Summary
The COP9 signalosome (CSN) is thought to maintain the stability of cullin-RING ubiquitin ligases (CRL) by limiting the autocatalytic destruction of substrate adapters such as F-box proteins (FBPs). CAND1, a protein associated with unneddylated CUL1, was proposed to assist in this role in an as yet unclear fashion. We found that only a subset of S. pombe FBPs, which feature a critical F-box proline that promotes their interaction with CUL1, required CSN for stability. Unlike the CRL3 adapter Btb3p, none of the CSN-sensitive FBPs were affected by deletion of ubp12. Contrary to current models, CAND1 does not control adapter stability, but maintains the cellular balance of CRL1 complexes by preventing rare FBPs from being out-competed for binding to CUL1 by more ample adapters. These findings were integrated into a refined model of CRL control where substrate availability toggles CRLs between independent CSN and CAND1 cycles.
Introduction
CRLs represent an extensive class of multisubunit E3 ubiquitin ligases each consisting of a core module containing a member of the cullin family and the RING domain protein RBX1 (= HRT1, ROC1), which recruits E2 ubiquitin conjugating enzymes to the ligases (reviewed in (Bosu and Kipreos, 2008; Petroski and Deshaies, 2005)). This core is joined by one of several hundred adapter proteins each of which appears to target a distinct array of substrates for ubiquitylation and proteasomal degradation. Whereas FBPs are tethered to the CUL1 core through the linker protein SKP1 to form SCF (or CRL1) complexes (Deshaies, 1999), CUL3 adapters are recruited into CRL3 complexes via their inherent BTB domains (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003; Xu et al., 2003). Several members of both adapter families are unstable proteins, due to autoubiquitylation by the intrinsic ubiquitin ligase activity of their associated core modules (Galan and Peter, 1999; Geyer et al., 2003; Luke-Glaser et al., 2007; Rouillon et al., 2000; Wirbelauer et al., 2000; Zhou and Howley, 1998) .
CRLs are stimulated through modification of cullins with the ubiquitin-related peptide NEDD8 (reviewed in (Pan et al., 2004). Cullin neddylation is reversed by the COP9 signalosome (CSN), a highly conserved protein complex that binds cullins (Lyapina et al., 2001; Schwechheimer et al., 2001; Wee et al., 2002; Zhou et al., 2001) and thereby exposes them to a deneddylating activity intrinsic to subunit 5 of the CSN (Cope et al., 2002). Consistent with neddylation being a stimulatory modification is the observation that purified CSN inhibits CRL activity in vitro. In addition, the CSN-associated deubiquitylating enzyme (DUB) Ubp12/USP15 acts to neutralize CRL activity when assayed in vitro (Groisman et al., 2003; Zhou et al., 2003).
These biochemical findings conflicted with genetic evidence that CSN is required for efficient CRL-dependent substrate degradation in vivo (Reviewed in (Cope and Deshaies, 2003; von Arnim, 2003; Wolf et al., 2003)). This so-called CSN paradox was resolved by the demonstration that CSN's inhibitory enzymatic activities revealed in vitro prevent the autocatalytic degradation of CRL substrate adapters in vivo thus promoting CRL activity. For example, the Cul3p adapter Btb3p is destabilized in fission yeast csn and ubp12 mutants (Wee et al., 2005). In addition, several FBPs are unstable in csn mutants of S. pombe (Zhou et al., 2003) and N. crassa (He et al., 2005), and in human CSN knockdown cells (Cope and Deshaies, 2006; Denti et al., 2006).
CAND1 is a highly conserved protein that binds to the unneddylated form of human and plant CUL1 and inhibits complex formation with SKP1-FBP modules (Hwang et al., 2003; Liu et al., 2002; Min et al., 2003; Oshikawa et al., 2003; Zheng et al., 2002a). By virtue of these properties, CAND1 inhibits CRL activity in vitro. However, CAND1 was also shown to be required for efficient CRL function in vivo (Chuang et al., 2004; Feng et al., 2004; Zheng et al., 2002a). This contradictory pattern reiterates the CSN paradox, and it was therefore suggested that CAND1 and CSN participate in the same pathway of CRL adapter stabilization (Cope and Deshaies, 2003; He et al., 2005; Min et al., 2005), although experimental proof is still outstanding. It also remained unclear whether the model is broadly applicable to all FBPs. In the present report, we address these questions by determining the regulation by CSN and CAND1 of a panel of eight FBPs from fission yeast.
Results
Differential effect of CSN on FBP levels
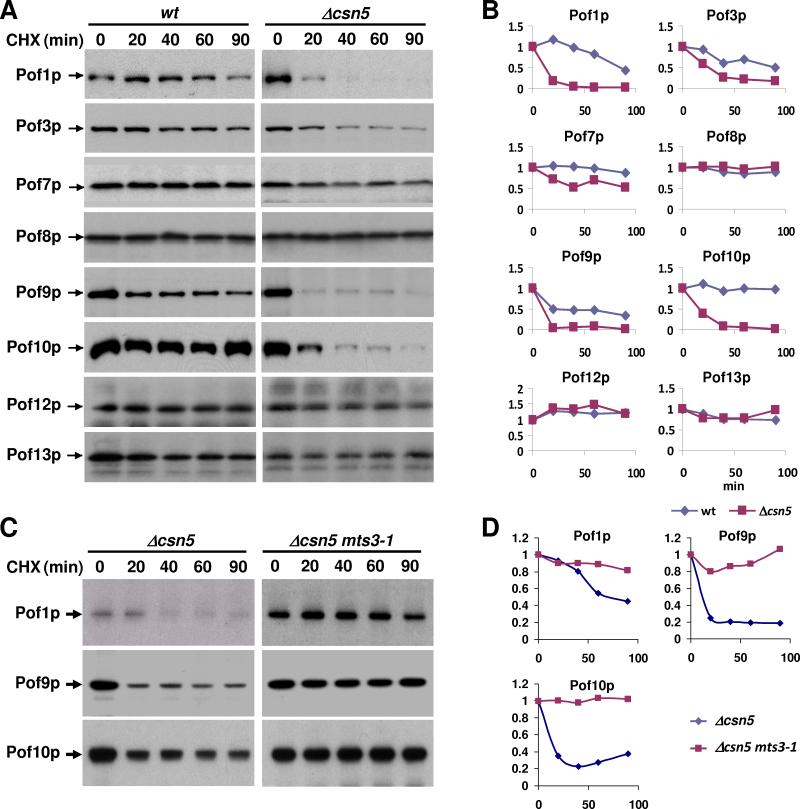
In previous studies we demonstrated that the stability of the CRL1 adapter Pop1p and the CRL3 adapter Btb3p is promoted by the CSN in vivo (Wee et al., 2005; Zhou et al., 2003). In addition, genetic interaction studies suggested that FBPs other than Pop1p are also subject to regulation by the CSN (Wee et al., 2005). The S. pombe genome encodes a minimum of 16 FBPs (named Pofs and Pops), but whether all are targets for regulation by the CSN is unknown. We compared the steady-state protein levels of eight randomly chosen FBPs in wild-type and csn5 mutant cells, utilizing a panel of strains harbouring FBPs modified at their endogenous genomic loci with C-terminal Myc-epitope tags (Lehmann et al., 2004). Pof1p, 3p, 7p, 9p, 10p, and 13p steady-state levels were reduced in csn5 mutants, whereas Pof8p and Pof12p levels were largely unaffected (Fig. 1A).
Fig. 1. Differential effect of CSN on FBP levels.
(A) Steady-state levels of Myc-tagged FBPs in csn5 deletion strains. Cdc2p and Cul1p are shown for reference in the lower panels. The mobility differences of Cul1p reflect the loss of deneddylation activity in csn5 mutants.
(B) Complementation of Pof10p-Myc levels. The indicated csn4 and csn5 deletion strains were transformed with plasmids driving the expression of Myc-tagged Csn4p or Csn5p, and Pof10p-Myc levels were determined by immunoblotting. Two independently derived strains are shown in the right panel.
(C) Dependence of the rescue of Pof10p-Myc levels on the JAMM motif of Csn5p. Myc-tagged wild-type Csn5p or the JAMM point mutant Csn5p-H118A were expressed in csn5 deletion strains, and the level of Pof10p-Myc was determined by immunoblotting. Two independently derived strains are shown.
The levels of the CSN-regulated Pof1p and Pof10p were also diminished in csn3 and csn4 mutants (Fig. 1B, Supplementary Fig. 1). Conversely, downregulation of Pof10p in csn5 mutants was complemented by providing wild-type csn5 from a plasmid (Fig. 1B). This rescue failed in csn4 csn5 double mutants (Fig. 1B). In addition, efficient rescue depended on the enzymatic function of the deneddylating enzyme Csn5p, since point mutants in the catalytic JAMM motif were unable to maintain Pof10p levels in csn5 mutants (Fig, 1C). These results suggested that the cullin deneddylation function of the entire CSN complex is required to maintain the steady-state expression levels of CSN-sensitive FBPs.
CSN regulates FBP protein stability
The levels of the mRNAs encoding Pof1p and Pof10p were unchanged in csn5 mutants as determined by semiquantitative RT-PCR (Supplementary Fig. 2A), suggesting that CSN-sensitive FBPs might be regulated at the level of protein stability. Cycloheximide (CHX) chase experiments confirmed that the CSN-sensitive FBPs Pof1p, Pof3p, Pof7p, Pof9p, and Pof10p were considerably destabilized in csn5 mutants (Fig. 2A, B). Destabilization of Pof1p, Pof9p, and Pof10p did not occur in csn5 mutants that carried the temperature-sensitive mts3-1 allele (Gordon et al., 1996), which encodes a mutant version of the proteasome subunit Rpn12p (Fig. 2C, D). Likewise, the downregulation of these FBPs in csn5 mutants was rescued in csn5 mts3-1 double mutants, indicating that proteasome activity is required for FBP destabilization in csn5 mutants (Supplementary Fig. 3). No destabilization was observed for Pof8p and Pof12p. Pof13p stability was also unaffected in csn5 mutants (Fig. 2A, B) despite the decrease in Pof13p steady-state levels apparent in Fig. 1A. Since pof13 mRNA was not downregulated in csn5 mutants as determined by quantitative real time PCR (Supplementary Fig. 2B), decreased Pof13p protein levels in csn5 mutants may reflect an unidentified role of CSN in facilitating Pof13p protein synthesis. It is well established that some CSN components have dual functions in proteolysis and protein synthesis (Luke-Glaser et al., 2007).
Fig. 2. Stability of FBPs in csn5 mutants.
(A). Strains expressing the indicated Myc-tagged FBPs in a wild-type or csn5 mutant background were employed in cycloheximide (CHX) chase experiments to determine their stability (see Materials and methods). Note that exposure times were adjusted such that comparable base line signals of the FBPs in wildtype and csn5 mutant cells were obtained for improved comparison.
(B) The levels of FBPs remaining in wild-type and csn5 mutants upon CHX chase were quantified. Quantification was done using IMAGEJ v1.40f analysis software. Mean intensities were background subtracted and normalized to the loading control Cdc2 (blots not shown).
(C) Strains expressing the indicated Myc-tagged FBPs in a csn5 or csn5 mts3-1 mutant background were employed in cycloheximide (CHX) chase experiments. Note that loading and exposure times were adjusted such that comparable base line signals of the FBPs in csn5 and csn5 mts3-1 mutant cells were obtained for improved comparison.
(D) The levels of FBPs remaining in csn5 and csn5 mts3-1 mutants upon CHX chase were quantified. Quantification was done using IMAGEJ analysis software. Mean intensities were background subtracted and normalized to the loading control Cdc2 (blots not shown).
The DUB Ubp12p maintains the stability of the CRL3 adapter Btb3p but not FBPs
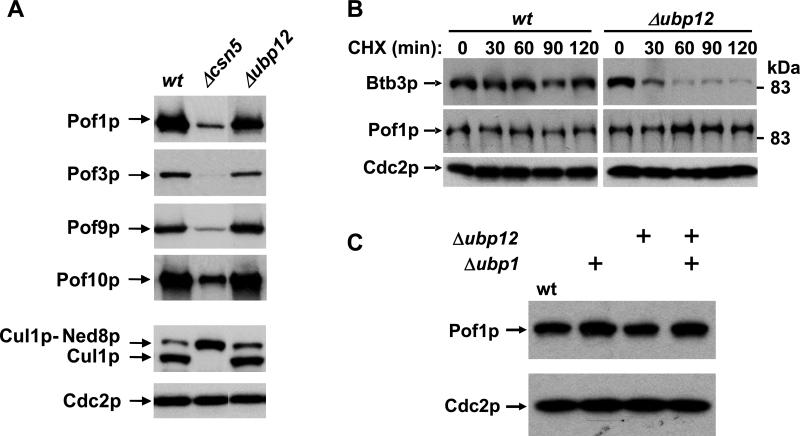
We previously showed that the level of the Cul3p adapter Btb3p is strongly reduced in cells lacking CSN deneddylation activity (Wee et al., 2005). An even more pronounced downregulation was observed in cells deficient of the CSN-associated DUB Ubp12p, and both effects were attributed to increased autocatalytic destruction of Btb3p by its associated CRL3 core module (Wee et al., 2005). Unlike with Btb3p, the steady-state levels of the CSN-regulated FBPs Pof1p, Pof3p, Pof9p, and Pof10p were only minimally altered in ubp12 mutants when compared to csn5 mutants (Fig. 3A), indicating that Ubp12p is not a major regulator of these adapters.
Fig. 3. Ubp12p maintains the stability of the CRL3 adapter Btb3p but not FBPs.
(A) Steady-state levels of the indicated CSN-sensitive FBPs in csn5 and ubp12 mutants.
(B) A strain coexpressing protein A-tagged Btb3p and Myc-tagged Pof1p from their respective endogenous genomic loci was employed in a CHX chase experiment to determine the stability of the adapters.
(C) Pof1p-Myc levels were determined in ubp1 and ubp12 single mutants, and in ubp1 ubp12 double mutants.
To illustrate the differential effect of Ubp12p on Btb3p and FBPs more rigorously, we generated a ubp12 deletion strain coexpressing protein A-tagged Btb3p and Myc-tagged Pof1p from their endogenous promoters. A CHX chase experiment revealed that Btb3p was drastically destabilized in ubp12 mutants as described (Wee et al., 2005), whereas Pof1p was entirely stable in the same cells (Fig. 3B).
To exclude the possibility that Ubp1p, a DUB sharing 48% amino acid similarity (32% identity) with Ubp12p and an overall identical domain structure and length (data not shown), maintained FBP stability in ubp12 mutants, we compared Pof1p levels in ubp1 and ubp12 single mutants and in ubp1 ubp12 double mutants. Pof1p levels were unchanged in either mutant (Fig. 3C). These data suggested that, unlike with Btb3p, the steady-state levels of CSN-sensitive FBPs were not affected by lack of CSN-associated DUB activity, although we cannot entirely exclude minor destabilization in ubp12 mutants as previously detected for Pop1p (Zhou et al., 2003).
CSN-insensitive FBPs are deficient in forming canonical CRL1 complexes
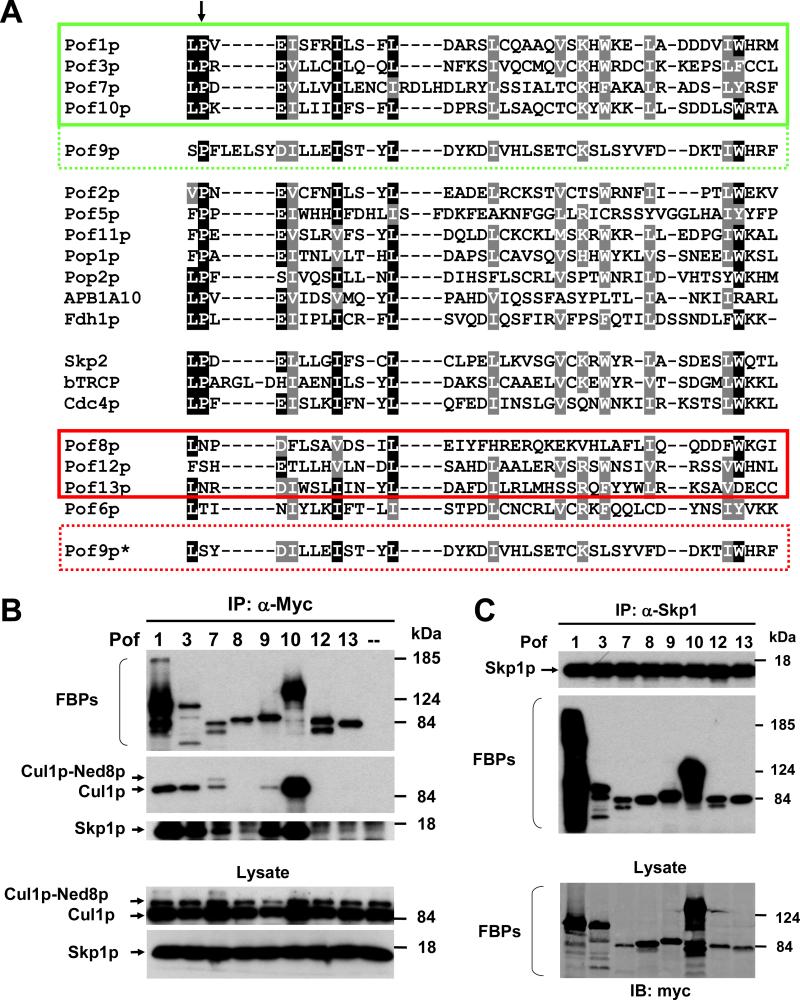
To understand why the stability of only five of the eight FBPs tested was regulated by the CSN, we searched for structural features setting the two groups apart. Since the F-box is the only motif shared by all of these proteins, we performed a ClustalW alignment of the F-boxes of all 16 fission yeast FBPs. Human SKP2 and β-TRCP1 as well as budding yeast Cdc4p were included for reference. The alignment revealed that all CSN-insensitive FBPs missed a conserved proline residue at the beginning of the F-box motif (Fig. 4A). None of the other signature residues of the F-box motif segregated consistently with CSN regulation. Since the proline is one of the most highly conserved amino acids of the F-box motif across all species, we reasoned that it might be important for CRL complex formation.
Fig. 4. CSN-insensitive FBPs are less efficient in forming CRL complexes.
(A) CLUSTALW alignment of the F-boxes of the indicated proteins. The conserved proline residue is highlighted by an arrow. Two different alignments are shown for Pof9p (stippled green and red boxes). Pof9p* denotes an alignment according to which the Pof9p F-box does not contain the conserved proline. The proline containing FBPs examined in this study are highlighted in a green box, the proline-less FBPs are in a red box.
(B) Interaction of FBPs with Cul1p and Skp1p. The indicated Myc-tagged FBPs were immunoprecipitated with Myc antibodies, followed by immunoblotting with Cul1p and Skp1 sera. The negative control (denoted “--“) represents cell lysate from a strain not expressing any Myc-tagged proteins. Total cell lysates are shown in the bottom panel.
(C) The same lysates as in (B) were subjected to immunoprecipitation with Skp1p antisera, followed by immunoblotting with Myc antibodies to detect co-purification of Myc-tagged FBPs. Total cell lysates are shown in the bottom panel.
To test this, Myc-tagged FBPs were immunoprecipitated with Myc antibodies, followed by immunoblotting with Cul1p and Skp1p antisera. Binding of Cul1p was only detected for the CSN-regulated FBPs (Fig. 4B). In contrast, Pof8p, 12p, and 13p showed no detectable interaction with Cul1p (Fig. 4B). Skp1p exhibited a binding pattern closely resembling that of Cul1p, although a low level of Skp1p was also retrieved in immunoprecipitates of Pof8p, 12p, and 13p (Fig. 4B), suggesting that these FBPs are principally capable of interacting with Skp1p. In fact, Skp1p immunoprecipitates prepared from the same lysates efficiently retrieved all eight FBPs (Fig. 4C). These results suggested that CSN-insensitive FBPs lacking the conserved proline residue in their F-boxes are impaired in binding Cul1p, and thus, in forming canonical CRL complexes.
The conserved proline residue determines CRL1 complex formation
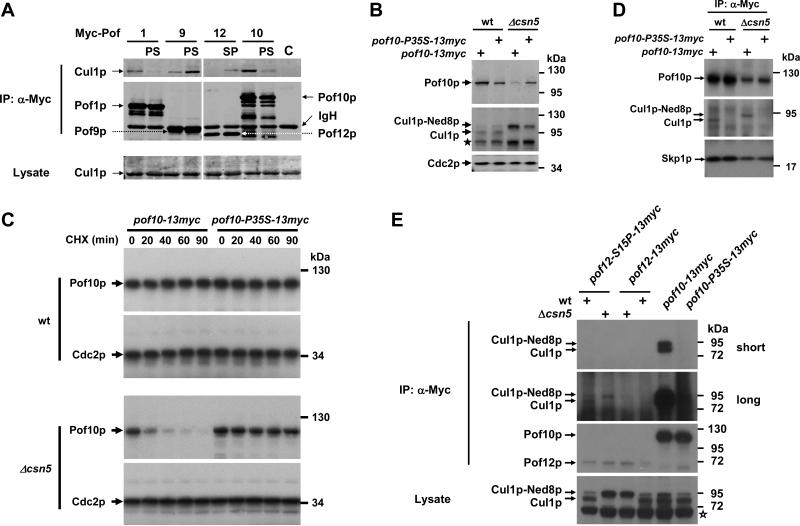
To directly address the role of the proline residue, we constructed a point mutant of the CSN-regulated Pof1p, exchanging proline 114 for serine (= Pof1p-P114S), which is found in the corresponding position in the F-box of the CSN-independent Pof12p (Fig. 4A). Similarly, we mutated the conserved proline 35 in Pof10p to serine (Pof10p-P35S). Myc-tagged wild-type and proline mutant FBPs were expressed from pRep81 plasmids driven by the low strength nmt1 promoter, and binding to endogenous Cul1p was determined by co-immunoprecipitation. The Pof1p-P114S and Pof10p-P35S mutants interacted with Cul1p much less efficiently than the respective wild-type proteins (Fig. 5A). For Pof1p-P114S, this deficiency was verified in both wildtype and csn5 mutant backgrounds, whereas binding of Skp1p was not affected by the proline mutation (Supplementary Fig. 3).
Fig. 5. The conserved proline residue determines FBP binding to Cul1p and regulation by the CSN.
(A) Interaction of plasmid derived wildtype and proline mutant FBPs with Cul1p. Myc-tagged FBPs were immunoprecipitated and copurification of Cul1p was assayed by immunoblotting. “C” denotes a specificity control (lysate from untransformed cells).
(B) Steady state levels of endogenously Myc-tagged wildtype Pof10p-Myc and Pof10p-P35S-Myc in csn5 mutants and in wildtype cells. Note that, in contrast to wildtype Pof10p, Pof10p-P35S is not downregulated in csn5 mutants. Cul1p and Cdc2p are shown for reference. The asterisk denotes an unspecific cross reactivity of the Cul1p sera.
(C) The stabilities of Pof10p and Pof10p-P35S in wildtype (top panel) csn5 mutants (bottom panel) were determined by CHX chase.
(D) Interaction of Pof10p and Pof10p-P35S with Cul1p and Skp1p. Lysates from wildtype and csn5 mutant cells harbouring endogenously Myc-tagged Pof10p and Pof10p-P35S were immunoprecipitated with Myc antibodies followed by immunoblotting with Cul1p, Skp1p, and Myc antisera. Whereas Pof10p-P35S is deficient in binding of Cul1p, its interaction with Skp1p is maintained.
(E) Interaction of Pof12p and Pof12p-S15P with Cul1p. Lysates from wildtype and csn5 mutant cells harbouring endogenously Myc-tagged Pof12p and Pof12p-S15P were immunoprecipitated with Myc antibodies followed by immunoblotting with Cul1p and Myc antisera. Whereas wildtype Pof12p is deficient in binding of Cul1p, the mutant Pof12p-S15P shows binding (strong exposure, second panel from top). The Pof10p-Cul1p interaction is shown for reference (weak and strong exposures, top two panels). The asterisk denotes an unspecific cross reactivity of the Cul1p sera.
We next asked whether the proline was sufficient to target an FBP into a CRL1 complex. To this end, we changed serine 15 of the Pof12p F-box to proline and determined binding to Cul1p. As with endogenous Pof12p (see Fig. 4B), plasmid-derived wildtype Pof12p was inefficient in binding Cul1p (Fig. 5A). In contrast, proline-containing Pof12p-S15P bound Cul1p (Fig. 5A). Thus, the F-box proline residue appears to be both required and sufficient to target FBPs into canonical CRL1 complexes.
The instability of Pof9p in csn5 mutants (Fig. 2A) and the binding of Pof9p to Cul1p (Fig. 4B) suggested that its F-box also contains the conserved proline. Indeed, under the parameters used, the ClustalW algorithm aligned proline 5, a residue near the beginning of the Pof9p F-box, with the conserved proline of other FBPs (Fig. 4A). The alignment also highlighted a short insertion following the proline that is shared by human β-TRCP but not by other S. pombe FBPs (Fig. 4A). Remarkably, unlike with Pof1p and Pof10p, mutation of proline 5 to serine did not reduce the binding of Pof9p to Cul1p but enhanced it instead (Fig. 5A). Manual editing of the sequence alignment revealed that Pof9p can also be aligned such that it features a serine in the position of the conserved proline, a maneuver that would place Pof9p into the group of CSN-independent FBPs (designated Pof9p* at the bottom of Fig. 4A).
To reconcile this apparent paradox, we considered the possibility that Pof9p may be targeted into CRL1 complexes independently of its F-box motif. Precedence for such a scenario was previously provided by the S. pombe FBP Pop2p, which can be recruited into functional CRL1 complexes in an F-box independent manner through dimerization with another FBP, Pop1p (Seibert et al., 2002). Truncated Pof9p lacking the N-terminal F-box retained efficient interaction with Cul1p similar in extent to the wildtype and the proline mutant proteins (Supplementary Fig. 5). In summary, these findings indicated that Pof9p, unlike most other FBPs, is targeted into CRL1 complexes and subjected to stability control by the CSN independently of the integrity of its F-box. Nevertheless, insertion of a serine upstream of the F-box further enhances Pof9p binding to Cul1p for reasons that are presently unclear (Fig. 5A).
Proline-dependent regulation of Pof10p stability by the CSN
Our data revealed a strict correlation between the ability of FBPs to form CRL1 complexes and their requirement for CSN to maintain stability. From this correlation, we predicted that proline mutant FBPs are no longer targets for stability control by CSN. To critically test this prediction, we replaced the endogenous copy of the non-essential pof10 gene with the proline mutant Pof10p-P35S in both wildtype as well as csn5 mutants, and determined its stability by CHX chase. Whereas wild-type Pof10p was downregulated and destabilized in csn5 mutants, Pof10p-P35S was as stable in the mutant as in wild-type cells (Fig. 5B,C). Like plasmid expressed Pof10p-P35S (Fig. 5A), the point mutant expressed from its endogenous genomic locus in csn5 mutants, was completely deficient in binding Cul1p, but maintained wild-type levels of interaction with Skp1p (Fig. 5D). These data establish that proline-dependent recruitment of FBPs into CRL1 complexes destines FBPs for stabilization by the CSN pathway.
We also created a proline knock-in mutation in the endogenous pof12 gene to generate a strain that expressed Pof12p-S15P in either a wildtype or csn5 mutant background. Unlike wildtype Pof12p, Pof12p-S15P expressed from its endogenous promoter gained the ability to interact with Cul1p, albeit at a low level (Fig. 5E). Whereas, as with overexpressed Pof12p-S15P (Fig. 5A), knock-in of the F-box proline was sufficient to target Pof12p into a CRL1 complex, the mutant protein was neither downregulated nor destabilized in csn5 mutants (Fig. 5E; Supplementary Fig. 6). This might be due to the low level of recruitment to Cul1p. In addition, the mutant protein, which is not normally destined for a CRL1 complex, may not bear lysine residues accessible to autocatalytic modification by its associated Cul1p core complex.
CAND1 is not required for maintaining the stability of CSN-regulated FBPs
CAND1 was proposed to participate in the same process of CRL adapter stabilization as the CSN (Cope and Deshaies, 2003; He et al., 2005; Min et al., 2005), although no supporting experimental evidence was provided. To address this proposition in fission yeast, we turned our attention to the uncharacterized open reading frame SPAC1565.07c, which we named knd1. This gene encodes a protein with an overall similarity of 43% (22% identity) with human CAND1 over its entire length of 1220 amino acids. In addition, 25 of the 27 HEAT repeats found in human CAND1, which are responsible for its hallmark solenoid structure (Goldenberg et al., 2004), are readily predicted from the primary sequence of S. pombe Knd1p (data not shown). These considerations suggested that Knd1p is the homolog of CAND1 identified in higher eukaryotes.
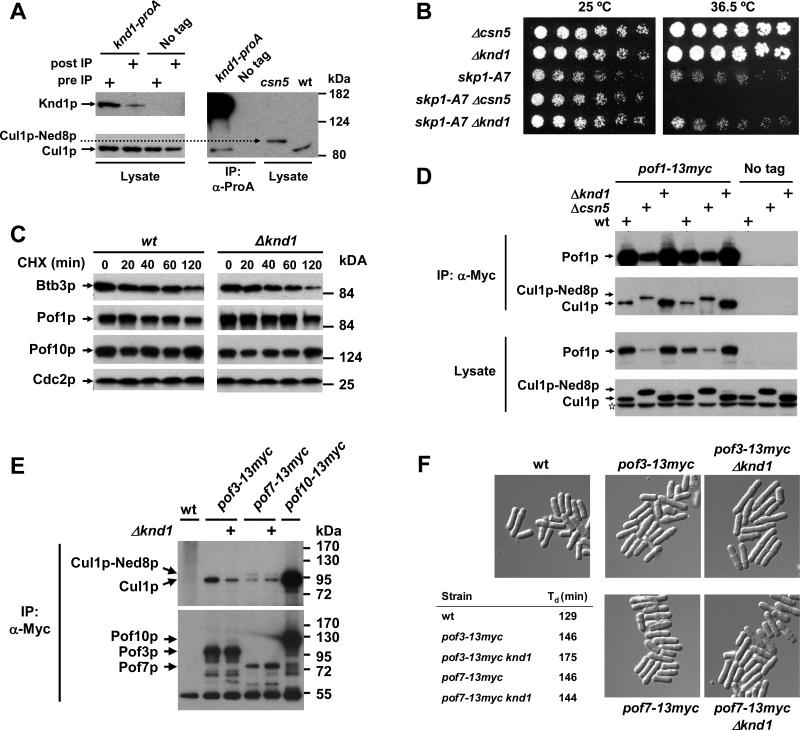
This was supported by the finding that Knd1 expressed from a plasmid specifically interacted with the unneddylated form of Cul1p but not with neddylated Cul1p present in csn5 mutants (Supplementary Fig. 4). Likewise, Knd1p modified with a single N-terminal protein A tag at the endogenous genomic locus co-immunoprecipitated the unneddylated form of Cul1p (Fig. 6A). Importantly, whereas >50% of ProA-Knd1p was depleted from the cell lysate upon absorption to IgG resin, the bulk of Cul1p was retained in the lysate, indicating that only a minor fraction of Cul1p was in a stable complex with Knd1p, at least under steady-state conditions (Fig. 6A).
Fig. 6. CAND1/Knd1p does not maintain FBP stability but regulates cellular CRL composition.
(A) Lysate from a strain expressing Knd1p modified with a single N-terminal protein A tag at the endogenous genomic locus was absorbed to IgG beads, followed by immunoblotting with Cul1p sera. Total cell lysates before and after chromatography on IgG resin are shown on the left to indicate the extent of the Knd1p-Cul1p interaction.
(B) Genetic interactions of knd1 and csn5 with the temperature-sensitive allele skp1-A7. Serial dilutions of the indicated strains were spotted onto YES plates and incubated at the indicated temperatures. Whereas csn5 shows synthetic interaction with skp1-A7, knd1 does not.
(C) The stability of Pof1p-Myc, Pof10p-Myc, and Btb3p-proA in knd1 mutants was determined by CHX chase. Cdc2p is shown for reference.
(D) Binding of Pof1p to Cul1p in csn5 and knd1 mutants. Lysates from csn5 or knd1 mutant cells harbouring endogenously Myc-tagged Pof1p were immunoprecipitated with Myc antibodies followed by immunoblotting with Cul1l antibodies. Duplicate experiments are shown. Csn5 and knd1 mutants not containing Pof1p-Myc are shown as negative controls. The asterisk in the Cul1p blot denotes an unspecific band.
(E) Binding of Pof3p and Pof7p to Cul1p in knd1 mutants. The experiment was performed as described in (D). Pof10p was included as reference for an abundant FBP.
(F) Cell elongation phenotype of pof3-13myc knd1 cells. The indicated strains were grown in liquid YES, fixed with formaldehyde, and visualized by digital interference contrast microscopy. The table shows the doubling times (Td) determined in mid log phase.
Like most csn and ubp12 deletions strains (Mundt et al., 2002; Zhou et al., 2001; Zhou et al., 2003), haploid cells lacking knd1 were viable and did not exhibit any gross morphological or growth phenotypes (data not shown). Unlike csn mutants, however, knd1 mutants did not display accumulation of Cul1p in the neddylated state (Fig. 6D). Nevertheless, since neddylated Cul1p can not interact with CAND1 (Hwang et al., 2003; Liu et al., 2002; Min et al., 2003; Oshikawa et al., 2003; Zheng et al., 2002a), and since Cul1p is fully neddylated in csn5 mutants, CSN deficiency might mimic CAND1 deficiency with respect to CRL1 activity
To test this idea, we performed a genetic assay using a strain containing a temperature-sensitive allele of skp1, which is specifically impaired in binding of FBPs (Lehmann et al., 2004), but not in binding of Cul1p (Wee et al., 2005). In this strain background, we previously demonstrated that CSN becomes essential for viability when adapter recruitment to CRL core complexes is compromised. Whereas skp1-ts csn5 mutants lost viability upon shift to the restrictive temperature as demonstrated before (Wee et al., 2005), skp1-ts knd1 double mutants were fully viable at 36.5 °C (Fig. 6B). Thus, unlike CSN, Knd1p is dispensable for viability, even when adapter recruitment to CRL1 core complexes is compromised.
This finding suggested that knd1 mutants do not suffer the same adapter instability as csn mutants. In fact, downregulation of Pof1p, Pof3p, and Pof7p steady state levels occurring in csn5 mutants was not observed in knd1 mutants (Fig. 6D, E). In addition, the stabilities of the CSN-regulated FBPs, Pof1p and Pof10p, as well as the CRL3 adapter Btb3p were unaffected (Fig. 6C). These findings indicated that CSN maintains CRL adapter stability independently of CAND1.
CAND1 controls the composition of cellular CRL1 complexes
Since, CAND1 deficiency did not phenocopy CSN deficiency with regards to the control of FBP levels and stability, we wondered whether it affected the recruitment of FBPs to Cul1p. Co-immunoprecipitation experiments revealed that binding of Pof1p to Cul1p was strongly enhanced in a knd1 mutants (Fig. 6D). Consistent with CAND1 being displaced from fully neddylated Cul1p, this interaction was also enhanced in a csn5 deletion strain, in particular when accounting for the low steady-state levels of Pof1p present in this mutant (Fig. 6D). Whereas the Pof7p-Cul1p interaction was unaffected by the absence of Knd1p, the Pof3p-Cul1p interaction was substantially decreased (Fig. 6E). Knd1 mutants carrying C-terminally tagged Pof3p showed the same slow growth and cell elongation phenotype as described for pof3 deletion strains (Katayama et al., 2002), indicating that they have a defect in SCFPof3p function (Fig. 6F). These phenotypes were neither observed in wildtype cells carrying tagged Pof3p nor in knd1 mutants harbouring tagged Pof7p (Fig. 6F), suggesting that the pof3-13myc allele is partially defective and therefore requires CAND1 to maintain a sufficient level of activity. Taken together, these findings show that loss of CAND1 differentially affects the composition of CRL1 complexes, presumably by allowing increased recruitment of some abundant FBPs such as Pof1p at the expense of less abundant adapters such as Pof3p.
Discussion
F-box directed CRL assembly and control by the CSN
The results of this study add further credence to our model that CSN's cullin deneddylation activity revealed in vitro serves to maintain the stability of CRL adapters thus promoting CRL activity in vivo (Wolf et al., 2003; Zhou et al., 2003). However, not all FBPs are subject to this mode of regulation, a surprising finding, considering that all FBPs examined here share a canonical F-box motif and interact with Skp1p (Lehmann et al., 2004). Yet, the CSN-insensitive FBPs are not efficiently incorporated into CRL1 complexes, because they lack a critical proline residue in their F-boxes, which is required for binding of Cul1p, but not Skp1p (Fig. 4). This proline is also missing from budding yeast Rcy1p, its fission yeast ortholog Pof6p, and from human Emi1, all of which were previously found to bind Skp1p, but not Cul1p (Galan et al., 2001; Hermand, 2006; Hermand et al., 2003; Seol et al., 2001); and Peter K. Jackson, personal communication).
These findings can be rationalized by the crystal structure of the human CUL1-RBX1-SKP1-SKP2 complex, which showed that the proline is the only conserved F-box residue within a cluster of three contiguous amino acids that makes side chain contacts with CUL1 (Zheng et al., 2002b). Despite its extensive interface with both CUL1 and the F-box, SKP1 is insufficient to recruit FBPs into CRL complexes; instead, this process is specified by proline-dependent F-box-CUL1 interactions. An interesting possibility is that recruitment of FBP-SKP1 dimers to the CRL1 core complex requires the induction of proline-dependent conformational changes within the N-terminus of CUL1.
Based on these data, we propose that all FBPs that lack the critical proline residue are not engaged in canonical CRL1 complexes and are hence not subject to stabilization by the CSN. Consistent with this interpretation is our demonstration that mutation of the conserved proline circumvents the CSN requirement for maintaining Pof10p stability (Fig. 5C). The same scenario would apply to at least 11 human FBPs that lack the proline. Conversely, CSN-mediated cullin deneddylation would assist in the assembly of proline-containing FBPs into CRL1 complexes by shielding them from autocatalytic inactivation. This mechanism provides a safe environment for the de novo assembly and maintenance of CRL complexes with labile FBPs (Wolf et al., 2003).
Distinct CAND1 and CSN cycles
CAND1 inhibits CRL activity in vitro (Hwang et al., 2003; Liu et al., 2002; Min et al., 2003; Oshikawa et al., 2003; Zheng et al., 2002a), but is required for full CRL activity in vivo (Chuang et al., 2004; Feng et al., 2004; Zheng et al., 2002a). This pattern reiterates the CSN paradox, and similar resolutions involving adapter stabilization were invoked for both the CSN and CAND1 paradoxes (Cope and Deshaies, 2003, 2006; He et al., 2005; Zheng et al., 2002a). This seemed attractive considering that inactivation of CSN is expected to phenocopy deletion of CAND1.
The results presented here contradict several key prediction of a model of adapter stabilization involving CAND1: First, only a miniscule fraction of unneddylated Cul1p was in a stable complex with Knd1p, although the vast majority of Cul1p was in the unneddylated state (Fig. 6A). Conversely, unneddylated Cul1p readily interacted with Skp1p and FBPs (Fig. 4B). Both findings are inconsistent with quantitative sequestration of unneddylated Cul1p into inert CAND1 complexes. Secondly, deletion of knd1 did not interfere with adapter stability (Fig. 6C). Likewise, the FBP COI1 is not stabilized in plant CAND1 mutants (Feng et al., 2004). Finally, unlike CSN, Knd1p was not rendered essential when adapter recruitment to CRL1 core complexes was compromised (Fig. 6D). Based on these findings, we conclude that CSN-dependent adapter stabilization principally functions independently of CAND1. CSN is sufficient to sustain this mode of regulation, apparently because the autocatalytic activity of CRL1 complexes is dominantly suppressed by CUL1 deneddylation.
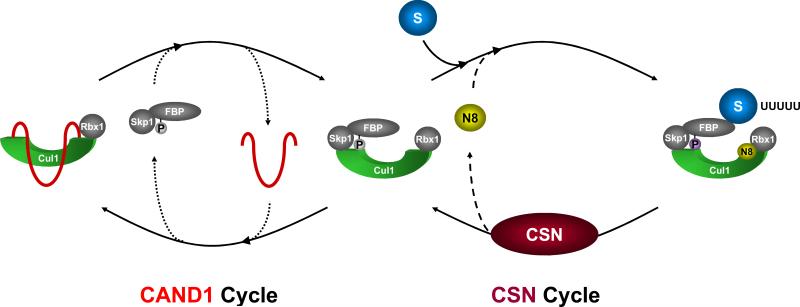
If CAND1 does not maintain adapter stability, what might its function be in promoting CRL activity? Our findings suggest a model according to which transient interaction of CAND1 with unneddylated CUL1-RBX1 core complexes promotes a continuous cycle of adapter recruitment and displacement (CAND1 cycle, Fig. 7). This cycle would assure rapid and constitutive turnover of a given CRL complex in the absence of its substrate. In support of this mechanism are biochemical studies that demonstrated that human SKP2-SKP1 can displace CAND1 from CUL1 (Bornstein et al., 2006). If substrate becomes available, the corresponding CRL-adapter complex would be removed from the CAND1 cycle by substrate driven CUL1 neddylation and transferred into an independent CSN cycle (Fig. 7). Once the substrate is consumed, the CRL will be deneddylated and redirected into the CAND1 cycle.
Fig. 7. Model for toggling of CRL complexes between the CAND1 and CSN cycles.
In the absence of substrate, CRL complexes undergo continuous and rapid adapter exchange in the CAND1 cycle (left). Upon substrate-induced neddylation, CRLs transition into the CSN cycle (right). Once substrate is consumed, CSN-mediated deneddylation toggles CRLs back into the CAND1 cycle for maintenance. CAND1 is symbolized by a red “hook”, NEDD8 by a yellow circle (N), and substrate by a blue circle (S).
This model can readily reconcile the biochemical phenotypes of csn and knd1 mutants: Although only a small fraction of Cul1p was engaged in a stable complex with Knd1p, a situation that is reiterated in A. thaliana (Feng et al., 2004), lack of Knd1p, nevertheless, caused a substantial increase in the recruitment of the abundant FBP, Pof1p, to Cul1p (Fig. 6B). This constellation can only be explained by a continuous and rapid adapter exchange cycle driven by transient Knd1p-Cul1p interactions. In the absence of CAND1, slow exchange would competitively disadvantage relatively rare adapters such as Pof3p thus disturbing the disposition of a subset of cellular CRL complexes. Indeed, in A. thaliana, an organism with an extensive FBP repertoire, CRL-dependent auxin response and photomorphogenesis are compromised in the absence of CAND1, albeit to a lower extent than in CSN mutants, which are presumed to be broadly deficient in all CRLs (Chuang et al., 2004; Feng et al., 2004). Likewise, in S. pombe, loss of knd1 does not show the same synthetic effects with skp1-ts as loss of csn5 (Fig. 6E), again suggesting that only a subset of CRLs is compromised in knd1 mutants.
The proposition that substrate availability transfers CRLs from the CAND1 to the CSN cycle is corroborated by the finding that the CRL substrate p27 can drive CUL1 neddylation and CAND1 displacement from SCFSKP2 in vitro (Bornstein et al., 2006). Likewise, CUL1 and CUL2 neddylation is stimulated by substrate in vivo (Chew and Hagen, 2007). Substrate-dependent toggling of CRLs between the CAND1 and CSN cycle thus provides a ready means of driving both the selection of corresponding CRLs and their recycling.
Role of Ubp12p in CRL regulation
Like Knd1p, the CSN-associated deubiquitylating enzyme Ubp12p was dispensable for the stability of the FBPs examined here. However, the Cul3p adapter Btb3p is strongly destabilized in the same ubp12 deleted cells, in which FBPs are largely stable ((Wee et al., 2005) and Fig. 3). Since we were unable to detect an interaction of Cul3p with Knd1p (data not shown) under conditions at which Knd1p bound Cul1p, CRL3 complexes may not be subject to the CAND1 cycle. The much reduced adapter repertoire for Cul3p – only three BTB adapters exist in fission yeast (Geyer et al., 2003) – may alleviate the need for constitutive exchange. Instead, BTB adapters stably associated with neddylated Cul3p appear to be protected from autocatalytic turn-over by CSN-bound Ubp12p. Additionally, Ubp12p may stabilize CRL core components as described for some organisms (He et al., 2005; Hetfeld et al., 2005; Wu et al., 2005), although we have yet to detect conditions under which similar regulation would occur in fission yeast.
Materials and methods
Yeast strains and plasmids
The knd1 and ubp1 deletion strains and the epitope-tagged strains were constructed by one-step gene replacement using PCR-generated fragments containing ura4 or kanamycin cassettes (Bahler et al., 1998). The ubp1 ubp12 double mutant was obtained by mating and confirmed by PCR. Strains containing Myc-tagged FBPs were provided by T. Toda and crossed into our wild-type background and into csn5 mutants. The skp1-ts knd1 mutant was created by mating, followed by verification of the recombinants by colony PCR. The strain was assayed for synthetic phenotypes by spotting serial dilutions exactly as described (Wee et al., 2005). N-terminally protein A tagged knd1 was constructed following a published protocol (Werler et al., 2003).
A pof10 disruptant was constructed by one-step gene replacement using a PCR-generated fragment containing the ura4 cassette and flanking regions thus inserting the ura4 cassette between codon 31 and 636 of the pof10 open reading frame. The pof10 disruptant was transformed with a PCR-generated fragment consisting of the coding sequence of full length pof10 carrying a mutation of serine 35 to proline. Transformants were grown in 10 ml liquid YES at 30 °C for 22 hours. Ten 10-fold serial dilutions of the liquid culture with fresh YES were prepared and 100μl of each dilution was plated on YES plates containing 0.1% 5-fluororotic acid (5-FOA). After 3-5 days, colonies growing on YES + 5-FOA were confirmed as containing the pof10-P35S mutation by colony-PCR and sequencing. Subsequently, the point mutant pof10 locus was modified with a 13Myc tag. The pof10-P35S Δcsn5 mutant was created by mating. The pof12-S15P and pof12-S15P csn5 mutants were created in an identical manner.
The plasmids expressing Csn5p JAMM mutants were described previously (Wee et al., 2005). FBP plasmids were prepared by amplifying the respective ORFs from Schizosaccharomyces pombe complementary DNA. PCR products were sequenced and then cloned into pRep3 plasmids, which drive the expression of amino-terminally Myc-epitope-tagged proteins from the thiamine-repressible nmt1 promoter. FBP point mutants were constructed with the QuickChange site-directed mutagenesis method (Stratagene) and verified by sequencing.
Immunological methods
Epitope-tagged proteins were detected by the monoclonal anti-Myc antibody 9E10 or with monoclonal anti-Protein A antibodies (Sigma). Cell lysates for immunoprecipitation were prepared as described in a buffer containing 50 mM Tris, pH 7.4, 50 mM NaCl, and 0.5% Triton X-100 (Zhou et al., 2001). Lysates were cleared by centrifugation, and proteins were precipitated with the respective antisera. Immunocomplexes were collected by binding to protein A beads, washed, and analyzed by immunoblotting as described (Zhou et al., 2001). Protein A-tagged proteins were precipitated using whole rabbit immunoglobulin absorbed to Dynabeads. Affinity-purified rabbit antisera against Cul1p, Skp1p, and Rbx1p were described before (Geyer et al., 2003; Seibert et al., 2002). For loading controls Cdc2 (PSTAIR, Santa Cruz) antibodies were used at a dilution of 1:500.
Cycloheximide chase experiments
To measure the stability of adapter proteins, strains containing epitope-tagged versions of FBPs and Btb3p were grown to an OD600 of 1.0 in 50 ml YES media. 100 ug/ml cycloheximide (CHX) was added, and cultures were incubated at 25 °C. 10 ml aliquots removed after the times indicated in the figures. NaN3 was added to a final concentration of 1 mM, in order to instantly kill the cells. Cell were harvested by centrifugation and frozen in liquid nitrogen. Once all samples of the chase period were obtained, cell lysates were prepared by standard bead lysis, and equal amounts of proteins were analyzed by immunoblotting.
CHX chase experiments with strains containing the temperature-sensitive mts3-1 allele were performed in the same way, except that strains were maintained at 37°C for 90 minutes prior to the addition of CHX.
RT/PCR
Total cellular RNA was isolated by cell lysis in hot RNAzol (TelTest, Inc.) and three cycles each of vortexing in the presence of beads for 5 minutes and heating to 65°C for 5 minutes. The RNA was extracted and precipitated according to the recommendations of the manufacturer. 2μg RNA was used in each RT/PCR reaction. Primer design and RT-PCR conditions were according to the manual supplied with the Platinum Quantitative RT-PCR Thermoscript kit (Invitrogen). The primer concentration for actin amplification was 0.25 μM. Other primers were used at 1 μM. Pof13 expression was assayed using a iQ Multiplex Powermix enzyme kit (BioRad) following the supplied protocol. Triplicate reactions were analyzed with a iCycler iQ PCR detection system (BioRad).
Supplementary Material
Acknowledgements
We are grateful to T. Toda for providing strains expressing tagged FBPs and to R. J. Deshaies for JAMM mutants. We wish to thank C. Hoffman for help with the creation of the pof1-P35S mutant. M.W.S is grateful to D. H. Wolf (University of Stuttgart) for support. We thank M. Petroski for helpful comments on the manuscript. This study was funded by NIH grant GM59780 to D.A.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proceedings of the National Academy of Sciences. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu D, Kipreos E. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Division. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew E-H, Hagen T. Substrate-mediated Regulation of Cullin Neddylation. J Biol Chem. 2007;282:17032–17040. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- Chuang H.-w., Zhang W, Gray WM. Arabidopsis ETA2, an Apparent Ortholog of the Human Cullin-Interacting Protein CAND1, Is Required for Auxin Responses Mediated by the SCFTIR1 Ubiquitin Ligase. Plant Cell. 2004;16:1883–1897. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope G, Suh GSB, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of Predicted Metalloprotease Motif of Jab1/Csn5 in Cleavage of NEDD8 from CUL1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti S, Fernandez-Sanchez ME, Rogge L, Bianchi E. The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J Biol Chem. 2006;281:32188–32196. doi: 10.1074/jbc.M604746200. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun T.-p., Deng XW. Arabidopsis CAND1, an Unmodified CUL1-Interacting Protein, Is Involved in Multiple Developmental Pathways Controlled by Ubiquitin/Proteasome-Mediated Protein Degradation. Plant Cell. 2004;16:1870–1882. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci U S A. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates JRI, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 Complex Reveals Regulatory Mechanisms for the Assembly of the Multisubunit Cullin-Dependent Ubiquitin Ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Wallace M, Hastie ND. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, He Q, Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermand D. F-box proteins: more than baits for the SCF? Cell Div. 2006;1:30. doi: 10.1186/1747-1028-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermand D, Bamps S, Tafforeau L, Vandenhaute J, Makela TP. Skp1 and the F-box Protein Pof6 Are Essential for Cell Separation in Fission Yeast. J Biol Chem. 2003;278:9671–9677. doi: 10.1074/jbc.M211358200. [DOI] [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–1221. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Hwang JW, Min KW, Tamura TA, Yoon JB. TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett. 2003;541:102–108. doi: 10.1016/s0014-5793(03)00321-1. [DOI] [PubMed] [Google Scholar]
- Katayama S, Kitamura K, Lehmann A, Nikaido O, Toda T. Fission Yeast F-box Protein Pof3 Is Required for Genome Integrity and Telomere Function. Mol Biol Cell. 2002;13:211–224. doi: 10.1091/mbc.01-07-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A, Katayama S, Harrison C, Dhut S, Kitamura K, McDonald N, Toda T. Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells. 2004;9:367–382. doi: 10.1111/j.1356-9597.2004.00730.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S, Roy M, Larsen B, Le Bihan T, Metalnikov P, Tyers M, Peter M, Pintard L. CIF-1, a Shared Subunit of the COP9/Signalosome and Eukaryotic Initiation Factor 3 Complexes, Regulates MEL-26 Levels in the Caenorhabditis elegans Embryo. Mol Cell Biol. 2007;27:4526–4540. doi: 10.1128/MCB.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Min KW, Hwang JW, Lee JS, Park Y, Tamura TA, Yoon JB. TIP120A Associates with Cullins and Modulates Ubiquitin Ligase Activity. J Biol Chem. 2003;278:15905–15910. doi: 10.1074/jbc.M213070200. [DOI] [PubMed] [Google Scholar]
- Min KW, Kwon MJ, Park HS, Park Y, Yoon SK, Yoon JB. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem Biophys Res Commun. 2005;334:867–874. doi: 10.1016/j.bbrc.2005.06.188. [DOI] [PubMed] [Google Scholar]
- Mundt KE, Liu C, Carr AM. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun. 2003;303:1209–1216. doi: 10.1016/s0006-291x(03)00501-1. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30 )complex. Embo J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- Seibert V, Prohl C, Schoultz I, Rhee E, Lopez R, Abderazzaq K, Zhou C, Wolf DA. Combinatorial diversity of fission yeast SCF ubiquitin ligases by homo- and heterooligomeric assemblies of the F-box proteins Pop1p and Pop2p. BMC Biochemistry. 2002;3:22. doi: 10.1186/1471-2091-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- von Arnim AG. On again - off again: COP9 signalosome turns the key on protein degradation. Current Opinion in Plant Biology. 2003;6:520–529. doi: 10.1016/j.pbi.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002;3:15. doi: 10.1186/1471-2156-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werler PJ, Hartsuiker E, Carr AM. A simple Cre-loxP method for chromosomal N-terminal tagging of essential and non-essential Schizosaccharomyces pombe genes. Gene. 2003;304:133–141. doi: 10.1016/s0378-1119(03)00402-5. [DOI] [PubMed] [Google Scholar]
- Wirbelauer C, Sutterluty H, Blondel M, Gstaiger M, Peter M, Reymond F, Krek W. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. Embo J. 2000;19:5362–5375. doi: 10.1093/emboj/19.20.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002a;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002b;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zhou C, Seibert V, Geyer R, Rhee E, Lyapina S, Cope G, Deshaies RJ, Wolf DA. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochemistry. 2001;2:7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission Yeast COP9/Signalosome Suppresses Cullin Activity through Recruitment of the Deubiquitylating Enzyme Ubp12p. Mol Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.