Abstract
By exploiting the cell-cycle-dependent proteolysis of two ubiquitination oscillators, human Cdt1 and geminin, which are the direct substrates of SCFSkp2 and APCCdh1 complexes, respectively, Fucci technique labels mammalian cell nuclei in G1 and S/G2/M phases with different colors. Transgenic mice expressing these G1 and S/G2/M markers offer a powerful means to investigate the coordination of the cell cycle with morphogenetic processes. We attempted to introduce these markers into zebrafish embryos to take advantage of their favorable optical properties. However, although the fundamental mechanisms for cell-cycle control appear to be well conserved among species, the G1 marker based on the SCFSkp2-mediated degradation of human Cdt1 did not work in fish cells, probably because the marker was not ubiquitinated properly by a fish E3 ligase complex. We describe here the generation of a Fucci derivative using zebrafish homologs of Cdt1 and geminin, which provides sweeping views of cell proliferation in whole fish embryos. Remarkably, we discovered two anterior-to-posterior waves of cell-cycle transitions, G1/S and M/G1, in the differentiating notochord. Our study demonstrates the effectiveness of using the Cul4Ddb1-mediated Cdt1 degradation pathway common to all metazoans for the development of a G1 marker that works in the nonmammalian animal model.
Keywords: cell cycle, fluorescent protein, imaging, ubiquitination
Eukaryotic cells ensure tight regulation of cell division by maintaining close control over the levels of cell-cycle proteins. For example, Cdt1 and geminin have opposite effects on DNA replication during S phase, and their levels fluctuate accordingly throughout the cell cycle (1, 2). Cdt1 levels are highest in G1 phase just before DNA replication and decrease as cells transition into S phase, whereas geminin levels rise during S phase and fall during G1 phase. Cells control Cdt1 and geminin activity at the protein level by ubiquitination, which precisely targets unwanted proteins for destruction.
We harnessed the regulation of cell-cycle-dependent ubiquitination to develop a genetically encoded indicator for cell-cycle progression: Fucci (fluorescent ubiquitination-based cell cycle indicator) (3). The original Fucci probe was generated by fusing mKO2 (monomeric Kusabira Orange2) and mAG (monomeric Azami Green) to the ubiquitination domains of human Cdt1 (hCdt1) and human geminin (hGem): hCdt1(30/120) and hGem(1/110), respectively. These two chimeric proteins, mKO2-hCdt1(30/120) and mAG-hGem(1/110) (Fig. 1 A and B), accumulate reciprocally in the nuclei of transfected mammalian cells during the cell cycle, labeling nuclei of G1 phase cells orange and those in S/G2/M phase green. Thus, these proteins function as effective G1 and S/G2/M markers. We also developed a S/G2/M marker, mAG-hGem(1/60), which accumulates in both the nucleus and cytoplasm (4) and reveals the morphology of individual cells that have undergone DNA replication. This permits cell proliferation to be monitored along with the morphological differentiation of various cell types.
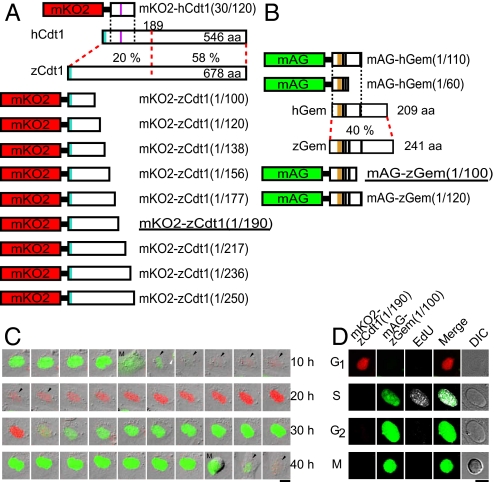
Fig. 1.
Development of fluorescent indicators for cell-cycle progression in fish cells (zFucci), and characterization of zFucci in transgenic fish (Cecyil) cells compared with Fucci in HeLa cells. (A) Structural domains of hCdt1 (human Cdt1) and zCdt1 (zebrafish Cdt1). Cyan box, PIP box or QXRVTDF motif (amino acids 3–9); violet box, Cy motif (amino acids 68–70). The N-terminal 189 aa of hCdt1 are sufficient for G1-specific accumulation of the protein (11). This regulatory region shows 20% sequence homology between hCdt1 and zCdt1. The other region, which contains the geminin and MCM6 binding domains, shows 58% homology between the two proteins. The G1 marker of the original Fucci, mKO2-hCdt1(30/120), is illustrated at the Top. Various constructs with concatenated mKO2 and deletion mutants of zCdt1 are shown at the Bottom. mKO2-zCdt1(1/190) is underlined. (B) Structural domains of hGem (human geminin) and zGem (zebrafish geminin). The S/G2/M markers of the original Fucci, mAG-hGem(1/110) and mAG-hGem(1/60), are illustrated at the Top. Two constructs with concatenated mAG and deletion mutants of hGem for labeling nuclei in S, G2, and M phases. mAG-zGem(1/100) is underlined. Orange box, D (destruction) box; black box, NLS (nuclear localization signal). (C) Cell-cycle-dependent changes in fluorescence of zFucci (mKO2-zCdt1(1/190) and mAG-zGem(1/100)) in Cecyil cells. Arrows indicate cells that were tracked. (Scale bar, 10 μm.) M, M phase. (D) Typical fluorescence images of Cecyil cells expressing zFucci (mKO2-zCdt1(1/190) and mAG-zGem(1/100)) and fluorescence from incorporated EdU (white) at G1, S, G2, and M phases. (Scale bar, 10 μm.)
Visualizing the cell-cycle behavior of individual cells within complex tissues presents an irresistible challenge to biologists studying multicellular structures. We previously generated transgenic mice that express Fucci in every cell, and characterized the cell-cycle behavior of embryonic neural progenitor cells (3). However, we wished to extend our studies even further by following the dynamic process of cell proliferation in whole embryos. We therefore turned to zebrafish embryos, whose external development and transparency provide good access to almost every stage of embryogenesis. Using the hspa8 promoter, we generated transgenic zebrafish lines that express mAG-hGem(1/110) or mKO2-hCdt1(30/120). Four mAG-hGem(1/110) lines (Tg(hspa8:mAG-hGem(1/110))rw0409a-d, Table S2) showed faithful green fluorescent labeling of nuclei in S/G2/M phases. However, orange fluorescence was observed throughout the cell cycle rather than just in G1 in two mKO2-hCdt1(30/120) lines (Tg(hspa8:mKO2-hCdt1(30/120))rw0401b,c, Table S1). These results suggest that geminin, but not Cdt1, is interchangeable between mammals and fish in terms of ubiquitin-mediated degradation (Fig. S1). Thus, application of Fucci technology in nonmammalian animals requires further study.
Here, we describe our development of zebrafish Fucci (zFucci), a powerful new tool to dissect cell-cycle behavior in vivo. We generated DNA constructs using the zebrafish homologs of Cdt1 (zCdt1) and geminin (zGem), characterized them using cultured fish cells, and constructed transgenic zebrafish lines. We were able to observe the dynamic patterns of cell-cycle progression in several parts of the embryo, including the retina and notochord.
Results
Construction of a G1 Marker for Fish Cells.
We redesigned the G1 marker for zebrafish cells using zCdt1. Regulated proteolysis of Cdt1 is the major mechanism behind preventing rereplication (5). Importantly, Cdt1 is also destroyed in response to DNA damage. A pathway of both the DNA replication- and damage-related Cdt1 destruction is well conserved among metazoans (6, 7). The pathway involves a common ubiquitin E3 ligase (Cul4Ddb1) and chromatin-bound proliferating cell nuclear antigen (PCNA); Cdt1 contains a conserved PCNA interaction protein motif (PIP box or QXRVTDF motif) (8, 9) (Fig. 1A). However, in addition to the Cul4Ddb1-mediated Cdt1 destruction, mammalian cells employ SCFSkp2-mediated Cdt1 destruction, which depends on the phosphorylation of Cdt1 by cyclin E/A-cyclin-dependent kinases; hCdt1 contains the Cy motif that binds to the SCFSkp2 E3 ligase (Fig. 1A). Experiments using mammalian cells have revealed that Cul4Ddb1 is required for Cdt1 proteolysis in response to both DNA replication and damage, whereas SCFSkp2 is mainly involved in DNA replication (6, 7). In the original work on Fucci (3), therefore, we engineered the hCdt1-based G1 marker to be unaffected by DNA damage; the marker, mKO2-hCdt1(30/120), carries the Cy motif but not the N terminus containing the PIP box.
Cdt1 consists of three domains: the N-terminal domain, which is poorly conserved among eukaryotes and is involved in the protein's degradation; the central domain, which contains a geminin binding site and is conserved among metazoans; and the C-terminal domain, which binds to minichromosome maintenance (MCM) 6 protein and is highly conserved among eukaryotes (10). Since the 189 N-terminal amino acids of hCdt1 are sufficient for S-phase-specific proteolysis (11), we focused on the N-terminal domain of zCdt1 (Fig. 1A). zCdt1 lacks the Cy motif, suggesting that the PIP box is required to develop a functional G1 maker in fish cells.
We generated numerous zCdt1 mutant constructs containing the PIP box but with varying degrees of C-terminal truncations. These were fused to mKO2 and evaluated for cell-cycle-dependent orange fluorescence in cultured fish cells by time-lapse imaging (Table S1). A permanent goldfish cell line (GEM-81) (12) was imaged after transient transfection with the DNA constructs (see SI Methods). Interestingly, zCdt1(1/100) and zCdt1(1/120), which were similar in size to hCdt1(30/120), produced a constant fluorescence signal throughout the cell cycle in both the nucleus and cytoplasm. In contrast, a series of longer mutant constructs, including zCdt1(1/138), zCdt1(1/156), zCdt1(1/177), zCdt1(1/190), zCdt1(1/217), zCdt1(1/236), and zCdt1(1/250) fused to mKO2 all showed clear fluctuation of orange fluorescence in the nucleus. Because the fluorescence was apparent in the first half of the cell cycle, the proteins likely accumulated specifically in G1 phase.
Next, these constructs with the EF1α promoter (13) were introduced into zebrafish as transgenes (see SI Methods). For each construct, multiple transgenic lines were generated and screened for fluorescence. Ten lines, Tg(EF1α:mKO2-zCdt1(1/138))rw0402a,b, Tg(EF1α:mKO2-zCdt1(1/177))rw0404a-c, Tg(EF1α:mKO2-zCdt1(1/190))rw0405a-d, and Tg(EF1α:mKO2-zCdt1(1/236))rw0407a, were selected for characterization of the spatiotemporal fluorescence patterns (Table S1). We examined when the mKO2 fluorescence was first detectable after fertilization. zFucci should produce sufficient signal to report cell-cycle progression after the midblastula transition (MBT), which leads to a lengthening of the cell cycle and an increase in RNA synthesis. In the lines expressing mKO2-zCdt1(1/138), mKO2-zCdt1(1/177), and mKO2-zCdt1(1/190), orange fluorescence became observable at 50% epiboly, during which the beginning of involution defines the onset of gastrulation. We noticed that the constructs encoding less truncated versions of zCdt1 showed later onset of fluorescence. For example, embryos expressing mKO2-zCdt1(1/236) started to fluoresce during the segmentation (10-somite stage). In contrast, constructs encoding more truncated versions of zCdt1 (mKO2-zCdt1(1/138)) exhibited unclear fluctuation of fluorescence, as was observed for mKO2-zCdt1(1/100) and mKO2-zCdt1(1/120) in GEM-81 cells. After investigation via time-lapse imaging experiments, we concluded that mKO2-zCdt1(1/190) had the best performance in vivo. We selected two lines, Tg(EF1α:mKO2-zCdt1(1/190))rw0405b,d, which expressed bright orange fluorescence reliably, turning on and off in every cell (Table S1). Importantly, their embryos seemed to grow normally, and the adults were fertile.
Construction of an S/G2/M Marker for Fish Cells.
Although transgenic lines expressing mAG-hGem(1/110) showed fluctuating green fluorescence in every nucleus after 50% epiboly, we used zGem to generate S/G2/M markers (Fig. 1B). A chimeric protein composed of mAG and the N-terminal 100 aa of zGem (mAG-zGem(1/100)) was found to label the nucleus of transfected GEM-81 cells in the second half of the cell cycle. mAG-zGem(1/120) gave the same results as mAG-zGem(1/100). We next made transgenic lines producing mAG-zGem(1/100) or mAG-zGem(1/120) using the EF1α promoter. Time-lapse fluorescence imaging was performed using four lines (Tg(EF1α:mAG-zGem(1/100))rw0410d,g,h and Tg(EF1α:mAG-zGem(1/120))rw0411b), and bright green nuclei were observed to blink after the 50% epiboly stage in every line (Table S2). Tg(EF1α:mAG-zGem(1/100))rw0410h was selected for further experiments.
mAG-hGem(1/60) is an S/G2/M marker distributed in both the nucleus and cytoplasm (4). We found that mAG-hGem(1/60) showed the same spatiotemporal pattern of fluorescence in GEM-81 cells and transgenic zebrafish lines as in mammalian cells (Fig. 1B, Table S2). Tg(EF1α:mAG-hGem(1/60))rw0412a was selected for further experiments.
zFucci Technology Established in Transgenic Zebrafish Lines.
We cross-bred Tg(EF1α:mKO2-zCdt1(1/190))rw0405b and Tg(EF1α:mAG-zGem(1/100))rw0410h to generate a zebrafish line producing zFucci. In this line, which we called Cecyil (cell cycle illuminated), every cell nucleus appeared to exhibit either orange or green fluorescence. To characterize the performance of zFucci in detail, we dissociated and cultured cells from a Cecyil embryo (10-somite stage) and performed time-lapse imaging (see SI Methods and Movie S1). In each cell, orange fluorescence alternated with green fluorescence in the nucleus (Fig. 1C). The intensities of orange and green signals were plotted against time (Fig. S2A). Using data from many (>20) cells, we characterized the temporal profile of zFucci as schematized in the inset. For comparison, a typical temporal profile of the original Fucci in HeLa cells and its schematic graph are also shown (Fig. S2B). There is a slight difference in the timing of the orange-to-green conversion between the two Fucci systems. In the mammalian system, orange and green fluorescence overlapped to yield a yellow nucleus (arrows, Fig. S2B). In contrast, zFucci did not produce a yellow nucleus, because the orange fluorescence dropped off quickly (arrows, Fig. S2A). This difference is likely because of the fact that fish use a Cul4Ddb1-mediated mechanism for Cdt1 destruction, whereas the mammals employ SCFSkp2. To examine whether the timing of the color conversion correlates with the onset of S phase, Cecyil cells were pulse labeled with 5-ethynyl-2′-deoxyuridine (EdU) (14) (Fig. 1D). None of the cells with orange nuclei showed EdU incorporation (Fig. S3). Cells with green nuclei were either in the S or G2 phase, and were distinguishable by nuclear EdU staining.
We also cross-bred Tg(EF1α:mKO2-zCdt1(1/190))rw0405d and Tg(EF1α:mAG-hGem(1/60))rw0412a to generate another transgenic line, Cecyil2, in which each cell exhibited orange fluorescence in G1 phase nuclei and green fluorescence in both the nucleus and cytoplasm of S/G2/M phase cells. This zFucci-S/G2/M(NC) line permits us to trace the silhouette of individual proliferating cells, and thus, to identify cell types and differentiation states by their characteristic morphologies.
Panoramic Views of Cell-Cycle Progression Inside the Developing Fish Embryo.
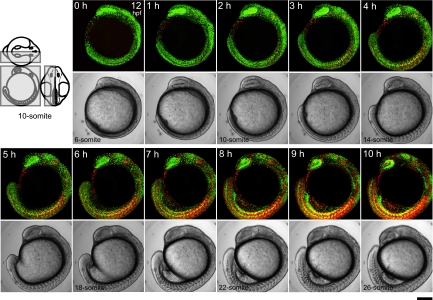
We observed zFucci fluorescence in a Cecyil embryo during segmentation. Three-dimensional time-lapse imaging was performed to collect fluorescence images from the left half of the embryo at 10-min intervals using an upright confocal microscope (Movie S2). Sagital (xy) images (every hour) are shown in Fig. 2. Because they were stacked along the z axis, which extended between the midplane and body surface (Fig. 2, three drawings), each image contains complete information about cell proliferation occurring in the embryo. Initially, the green signal predominated over the orange signal almost everywhere, indicating rapid mitotic cycling. The overall ratio of green-to-orange signal decreased as the embryo grew. By the end of segmentation, the color balance was reversed. While green signal was concentrated in several organs, including the retina and brain, orange signal strongly highlighted well-differentiated cells, such as postmitotic neurons and muscle cells.
Fig. 2.
Time-lapse imaging of a Cecyil embryo during segmentation. Three-dimensional time-lapse imaging was performed to collect fluorescence images from the left half of the embryo at 10-min intervals using an Olympus FV1000 upright confocal microscope equipped with an objective lens (×10 N.A. 0.3). A dechorionated embryo in the early segmentation period at 12 hpf (6-somite stage) was mounted with the left side up in a chamber containing 0.3% agar. Since the sample was kept at less than 28 °C during observation, developmental stages cannot be accurately expressed in hpf. Due to z-stacking, green and orange signals at different z-positions merge to generate yellow signal. Note that zFucci does not yield yellow fluorescence at the G1/S transition, whereas the original Fucci in mammalian cells does. The scanned region is indicated by the gray box on the three views of an embryo at the 10-somite stage. (Scale bar, 200 μm.)
Time-Lapse Imaging of Interkinetic Nuclear Migration in the Early Developing Retina.
During retinogenesis, multipotent retinal progenitor cells exit the cell cycle to differentiate into all retinal cell types. At 28 hours postfertilization (hpf), a group of cells in the ventronasal region first exit the cell cycle and differentiate into ganglion cells (15). Many studies have focused on zebrafish retinogenesis after 28 hpf to examine how cell proliferation, cell cycle exit, and neurogenesis are coordinated (16–18). We instead focused on early developmental stages (22–27 hpf), when all retinal neuroepithelial cells are mitotic. To verify that zFucci permits in vivo visualization of interkinetic nuclear migration (18), we collected optical z-sections of the developing retina of Cecyil embryos at 3.5-μm steps every 10 min for 6 h (Movie S3). During this observation period, green signal predominated over orange in the neuroepithelial sheet, indicating rapid cycling of neuronal progenitor cells. It was possible to track green nuclei migrating from the basal to the apical side (Fig. S4A, yellow arrowheads) and orange nuclei migrating in the opposite direction (Fig. S4A, white arrows).
A similar experiment was performed using Cecyil2 embryos. Retinal neuroepithelial cells expressing zFucci-S/G2/M(NC) were imaged beginning at 22 hpf (Fig. S4B). The nuclei of green cells at S/G2/M phases could be distinguished from the cytoplasm by the more intense nuclear fluorescence. While progressing through the cell cycle, green progenitor cells extended processes and displaced their nuclei from the basal to the apical side.
Two Waves of Cell-Cycle Transitions Traveling from the Anterior to the Posterior of the Notochord.
The notochord is the first organ to fully differentiate during embryogenesis. Although this slender rod of cells is essential for the patterning of surrounding tissues, such as the neuroectoderm and paraxial mesoderm (19, 20), little is known about how the cell cycle progresses within this organ in coordination with its differentiation. Because the notochord is in the center of the trunk and difficult to access, and the high mobility of embryos makes it difficult to keep a large portion of the notochord within a single focal plane for a sufficient length of time, time-lapse imaging of this organ in developing embryos is a formidable task. Thus, we used multiple fixed embryos to examine the spatiotemporal patterns of cell-cycle progression in the notochord.
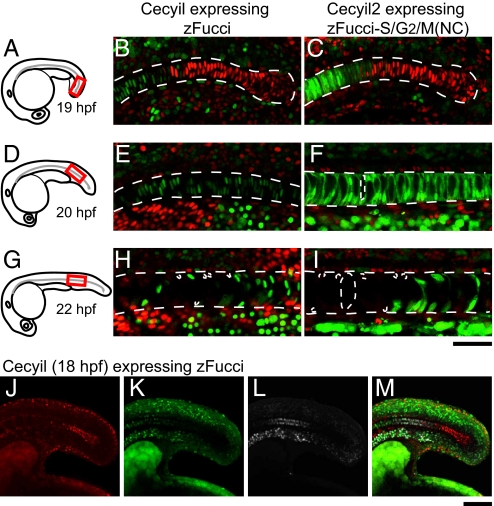
As observed in the tail region of the notochord of a Cecyil embryo at 19 hpf (Fig. 3A, red box), there was a discernible boundary between two cell populations (Fig. 3B). Orange nuclei were on the posterior side of the boundary, whereas green nuclei were on the anterior side. As the notochord elongated further, the G1/S boundary moved toward the posterior. An embryo at 20 hpf was used to observe the same region relative to the head (Fig. 3D, red box). All of the cells showed green nuclei (Fig. 3E), indicating that they had undergone the G1/S transition. Then, after remaining in G2 phase for some time, the notochordal cells entered M phase. This M/G1 transition boundary also moved from anterior to posterior. At 22 hpf, dim orange G1 nuclei were observed on the anterior side of the boundary, and green G2 or M nuclei were on the posterior side (Fig. 3H). Interestingly, mitosis in the notochord was accompanied by extreme vacuolization, such that the orange nuclei of new daughter cells were pushed to one side of the cell (21).
Fig. 3.
Cell-cycle transition waves in the differentiating notochord. Dechorionated embryos at various stages were fixed in 4% paraformaldehyde (PFA) solution, then each sample was mounted in 0.3% agar so that a confocal image of the posterior region of the notochord could be obtained. (A) A schematic drawing of an embryo at 19 hpf. (B) A fluorescence image of the posterior region (indicated in A) of the notochord of a Cecyil embryo at 19 hpf. (C) A fluorescence image of the posterior region (indicated in A) of the notochord of a Cecyil2 embryo at 19 hpf. (D) A schematic drawing of an embryo at 20 hpf. (E) A fluorescence image of the posterior region (indicated in D) of the notochord of a Cecyil embryo at 20 hpf. (F) A fluorescence image of the posterior region (indicated in D) of the notochord of a Cecyil2 embryo at 20 hpf. (G) A schematic drawing of an embryo at 22 hpf. (H) A fluorescence image of the posterior region (indicated in G) of the notochord of a Cecyil embryo at 22 hpf. (I) A fluorescence image of the posterior region (indicated in G) of the notochord of a Cecyil2 embryo at 22 hpf. (J–M) A Cecyil embryo at 18 hpf was treated with 400 μM EdU for 1 h and then fixed with 4% PFA. Alexa647-azide was used to visualize EdU incorporation. Fluorescence images of the notochord of a Cecyil embryo at 18 hpf for G1 marker (red) (J), S/G2/M marker (green) (K), incorporated EdU (white) (L), and their merge (M). [Scale bar, 50 μm (A–I); 100 μm (J–M.]
We identified two waves of cell-cycle transitions: G1/S and M/G1, both of which traveled from anterior to posterior in the differentiating notochord. The discovery of a G1/S wave suggests a strong coordination between DNA replication and differentiation of the notochord. To map S-phase entry along this organ, we pulse labeled a Cecyil embryo (18 hpf) with EdU (Fig. 3 J–M). Whereas EdU localized to green nuclei throughout the embryo, EdU-positive green nuclei in the notochord were confined to the posterior end. EdU-negative green nuclei were likely in G2 phase. Thus, notochordal cells were arranged in the order of cell-cycle phase (G1, S, and G2 phases) from posterior to anterior.
The two waves of cell-cycle transitions in the notochord were also observed in Cecyil2 embryos, but additional features of these cells were revealed. Although the G1/S boundary was in the same region and at the same stage as in Cecyil embryos (Fig. 3B), cells immediately behind the G1/S wave had no vacuoles (Fig. 3C). In contrast, notochordal cells that were probably in G2 phase were slightly polarized and contained some vacuoles (Fig. 3F). Furthermore, the M/G1 boundary was highlighted clearly, with both the nucleus and the cytoplasm filled with green fluorescence during vacuolization and cell division (Fig. 3I).
Discussion
Because it is generally thought that the fundamental mechanisms for controlling the cell cycle are well conserved among species, it seemed likely that the original Fucci technique designed for mammalian cells would be fully applicable to nonmammalian species. However, the G1 marker based on hCdt1 does not function in zebrafish, probably because hCdt1 is not ubiquitinated properly by a fish E3 ligase complex. Differences between the mammalian and nonmammalian mechanisms of Cdt1 degradation likely account for this; in addition to the Cul4Ddb1-mediated Cdt1 destruction pathway common to all metazoans, mammalian cells employ a SCFSkp2-mediated Cdt1 destruction pathway. We used the Cy motif of hCdt1, which binds to the SCFSkp2 E3 ligase, to develop the G1 marker for mammalian cells (3). However, because zCdt1 lacks the Cy motif, we instead used the PIP box, which is involved in Cul4Ddb1-mediated Cdt1 destruction, to develop the G1 marker for fish cells. mKO2-zCdt(1/190) was able to label the G1 nucleus faithfully in cultured fish cells and transgenic zebrafish lines. Together with the S/G2/M markers based on hGem or zGem, mKO2-zCdt(1/190) was used to establish the zFucci system. The transgenic lines Cecyil and Cecyil2, which express zFucci and zFucci-S/G2/M(NC), respectively, exhibit normal development, confirming that the indicators do not affect normal cell-cycle progression of fish cells.
Although the original Fucci causes nuclei to turn yellow at the G1/S transition, zFucci does not. There is a very short time gap between the green and orange fluorescence in zFucci, which makes it slightly difficult to track the G1/S transition of a migrating cell in three-dimensional space. One solution is to image cells at short time intervals to get smooth trajectories. Alternatively, limiting expression of zFucci to a small fraction of cells may be helpful. Transplantation of fluorescent cells from Cecyil or Cecyil2 embryos into nonfluorescent recipient embryos can generate mosaic animals in which individual cells expressing zFucci can be easily identified within a tissue. Using this approach, wide-field observation using an objective lens of low numerical aperture will allow for efficient tracking of cell-cycle progression of moving cells. This should be particularly effective for cells expressing zFucci-S/G2/M(NC), which reveals the morphology of cells at S/G2/M phases. Another method of restricting expression of zFucci is the use of tissue-specific promoters. For instance, live imaging of the notochord in an intact embryo could be achieved by using a notochord-specific promoter (22). Cecyil and Cecyil2 will be particularly powerful when used in conjunction with additional markers of different colors. For example, although we focused on early stages of zebrafish retina development to observe interkinetic nuclear migration, the addition of fluorescent markers such as huc:RFP (23) at later developmental stages will allow us to study how cell proliferation and cell-cycle exit during neurogenesis are coordinated.
Although the transition from G1 to S is difficult to observe in live samples, the spatial pattern of the G1/S transition occurring within the notochord could be directly visualized using zFucci technology. Our experiments using fixed embryos at multiple stages revealed an anterior to posterior wave of cells undergoing the G1/S transition. The M/G1 transition can generally be monitored by morphological changes, but this is also difficult in the notochord due to the extensive vacuolization accompanying cell division. By labeling the M- and G1-phase nuclei with green and orange fluorescence, respectively, we were able to distinguish between these two cell populations.
Two intriguing questions arise from our data. First, what regulates the occurrence and speed of cell-cycle transition waves? Second, what are the developmental consequences of these waves? These questions will be addressed by time-lapse imaging of notochord cells expressing zFucci. Moreover, Fucci technology will be useful for addressing evo-devo questions (24), such as how development of the notochord, which is found in embryos of all chordates, was modified during evolution (25, 26). Whereas the notochord is replaced by the vertebral column in higher vertebrates, it persists throughout life as the main axial support of the body in lower vertebrates (27). We plan to expand Fucci technology to urochordates, such as Ciona intestinalis.
Although the fish embryo is transparent during early development, special technologies may be required to perform fluorescence imaging in a large three-dimensional space. For example, selective plane illumination microscopy (28) and digital scanned laser light sheet fluorescence microscopy (29) have been developed for high-speed in vivo observation of embryonic development at subcellular resolution. These systems enable tracking of individual cells in the whole fish embryo to provide a “digital embryo,” a three-dimensional reconstruction of early cell division patterns. While these studies used histone-2B-EGFP for labeling the nucleus (29), substitution of zFucci will add DNA replication information to these digital embryos.
Descriptive embryology using zFucci will permit the construction of comprehensive databases of cell proliferation, differentiation, and movement during zebrafish development. In addition, combining zFucci with genetic approaches will be fruitful. For example, crossing zFucci-expressing lines to the many mutant lines isolated by forward genetic screens or to the numerous transgenic lines expressing fluorescent markers under the control of cell type- or stage-specific promoters via Cre/lox or UAS/Gal4 technology (30) will be interesting. Finally, because the external development and transparency of zebrafish embryos favor experimental embryology, zFucci-expresing cells can be used for tissue recombination and interchimeric transplantation, or grown in isolation to study cell–cell communication. Thus, zFucci technology will facilitate a number of marriages and crossovers between various fields of embryology, allowing us to decipher the underlying rules of cell proliferation in morphogenetic processes.
Methods
In Vivo Time-Lapse Imaging.
A glass bead (Iuchi BZ-1) was placed on a coverslip, onto which 1% agarose (Takara L03) solution in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.4 mM CaCl2, and 0.16 mM MgSO4) was poured and allowed to harden. Then the glass bead was removed to generate a round chamber. An embryo, anesthetized with Tricaine at >22 hpf, was placed in the chamber and covered with 0.3% agarose in E3 solution. The chamber was submerged in E3 solution containing Tricaine and PTU. Time-lapse 3D imaging was performed in the xyz-t mode using an FV1000 (Olympus) confocal upright microscope system equipped with a water-immersion 20× objective (N.A. 0.9). Two laser lines, 473 nm and 559 nm were used. The recording interval was 5 min. At each time point, 40 confocal images along the z axis were acquired. To avoid cross-detection of green and orange signals, images were acquired sequentially at 473 nm and 559 nm. Proper alignment and correct image registration of two laser lines and detection channels were verified using double-labeled fluorescent beads (TetraSpeck Fluorescent Microsphere Standards, 0.5 μm in diameter, Molecular Probes). Data analysis was performed using Volocity software (Improvision) and MetaMorph software.
Imaging of the Notochord.
Dechorionated embryos (19–22 hpf) were fixed in 4% PFA (pH 7.4) in PBS for 2 h at room temperature. EdU labeling was carried out using the Click-iTTMEdU Alexa Fluor imaging kit (Molecular Probes) according to the manufacturer's instruction, with some modifications. Briefly, dechorionated embryos were incubated with 400 μM EdU for 1 h at room temperature. After fixation, they were treated with reagent containing Alexa647-azide for detection. Image acquisition was performed using an FV1000 (Olympus) confocal upright microscope system equipped with 473 nm, 559 nm, and 633 nm laser lines, or an EZ-S1 (Nikon) confocal upright microscope system equipped with 488 nm, 543 nm, and 647 nm laser lines.
Distribution of Materials.
Transgenic zebrafish lines: Tg(EF1α:mKO2-zCdt1(1/190))rw0405b,d, Tg(EF1α:mAG-zGem(1/100))rw0410h, and Tg(EF1α:mAG-hGem(1/60))rw0412a (see Table S1 and Table S2) will be distributed from the National BioResource Project, Zebrafish (http://www.shigen.nig.ac.jp/zebra/index_en.html).
Supplementary Material
Acknowledgments.
The authors thank Hiroshi Kurokawa, Akiko Ishioka, Reiko Sato, and Yoshiko Wada for technical assistance; Dr. Hideaki Mizuno, Dr. Ichiro Masai, Dr. Kaoru Sugimura, Dr. David Mou, and Dr. Hiroyuki Takeda for valuable advice. This work was partly supported by grants from Japan MEXT Grant-in-Aid for Scientific Research on priority areas and the Human Frontier Science Program. None of the authors have a financial interest related to this work. M.S. was supported by RIKEN′s Junior Research Associate Program. T.I. was supported by the grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan for Global Center of Excellence Program, “International Research Center for Molecular Science in Tooth and Bone Diseases,” and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (21659426).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences for mKO2-zCdt1(1/190), mAG-zGeminin(1/100), and mAG-hGeminin(1/60) reported in this paper have been deposited in the DDBJ/EMBL/GenBank databases (accession nos. AB505859, AB505860, and AB505861, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906464106/DCSupplemental.
References
- 1.Ang XL, Harper JW. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci STKE. 2004;242:pe31. doi: 10.1126/stke.2422004pe31. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama KI, Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 3.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 4.Sakaue-Sawano A, et al. Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem Biol. 2008;15:1243–1248. doi: 10.1016/j.chembiol.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y, Kipreos ET. Cdt1 degradation to prevent DNA re-replication: Conserved and non-conserved pathways. Cell Div. 2007;2:18. doi: 10.1186/1747-1028-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connell B, Harper JW. Ubiquitin proteasome system (UPS): What can chromatin do for you? Curr Opin Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Arias EE, Walter JC. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol. 2005;8:84–90. doi: 10.1038/ncb1346. [DOI] [PubMed] [Google Scholar]
- 9.Nishitani H, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensind factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- 11.Nishitani H, Lygerou Z, Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of Geminin through its N-terminal region. J Biol Chem. 2004;279:30807–30816. doi: 10.1074/jbc.M312644200. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto J, Ishikawa T, Masahito P, Takayama S. Permanent cell lines from erythrophomas in goldfish (Carassius auratus) J Natl Cancer Inst. 1980;64:879–890. [PubMed] [Google Scholar]
- 13.Urasaki A, Morva G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zolessi FR, et al. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–263. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 17.Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/s0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 18.Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodd J, Jessell TM, Placzek M. The when and where of floor plate induction. Science. 1998;282:1654–1657. doi: 10.1126/science.282.5394.1654. [DOI] [PubMed] [Google Scholar]
- 20.Hirsinger E, Jouve C, Dubrulle J, Pourquié O. Somite formation and patterning. Int Rev Cytol. 2000;198:1–65. doi: 10.1016/s0074-7696(00)98002-1. [DOI] [PubMed] [Google Scholar]
- 21.Scott A, Stemple DL. Zebrafish notochordal basement membrane: Signaling and structure. Curr Top Dev Biol. 2005;65:229–253. doi: 10.1016/S0070-2153(04)65009-5. [DOI] [PubMed] [Google Scholar]
- 22.Du SJ, Dienhart M. Zebrafish tiggy-winkle hedgehog promoter directs notochord and floor plate green fluorescence protein expression in transgenic zebrafish embryos. Dev Dyn. 2001;222:655–666. doi: 10.1002/dvdy.1219. [DOI] [PubMed] [Google Scholar]
- 23.Higashijima S, Masino MA, Mandel G, Fetcho JR. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol. 2003;90:3986–3997. doi: 10.1152/jn.00576.2003. [DOI] [PubMed] [Google Scholar]
- 24.Müller GB. Evo-devo: Extending the evolutionary synthesis. Nat Rev Genet. 2007;8:943–949. doi: 10.1038/nrg2219. [DOI] [PubMed] [Google Scholar]
- 25.Satoh N, Jeffery WR. Chasing tails in ascidians: Developmental insights into the origin and evolution of chordates. Trends Genet. 1995;11:354–359. doi: 10.1016/s0168-9525(00)89106-4. [DOI] [PubMed] [Google Scholar]
- 26.Heisenberg CP, Solnica-Krezel L. Back and forth between cell fate specification and movement during vertebrate gastrulation. Curr Opin Genet Dev. 2008;18:311–316. doi: 10.1016/j.gde.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming A, Keynes R, Tannahill D. A central role for the notochord in vertebral patterning. Development. 2004;131:873–880. doi: 10.1242/dev.00952. [DOI] [PubMed] [Google Scholar]
- 28.Huisken J, et al. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 29.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 30.Dong J, Stuart GW. Transgene manipulation in zebrafish by using recombinases. Methods Cell Biol. 2004;77:363–379. doi: 10.1016/s0091-679x(04)77020-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.