Abstract
Many splicing factors interact with both mRNA and pre-mRNA. The identification of these interactions has been greatly improved by the development of in vivo cross-linking immunoprecipitation. However, the output carries a strong sampling bias in favor of RNPs that form on more abundant RNA species like mRNA. We have developed a novel in vitro approach for surveying binding on pre-mRNA, without cross-linking or sampling bias. Briefly, this approach entails specifically designed oligonucleotide pools that tile through a pre-mRNA sequence. The pool is then partitioned into bound and unbound fractions, which are quantified by a two-color microarray. We applied this approach to locating splicing factor binding sites in and around ∼4000 exons. We also quantified the effect of secondary structure on binding. The method is validated by the finding that U1snRNP binds at the 5′ splice site (5′ss) with a specificity that is nearly identical to the splice donor motif. In agreement with prior reports, we also show that U1snRNP appears to have some affinity for intronic G triplets that are proximal to the 5′ss. Both U1snRNP and the polypyrimidine tract binding protein (PTB) avoid exonic binding, and the PTB binding map shows increased enrichment at the polypyrimidine tract. For PTB, we confirm polypyrimidine specificity and are also able to identify structural determinants of PTB binding. We detect multiple binding motifs enriched in the PTB bound fraction of oligonucleotides. These motif combinations augment binding in vitro and are also enriched in the vicinity of exons that have been determined to be in vivo targets of PTB.
Keywords: PTB, SELEX, U1snRNP, genomics, splicing
INTRODUCTION
Splice site selection occurs through the coordinated recognition of multiple cis-elements: the branch point, the 5′ splice site (5′ss), the polypyrimidine tract, the 3′ splice site (3′ss), and a variety of auxiliary elements (Fairbrother and Chasin 2000; Fairbrother et al. 2002). The binding of some factors such as the snRNPs is thought to be well understood. Through a mechanism of RNA:RNA base-pairing, U1snRNP predominantly recognizes the 5′ss (Mount et al. 1983), but there have also been reports of binding outside the 5′ss (Puig et al. 1999; McCullough and Berget 2000), and also reports of U1snRNP performing nonsplicing functions (Furth et al. 1994; Abad et al. 2008). In addition to cooperation between the factors that recognize these core elements, there are a variety of other factors that act as splicing activators and repressors by binding pre-mRNA to recruit or to modulate the recruitment of other components of the splicing machinery to the splice sites.
One such repressor is the abundant polypyrimidine tract binding protein (PTB), a well-studied member of the hnRNP family (Perez et al. 1997a; Liu et al. 2002; Shen et al. 2004; Wollerton et al. 2004; Amir-Ahmady et al. 2005; Sharma et al. 2005; Spellman and Smith 2006; Boutz et al. 2007; Matlin et al. 2007; Sawicka et al. 2008). The simplest model of PTB-mediated silencing is through competition with U2AF65 for the binding of the polypyrimidine tract (Singh et al. 1995; Liu et al. 2002; Sauliere et al. 2006; Matlin et al. 2007). As U2AF65 and PTB possess similar but not identical binding specificities (Singh et al. 1995), and U2AF binding is required for 3′ss recognition (Ruskin et al. 1988; Zamore and Green 1991), slight differences in sequence could shift the balance toward or away from PTB binding, perhaps explaining why only a subset of exons is repressed by PTB. PTB binding sites are often found flanking regulated exons, and the disruption of one binding site has been shown to interfere with the binding of PTB at distal sites, demonstrating the potential for more complex interactions (Ruskin et al. 1988; Zamore and Green 1991; Chou et al. 2000). Other models of PTB repression propose exon looping (Wagner and Garcia-Blanco 2001), interference with 5′ss definition (Izquierdo et al. 2005; Sharma et al. 2005), and participation of co-repressors such as Raver1 (Huttelmaier et al. 2001; Gromak et al. 2003; Spellman and Smith 2006; Auweter and Allain 2008; Fairbrother and Lipscombe 2008). As each of these mechanisms is built from a relatively small number of cases, it is clear that a global view of PTB binding specificity will be necessary to allow a more comprehensive picture of PTB function to emerge.
Various methods exist to discover the RNA binding specificity of proteins. Systematic evolution of ligands by exponential enrichment (SELEX) is an iterative selection technique that selects high affinity ligands from a random pool (Tuerk and Gold 1990). The number of SELEX cycles needed varies depending on the study, sometimes greatly affecting the consensus. Separate determinations of PTB binding specificity identified G-rich sequences flanking short runs of pyrimidines after seven rounds of SELEX and the motif UCUUC after 11 rounds of SELEX (Singh et al. 1995; Perez et al. 1997b). In light of early observations that splicing factors that bind too tightly are inhibitive (Staley and Guthrie 1999), it is not clear that the highest affinity site is more biologically relevant than the more moderately bound sites. It may also be true that the highest affinity ligand has an even higher affinity for another factor, as was the case with sequences identified by SF2/ASF SELEX, which were found to be bound by tra2 β in extract (Tacke et al. 1998). This problem of physiological significance is compounded by the fact that many of the sequences in the initial, random pool do not exist in the genome and so oligonucleotides enriched in the bound fraction often do not map back to genomic sequence. Finally, SELEX studies are often performed with purified protein (PTB), thus neglecting cofactors such as Raver1, and competitors, like U2AF65, which are normally present in the cell (Singh et al. 1995; Rideau et al. 2006; Sauliere et al. 2006; Sickmier et al. 2006; Boutz et al. 2007; Matlin et al. 2007).
To determine global binding specificity in a more physiological context, RNPs formed in vivo can be isolated by immunoprecipitation (IP) for further analysis. In vivo location protocols to map RNPs vary from RNA adaptations of DNA ChIP protocols to UV cross-linking coupled to high throughput sequencing (cross-linking immunoprecipitation [CLIP]) (Niranjanakumari et al. 2002; Ule et al. 2003; Keene et al. 2006). CLIP has been very successful in mapping the binding of pre-mRNA splicing factors, NOVA and Fox, to intronic regions (Ule et al. 2003; Auweter et al. 2006; Yeo et al. 2009). However, all validated PTB and U2AF65 enrichments from in vivo location studies were shown to be occurring on mRNA and not pre-mRNA (Gama-Carvalho et al. 2006). Both PTB and U2AF65 are implicated in additional cellular functions that bring them into contact with mRNA (Cote et al. 1999; Tillmar and Welsh 2002; Tillmar et al. 2002; Zolotukhin et al. 2002; Hamilton et al. 2003; Kosinski et al. 2003; Castelo-Branco et al. 2004; Coles et al. 2004; Knoch et al. 2004; Le Sommer et al. 2005; Pautz et al. 2006; Kuwahata et al. 2007; Ma et al. 2007; Xu and Hecht 2007). While CLIP obviously provides invaluable insight into in vivo binding events, there exist certain biases in CLIP output that may influence certain downstream analysis. As the cellular level of mRNA is a thousand times higher than pre-mRNA, it is reasonable to expect CLIP studies to preferentially return mRNPs because the RNA upon which these complexes form persists longer in the cell than pre-mRNPs (Supplemental Fig. S1, and citations within). While it is clear that CLIP provides invaluable in vivo insight, subsequent analysis such as motif finding may suffer from inappropriately weighted input. In other words, binding sites on exons may be sampled at a higher rate that sites on introns, not because they are more tightly bound but because mRNA has a much longer half-life in the cell and therefore RNPs persist longer on mRNA.
To record the affinity of splicing factors for pre-mRNA in a cellular extract in a manner that circumvents this bias, we have adapted the MEGAshift protocol to identify RNA binding events around alternatively spliced exons that have been re-synthesized as a tiled oligonucleotide pool (Tantin et al. 2008). Similar to SELEX, this approach partitions complex oligonucleotide libraries into a bound and unbound fraction by utilizing gel shift and co-immunoprecipitation (co-IP). We follow the level of U1snRNP and then PTB occupancy on each oligonucleotide by two-color microarray and use the resulting enrichments to guide motif discovery. Importantly, unlike CLIP or SELEX, the output is not a sample of “winners” but a measurement of enrichment on a large set of potential ligands and so downstream analysis can also incorporate information from “losers” (i.e., sequences that do not bind well). We discover multiple binding motifs after only a single round of binding enrichment and find that particular combinations of motifs are highly enriched in the PTB bound fraction. The resulting enrichment values were also used to discover a relationship between secondary structure and PTB binding.
RESULTS
Experimental design and synthesis of oligonucleotide pool
To determine the pre-mRNA binding sites of splicing factors such as U1snRNP and PTB, we first assessed the utility of the CLIP methodology. Our interest was to incorporate secondary structure determinants into a model of RNA/splicing factor interactions. However, concerns arose about the expression bias of CLIP (Supplemental Fig. S1; Yeo et al. 2009), cross-linking biases, and selecting the appropriate folding window in pre-mRNA to predict the secondary structure around binding elements (Supplemental Fig. S2). Therefore, we developed a SELEX-based approach targeted to searching the pre-mRNA regions around alternatively and constitutively spliced exons that utilized oligonucleotides of a defined length. The exon set included pre-mRNA that demonstrated alternative splicing in both mouse and human (Holste et al. 2006), predicted alternatively spliced exons (Yeo et al. 2005), and arbitrarily chosen exons. The complete set is annotated in a custom UCSC Genome Browser tract that is available to download (http://fairbrother.biomed.brown.edu/data/SelexMap/).
These regions were resynthesized as an oligonucleotide pool that tiled a sequence window of 30 nucleotides (nt) in length by 10-nt increments through ∼4000 genomic regions centered around exons but encompassing 200 nt of their intronic flanks (Fig. 1A). Universal primer binding sites were appended to each end of this 30-nt window, and the resulting 60-mer were synthesized as a probe feature onto a custom oligonucleotide array.
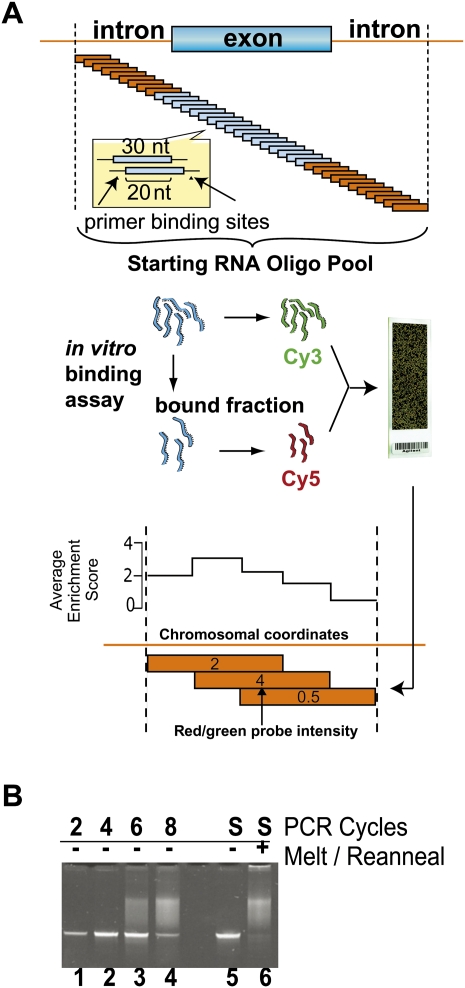
FIGURE 1.
Experimental scheme for mapping splicing factors to pre-mRNA. (A) The oligonucleotide pool was designed by tiling a length of 30 oligonucleotides in 10-nt increments across approximately 4000 genomic regions. A total of 241,347 experimental and 90 validation sequences were flanked by common primers and ordered as features on a custom microarray. Features were recovered from the array surface by scouring (Materials and Methods) and PCR-amplified using the common, T7-promoter-tailed primers. After T7 transcription, the amplified pool was partitioned into bound and unbound fractions by EMSA with U1snRNP preparations (see Fig. 2) or by co-immunoprecipitation from HeLa nuclear extract using αPTB mab BB7 (Fig. 3). Next, the starting pool was internally labeled with Cy3 dye, and the bound fraction was labeled with Cy5. These two RNA pools were mixed so oligonucleotides competed for binding on a two-color microarray, resulting in enrichment data. The array data were mapped to genomic coordinates, and the scores at each location were averaged and converted to base-10 log. An illustration of this averaging step is given for three theoretical overlapping 30-nt oligonucleotides with scores of 2, 4, and 0.5, where the average enrichment score for each 10-nt window is graphed above. (B) Pool overamplification was checked by electrophoresis. Acrylamide gel lanes 1–4 represent the PCR-amplified pool after an additional two, four, six, and eight cycles of PCR beyond the optimal amplification. Heteroduplexes composed of unextended template are presumed to comprise the low mobiliy smear in lanes 3 and 4. The optimally amplified starting material (S, lane 5) was melted and reannealed creating a mixed population of heteroduplexes that migrates as a similar low mobility smear (lane 6).
The oligonucleotide library was commercially synthesized as a custom oligonucleotide array, liberated from the slide, and then amplified via the universal primer binding sites at low-cycle polymerase chain reaction (PCR). PCR amplifications that exceeded the log linear range were identified by the presence of a low-mobility smeared product in the acrylamide gel (Fig. 1B, gel lanes 1–4). In melting and re-annealing experiments, the common flanks facilitated imperfect duplex formation between different members of the pool, resulting in a diffuse product that greatly resembled the smear observed from overamplification (Fig. 1B, gel lanes 5,6). We conclude that this diffuse product represented an imperfect duplex that formed from heterogeneous single-stranded DNA that failed to extend in the final PCR cycle. The oligonucleotide pool was transcribed into internally radiolabeled RNA via the T7 RNA polymerase promoter introduced into the forward primer.
This RNA pool was then subjected to gel shift, filter binding assay or co-immunoprecipitation to isolated RNPs. The RNA oligonucleotides upon which these complexes form are extracted, reverse-transcribed, and labeled with Cy3 or Cy5. The representation of each oligonucleotide in the bound fraction was compared to its abundance in the starting pool by Cy labeling the starting pool and allowing both sets of differentially labeled oligonucleotides to hybridize to a detection array. Enrichment was then measured as the ratio of oligonucleotide in the bound fraction versus that in the starting pool. After this process, the resulting red/green ratio for a particular oligonucleotide was then proportional to the fraction of that oligonucleotide species in bound state under experimental conditions. The degree of binding is projected onto genomic coordinates by averaging the red/green ratios of all probe features in each column of the alignment (Fig. 1A). This averaged ratio is logged such that a positive value indicates enrichment in the oligonucleotide's representation in the bound fraction of the pool.
U1snRNP predominantly binds the 5′ss but also contacts 5′ss proximal intronic regions and the 3′ss
To test the feasibility of mapping splicing factors within the regions of alternative splicing in pre-mRNA, we initially selected U1snRNP and isolated the oligonucleotides that were gel shifted upon incubation with purified U1snRNP. While not all splice sites may be dependant on U1snRNP (Crispino et al. 1996) and U1snRNP has been shown to function outside the splice sites (McCullough and Berget 2000), we reasoned that U1snRNP was a good choice for initial mapping because it binds annotated 5′ss and is therefore one of the few splicing factors with a large set of known ligands. Under experimental conditions, 63% of 5′ss that map to the central region of the oligonucleotide are enriched in the U1snRNP shifted fraction in this study.
To test the affinity of each RNA oligonucleotide for U1snRNP, we incubated the oligonucleotide pool with a purified fraction of human U1snRNP (Gunderson et al. 1998; Abad et al. 2008). After verifying that the U1snRNP prep was competent to bind the control BPV 5′ss probes, we adjusted the ratio of probe to protein such that ∼5% of the oligonucleotide pool was shifted (Fig. 2A, lanes 2,4). Increasing amounts of unlabeled pool were used to establish that U1snRNP was not present in great excess—the shifted band in lane 4 of Figure 2A could be competed away in lanes 5 and 6 by increasing amounts of unlabeled pool. The region of the gel corresponding to the shifted U1snRNP complexes was excised, amplified, and analyzed via microarray.
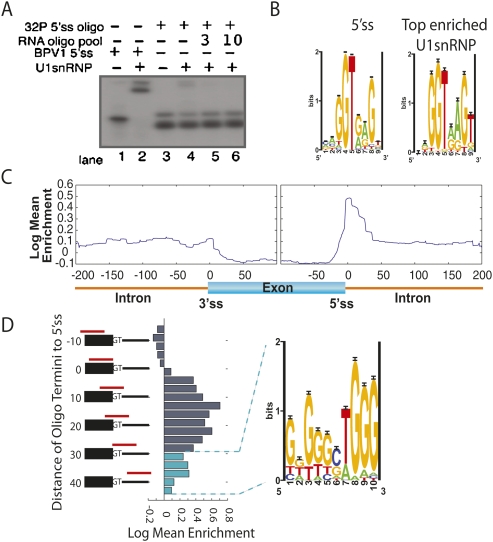
FIGURE 2.
Annotating pre-mRNA with locations of U1snRNP binding. (A) The parameters of U1snRNP gel shift were established with radiolabeled BPV 5′ss positive control (lane 2) and radiolabeled RNA oligonucleotide pool to achieve a 1:20 shifted-to-unshifted ratio of labeled RNA oligonucleotide pool (cf. lane 3 and lane 4). An additional 3 μg and 10 μg of cold RNA oligonucleotide pool established that the probe was in excess (lanes 5,6). (B) Oligonucleotides were ranked according to their ability to bind U1snRNP. (Left panel) The motif returned from Gibbs sampling on the 5′ss; (right panel) the motif returned from the top 100 U1snRNP enriched oligonucleotides. (C) A generalized RNA map for U1snRNP enrichment data was made by compiling the information from all ∼4000 regions into one map. The enrichment scores for each oligonucleotide were sorted based on their distance from splice sites (x-axis). An average enrichment score (y-axis) was calculated for each position 200 bases into the intron and 100 bases into the exon from each splice site and plotted, resulting in a single map encompassing the entire enrichment data set. (D) Oligonucleotide enrichment in the U1snRNP fraction was compared to the 5′ss location. The registry of the 5′ss with respect the 3′-end of the oligonucleotide (red bars in alignment) is listed on the vertical axis. Enrichment was plotted as a histogram. Oligonucleotides used to calculate cyan histogram bars were selected for motif finding analysis.
Ranking the oligonucleotides by the array measurement of enrichment in the U1snRNP bound fraction led to the identification of a U1snRNP binding specificity that was nearly identical to the canonical donor site (Fig. 2B). Furthermore, plotting the average binding enrichment as a function of distance from the splice site reveals a strong peak at the 5′ss (Fig. 2C). This peak around the 5′ss was asymmetric—falling sharply on the exonic side and more gradually into the intron. To gain better resolution of U1snRNP binding at the 5′ss, we re-graphed the average enrichment in a manner that preserved the registry of the oligonucleotide at the 5′ss. We reasoned that this analysis would reveal whether any additional information that U1snRNP might require for robust binding would reside predominantly on the exonic or intronic side of the 5′ss. However, the location of the 5′ss within the oligonucleotide did not seem to affect U1snRNP binding—oligonucleotides with a donor site and predominantly upstream exonic flank appeared with the same average enrichment as oligonucleotides with predominantly downstream intronic flank (Fig. 2D). Oligonucleotides lacking the intronic portion of the 5′ss were not enriched in the U1snRNP bound fraction, whereas oligonucleotides lacking the exonic portion were enriched, but to a lesser degree than oligonucleotides with intact 5′ss.
While the average enrichment of purely exonic sequences was negative in the U1snRNP bound fraction, we observed a residual level of U1snRNP binding in the intronic regions immediately downstream from the 5′ splice site (Fig. 2D). Motif finding algorithms run on the top 100 most enriched oligonucleotides extracted from these downstream regions returned a G triplet motif that has been previously identified as an intronic enhancer that was proposed to directly bind U1snRNP through base-pairing interactions (McCullough and Berget 2000).
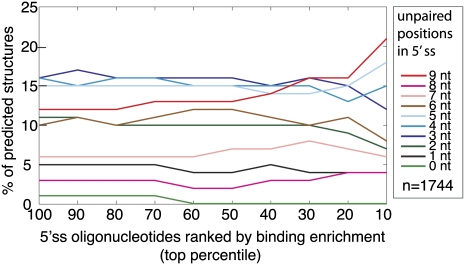
Analyzing the oligonucleotides that contain splice sites with the RNA folding program, Sfold, reveals that the 5′ss regions within the individual oligonucleotides are engaged in RNA secondary structure to varying degrees. It appears that U1snRNP binds preferentially to single-stranded splice sites. After ranking the oligonucleotides according to their enrichment in the shifted fraction, we observe that the top 10% is 1.75 times more likely to be completely unpaired than the pool-wide average (Fig. 3). Conversely, none of oligonucleotides that contain 5′ss predicted to be completely sequestered by secondary structure fall within the top 60% of the pool. This trend of favoring unstructured splice sites in the shifted fraction and sequestered sites in the unshifted fraction becomes less pronounced for intermediate degrees of structure (Fig. 3). The direct annotation of U1snRNP on pre-mRNA was written as a custom annotation track for the UCSC Genome Browser and is available to download (http://fairbrother.biomed.brown.edu/data/SelexMap/).
FIGURE 3.
Comparison of secondary structure with U1snRNP binding. Secondary structure was modeled using Sfold for each of the 1744 oligonucleotides that contained an annotated 5′ splice site (Ding et al. 2005). The structures are classified according to the degree of predicted base-pairing over the windows that encompass the 9-nt 5′ss. The y-axis plots the average number of predicted structures in each category for the particular bin of oligonucleotides defined by the x-axis. The x-axis defines the bins as top cumulative percentile ranks where oligonucleotides are ranked according to their enrichment in the U1snRNP shifted fraction.
Selection of high affinity PTB ligands
After concluding that the overlap between U1snRNP-enriched oligonucleotides and 5′ss supported the general validity of the method, we applied this approach to mapping the binding sites of PTB. While PTB has been determined to bind short tracts of polypyrimidines, these motifs are too numerous to be useful in describing the specificity of PTB. PTB, unlike U1snRNP, is not associated with a large set of known ligands. In order to physically isolate the PTB-bound fraction of the library, we co-immunoprecipitated PTB ligands with PTB from HeLa nuclear extract. To establish that the PTB immunoprecipitation was also precipitating PTB-bound RNA, we demonstrated by semi-quantitative PCR that the partition of the total oligonucleotide pool into the immunoprecipitated fraction is dependent on both the HeLa nuclear extract and the αPTB mabBB7 (Fig. 4A). The bound and starting fraction were reverse transcribed, differentially labeled with Cy5 and Cy3, and then hybridized to the detection array. Enrichment was then measured as the ratio of oligonucleotide in the bound fraction versus that in the starting pool as described above. While we utilize the term “PTB bound” to describe the enriched fraction, the RNA is incubated in extract so some cases of binding may be indirect.
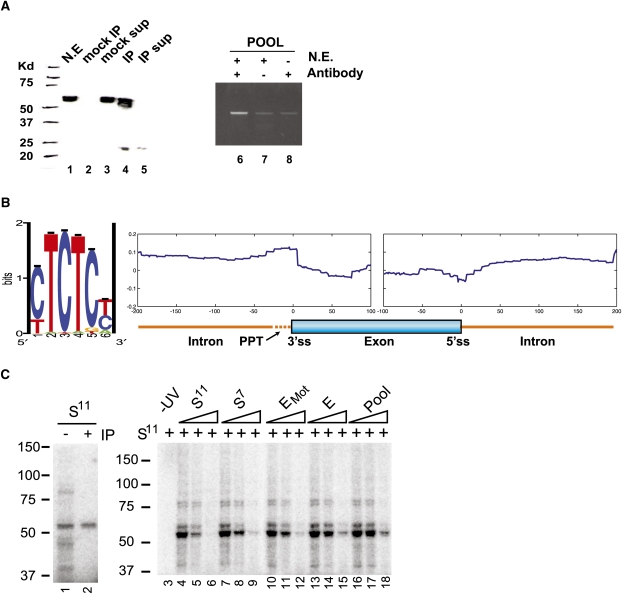
FIGURE 4.
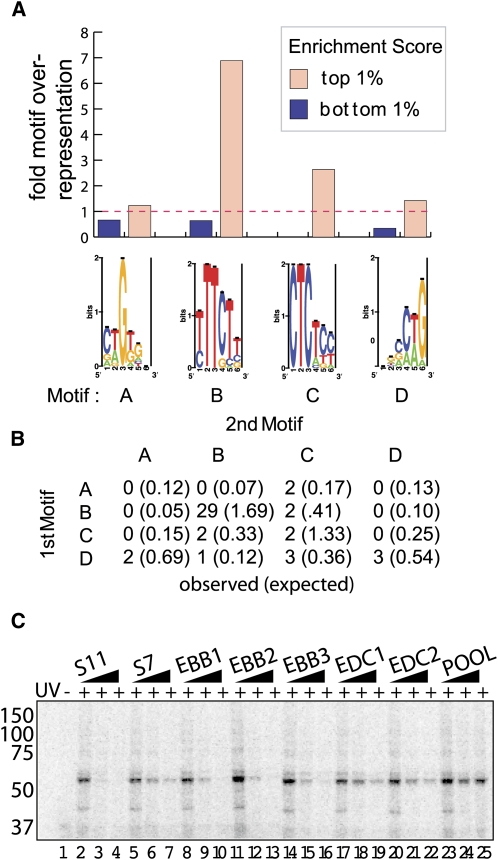
Discovering and validating PTB binding specificity. (A, lanes 1–5) A Western blot of PTB immunoprecipitated from HeLa nuclear extract (N.E.) with (lanes 4,5) or without (lanes 2,3) mab BB7. (sup) Lanes containing IP or mock IP supernatant. (Lanes 6–8) Semi-quantitative PCR of oligonucleotides from the pool that co-immunoprecipitated with PTB. (Lanes 7,8) Oligonucleotides co-IP'ed in the absence of mab BB7 antibody or HeLa nuclear extract, respectively. (B) Motif finding on the top 1% of enriched oligonucleotides, using Gibbs sampling (Materials and Methods), resulted in a PTB consensus motif. (C, lane 1) UV-cross-linking to a high affinity ligand isolated from 11 rounds of SELEX (S11) was performed on HeLa extract (Perez et al. 1997b). (Lane 2) The cross-linked extract was then immunoprecipitated using mab BB7 αPTB antibody. Samples were separated by PAGE. (Lanes 3–18) UV-crosslinking was performed using radiolabeled S11 as a probe in the presence of increasing amounts (0-, 5-, or 50-fold molar excess) of unlabeled competitors as indicated. S7 is a PTB ligand isolated from seven rounds of SELEX (Singh et al. 1995). (E and EMot) Oligonucleotides associated with high enrichment scores; (EMot) also enriched for the motif described above; (Pool) the unenriched oligonucleotide pool.
In order to make a useful generalization about where a splicing factor binds pre-mRNA, the distances of all 60,592 probes to the nearest splice sites were calculated, and their genomic average enrichment values were plotted as a function of distance to the splice site (Fig. 4B). The largest region of enrichment was immediately upstream of the 3′ss in the vicinity of the polypyrimidine tract. PTB sites could also be seen as reduced in abundance around the 5′ss and occurred less frequently in exons than introns (Fig. 4B).
We used Gibbs sampling to discover sequence motifs overrepresented within the PTB-enriched pool of our experiment. As in previous studies (Tantin et al. 2008), ranking the oligonucleotides by enrichment score and then analyzing the top 1% of the data set resulted in the identification of the motif CUCUC, similar to the in vivo identified UCUCU motif enriched upstream of regulated cassette exons (Fig. 4B; Castle et al. 2008) and more importantly, identical to the oligonucleotide ligands used in the PTB-ligand co-crystal structure (Oberstrass et al. 2005).
Biochemical validation of array data
To confirm array predictions and assess how well RNA oligonucleotides that correspond to natural genomic sequence bind PTB, we selected several oligonucleotides for UV cross-linking analysis. Two oligonucleotides selected from the PTB-enriched fraction both cross-linked to a 58-kDa protein (data not shown). As the efficiency of UV cross-linking can vary with sequence, these measurements were not considered quantitative. To better quantify PTB binding, we established a standard UV cross-linking reaction on a well-studied ligand and assayed oligonucleotides for their ability to compete in trans. The radiolabeled PTB ligand isolated from 11 rounds of SELEX (Fig. 4C, S11) bound efficiently to a 58-kDa protein in HeLa nuclear extract (Fig. 4C, lane 1). This product efficiently immunoprecipitated with the PTB monoclonal antibody, mab BB7 (Fig. 4C, lane 2; Perez et al. 1997b). This interaction was decreased and then lost upon addition of an increasing molar excess of unlabeled S11 ligand (Fig. 4C, lanes 3–5). Using S11 as a standard, we shifted our analysis to repeating a single cross-linking assay in the presence of a series of unlabeled competitors (Fig. 4C, lanes 3–18).
With this approach, we find that the PTB aptamer selected after 11 rounds of SELEX (S11) bound better than the aptamer selected after seven rounds (S7) (Fig. 4C, lanes 4–6 vs. lanes 7–9). The natural sequences enriched in the PTB bound fraction of our pool were scored for their similarity to the CUCUC motif returned from the Gibbs sampling of the enriched set (Fig. 4B). Two oligonucleotides from the top 1% of array enrichment data were compared for their ability to bind PTB. Considering the top five match windows, one oligonucleotide (EMot) was enriched for the PTB binding motif relative to a second oligonucleotide (E) that was not enriched for the PTB binding motif (Fig. 4C). While both these sequences bound PTB with a higher affinity than the unselected pool (Fig. 4C, lanes 16–18), the natural oligonucleotide that also contained high scoring matches to the enriched motif appeared stronger (Fig. 4C, cf. lanes 10–12 and lanes 13–15). Targets of PTB defined in this manner appeared to share a similar level of affinity for PTB as the aptamer selected after seven rounds of SELEX. This set of sequences that both fell within the enriched set and contained high scoring PTB motifs is listed in Supplemental Table S1.
The role of secondary structure in PTB binding
While the experimental validation and the general agreement between array-derived and published motifs suggests that the co-IP approach largely selects for PTB ligands, we considered the scenario in which secondary structure influences PTB binding its ligand. Crystallographic and SELEX studies have not detected a structural component to PTB binding; however, there have been reports of PTB binding structured regions of IRES (Kolupaeva et al. 1996; Song et al. 2005).
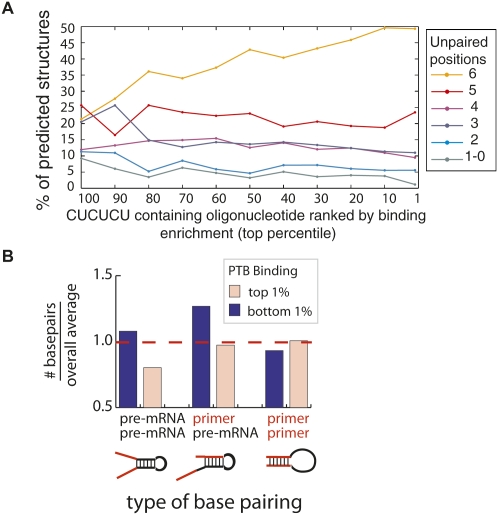
To test whether predicted RNA secondary structure significantly affected PTB binding, we used the RNA structure prediction program Sfold to fold all 60,592 oligonucleotides (Ding et al. 2005). Unlike lowest free energy prediction, Sfold returns an ensemble of 1000 structures that were sampled with replacement from all possible structures with probabilities derived from their predicted energies. In this way, lower-energy structures are more likely to be included, possibly multiple times. We ranked the oligonucleotides according to their binding enrichment and then separated the 60 million predicted structures into six categories according to their degree of predicted secondary structure. If PTB binding was significantly affected by secondary structure, we hypothesized that the highly enriched (PTB bound) set would have less secondary structure. Indeed, an increasing fraction of oligonucleotides fell into the unstructured category as enrichment in the PTB bound set increased (Supplemental Fig. S3). In addition, the more structured oligonucleotides appeared to occur less frequently in the PTB-enriched set (blue and cyan lines curve down in Supplemental Fig. S3). As we have determined a PTB binding motif that is over-represented in the enriched set, we repeated this analysis considering only the 6-nt window that best fit the PTB binding model. In other words, we limited our analysis to the set of 2879 oligonucleotides that contained CUCUCU and examined their predicted structures as a function of array enrichment (Fig. 5A). When we considered only the predicted binding site, we observed a more pronounced association between enrichment and an open structure over the spectrum of PTB oligonucleotide enrichment—there was a twofold increase in completely unstructured PTB motifs in the PTB-enriched set (Fig. 5A).
FIGURE 5.
Comparison of secondary structure with PTB binding. Secondary structure was modeled using Sfold for each of the 2879 oligonucleotides that contained CUCUCU, the highest possible scoring match to the PTB motif (Ding et al. 2005). (A) The predicted structures are classified according to the degree of predicted base-pairing over the windows that encompass the 6-nt PTB motif, CUCUCU. The y-axis plots the average number of predicted structures in each category for the particular bin of oligonucleotides defined by the x-axis. The x-axis represents 10-nt bins of percentile ranks (100–91, 90–89, …), where oligonucleotides are ranked according to enrichment in the PTB bound fraction. (B) Compares types of base-pairing observed. Predicted base pairs were grouped into three categories: pre-mRNA/pre-mRNA, primer/pre-mRNA, and primer/primer. For each category, the number of predicted base pairs in the (pink bar) top and (blue bar) bottom 1% of oligonucleotides ranked by array enrichment was normalized to the pool-wide average and plotted as a histogram.
Finally, we sought a qualitative description of the structure predicted on these oligonucleotides. Referring to the primer binding regions as “primer,” we classified predicted base pairs in the oligonucleotide pool into three similarly sized groups: pre-mRNA/pre-mRNA (30.4%), primer/pre-mRNA (34.5%), and primer/primer (35.1%) (Fig. 5B). By comparing the top and bottom 1% of oligonucleotide enrichment, we determined that primer/primer base-pairing had a negligible effect on PTB enrichment (Fig. 5B, right pair, similar bars). A lack of inhibitory primer/pre-mRNA interactions also did not seem to explain the top 1% of PTB enrichment (Fig. 5B, middle pair, pink bar is similar to average), although this pairing did explain a few cases of poor PTB enrichment (Fig. 5B, middle pair, blue bar). The category of structure that best discriminates the enriched class from the background was the pre-mRNA/pre-mRNA structure. In the top 1% there is 20% less pre-mRNA/pre-mRNA structure than observed in the overall pool. These results demonstrate PTB's strong preference for single-stranded occurrences of the polypyrimidine motifs. High affinity binding sites appear to contain less pre-mRNA/pre-mRNA structure than expected. This structure is not an artifact of the oligonucleotide design but rather comes entirely from endogenous sequence. In the Discussion section, we formalize how this structural element can be incorporated into a predictive model for detecting PTB binding sites in RNA. However, the utility of such a predictive scheme will be limited by our ability to accurately predict secondary structure in pre-mRNAs—a task that is considerably more challenging in longer sequences.
Modeling PTB binding with multiple motifs
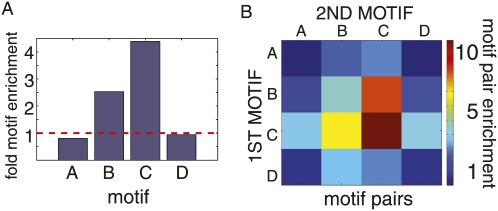
Another aspect of PTB binding that emerged from structural studies was the possibility that PTB binds RNA semi-independently with its four RRMs (Conte et al. 2000; Simpson et al. 2004; Oberstrass et al. 2005; Petoukhov et al. 2006). In order to explore the possibility of multiple binding models, we reanalyzed the oligonucleotide pool by Gibbs sampling, allowing up to four binding motifs, which we labeled motifs A–D (Fig. 6A). The CU repeats identified as the dominant motif were returned (Fig. 6A, motif C) as well as a UUUCU motif (Fig. 6A, motif B) similar to the pattern discovered in previous SELEX results (Perez et al. 1997b) and two novel CTG motifs (Fig. 6A, motifs A,D). Annotating the entire oligonucleotide set with these four motifs allowed us to compare the distribution of PTB motifs in the set of oligonucleotides with the highest enrichment scores versus the set with the lowest. All four motifs were over-represented in the top 1% of oligonucleotides ranked by enrichment score and under-represented in the bottom 1% (Fig. 6A). The pyrimidine motifs associated more with highly enriched oligonucleotides than the g-rich motif. Interestingly, the motif returned from 11 rounds of SELEX and presumed to be the strongest PTB binder has the largest (sevenfold) over-representation in the top 1% of oligonucleotides ranked by enrichment. We also considered a scenario in which certain combinations of motifs co-occurring within a single oligonucleotide functioned synergistically. While multiple motifs frequently co-occurred in the top 1% of oligonucleotides ranked by enrichment, there were no cases of co-occurrence in the bottom 1% (data not shown). Particular combinations of motifs were observed more frequently in highly enriched oligonucleotides than in the entire pool (P-value = 0.004), indicating that it was not just the number but the identity and order of the motifs that influenced PTB binding (Fig. 6B). One such combination was a motif B pair, which occurred 169 times in the entire oligonucleotide pool, with 17% of these occurrences concentrated within the top 1% of oligonucleotides ranked by enrichment (Fig. 6B).
FIGURE 6.
Particular multi-motif combinations enhance PTB binding. PTB contains four RRMs that have been shown to interact with RNA semi-independently. (A) Motif finding was repeated allowing up to four binding models. The resulting motifs (A–D) were used to annotate the pool. The average number of annotated sites was used to calculate the over- or under-representation of motifs in the top and bottom 1% of oligonucleotides ranked by enrichment (histogram bars over each motif). (B) Co-occurrences of motifs were analyzed in the top 1% of oligonucleotides ranked by enrichment. The number of pairs that occur in the specified order were counted in the top 1% of the data. The value in parentheses represents the number of co-occurrences expected in the top 1% under a null model of no motif enrichment. (C) Oligonucleotides containing motif combinations were selected from the top one percentile for validation. UV-cross-linking was performed as before using radiolabeled S11 as a probe in the presence of increasing amounts (0-, 5-, or 50-fold molar excess) of unlabeled competitors as indicated.
To test potential synergy between co-occurrences of motifs, we selected five additional oligonucleotides that represented examples of multiple motif combinations. As has been observed in previous studies, bona fide PTB targets contain multiple clustered PTB sites (Chou et al. 2000; Amir-Ahmady et al. 2005; Matlin et al. 2007). To compare B–B and D–C motif combinations to the PTB ligands established by SELEX, we repeated the UV cross-linking/competition assay with increasing concentrations of motif competitor (Fig. 6C). The combinations of B-B bound PTB with an affinity comparable to the S11 substrate that was produced after 11 rounds of SELEX. All tested D–C motif combinations bind PTB with an affinity comparable to seven rounds of SELEX (Fig. 6C). The 29 oligonucleotides that contain multiple matches to motif B and fell within the top 1% of oligonucleotides ranked by PTB enrichment score are listed in Supplemental Table S2.
PTB motifs (pairs) are enriched around in vivo targets of PTB regulation
To determine if the binding model developed with the high throughput oligonucleotide binding assay accurately reflects in vivo binding, we analyzed a set of PTB regulated exons for overrepresentation of PTB motifs. These 11 exon–intron regions were identified by the Smith lab on the basis of their significant difference in splicing following dual PTB/nPTB RNAi knockdown (Spellman et al. 2007). To compare the number of PTB motifs in this set of in vivo targets to a baseline value, we employed a sampling approach. Briefly, we compared the number of annotated motifs in the set of PTB targets to the number of annotated motifs in randomly selected sets of 11 exon–intron regions. We performed this analysis with 1000 trials, where the random sets were selected to preserve the size characteristics of the set of PTB targets. Motifs B and C were both enriched (both P-values = 0); however, motifs A and D were not enriched (P-value = 0.81 and 0.57) (Fig. 7A). Motif C was the dominant motif in the in vivo targets of PTB (Fig. 7A). Motif combinations were scored in 30-nt windows and, again, compared to random draws of 11 exon–intron regions. Motifs B–B and D–C were both over-represented in the in vivo target set; however, the over-representation of motif BB was significantly higher than D–C, again consistent with the oligonucleotide binding study (Figs. 7B, 6C). Motif C and motif pair C–C predominated in the set of in vivo targets (Fig. 7B).
FIGURE 7.
The distribution of PTB motifs in endogenous PTB pre-mRNA targets. A set of exons regulated by PTB in HeLa cells was obtained from the literature (Spellman et al. 2007) . Motifs A–D were annotated in the intronic and exonic regions around PTB regulated splice sites. The number of sites found around splice sites that were targets of PTB regulation was compared to the number of sites found around randomly selected splice sites. A sampling strategy was utilized to make this comparison. Control data sets were constructed by drawing randomly selected pre-mRNA regions from refseq pre-mRNAs such that each control set was equal in number and size and arrangement (exonic and intronic portions) to the PTB regulated set. (A) Histogram bars record the fold over-representation of each motif to the average counted in the 1000 control sets. (B) Pairs of motifs were searched with a 30-nt window as in Figure 6B. A heat map conveys the over-representation of each combination in the PTB targets relative to the average tallied over 1000 control sets.
DISCUSSION
We used Gibbs sampling to identify CUCUC and UUUCU and two CUG-type motifs as the most commonly occurring motifs within the set of PTB-bound sequences (Figs. 4B, 6A). Previous SELEX experiments identified UCUUC, similar to our motif B, as the highest affinity PTB binding site after 11 rounds of selection (Perez et al. 1997b). It is interesting to note that motif B, which is present in the strongest ligand, enjoys the highest fold enrichment in our experiment, about a sevenfold over-representation in the top-scoring enrichment set (Fig. 6A). A similar SELEX experiment performed with fewer (seven) rounds of enrichment identified a more degenerate short run of pyrimidines flanked by a G-rich sequence (similar to our motifs A, D), and structural studies of PTB in complex with RNA were solved using poly(CU) (Fig. 6A, motif C) (Singh et al. 1995; Oberstrass et al. 2005). Indeed, these results are supported by our cross-linking experiments in which an oligonucleotide enriched for PTB binding and also containing the CUCUC motif competed for PTB binding as well as the seven-round SELEX result (Fig. 4C).
In addition, we investigated the role of secondary structure in U1snRNP and PTB binding RNA. Both factors demonstrate a clear preference for a single-stranded substrate and a clear avoidance of a site completely sequestered in secondary structure. Perhaps against expectation, PTB binding appears slightly more affected by predicted secondary structure. PTB is a single-stranded RNA binding protein that binds CU repeats. We observe that oligonucleotides that bind PTB strongly tend to be unpaired in the window that encompasses the best match to the CU motif. The best relationship between binding and reduced structure appears to be found in the structure that is confined entirely to the pre-mRNA region and does not include the primer binding site. While a SELEX study revealed NOVA-1 bound preferentially to single-stranded motifs within hairpin structures, to our knowledge this study is the first attempt to incorporate structure into a binding model for a splicing factor (Song et al. 2005). Our future efforts will be to extend this analysis incorporating structure and motif combinations into a predictive model. One form that such a predictive model could take is to extend the model used by pattern search programs such as Patser (Hertz and Stormo 1999) with an additional scoring component related to the degree of single strandedness of the target site. While the implementation of such a model awaits improvements in RNA structural prediction of long pre-mRNAs, the model and relevant data are included in Supplemental Table S3. To our knowledge, this represents the first binding model that incorporates both sequence and structural parameters.
In closing, this method is similar to SELEX performed on a subset of sequences selected by the researcher. In this case, we utilized real pre-mRNA sequences around alternatively spliced exons; however, mutations or polymorphisms or random sequences could be used. As high affinity binding sites are not always the physiological ligand of an RNA binding protein, there is an inherent advantage to restricting the search to real sequences. Furthermore, such an approach does not sample enriched oligos but returns data for each oligonucleotide allowing for information to be gleaned from poorly binding sequences as well as highly bound sequences. One potential drawback is a failure to distinguish between direct and indirect binding events and the possibility of false negatives on cooperative binding events that depend upon long-range combinations of cis elements that exceed the oligo window size. It is unclear whether this is an issue with PTB, as modules were discovered and validated with the oligonucleotide widths used in this study. However, both these issues can be remedied by repeating the method with recombinant PTB on longer oligonucleotides.
MATERIALS AND METHODS
Design of the array library and pool recovery
The total library consisted of 241,347 oligonucleotides tiled through ∼4000 exons downloaded from the UCSC Genome Browser. Tiling of the 30-mers by 10-nt increments extends 100 nt into the exonic and 200 nt into the intronic region. DNA was recovered from synthesis arrays by adding 500 μL of dH2O to the surface of the array, thoroughly scouring and resuspending using a 25-gauge needle, sonicating, and PCR amplifying.
U1snRNP EMSA
Electrophoretic mobility shift assay (EMSA) was done as previously described (Gunderson et al. 1998) and contained either 20 ng (0.03 μM final) of 100-nt 32P-labeled BPV1-RNA derived from BPV1 that contains a 9-nt U1snRNP binding site (Gunderson et al. 1998) or 1 μg (3 μM) of 32P-labeled RNA oligonucleotide pool. Two samples also contained 9 μg and 30 μg of unlabeled oligonucleotide pool as a competitor. Binding reactions (final volume of 20 μL) contained 1 mM MgCl2, 60 mM KCl, 0.1% Triton X-100, 8% glycerol, 1 μg of total yeast tRNA, 10 mM DTT, 20 mM Tris-HCl (pH 7.5), 3 units of RNasin (Promega), 0.1 mM EDTA, and 3 μg of bovine serum albumin. Two micrograms of U1snRNP was added last (final concentration of 0.3 μM), and the reaction was incubated for 5 min at room temperature prior to loading a 6% (60:1) polyacrylamide gel run in Tris-Borate-EDTA buffer. The amount of U1snRNP was varied across a 20-fold range to ensure the linear range of the assay and to maintain specificity of binding as defined by use of a BPV1-RNA containing a mutated U1snRNP binding site (Gunderson et al. 1998). Electrophoresis was for 3 h at 20 V/cm. The purification of U1snRNP (judged to be >98% pure) from HeLa cells is described in Abad et al. (2008).
After autoradiography and phosphoimage analysis, the U1snRNP bound-RNA was excised from the gel, passively eluted in SDS buffer overnight, and the RNA was collected by phenol-chloroform extraction and ethanol precipitation and subjected to microarray analysis as described below.
Array hybridization
RNA that precipitated with ASF/SF2 as well as the pre-enrichment starting pool was transcribed from the cDNA using the common flanking primers containing a T7 polymerase promoter.
The oligonucleotides were labeled with Cy5 and Cy3 dyes, respectively. The MEGAshortscript transcription kit was used (Ambion), using 1 μL of 5-(3-aminoallyl)-UTP (Ambion) and no regular UTP. Monoreactive Cy3 and Cy5 dyes (GE Healthcare) were prepared by mixing them with 45 μL of DMSO. To the RNA product, 4.5 μL of Coupling Buffer (0.1 M Na2CO3), 2.5 μL of H2O, and 3 μL of prepared dye were added. The mixture was incubated for 1 h at room temperature; the reaction was terminated by incubating it with 6 μL of 4 M hydroxylamine for 15 min. The RNA was then extracted by phenol-chloroform and ethanol precipitation. The following was used as a hybridization solution: 50 μL of blocking buffer, 30 μL of starting RNA/45 μL of elution RNA (corresponding to 750 ng of RNA), 10 μL of 25× fragmentation buffer, 250 μL of 2× hybridization buffer, and H2O up to 500 μL (all buffers by Agilent). This was then injected in the array chamber and incubated for 3 h at 50°C. The array was then gridded and subjected to signal-to-noise filter—only data points that, in the starting pool, scored higher than 2.6 standard deviations above background were analyzed.
PTB binding and protein analysis
A 1:1 mixture of magnetic Protein A and Protein G Dynabeads (Invitrogen) was incubated with 5 μL of mab BB7 hybridoma supernatant (ATCC# CRL-2501). Fifty microliters of this slurry were added to 120 μg of HeLa nuclear extract and 200 ng of RNA pool (4°C, 1 h). RNA was recovered by boiling in 1% SDS. Enrichment was quantified by two-color microarray analysis using standard protocols. For Western blotting, 15 μL of mab BB7 were incubated overnight on the membrane and imaged with HRP-conjugated anti-mouse secondary.
UV-cross-linking
Unlabeled cold competitors were prepared following standard protocols. Radiolabeled probe was prepared using 32P-UTP to a final concentration of 10 μCi. All samples were visualized on 7 M urea polyacrylamide gels and quantified via RiboGreen (Invitrogen), phosphorimaging (ImageQuant; GE Healthcare), and/or UV spectrometry (Nanodrop).
For each reaction, 200 ng (0.01 nM) of radiolabeled probe were added to 30 μg of HeLa cell nuclear extract and an increasing amount (0-, 5-, or 50-fold molar excess) of competitor RNA and incubated for 30 min at 25°C. Reactions were exposed to UV for 15 min at 120 mJ/cm2 5 cm from the source. RNA was digested using RNAse A/T1 mix for 1 h at 37°C. For cross-linking-IP, a portion of the reaction was incubated with the bead mixture prepared above for 1 h at 4°C. All samples were separated by SDS-PAGE and imaged.
Data visualization
The enrichment values were visualized using the UCSC Genome Browser. A wiggle-format custom track calculates the logged average enrichment score for overlapping oligonucleotides. The data, scripts, and documentation for this project are available for download (http://fairbrother.biomed.brown.edu/data/SelexMap/).
Genomic intron and exon positions were obtained using the Known Genes track from the UCSC Genome Browser (Karolchik et al. 2008). BLAST (Altschul et al. 1997) was used to map each oligonucleotide to its nearest splice site. An average enrichment score was calculated from overlapping oligonucleotides for each of the 100 exonic and 200 intronic bases.
Motif finding, annotation, and secondary structure prediction
Binding motifs were identified in the top 1% of oligonucleotides ranked by enrichment using the Gibbs sampler (V 3.04.006) (Thompson et al. 2007). For multiple motifs, oligonucleotides containing splice sites were omitted. Oligonucleotides were annotated using Patser v3e (Hertz and Stormo 1999). Patser was used to obtain and sum the scores of the top five CUCUC (Supplemental Table S1, EMot) or UUUCU (Supplemental Table S2) motifs per oligonucleotide. Motifs were visualized using SeqLogo (Schneider and Stephens 1990). Structures were sampled using Sfold 2.0 (Ding et al. 2005). The top scoring motif site in each oligonucleotide (Fig. 4A) was determined using Patser v3e (Hertz and Stormo 1999). The number of nonpaired positions in each of the top scoring sites was plotted against the oligo's enrichment score. The P-value for the enrichment of pairs in the top 1% versus the entire pool (Fig. 6B) was calculated by simulating 1,000,000 draws from the null distribution.
Motif analysis in in vivo targets of PTB
For the 11 intron–exon regions identified by the Smith lab, Patser v3e (GZ Hertz and GD Stormo, unpubl.) was used to determine the top-scoring motif sites in these sequences, for each of the four motifs. Score cutoffs for each motif were determined by selecting the top 2% of scores. To determine the distribution of motif counts in the rest of the genome, 1000 random sets of 11 intron–exon pairs from the genome were also run through Patser v3e using each of the four motifs, and scores above the aforementioned cutoffs were recorded, yielding a distribution for each motif, from which means were calculated. A fold-enrichment score was then calculated as the ratio of the number of motif scores in the top 2% of the Smith data to the mean number of motif scores above that 2% cutoff in the randomly sampled data.
To examine motif combinations, two motifs were considered to be a pair if their first positions were within 30 nt of each other. Pairs were considered only when both of the annotated motifs had scores falling above the 2% cutoff. For each of the 16 ordered pairs, counts of the pairs were tabulated for both the Smith sequences and the 1000 random sets of sequences, and a mean count value for the random data was calculated. A fold-enrichment score was calculated in the same way as above, as the ratio of the number of pairs in the Smith data to the mean number of pairs in the randomly sampled data.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Matthew Gemberling for pool generation and validation, Luciana Ferraris for expert technical assistance, Jonathan Levin and Eric Lim for computational expertise, and Wendy Virgadamo for administrative assistance. We especially thank Kim Mowry for her support and critical review of this manuscript. The authors thank Martin Maxey and the NSF UBM-Group for summer support to B.L.C. (DUE-0734234), for critical comments, and for useful discussions. This work was partially supported by the UTRA program (to B.L.C.), a CCMB Scholarship Award (to W.G.F.), an NIH RO1 57286 (to S.I.G.), and an NIH COBRE award (to W.A.T.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1821809.
REFERENCES
- Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res. 2008;36:2338–2352. doi: 10.1093/nar/gkn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Allain FH. Structure–function relationships of the polypyrimidine tract binding protein. Cell Mol Life Sci. 2008;65:516–527. doi: 10.1007/s00018-007-7378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FHT. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Underwood JG, Nikolic J, Luu MH, Black DL. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- Coles LS, Bartley MA, Bert A, Hunter J, Polyak S, Diamond P, Vadas MA, Goodall GJ. A multi-protein complex containing cold shock domain (Y-box) and polypyrimidine tract binding proteins forms on the vascular endothelial growth factor mRNA. Potential role in mRNA stabilization. Eur J Biochem. 2004;271:648–660. doi: 10.1111/j.1432-1033.2003.03968.x. [DOI] [PubMed] [Google Scholar]
- Conte MR, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Mermoud JE, Lamond AI, Sharp PA. Cis-acting elements distinct from the 5′ splice site promote U1-independent pre-mRNA splicing. RNA. 1996;2:664–673. [PMC free article] [PubMed] [Google Scholar]
- Ding YE, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Chasin LA. Human genomic sequences that inhibit splicing. Mol Cell Biol. 2000;20:6816–6825. doi: 10.1128/mcb.20.18.6816-6825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother W, Lipscombe D. Repressing the neuron within. Bioessays. 2008;30:1–4. doi: 10.1002/bies.20696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, Rideau A, Southby J, Scadden AD, Gooding C, Huttelmaier S, Singer RH, Smith CW. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol Cell Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Holste D, Huo G, Tung V, Burge CB. HOLLYWOOD: A comparative relational database of alternative splicing. Nucleic Acids Res. 2006;34:D56–D62. doi: 10.1093/nar/gkj048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Hellen CU, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- Kosinski PA, Laughlin J, Singh K, Covey LR. A complex containing polypyrimidine tract-binding protein is involved in regulating the stability of CD40 ligand (CD154) mRNA. J Immunol. 2003;170:979–988. doi: 10.4049/jimmunol.170.2.979. [DOI] [PubMed] [Google Scholar]
- Kuwahata M, Tomoe Y, Harada N, Amano S, Segawa H, Tatsumi S, Ito M, Oka T, Miyamoto K. Characterization of the molecular mechanisms involved in the increased insulin secretion in rats with acute liver failure. Biochim Biophys Acta. 2007;1772:60–65. doi: 10.1016/j.bbadis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Le Sommer C, Lesimple M, Mereau A, Menoret S, Allo MR, Hardy S. PTB regulates the processing of a 3′-terminal exon by repressing both splicing and polyadenylation. Mol Cell Biol. 2005;25:9595–9607. doi: 10.1128/MCB.25.21.9595-9607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang W, Reed RB, Liu W, Grabowski PJ. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA. 2002;8:137–149. doi: 10.1017/s1355838202015029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Liu G, Sun Y, Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim Biophys Acta. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Southby J, Gooding C, Smith CW. Repression of α-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA. 2007;13:1214–1223. doi: 10.1261/rna.219607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AJ, Berget SM. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol Cell Biol. 2000;20:9225–9235. doi: 10.1128/mcb.20.24.9225-9235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell. 1983;33:509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J Biol Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997a;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Perez I, McAfee JG, Patton JG. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997b;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Monie TP, Allain FH, Matthews S, Curry S, Svergun DI. Conformation of polypyrimidine tract binding protein in solution. Structure. 2006;14:1021–1027. doi: 10.1016/j.str.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes & Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat Struct Mol Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the β-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol Cell Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Kan JL, Ghigna C, Biamonti G, Green MR. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA. 2004;10:787–794. doi: 10.1261/rna.5229704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, et al. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12:1631–1643. doi: 10.1016/j.str.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Cross-regulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Tantin D, Gemberling M, Callister C, Fairbrother W. High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes. Genome Res. 2008;18:631–639. doi: 10.1101/gr.072942.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WA, Newberg LA, Conlan S, McCue LA, Lawrence CE. The Gibbs centroid sampler. Nucleic Acids Res. 2007;35:W232–W237. doi: 10.1093/nar/gkm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmar L, Welsh N. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol Med. 2002;8:263–272. [PMC free article] [PubMed] [Google Scholar]
- Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies NOVA-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Xu M, Hecht NB. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3′UTR. Biol Reprod. 2007;76:1025–1033. doi: 10.1095/biolreprod.107.060079. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: An essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin AS, Tan W, Bear J, Smulevitch S, Felber BK. U2AF participates in the binding of TAP (NXF1) to mRNA. J Biol Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]