, and various methods have been developed to formally test whether such patterns are compatible with evolution under a neutral model. Other effects, such as geographical structure, population admixture, non-random mating, and varying population sizes, can also give rise to a departure from the neutral model, thereby confounding this analysis. To address this problem, here we use data from recent large-scale surveys of population variation that allow us to compare our observations to empirical, genomic distributions rather than to model-based predictions. This approach accounts for non-local genomic effects such as population structure and growth, at the expense of some loss of power.
Tajima's D values were acquired from the UCSC genome browser for American individuals of African, European and Asian ancestry populations [40]. These were computed at 10 kb intervals, each using 100 kb of data. Since both PRDM7 and PRDM9 span about 20 kb, we took the average of two neighbouring values. For the background distribution, averages were similarly computed for all neighbours.
To assess the existence of long haplotype blocks, we used HapMap data (public release 26). We computed derived allele frequencies (DAF) by polarizing using the chimpanzee genome. To avoid miscalls, we removed all potential CpG SNPs. Finally, we used r-squared values computed for SNPs at a minimum distance of 50 kb, as including more proximal SNPs which are often in strong LD would further reduce power. For any locus, we identified all pairs of SNPs spanning the locus that satisfied these filters; the maximum r-squared value among these pairs was taken as the observable for that locus. We computed this value for all genomic loci to create the empirical distribution. The entire procedure was done separately for each of the HapMap populations.
Calculations of zinc finger sequence identities in the mouse genome
Clusters of zinc finger repeats (Figure 2B) were identified in each of six possible reading frames of the mouse genome using the hmmsearch programme [80] and a hidden Markov model derived from the SMART domain resource [81]. We discarded all zinc finger clusters which show frameshift or stop codon disruptions, giving 473 putative open reading frames (ORFs). Within each ORF, zinc fingers which do not possess the canonical zinc finger Cys2His2 structure were excluded from subsequent comparisons. A multiple alignment of conceptual cDNA zinc finger sequences was constructed from peptide alignments using the MUSCLE programme [82]. Pairwise cDNA sequence alignments were calculated and the proportions of pairs which were higher than a given threshold calculated. Mouse Prdm9 was an extreme outlier for zinc finger pairwise sequence identities greater than 90%, and also for other thresholds (data not shown).
Supporting Information
Phylogenetic tree of rodent Prdm9 zinc finger nucleotide sequences as inferred by phyml [76] version 3.0 (http://www.atgc-montpellier.fr/phyml/) and drawn using FigTree (http://tree.bio.ed.ac.uk/software/figtree). Zinc fingers are numbered sequentially from the C-terminal array. The exceptions are zinc fingers from the mouse and rat, whose numbers start from the first Prdm9 zinc finger in exon 11. For species names see legend to Figure 6. Branches with Approximate Likelihood-Ratio Test values of over 0.75 are indicated with bold lines.
(6.18 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of Prdm9 zinc finger sequences from the water vole (Arvicola terrestris) and field vole (Microtus agrestis).
(2.75 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of Prdm9 zinc finger sequences from the sea anemone, Nematostella vectensis. These form part of a predicted gene (NEMVEDRAFT_v1g113856) that has been predicted from scaffold_120 of the N. vectensis v.1.0 genome assembly (Joint Genome Institute).
(2.64 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of 23 Lottia gigantea Prdm9 zinc finger sequences. These have been predicted from scaffold 11 (bases 1507994-1510370) of the Lottia genome assembly (v1.0) produced by the Joint Genome Institute. This gene prediction is supported by an expressed sequence tag from L. gigantea male gonad (accession code FC692069).
(2.84 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of 11 Capitella sp.I Prdm9 zinc finger sequences. These have been predicted from scaffold_236 of the Capitella sp.I v.1.0 genome assembly (Joint Genome Institute).
(2.95 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of Prdm9 zinc finger sequences from the Atlantic salmon (Salmo salar; accession ACN10800, supported by ESTs CX352799, GE785155, EG785159, and EG785158) and from the Pacific Ocean rainbow trout (Oncorhynchus mykiss; contig generated from ESTs CR372724, CX253406 1305997, CX253405, CX252076, CX251898, CX251897, and CX252077).
(2.67 MB EPS)
Multiple sequence alignment of Prdm9 zinc fingers from all analysed species in FASTA format.
(0.09 MB PDF)
Genbank accessions for rodent and human sequences
(0.04 MB DOC)
Footnotes
The authors have declared that no competing interests exist.
PLO, LG, ZB, GL, and CPP gratefully acknowledge funding from the UK Medical Research Council. KCR is supported by an NSF fellowship, JJB in part by NIH predoctoral training grant T32 GM07270, and NP and HSM are supported by grant R01-GM74108 from the US National Institutes of Health and funds from FHCRC. SAB is supported by an ARC Australian Research Fellowship (DP0881247). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Darwin CR. London: John Murray; 1859. On the origin of species by means of natural selection or the preservation of favoured races in the struggle for life. [PMC free article] [PubMed] [Google Scholar]
- 2.Muller HJ. Isolating mechanisms, evolution, and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 3.Coyne J, Orr HA. Speciation: Sinauer Associates, Sunderland MA 2004 [Google Scholar]
- 4.Orr HA, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;55:1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 5.Laurie CC. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics. 1997;147:937–951. doi: 10.1093/genetics/147.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Good JM, Dean MD, Nachman MW. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics. 2008;179:2213–2228. doi: 10.1534/genetics.107.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez DE, Wu CI. Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics. 1995;140:201–206. doi: 10.1093/genetics/140.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 10.Sun S, Ting CT, Wu CI. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science. 2004;305:81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- 11.Ting CT, Tsaur SC, Sun S, Browne WE, Chen YC, et al. Gene duplication and speciation in Drosophila: evidence from the Odysseus locus. Proc Natl Acad Sci U S A. 2004;101:12232–12235. doi: 10.1073/pnas.0401975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masly JP, Jones CD, Noor MA, Locke J, Orr HA. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 13.Phadnis N, Orr HA. A Single Gene Causes Both Male Sterility and Segregation Distortion in Drosophila Hybrids. Science. 2008 doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storchova R, Gregorova S, Buckiova D, Kyselova V, Divina P, et al. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- 15.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A Mouse Speciation Gene Encodes a Meiotic Histone H3 Methyltransferase. Science. 2008 doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 16.Vyskocilova M, Prazanova G, Pialek J. Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mamm Genome. 2009;20:83–91. doi: 10.1007/s00335-008-9164-3. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 18.Irie S, Tsujimura A, Miyagawa Y, Ueda T, Matsuoka Y, et al. Single Nucleotide Polymorphisms in PRDM9 (MEISETZ) in Patients with Nonobstructive Azoospermia. J Androl. 2009 doi: 10.2164/jandrol.108.006262. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto T, Koh E, Sakugawa N, Sato H, Hayashi H, et al. Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008;25:553–557. doi: 10.1007/s10815-008-9270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- 21.Wade CM, Kulbokas EJ, 3rd, Kirby AW, Zody MC, Mullikin JC, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 22.Trachtulec Z, Vlcek C, Mihola O, Gregorova S, Fotopulosova V, et al. Fine haplotype structure of a chromosome 17 region in the laboratory and wild mouse. Genetics. 2008;178:1777–1784. doi: 10.1534/genetics.107.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohta T. How gene families evolve. Theor Popul Biol. 1990;37:213–219. doi: 10.1016/0040-5809(90)90036-u. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, et al. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006;16:584–594. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaline J, Graf J-D. Phylogeny of the Arvicolidae (Rodentia): Biochemical and Paleontological Evidence. J Mammalogy. 1988;69:22–33. [Google Scholar]
- 27.Massingham T, Goldman N. Detecting amino acid sites under positive selection and purifying selection. Genetics. 2005;169:1753–1762. doi: 10.1534/genetics.104.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt D, Durrett R. Adaptive evolution drives the diversification of zinc-finger binding domains. Mol Biol Evol. 2004;21:2326–2339. doi: 10.1093/molbev/msh246. [DOI] [PubMed] [Google Scholar]
- 30.Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choo Y, Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc Natl Acad Sci U S A. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choo Y, Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc Natl Acad Sci U S A. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr Opin Struct Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 34.Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, et al. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007;7:187. doi: 10.1186/1471-2148-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun XJ, Xu PF, Zhou T, Hu M, Fu CT, et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS ONE. 2008;3:e1499. doi: 10.1371/journal.pone.0001499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 37.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 38.Zhai W, Nielsen R, Slatkin M. An investigation of the statistical power of neutrality tests based on comparative and population genetic data. Mol Biol Evol. 2009;26:273–283. doi: 10.1093/molbev/msn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 41.Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 43.Devlin RH. Sequence of sockeye salmon type 1 and 2 growth hormone genes and the relationship of rainbow trout with Atlantic and Pacific Salmon. Canadian Journal of Fisheries and Aquatic Sciences. 1993;50:1738–1748. [Google Scholar]
- 44.Birtle Z, Ponting CP. Meisetz and the birth of the KRAB motif. Bioinformatics. 2006;22:2841–2845. doi: 10.1093/bioinformatics/btl498. [DOI] [PubMed] [Google Scholar]
- 45.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 49.Dover GA, Strachan T, Coen ES, Brown SD. Molecular drive. Science. 1982;218:1069. doi: 10.1126/science.7146895. [DOI] [PubMed] [Google Scholar]
- 50.Rudd MK, Schueler MG, Willard HF. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol. 2003;68:141–149. doi: 10.1101/sqb.2003.68.141. [DOI] [PubMed] [Google Scholar]
- 51.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 52.Lee HR, Zhang W, Langdon T, Jin W, Yan H, et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A. 2005;102:11793–11798. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 54.Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- 55.Yu HG, Hiatt EN, Chan A, Sweeney M, Dawe RK. Neocentromere-mediated chromosome movement in maize. J Cell Biol. 1997;139:831–840. doi: 10.1083/jcb.139.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science (in press) 2008 doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 57.Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pardo-Manuel de Villena F, Sapienza C. Transmission ratio distortion in offspring of heterozygous female carriers of Robertsonian translocations. Hum Genet. 2001;108:31–36. doi: 10.1007/s004390000437. [DOI] [PubMed] [Google Scholar]
- 59.Daniel A. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A). Am J Med Genet. 2002;111:450–452. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- 60.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 61.Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell (submitted) 2009 doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 64.Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science in press. 2009 doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nei M, Zhang J. Molecular origin of species. Science. 1998;282:1428–1429. doi: 10.1126/science.282.5393.1428. [DOI] [PubMed] [Google Scholar]
- 66.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 68.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 69.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 70.Frank SA. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1990;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 71.Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A. 2001;98:13183–13188. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 74.Sawamura K, Yamamoto MT. Cytogenetical localization of Zygotic hybrid rescue (Zhr), a Drosophila melanogaster gene that rescues interspecific hybrids from embryonic lethality. Mol Gen Genet. 1993;239:441–449. doi: 10.1007/BF00276943. [DOI] [PubMed] [Google Scholar]
- 75.Mayr E. Cambridge: Harvard University Press; 1942. Systematics and the origin of species, from the viewpoint of a zoologist. [Google Scholar]
- 76.Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 77.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 78.Goodstadt L, Ponting CP. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. doi: 10.1371/journal.pcbi.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 80.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 81.Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, et al. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conroy CJ, Cook JA. Phylogeography of a post-glacial colonizer: Microtus longicaudus (Rodentia: muridae). Mol Ecol. 2000;9:165–175. doi: 10.1046/j.1365-294x.2000.00846.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of rodent Prdm9 zinc finger nucleotide sequences as inferred by phyml [76] version 3.0 (http://www.atgc-montpellier.fr/phyml/) and drawn using FigTree (http://tree.bio.ed.ac.uk/software/figtree). Zinc fingers are numbered sequentially from the C-terminal array. The exceptions are zinc fingers from the mouse and rat, whose numbers start from the first Prdm9 zinc finger in exon 11. For species names see legend to Figure 6. Branches with Approximate Likelihood-Ratio Test values of over 0.75 are indicated with bold lines.
(6.18 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of Prdm9 zinc finger sequences from the water vole (Arvicola terrestris) and field vole (Microtus agrestis).
(2.75 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of Prdm9 zinc finger sequences from the sea anemone, Nematostella vectensis. These form part of a predicted gene (NEMVEDRAFT_v1g113856) that has been predicted from scaffold_120 of the N. vectensis v.1.0 genome assembly (Joint Genome Institute).
(2.64 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of 23 Lottia gigantea Prdm9 zinc finger sequences. These have been predicted from scaffold 11 (bases 1507994-1510370) of the Lottia genome assembly (v1.0) produced by the Joint Genome Institute. This gene prediction is supported by an expressed sequence tag from L. gigantea male gonad (accession code FC692069).
(2.84 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of 11 Capitella sp.I Prdm9 zinc finger sequences. These have been predicted from scaffold_236 of the Capitella sp.I v.1.0 genome assembly (Joint Genome Institute).
(2.95 MB EPS)
Multiple sequence alignment (shaded according to a 90% consensus) of Prdm9 zinc finger sequences from the Atlantic salmon (Salmo salar; accession ACN10800, supported by ESTs CX352799, GE785155, EG785159, and EG785158) and from the Pacific Ocean rainbow trout (Oncorhynchus mykiss; contig generated from ESTs CR372724, CX253406 1305997, CX253405, CX252076, CX251898, CX251897, and CX252077).
(2.67 MB EPS)
Multiple sequence alignment of Prdm9 zinc fingers from all analysed species in FASTA format.
(0.09 MB PDF)
Genbank accessions for rodent and human sequences
(0.04 MB DOC)