Abstract
Mitochondria (mt) in plants house about 20 group-II introns, which lie within protein-coding genes required in both organellar genome expression and respiration activities. While in nonplant systems the splicing of group-II introns is mediated by proteins encoded within the introns themselves (known as “maturases”), only a single maturase ORF (matR) has retained in the mitochondrial genomes in plants; however, its putative role(s) in the splicing of organellar introns is yet to be established. Clues to other proteins are scarce, but these are likely encoded within the nucleus as there are no obvious candidates among the remaining ORFs within the mtDNA. Intriguingly, higher plants genomes contain four maturase-related genes, which exist in the nucleus as self-standing ORFs, out of the context of their evolutionary-related group-II introns “hosts.” These are all predicted to reside within mitochondria and may therefore act “in-trans” in the splicing of organellar-encoded introns. Here, we analyzed the intracellular locations of the four nuclear-encoded maturases in Arabidopsis and established the roles of one of these genes, At5g46920 (AtnMat2), in the splicing of several mitochondrial introns, including the single intron within cox2, nad1 intron2, and nad7 intron2.
Keywords: maturases, group-II introns, splicing, mitochondria, plant
INTRODUCTION
Higher plants' mitochondria contain their own genomes, which encode for a small number of genes (up to 60) required in both organellar genome expression and respiration activities (for review, see Brennicke and Leaver 2007). The expression of the mtDNA in plants is complex, particularly at the post-transcriptional level (Mackenzie and McIntosh 1999; Bonen and Vogel 2001; Binder and Brennicke 2003; Barkan 2004; Knoop 2004; Gagliardi and Binder 2007; Bonen 2008). RNA processing events that contribute to organellar genome expression include numerous editing events (see Takenaka et al. 2008) and the splicing of ∼25 introns, which lie mainly within complex I (NADH:ubiquinone oxidoreductase) subunits, but also disrupt the coding regions of genes required in both organellar genome expression and respiration activities (reviewed in Bonen 2008).
According to conserved features of their primary sequences, the mitochondrial introns in plants are classified as “group-II” introns (Michel et al. 1989; Bonen and Vogel 2001; Knoop 2004; Li-Pook-Than and Bonen 2006; Gagliardi and Binder 2007; Bonen 2008). Although few introns in this class are able to catalyze their own excision in vitro, the mitochondrial introns in plants are highly degenerated and seem to lose their ability to “self-splice.” Some become fragmented, such that they are transcribed in pieces and are then spliced “in trans” (for review, see Bonen 2008). The removal of these introns from the coding sequences they interrupt is therefore essential for organellar function and is dependent upon the activity of various protein cofactors (for review, see Bonen and Vogel 2001; Barkan 2004; Gagliardi and Binder 2007; Bonen 2008).
The splicing of group-II introns in bacteria and yeast mitochondria is facilitated by proteins encoded within the introns themselves (intron-encoded “maturases”), which act specifically in the splicing of the intron in which they are encoded from (Cousineau et al. 1998, 2000; Wank et al. 1999; Matsuura et al. 2001; Singh et al. 2002; Noah and Lambowitz 2003; Lambowitz and Zimmerly 2004; Blocker et al. 2005; Robart and Zimmerly 2005). Yet, the vast majority of the organellar introns in plants lost their intron-encoded ORFs (Bonen and Vogel 2001; Toor et al. 2001; Knoop 2004; Ahlert et al. 2006). In fact, only a single maturase was retained in the mitochondrial genome in plants (matR); however, its putative role(s) in the splicing of organellar introns is yet to be determined. The diversity of group-II introns within plant mitochondria suggests the involvement of multiple splicing factors. These are presumably encoded within the nucleus, as there are no obvious candidates among the remaining ORFs within the mtDNA in plants.
Although considerable progress has been made in the identification and characterization of numerous factors required for the splicing of many of the introns found in the plastid genomes in algae and plants (Goldschmidt-Clermont et al. 1990; Jenkins et al. 1997; Jenkins and Barkan 2001; Ostheimer et al. 2003; Perron et al. 2004; Ostersetzer et al. 2005; Asakura and Barkan 2006; Merendino et al. 2006; Schmitz-Linneweber et al. 2006; Asakura and Barkan 2007; Watkins et al. 2007; Asakura et al. 2008; Keren et al. 2008; Prikryl et al. 2008; Williams-Carrier et al. 2008; Kroeger et al. 2009), such information is only starting to emerge for mitochondrial introns in plants.
A nuclear mutation in Nicotiana sylvestris (nms1) was shown to disrupt the splicing of the first intron in the nad4 transcript (Brangeon et al. 2000), however the identity of this gene remains unknown. Recently, a pentatricopeptide repeat protein (OPT43) was found to function specifically in the trans-splicing activity of nad1 intron1 in Arabidopsis mitochondria (de Longevialle et al. 2007).
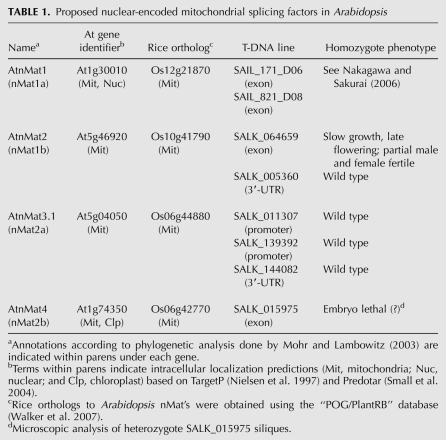
Intriguingly, analyses of the complete genomes of Arabidopsis and rice revealed the existence of four nuclear genes, which are closely related to group-II intron-encoded maturases identified in other systems (Mohr and Lambowitz 2003). According to conserved features of their N-termini sequences, these are all predicted to reside within mitochondria and are therefore expected to act in trans in the splicing of organellar introns. Indeed, while trying to elucidate the mechanism of cellulose synthesis in Arabidopsis, Nakagawa and Sakurai (2006) established a role for one of these genes, AtnMat1a (At1g30010), in the maturation of nad4 RNA in Arabidopsis mitochondria. However, the specific intron target in nad4 and whether AtnMat1 may also function in the splicing of additional introns are currently unknown. No data are available for the remaining nuclear-encoded maturase homologs in plants.
Here, we analyzed the intracellular locations of the four nuclear-encoded maturases in Arabidopsis and established the roles of another maturase-related gene, At5g46920 (annotated here as AtnMat2), in the splicing of several mitochondrial introns in Arabidopsis. Transient expression assays with a green fluorescent protein (GFP) tag and immunoblot analysis localized AtnMat2 to mitochondria. Fractionation of native organellar extracts indicated that nMat2 is found in a large RNA-associated particle(s) in vivo. Analysis of the phenotypes associated with a homozygotic T-DNA insertion line in the AtnMat2 gene established the roles of AtnMat2 in the splicing of at least three mitochondrial introns: the single intron within cox2 gene and the second introns within nad1 and nad7 intron2. nMat2 plants demonstrated growth to flowering defect phenotypes, which were tightly correlated with impaired organellar activity in the mutant.
RESULTS
Intracellular locations of the four nuclear-encoded maturases in Arabidopsis
The nuclear genome in Arabidopsis harbors four genes that are closely related to group-II intron-encoded maturases found in bacterial and yeast mitochondrial genomes (see Mohr and Lambowitz 2003). The genomic context of these genes differs from “model” maturases, as they are all found in the nucleus as self-standing ORFs, outside the context of their evolutionary-related group-II intron “hosts.” Analysis of their expression profiles, available in EST databases and the “microarray database and analysis toolbox” (Hruz et al. 2008), suggest that the four nuclear-encoded maturases are expressed at low levels in all tissues, throughout a plant's development (data not shown).
Group-II maturases, typically, are characterized by three domains: (1) an N-terminal reverse transcriptase (RT) domain; (2) a unique maturase RNA-binding motif (domain X); and (3) a C-terminal DNA endonuclease (En) domain (Mohr et al. 1993). Based on phylogenetic analysis, the nuclear-encoded maturases in plants were divided into two main categories (Mohr and Lambowitz, 2003): (1) genes encoding maturase proteins containing the “RT” and “X” domains, but lacking an “En” motif (in Arabidopsis these include At1g30010 and At5g46920); and (2) genes encoding maturase ORFs with all three domains typical in group-II intron maturases (At1g74350 and At5g04050 in Arabidopsis). However, At5g04050, alternatively, may be spliced into two putative isoforms: At5g04050.1, encoding a protein, which lacks the endonuclease domain; and At5g04050.2, which contains all three domains. RT-PCR followed by sequencing analysis (data not shown) and EST databases (NM_120487.6), supported only the presence of a transcript corresponding to At5g04050.1 mRNA.
Here, we changed the previous annotations of the four nuclear-encoded maturases in Arabidopsis (i.e., nMat 1a, 1b, 2a, and 2b) (see Mohr and Lambowitz 2003) into AtnMat 1 to 4 (see Table 1) to reflect the differences in their intracellular locations (AtnMat 1, 2, and 3.1 are localized to mitochondria, while AtnMat4 is localized to both mitochondria and plastids), as indicated by GFP localization and immunoblot analyses (see below).
TABLE 1.
Proposed nuclear-encoded mitochondrial splicing factors in Arabidopsis
Targeting prediction programs suggest that the four nuclear-encoded maturases in Arabidopsis, as their rice orthologs, are localized to mitochondria (Table 1; Mohr and Lambowitz 2003). Yet, despite their predictive power, these programs are still inaccurate, ascribing many nonmitochondrial proteins to that organelle (Heazlewood et al. 2005). Moreover, none of the maturases, including MatR and the four nuclear-encoded maturases, are found among proteins identified in mass-spectrometry analyses of plant organellar fractions, which also include numerous proteins with predicted RNA-mediated (or DNA-mediated) functions (Peltier et al. 2000, 2002, 2004; Baginsky et al. 2004, 2007; Heazlewood et al. 2005, 2007; Phinney and Thelen 2005; Kleffmann et al. 2007; Sun et al. 2009). However, these may have escaped our notice due to their predicted low expression profiles.
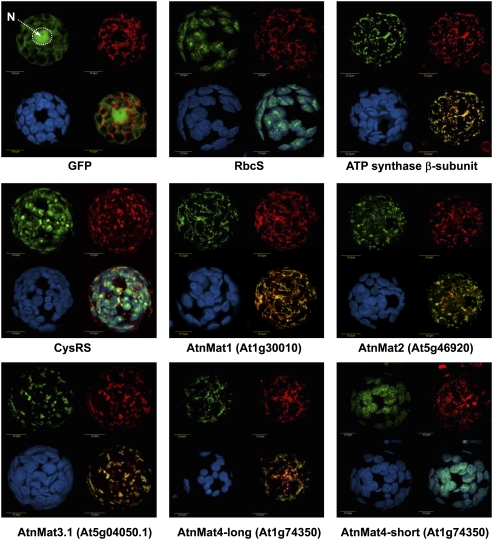
To establish their intracellular locations in vivo, constructs encoding the N-terminal region (about 150 amino acids) of each of the four nuclear-encoded maturases in Arabidopsis were cloned in-frame to GFP, introduced into tobacco protoplasts, and the location of each GFP-fusion protein was determined by confocal microscopy (Fig. 1). 5′ RACE and “in silico” analyses were used to ensure the integrity of the start codon in each maturase gene in Arabidopsis.
FIGURE 1.
Analysis of the intracellular locations of the four Arabidopsis nuclear-encoded maturase proteins in tobacco protoplasts. Tobacco protoplasts were transformed with GFP alone or GFP fused to the N-termini regions (about 150 amino acids) of: Rubisco small subunit (RbcS), ATP synthase β-subunit, cysteinyl-tRNA synthase (CysRS), AtnMat1 (At1g30010), AtnMat2 (At5g46920), AtnMat3.1 (At5g04050.1), and the N-termini regions of the two putative isoforms of AtnMat4 (At1g74350 “long” and “short”). The following substitutions were made for the GFP cloning: AtnMat1 K2V, AtnMat2 R2G, and AtnMat4-full, F2V. GFP signals (green, upper left), MitoTracker marker (red, upper right), chlorophyll autofluorescence (blue, lower left) and merged images (lower right), are outlined in each panel. Merged images in GFP, ATP synthase β-subunit, AtnMat 1, 2, 3.1 and 4-long represent GFP and MitoTracker signals; CysRS merged image represents GFP, MitoTracker, and chlorophyll autofluorescence signals; the merged RbcS and AtnMat4-short images correspond to GFP and chlorophyll autofluorescence signals. The position of the nucleus (N) is indicated in the “GFP” panel.
GFP alone showed a cytosolic distribution of the GFP signal, with considerable diffusion into the nucleus; whereas the signals of the N-terminal regions of the Rubisco small subunit (RbcS) and ATP synthase β-subunit fused to GFP, colocalized with chlorophyll autoflorescence and the MitoTracker marker, a mitochondrion-specific fluorescent probe, respectively. The signal obtained from the N-termini of the cysteinyl-tRNA synthetase (CysRS) protein was observed in both plastids and mitochondria, as expected for its dual localization to both these organelles (Peeters et al. 2000). The signals were clearly distinct from peroxisomes, as indicated in colocalization analysis with a peroxisomal, Cherry-PTS1 (Avisar et al. 2008), marker (data not shown).
When protoplasts were transfected with the N-termini of AtnMat1 fused to GFP, the signal was exclusively detected as rod-shaped granules colocalizing with those of MitoTracker (Fig. 1). These results coincide with the predictions of AtnMat1 to that organelle (Table 1) and previous observations, which indicated a role for this protein in the splicing of nad4 in Arabidopsis mitochondria (Nakagawa and Sakurai 2006). Similar to AtnMat1, the signals obtained from the N-termini regions of AtnMat2 and AtnMat3 colocalized with those of the MitoTracker marker, with no other signal observed elsewhere in the cell (Fig. 1).
However, the localization of the fourth paralog was less clear. 5′-RACE analysis of At1g74350 (AtnMat4) indicated two possible start codons, in which the putative 5′-proximal initiation site (UUGAUGUU) is found 138 nucleotides (nt) upstream (in-frame) of the NCBI-annotated AUG start codon (GAAAUGGC). Interestingly, the intracellular locations of these two putative isoforms of AtnMat4 differed markedly: while the signal obtained with the “long” N-terminal region of AtnMat4 (amino acids 1–196) was observed in the mitochondria, the “short” N-terminal region, containing only the “second” methionine (amino acids 46–196), colocalized with chlorophyll autoflorescence (Fig. 1). As the second “AUG” also shares a high similarity with the consensus translation initiation site in plants, (A/G)(A/C)AAUGGC (Joshi 1987; Lütcke et al. 1987; Rangan et al. 2008), these results may indicate that chloroplasts in Arabidopsis are also represented by at least one nuclear-encoded maturase paralog.
AtnMat2 mutants display defective vegetative to floral meristem developmental phenotypes
To examine their roles in organellar biogenesis, we screened available Arabidopsis T-DNA lines (Alonso et al. 2003) found within the promoter, untranslated regions (UTRs) and coding regions of AtnMat 2, 3, and 4 genes (Table 1). Insertions within the promoter region or the 3′-UTR of AtnMat3.1 had no obvious effect on the expression of the gene. No homozygotes were obtained for a T-DNA insertion line within the coding region of the AtnMat4 gene (Table 1, SALK-015975).
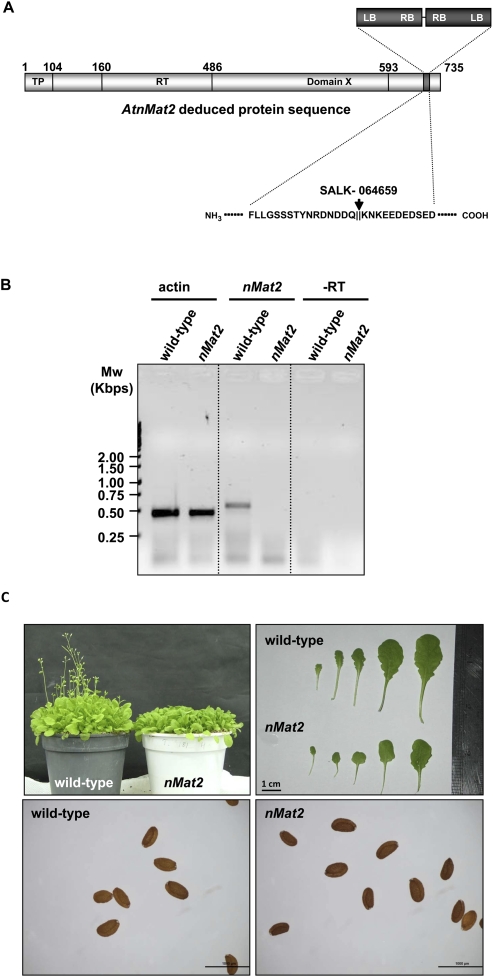
Although an insertion within the 3′-UTR of the At5g46920 gene (SALK-005360 line) had no effect on AtnMat2 expression, the mRNA level was largely reduced (undetectable levels) in the SALK-064659 line, where the T-DNA insertion is found within the AtnMat2 coding region (see Fig. 2). PCR, followed by DNA sequencing analysis, suggested a “double-inverted” insertion event, in which two T-DNAs are inserted in opposite directions (left borders facing out) within the C-termini of AtnMat2, in proximity to the RNA-binding motif (domain X) (Fig. 2A).
FIGURE 2.
Plant phenotypes associated with a T-DNA insertion line in AtnMat2. (A) Schematic representation of AtnMat2 (At5g46920) deduced sequence. The putative N-terminal organellar targeting region (TP), reverse transcriptase domain (RT), and the RNA-binding motif (domain X) are highlighted within the sequence according to Mohr and Lambowitz (2003). The precise location of the T-DNA insertion site (SALK_line 064659) is outlined within and below. Genotypes were confirmed by PCR and sequencing. (B) Analysis of relative AtnMat2 mRNA transcript accumulation by RT-PCR in wild-type and nMat2 plants; At-actin2 (At3g18780) was used as a control in the reaction. Primers are listed in Supplemental Table S3. (C) The effects of nMat2 knockout on seeds, flowering, and leaf morphology of 5-wk-old Arabidopsis thaliana wild-type and nMat2.
Homozygotic nMat2 mutants (SALK-064659) were able to grow, flower, and set normal viable seeds, thus allowing us to obtain large quantities of plant material required for analysis of their associated phenotypes. nMat2 plants, grown under long- or short-day conditions, displayed low germination, slow growth, and late flowering (about 5 wk to flowering in wild-type under long-day conditions, versus 7–8 wk in nMat2 mutants), and were partially sterile for both male and female gametes (data not shown). nMat2 leaves were smaller, round-shaped, and somewhat paler when compared with those of wild type (Fig. 2C). The insertion appeared tightly linked to these phenotypes, as all nMat2 plants demonstrated the same phenotypes.
nMat2 is found in a large ribonucleoprotein complex in the mitochondria that cofractionates with group-II intron RNAs
To more carefully establish the localization of AtnMat2 to mitochondria, we performed immunoblot analysis of Arabidopsis organellar extracts, using affinity-purified polyclonal antibodies raised against a unique region in the C-termini of AtnMat2 (amino acids 612–735, precursor protein). Antibodies to hydroxymethyltransferase (SHMT), cytochrome oxidase subunit 2 (COX2), NADH dehydrogenase subunit 9 (NAD9) (Lamattina et al. 1993), and the plastidic 33-kDa subunit of the photosystem II oxygen-evolving enzyme (PsbO) confirmed that the organelle preparations were reasonably pure (Fig. 3A).
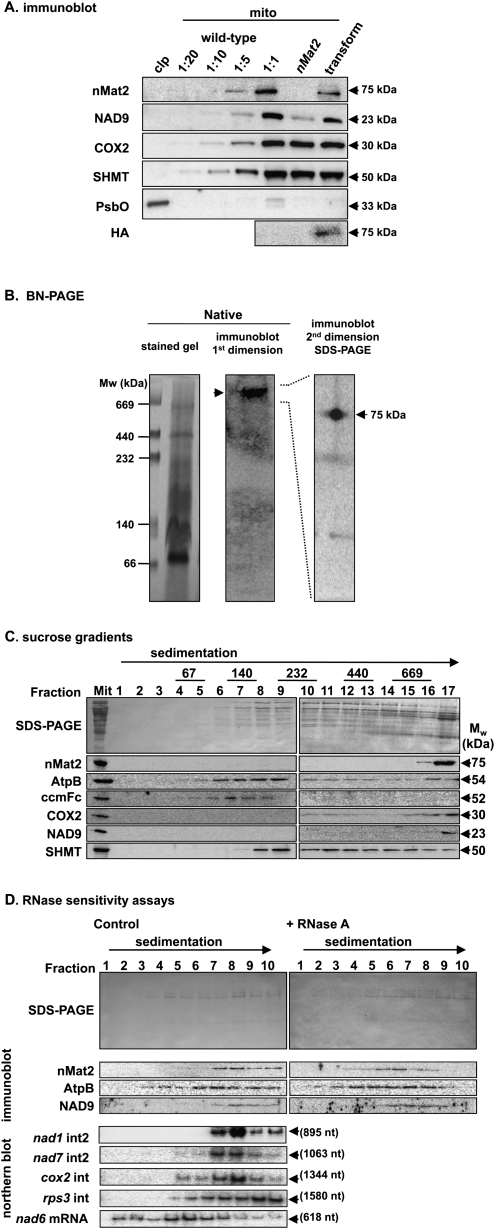
FIGURE 3.
nMat2 is identified in a large ribonucleoprotein particle in higher plants mitochondria. (A) Immunoblots with total proteins (about 50 μg) extracted from 3-wk-old rosette leaves of wild-type, homozygotic nMat2 plants, and 35S∷AtnMat2-HA transformant nMat2 plants. The blots were probed with polyclonal antibodies raised to AtnMat2, cytochrome oxidase subunit 2 (COX2), hydroxymethyltransferase (SHMT) protein, NADH-oxidoreductase subunit 9 (NAD9), plastid 33-kDa subunit of the oxygen evolving complex (PsbO), and monoclonal antibodies raised to hemagglutinin (HA). Detection was carried out by chemiluminescence assay after incubation with HRP-conjugated secondary antibody. (B) BN-PAGE and two-dimensional blue-native/SDS-PAGE of digitonin-treated cauliflower mitochondria. (C) Fractionation of solubilized cauliflower mitochondria by sucrose gradient centrifugation. Aliquots of total organellar protein extract (Mit) and sucrose gradient fractions (1–17) were subjected to immunoblot analysis with different antibodies, as indicated in each blot. High molecular mass standards (GE Healthcare) sizes are given in kDa. (D) nMat2-enriched fractions (16 and 17) were treated with RNase inhibitor (control) or RNase A prior to their fractionation by a second sucrose gradient centrifugation. Aliquots of total organellar extracts (Mit) and sucrose gradient fractions (1–10) were subjected to immunoblot, and Northern blot analyses with RNA probes raised to domains V and VI of each intron (see Supplemental Table S6).
As anticipated by its organellar prediction (Table 1) and GFP localization (Fig. 1), anti-AtnMat2 antibodies cross-reacted with a ∼75 kDa mitochondrial protein, in the expected size for a mature nMat2 protein (Fig. 3A). No signal was observed in crude mitochondrial extracts obtained from nMat2 plants (Fig. 3A), confirming that nMat2 was largely reduced in the mutant. While COX2 and SHMT accumulated to similar levels in wild-type and nMat2 plants, immunoblot analysis with antibodies raised in wheat NAD9 protein (Lamattina et al. 1993) indicated that the protein was reduced by several folds in the mutant (Fig. 3A, NAD9). As the accumulation of mRNA transcripts corresponding to nad9 apparently was unaffected in nMat2 (data not shown), these results may indicate that the NAD9 protein is unstable in the mutant.
For analysis of native organellar complexes we used mitochondria isolated from cauliflower (Brassica oleracea) inflorescences, as cauliflower is closely related to Arabidopsis thaliana and also enables the purification of large quantities of relatively pure mitochondria from plants (Neuwirt et al. 2005). Analysis of EST databases revealed the presence of several nMat2 orthologs in Brassicaceae, including a highly homologous sequence in cauliflower (AM395329). Immunoblot analysis confirmed that the antibodies raised in the Arabidopsis nMat2 protein also recognize its ortholog in cauliflower mitochondria (Fig. 3).
For BN-PAGE analysis, the equivalent of 600 μg cauliflower mitochondria protein was solubilized with digitonin, and native organellar complexes were separated on preparative blue-native gels (BN-PAGE) (Fig. 3B). nMat2 particle size was estimated from its relative mobility on the gel (Fig. 3B, middle panel), in comparison with those of native high-molecular-mass standards (Fig. 3B, left panel). The calculated mass of nMat2 complex, at ∼900 kDa, is substantially larger than the expected size for a monomeric nMat2 protein (∼75 kDa), suggesting that the nMat2 complex may involve multiple protein and/or RNA components.
For the second dimension, a gel lane from a native gel was excised and layered onto a linear 12% SDS-polyacrylamide gel. Following electrophoresis, the proteins were transferred to a nitrocellulose membrane and blotted with anti-AtnMat2 antibodies. Under denatured conditions, the nMat2 signal was identified as a ∼75 kDa protein (Fig. 3B, right panel), further supporting that the high-molecular-weight signal in the BN-PAGE indeed corresponded to organellar particles containing the nMat2 protein.
To analyze whether nMat2 is associated with organellar transcripts in vivo, about 1.5 mg of solubilized cauliflower mitochondria were fractionated by velocity centrifugation sedimentation throughout sucrose gradients, and each fraction was analyzed by immunoblot analysis with antibodies raised in different mitochondrial proteins (Fig. 3C). The nMat2 particles migrated through the sucrose gradients toward the bottom of the tube (fractions 16 and 17), further indicating that nMat2 is found in a large organellar complex in vivo (Fig. 3C).
For RNase sensitivity experiments, nMat2-enriched fractions were collected and treated with RiboLock RNase-inhibitor (control) or ribonuclease A (+RNase A), and were then applied to a second sucrose gradient. As indicated in Figure 3D, the nMat2 sedimentation rate was reduced in the presence of RNase A, suggesting that the nMat2 complex contains organellar transcripts in vivo. Northern blot analysis indicated that nMat2 cofractionated with several group-II introns, including nad1 intron2, nad7 intron2, and cox2 intron. In contrast, the sedimentation rate of nMat2 differs from those of rps3 intron RNA and nad6 mRNA after centrifugation. No signal was observed in fractions obtained from RNase A-treated fractions (data not shown). Together, these results strongly supported that nMat2 is found in a stable and specific complex with various group-II intron RNAs in mitochondria.
AtnMat2 is required for the efficient splicing of at least three organellar introns, including cox2 intron, nad1 intron2, and nad7 intron2
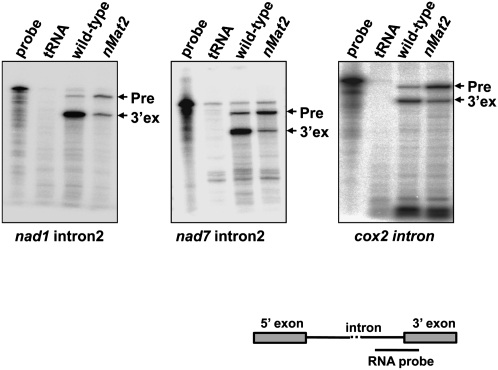
The high similarity of nMat2 to “model” maturases (Mohr and Lambowitz 2003; Robart and Zimmerly 2005) combined with its association with organellar transcripts in vivo (Fig. 3D), suggests that nMat2 may act in the splicing of at least one of the introns in the mtDNA in plants. To test this possibility, we analyzed the splicing of each of the 23 introns in Arabidopsis mitochondria (Unseld et al. 1997) in wild-type and nMat2 plants, using ribonuclease-protection assays (Fig. 4; Supplemental Fig. S1), Northern blot (Fig. 5), and RT-PCR (Supplemental Fig. S2).
FIGURE 4.
The splicing of cox2 intron, nad1 intron2, and nad7 intron2 are altered nMat2 mutant. Accumulation of precursor (Pre) and mature (3′ ex) transcripts was analyzed by RNase (RNase-T1) protection assay with 32P-labeled antisense RNA probes, which span the splice junctions (3′-exons and introns region; see illustration) of cox2 intron, nad1 intron2, and nad7 intron2. Total leaf RNA (20 μg) was analyzed in each assay. Protection of the probe by unspliced or spliced RNAs yield 300- and 100- nt products, respectively. The left-most lane in each panel contains a 1/1000 dilution of the probe used in the RNase-protection assay, while the lane labeled “tRNA” shows the results when leaf RNA was substituted with yeast tRNA (30 μg).
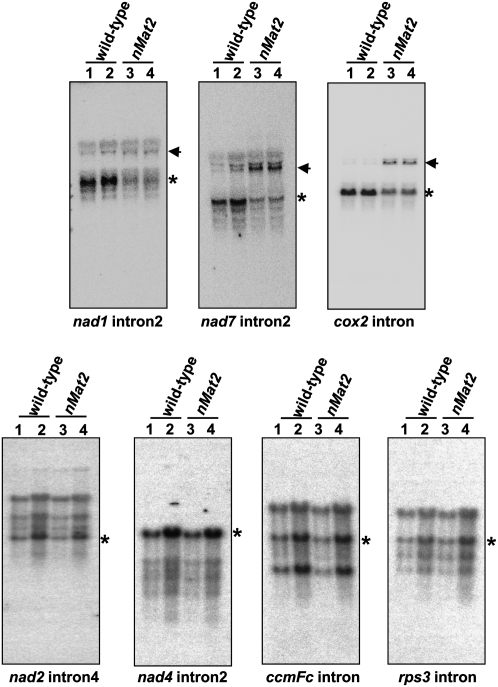
FIGURE 5.
Northern blots of mitochondrial transcripts. RNA gel-blot hybridizations showing specific organellar transcripts were assayed with either 5 μg (lanes 1,3) or 10 μg (lanes 2,4) total RNA obtained from wild-type (lanes 1,2) and nMat2 (lanes 3,4). Blots were probed with [32P]-“body-labeled” antisense RNA probes, which span the intron–exon boundaries of different organellar transcripts, as indicated below each blot. Asterisks mark bands corresponding to mature transcripts, while arrows point to predicted precursor RNAs. The sizes for the organellar transcript are: 2127 (pre-mRNA) and 783 nt (mRNA) for cox2; 2248 (pre-mRNA) and 1,185 (mRNA) nt for nad7; 3320 (pre-mRNA) and 1501 (mRNA) nt for nad2; 4564 (pre-mRNA) and 1484 (mRNA) nt for nad4; 2319 (pre-mRNA) and 1359 (mRNA) nt for ccmFc; and 3248 (pre-mRNA) and 1668 (mRNA) nt for rps3.
An RNase-protection assay was preformed with in vitro transcribed “body-labeled” RNA fragments, complementary to the intron–exon boundaries in each gene. The probes were incubated with total RNA isolated from wild-type and nMat2 plants, and subjected to RNase-T1 digestion (Fig. 4; Supplemental Fig. S1). The intensities of individual bands, corresponding to precursor (“Pre”) or mature (5′ or 3′ exons) transcripts, were quantified; and the ratio of precursor to mature RNA was calculated for each of the introns and normalized with “nonrelated” bands (Fig. 5, marked with asterisks). To identify potential artifacts that may arise by this method, the reaction was performed several times with different batches of RNA, and we also included a tRNA control that was used as a substitute for “total leaf” RNA in each assay. Supplemental Table S1 summarizes the splicing activity of each of the introns in wild-type and nMat2 plants, based on quantification of the RNase-protection assays.
As demonstrated in Figure 4, notable splicing defects (i.e., accumulation of pre-mRNAs that are correlated with reduced mRNA levels in the mutant) were observed for three individual introns: the single “intron2-type” within the cox2 transcript (Bonen 2008), and the second introns in nad1 (nad1 intron2) and nad7 intron2. Specific defects in the splicing of these three introns in nMat2 were further supported by Northern blot analysis, with RNA probes complementary to the intron–exon boundaries in each gene (Fig. 5). Similar to the RNase-protection assay results, cox2, nad1, and nad7 mRNA levels (Fig. 5, marked with asterisks) were significantly reduced in nMat2, while their corresponding pre-mRNAs (Fig. 5, marked with arrows) accumulated to higher levels in the mutant.
Accumulation of pre-mRNAs in nMat2, in the RNase-protection assays, was also apparent in the case of several other introns; these included nad2 introns 1, 3, and 4; nad4 intron2; and nad7 introns 2 and 3 (Supplemental Fig. S1). However, nad2 maturation also seemed impaired in Arabidopsis mutants altered in the splicing of various other mitochondrial introns (I Small, pers. comm.), while defects in the splicing of nad4 intron2 and nad7 introns 3 and 4 could not be supported by different methodologies (Figs. 4, 5; Supplemental Figs. S1, S2). Thus, at this point, we could not draw any firm conclusions regarding the putative roles of nMat2 in the splicing of either of these introns.
No significant differences in the accumulation of transcripts corresponding to nad1 intron4; nad4 introns 1 and 3; nad5 intron1; and nad7 introns 1, 3, and 4 were observed between wild-type and nMat2 plants (Figs. 4, 5; Supplemental Figs.S1, S2). Also, the splicing of cytochrome-c maturation factor (ccmFc), ribosomal protein L2 (rpl2), and ribosomal protein S3 (rps3) was unaffected in the mutant (Fig. 5; Supplemental Fig. S1). No differences in transcript accumulation of the five trans-spliced introns in Arabidopsis mitochondria were supported by RT-PCR; these included nad1 introns 1 and 3, nad2 intron2, and nad5 introns 2 and 3 (Unseld et al. 1997; Supplemental Fig. S2). The splicing of these introns is, therefore, unlikely to be dependent upon nMat2 activity, and is likely to be facilitated by other proteinacous cofactors.
The insertion of AtnMat2 into nMat2 mutants restored the growth phenotypes and splicing defects associated with the mutant
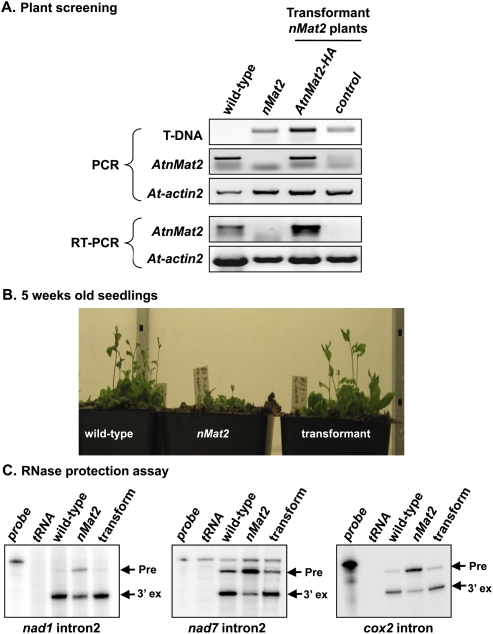
To establish whether the growth phenotypes and organellar splicing defects in nMat2 plants indeed result from disruption of At5g46920 loci, we performed a series of segregation tests and also introduced AtnMat2 into homozygotic nMat2 mutants. Due to their semisterile nature, reciprocal crosses between wild-type and nMat2 plants yielded only a few seeds, none of which were found to be heterozygotes. To address whether the insertion of AtnMat2 into nMat2 would restore the growth phenotypes and splicing defects associated with the mutant, the AtnMat2 gene was fused to a hemagglutinin (HA) tag at its carboxyl terminus, cloned into the binary pML-BART vector under a 35S promoter (pML-AtnMat2-HA) and introduced into nMat2 plants by the floral-dip method (Clough and Bent 1998).
Basta-resistant T1 transformants were selected, and the T2 progeny was analyzed for growth phenotypes and splicing defects. The expression of recombinant AtnMat2-HA in F2 progeny was analyzed by immunoblot (Fig. 3A) and RT-PCR (Fig. 6A) in wild-type, homozygotic nMat2, and transformant nMat2 plants with either a vector control (cont) or pML-BART containing AtnMat2-HA under 35S promoter. Oligonucleotides to SALK T-DNA and actin2 (At3g18780) were used as controls in the RT-PCR analysis (Fig. 6A).
FIGURE 6.
Insertion of AtnMat2 into nMat2 plants restored both the growth phenotypes and splicing defects associated with the mutant. AtnMat2, containing in its C-termini an HA-tag, was cloned into the binary pML-BART vector under the control of 35S promoter and introduced into homozygotic nMat2 plants by the floral-dip method (Clough and Bent 1998). (A) PCR and RT-PCR analyses of wild-type, nMat2, and transformant nMat2 plants with a vector control (cont) or 35S∷AtnMat2-HA; oligonucleotides to SALK T-DNA and At-actin2 gene (At3g18780) were used as control. (B) Growth to flowering phenotypes of 5-wk-old seedlings (F2 progeny). (C) Splicing activity in wild-type, nMat2, and transformant nMat2 plants was assayed by RNase protection assays, as described in more detail in Figure 4.
As indicated in Figures 3A and 6A, an individual 35S∷AtnMat2-HA transformant nMat2 plant was found to express AtnMat2-HA mRNA at high levels. Immunoblot analysis with anti-AtnMat2 antibodies and monoclonal anti-HA antibodies confirmed that the recombinant AtnMat2-HA protein accumulates in trasnformant nMat2 plants (Fig. 3A).
Analyses of the growth phenotypes of wild-type, nMat2, and 35S∷AtnMat2-HA transformant nMat2 plants demonstrated that vegetative and floral meristem development was similar in wild-type and transformant nMat2 plants (Fig. 6B), whereas transformant plants with a vector control (nMat2-cont) failed to restore the growth phenotypes associated with the nMat2 mutant (data not shown). Moreover, RNase-protection assays with total RNA, obtained from wild-type, nMat2, and 35S∷AtnMat2-HA transformant plants, indicated that cox2, nad1, and nad7 splicing activity was recovered upon the insertion and expression of a recombinant nMat2 protein in nMat2 mutants (Fig. 6C). Also, the NAD9 protein, which was reduced in nMat2 plants, accumulated at higher levels in transformant nMat2 plants expressing AtnMat2-HA (Fig. 3A).
DISCUSSION
The mitochondrial genome in Arabidopsis contains 23 group-II introns, which are found mainly within complex I subunits, but also disrupt the coding regions of ccmFc, cox2, rpl2, and rps3 genes (Unseld et al. 1997; Supplemental Table S1). Their degeneracy, the lack of intron-encoded ORFs, and the fact that none of these introns have been shown to self-splice in vitro, suggest that the splicing of these introns is facilitated by nuclear-encoded proteinacous cofactors in vivo (for review, see Binder and Brennicke 2003; Ahlert et al. 2006; Gagliardi and Binder 2007; Bonen 2008).
The nuclear genomes of Arabidopsis and rice contain four genes (denoted here as nMat 1–4) (Table 1), which are closely related to the group-II intron-encoded maturases identified in other systems (see Mohr and Lambowitz 2003; Robart and Zimmerly 2005). Based on their N-termini sequences, these are all predicted to reside within mitochondria and/or plastids (Table 1), and are thus predicted to function in the splicing of group-II introns in the organelle(s) to which they are localized. Indeed, a mutation in one of these genes, AtnMat1 (At1g30010), was recently shown to affect the accumulation of nad4 mRNA (Nakagawa and Sakurai 2006). Here, we provide evidence that strongly supports the roles of another nuclear-encoded maturase, AtnMat2 (At5g46920), in the splicing of several introns in Arabidopsis mitochondria.
GFP localizations (Fig. 1) and immunoblot (Fig. 3A) analyses localized nMat2 to mitochondria. Analysis of the RNA profiles in wild-type and nMat2 plants indicated that AtnMat2 is required for the efficient splicing of at least three mitochondrial introns: the single intron within cox2, nad1 intron2, and nad7 intron2 (Figs. 4, 5). In each case, a significant reduction in mRNA level was followed by accumulation of pre-mRNAs corresponding to each of these transcripts in the mutant. nMat2 may also function in the splicing of several other introns, including nad2 introns 1, 3, and 4; nad4 intron2; and nad7 introns 2 and 3. However, defects in the maturation of these introns in nMat2 were not as clear as in the cases of the cox2, nad1, and nad7 introns, and thus any speculation regarding the putative roles of AtnMat2 in the splicing of these introns must be considered with care.
Analysis of native organellar extracts indicated that nMat2 exists in a large (about 900 kDa) ribonucleoprotein complex (Fig. 3). The roles of nMat2 in the splicing of several mitochondrial introns suggest that the RNA components of this complex include group-II introns. To address this possibility, the sedimentation rate of nMat2 was compared with that of individual intron RNAs in fractionation of solubilized mitochondria through sucrose gradients. Indeed, cox2 intron, nad1 intron2, and nad7 intron2—the three genetically identified intron targets of nMat2—were found to cofractionate with nMat2, whereas the sedimentation rates of rps3 intron, the splicing of which is independent of nMat2 activity (Fig. 5; Supplemental Fig. S1), or nad6 mRNA did not match with those of nMat2 (Fig. 3D).
Although cox2, nad1, and nad7 splicing was altered, and their corresponding mature transcripts were significantly reduced, mRNA was still observed for each of these genes in nMat2 plants (Figs. 4, 5; Supplemental Fig. S2). Sequencing analysis confirmed their integrity and indicated that each of these transcripts is correctly spliced in the mutant (data not shown). Thus, we speculate that multiple factors may function in the splicing of each of these transcripts in mitochondria, as was also evident in the cases of many of the plastidic introns in plants (see Kroeger et al. 2009). Such proteins (e.g., MatR, nuclear-encoded maturases, PPR proteins, and other nuclear-encoded factors) may partially complement nMat2 activity in the absence of the protein.
The roles of AtnMat2 in the splicing of several individual introns in plant mitochondria differ from that of “model” maturases, which act specifically to promote the splicing of the introns from which they are encoded (Wank et al. 1999; Matsuura et al. 2001; Noah and Lambowitz 2003), although in rare cases may also function in the splicing of closely related group-II introns (Meng et al. 2005). It is possible that the cox2 intron, nad1 intron2, and nad7 intron2 are evolutionarily related to one another (i.e., share a common group-II intron ancestor), and thus remained as conserved “targets” of a single maturase protein. Yet, phylogenetic analysis of the group-II intron family in Arabidopsis mitochondria failed to agree with the maximum parsimony assumption of evolution (i.e., there is no single event in evolution that could explain the functionality of the three introns as targets of nMat2, which differentiate them from other organellar introns) (data not shown). However, it still remains possible that these introns retained short intronic sequences that may have escaped our notice. Small regions, particularly within group-II introns domain's I and IV, were previously shown to play an important role in the specific association of splicing factors with their group-II intron RNA targets (Matsuura et al. 2001; Ostersetzer et al. 2005; Keren et al. 2008). Detailed biochemical analysis of nMat2 binding to its genetically identified RNA targets, and characterization of the roles of orthologous nMat2 genes in the splicing of mitochondrial introns in other plant species, are required to address any of these speculations.
Immunoblot analysis indicated that NAD9 was reduced by several folds in the nMat2 mutant (Fig. 3A). As nad9 mRNA accumulated at similar levels in wild-type and nMat2 plants (data not shown), these results may indicate that complex I was affected in the mutant, possibly due to a lower availability of complex I subunits, where splicing defects were observed (Figs. 4, 5). Indeed, analysis of the respiration activity of crude mitochondrial extracts obtained from wild-type and nMat2 plants indicated that O2 uptake was reduced in the mutant (see Supplemental Fig. S3). Moreover, nMat2 respiration activity was found to be less susceptible to inhibition by rotenone, a specific inhibitor of complex I electron transport (Supplemental Fig. S3, +ROT), further indicating that complex I was impaired in the mutant.
Complex I dysfunctions have been previously associated with plants having growth to flowering developmental defects (Gutierres et al. 1997; Brangeon et al. 2000; Sabar et al. 2000; Pineau et al. 2005; Garmier et al. 2008). Accordingly, nMat2 plants demonstrated slow growth, late flowering, and were partially sterile (Fig. 2C; data not shown). These phenotypes are tightly associated with this mutation, as all homozygotic nMat2 plants demonstrated the same phenotypes. Moreover, insertion of AtnMat2-HA into nMat2 plants restored both the growth phenotypes and impaired organellar activities, associated with the nMat2 mutant (Figs. 3A, 6; Supplemental Fig. S3). However, a more comprehensive biochemical analysis of nMat2 mitochondria is required to determine whether complex I was functionally altered in the mutant.
In summary, our results provide the first detailed analysis of the roles of a nuclear-encoded maturase protein in the splicing of mitochondrial group-II introns in plants. Here, we demonstrated that AtnMat2 is localized to the mitochondria, where it facilitates the splicing of at least three mitochondrial introns: the single intron in cox2, nad1 intron2, and nad7 intron2. Homozygotic nMat2 mutants demonstrated slow growth, late flowering, and were partially fertile; these phenotypes may correlate with impaired complex I activity in the mutant.
MATERIALS AND METHODS
Protoplast preparation and GFP-based transient assay
A fragment (∼150 amino acids long) of the N terminus of each maturase was fused in-frame to GFP by cloning each fragment with NcoI and XhoI sites into a pTEX-GFP vector (gift of Dr. Yoram Eyal, Agricultural Research Organization). The specific primers used in the reactions are listed in Supplemental Table S2. Mesophyll protoplasts were isolated from tobacco (Nicotiana tabacum cv Samsun NN) plants grown under sterile conditions (Draper et al. 1988). Electroporation of ∼5 × 105 protoplasts per construct was performed in a prechilled electroporation medium (Fromm et al. 1985) using 1–5 μg of plasmid DNA. After electroporation, the protoplasts were transferred into a growth medium, incubated in darkness at 27°C for 48 h, and were then analyzed by confocal microscopy (Olympus IX 81, Fluoview 500). To visualize the mitochondria, protoplasts were treated with 0.4 μM MitoTracker (Invitrogen) for 15 min at room temperature. Images were color-coded green for GFP, red for MitoTracker, and blue for chlorophyll autofluorescence.
Plant material and growth conditions
Arabidopsis thaliana (ecotype Columbia) was used in all experiments. Wild-type and individual Arabidopsis mutants were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Prior to germination, seeds of wild-type and mutant lines were surface sterilized with 10% bleach (sodium hypochlorite) solution and 70% ethanol, and seeded on solid MS-agar plates containing 3% (w/v) sucrose. The plates were kept in the dark for 2 d at 4°C and then transferred to a controlled temperature and light growth chamber (22°C, 16/8 h day/night, 100–150 μE m−2/sec−1). After 10 d, the seedlings were transferred to soil, and grown under the same temperature and light regimes. PCR with gene-specific and/or T-DNA oligonucleotides (Supplemental Table S3) was used to screen the plant collection and verify the insertion integrity of each individual line. Sequencing of specific PCR products was used to analyze the precise insertion sites in each T-DNA line.
Antisera production
AtnMat2 fragment (amino acids 612–735) was obtained by RT-PCR of total RNA extracted from 3-wk-old rosette leaves and cloned into pET-28a vector (Novagen), yielding a 16-kDa translation product with a C-terminal histidine (6×) tag. The recombinant plasmid pET28-fnMat2-His was transformed to expression host E. coli BL21 (DE3) pLysS cells. The fusion protein was purified on a nickel column and verified for its integrity by mass-spectrometry analysis (Smoler Protein Center). Peptides identified in the MS–MS are listed in Supplemental Table S4. The purified protein was dialyzed against buffer containing 20 mM Tris-HCl at pH 6.8, and injected into rabbits for the production of polyclonal antisera (Genemed Synthesis Inc.). Specific anti-AtnMat2 antibodies were affinity purified against the antigen and used in all immunoblot experiments.
Protein extraction and analysis
Total leaf protein was extracted from 3-wk-old Arabidopsis leaves by the borate/ammonium acetate method (Maayan et al. 2008). Protein concentration was determined by the Bradford method (Bio-Rad) according to the manufacturer's protocol, with bovine serum albumin (BSA) used as a calibrator. Approximately 20 μg total leaf protein were mixed with an equal volume of 3× Laemmli sample loading buffer (Laemmli 1970) and subjected to 12% SDS-PAGE (at a constant 100 V). Following electrophoresis, the proteins were transferred to a nitrocellulose membrane (Whatman) and blotted overnight at 4°C with specific primary antibodies. Detection was carried out by chemiluminescence assay after incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Sigma or Santa Cruz).
Blue-native gel electrophoresis
Cauliflower mitochondrial samples were prepared as described previously (Takenaka et al. 2007). In general, about 500 g of cauliflower inflorescences (Brassica oleracea var. botrytis; purchased at local markets) were grinded, fractionated onto percoll gradients, yielding, on average, 10–15 mg total organellar protein per purification. One-dimensional BN-PAGE and two-dimensional BN/SDS-PAGE were carried out essentially as described previously (Wittig et al. 2006). About 600 μg cauliflower mitochondria were resuspended in 50 μL buffer containing 30 mM Hepes-KOH and 150 mM KOAc (pH 7.4), and solubilized with digitonin (10 g/g, detergent-to-protein ratio). The organellar extract was incubated for 30 min on ice and centrifuged for 30 min at 18,000g (4°C) to remove insoluble materials. Three microliters of Coomassie Blue G-250 solution (5% w/v solution) were added to the supernatant, and the native organellar complexes were loaded onto a linear 5%–15% native page and separated at 4°C for 3–4 h at 100–400 V.
Sucrose gradient centrifugation
Separation of mitochondrial protein complexes by sucrose gradient ultracentrifugation was performed according to the method of Zsigmond et al. (2008). Freshly prepared cauliflower mitochondria (1.5 mg) were solubilized in 200 μL buffer containing 30 mM Hepes-KOH, 150 mM KOAc (pH 7.4), and 5% digitonine (w/v). The organellar extract was fractionated by sucrose gradient (10%–50%) centrifugation (150,000g for 20 h at 4°C). For the RNA destabilization experiments (Uyttewaal et al. 2008), nMat2-enriched fractions were treated with 1400 U/mL RNase A (Sigma), or 40 U/mL RiboLock RNase-inhibitor (Fermentas), and were then fractionated by a second sucrose gradient (10%–50%, 2 h at 155,000g). Fractions 1–10 (top to bottom) were collected and analyzed by immunoblot assay with antibodies to various organellar proteins.
RNA extraction and analysis
RNA was extracted from 3-wk-old Arabidopsis rosette leaves (1 g) by Tri-Reagent (Sigma), according to the manufacturer's instructions. The RNA was treated with RNase-free RQ1-DNase (Promega) prior to its use in the assays. For reverse transcription, 2 μg total leaf RNA were incubated with gene-specific “reverse” oligonucleotides (Supplemental Table S5), and the complementary DNA fragment was generated with 5 U of AMV reverse transcriptase (Promega) in a reaction buffer containing 10 mM Tris-HCl at pH 9.0, 5 mM MgCl2, 5 mM dithiothreitol (DTT), 2.5 mM dNTP mix, and 10 U of ribonuclease inhibitor, at 42°C for 1 h. For the PCR amplification step, 1.0 μL of the reverse-transcription reaction were used in 25 μL reactions with an appropriate amount of reverse and forward primers (Supplemental Table S5).
For the ribonuclease protection assay, 32P-labeled RNA fragments (about 150,000 cpm), which unequally spanning the intron–exon boundaries (200 nt exons and 100 nt introns), were annealed to 20 μg total leaf RNA (16 h at 50°C) and subjected to RNA cleavage by RNase-T1 (100 U; Ambion). The RNA mixture was loaded onto a 10% polyacrylamide/7 M urea gel. Bands corresponding to spliced (mRNA) and unspliced (pre-mRNA) transcripts were visualized by a PhosphorImager (Fuji-Film FLA-5000) and analyzed by ImageQuant software (version 5.1, Molecular Dynamics). The Oligonucleotides of different introns/exons are listed in Supplemental Table S6.
For Northern blot analysis, total leaf RNA (5–10 μg) was loaded onto 1.3% agarose gels containing 15% formaldehyde in 1× MOPS buffer (20 mM MOPS at pH 7.0, 8 mM sodium acetate, 1 mM ethylenediaminetetraacetic acid [EDTA]). Following electrophoresis in 1× MOPS buffer (about 3 h at a constant 80 V), the RNA was transferred from the gel to a nylon-charged nitrocellulose membrane (Nytran SPC; Whatman) using a standard capillary technique. Hybridizations with each membrane were preformed with 32P-labeled (50,000 cpm) in vitro-transcribed antisense RNA probes (see Supplemental Table S6) at 68°C in “Church-hybridization” buffer (0.5 M Na2PO4 at pH 7.1, 7% w/v SDS, 2 mM EDTA). The membranes were washed twice with 0.1× SSC, 0.2% SDS, and twice with 0.01× SSC, 0.2% SDS. Bands corresponding to specific transcripts were visualized by a PhosphorImager. Specific signals were normalized with rRNA visualized by methylene blue staining.
Transformation to nMat2 plants
The complete coding region of the AtnMat2 (At5g46920) gene was generated by RT-PCR. PCR-based reactions were used to generate an HA-tag in-frame to the C-termini of AtnMat2. AtnMat2-HA was cloned into pART7 (Gleave 1992) using the EcoRI and XbaI cloning sites under control of the viral 35S promoter, and the expression cassette present in the vector was isolated as a NotI fragment and cloned into the binary pML-BART vector. The construct was then introduced into homozygous nMat2 plants by the floral-dip method (Clough and Bent 1998); Basta-resistant T1 transformants were selected, and the F2 progeny was analyzed for growth and flowering phenotypes, respiration activity, and RNA and protein profiles.
Respiration activity
Crude mitochondria extracts were obtained from 3-wk-old Arabidopsis seedlings, generally as described previously (Keech et al. 2005). About 2 mg of Arabidopsis leaves were homogenized in 10 mL buffer containing 50 mM N-Tris (hydroxymethyl)-methyl-2-aminoethanesulphonic acid (TES), 10 mM EDTA, 10 mM KH2PO4, 25 mM tetrasodium pyrophosphate, 1 mM glycine, 0.3 M sucrose, and 1% (w/v) polyvinylpyrrolidone (PVP)-40, 1% (w/v) BSA (pH 7.5). The extract was filtered through four layers of cheesecloth, and thylakoid membranes and other contaminants were removed by 15-min centrifugation at 2500g (4°C). Mitochondria were obtained by centrifugation of the clear supernatant at 18,000g for 30 min (4°C). O2 uptake was analyzed with a Clark-type electrode (Yellow Springs Instrument Co.), connected to a 1 mV recorder (Electronik, Honeywell Controls Ltd.). For each assay, the equivalent of 200 μg crude mitochondria protein was resuspended in 2 mL buffer, containing 25 mM TES, 10 mM KCl, 2 mM MgSO4, 5 mM KH2PO4, 0.3 M sucrose, and 0.1% BSA (pH 7.5), applied to the electrode in a sealed glass chamber. Respiration was initiated by the addition of 10 mM glycine and measured in the presence or absence of 50 μM rotenone.
In-silico analysis
Available T-DNA insertion lines in Arabidopsis and DNA clones were selected by the SIGnAL database (http://signal.salk.edu/cgi-bin/tdnaexpress). ESTs were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and Dana Farber Cancer Institute (http://compbio.dfci.harvard.edu/tgi) databases. Rice orthologs to Arabidopsis maturases were analyzed in the “POG/PlantRB” database (Walker et al. 2007). Intracellular localization predictions were analyzed by TargetP (Nielsen et al. 1997) and Predotar (Small et al. 2004) software. Multiple sequence alignments were performed by ProbCons (Do et al. 2005) and validated by the HoT method (Landan and Graur 2007). The alignments were then subjected to PHYLIP (Phylogeny Inference Package) to perform a maximum likelihood tree. The topology of the tree was statistically tested by a bootstrap method (1000× replicates).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Professor Ian Small (University of Western Australia), Professor Valerian V. Dolja (Oregon State University), and Dr. Yoram Eyal (Volcani Center, Agricultural Research Organization) for allowing us to use their GFP-CysRS, Cherry-PTS1, and GFP-RbcS markers, respectively; Dr. Yuval Eshed (Weizmann Institute) for providing us with the binary pML-BART vector system; and Professor J.M Grienenberger, Dr. Philippe Giegé (CNRS, Strasbourg), and Professor Zach Adam (the Hebrew University) for providing us with the antibodies to NAD9, ccmFc, and PsbO, respectively. We also wish to thank Mrs. Leah Naveh (the Hebrew University) for help with respiration analysis and Dr. Ron Ophir (Volcani center, Agricultural Research Organization) for his assistance with phylogenetic analysis. This work was supported by grants from the U.S–Israel Binational Agricultural Research and Development Fund (BARD IS-374-05), the U.S–Israel Binational Science Foundation (BSF 2007200), and in part by the Israeli Science Foundation (ISF 1176/07).
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1776409.
REFERENCES
- Ahlert D, Piepenburg K, Kudla J, Bock R. Evolutionary origin of a plant mitochondrial group II intron from a reverse transcriptase/maturase-encoding ancestor. J Plant Res. 2006;119:363–371. doi: 10.1007/s10265-006-0284-0. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19:3864–3875. doi: 10.1105/tpc.107.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Bayraktar OA, Barkan A. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA. 2008;14:2319–2332. doi: 10.1261/rna.1223708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. Myosin XI-K is required for rapid trafficking of golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 2008;146:1098–1108. doi: 10.1104/pp.107.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Siddique A, Gruissem W. Proteome analysis of tobacco Bright Yellow-2 (BY-2) cell culture plastids as a model for undifferentiated heterotrophic plastids. J Proteome Res. 2004;3:1128–1137. doi: 10.1021/pr0499186. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Grossmann J, Gruissem W. Proteome analysis of chloroplast mRNA processing and degradation. J Proteome Res. 2007;6:809–820. doi: 10.1021/pr060473q. [DOI] [PubMed] [Google Scholar]
- Barkan A. Intron splicing in plant organelles. In: Daniell H, Chase C, editors. Molecular biology and biotechnology of plant organelles. Kluwer Academic; Dordrecht, The Netherlands: 2004. pp. 281–308. [Google Scholar]
- Binder S, Brennicke A. Gene expression in plant mitochondria: Transcriptional and post-transcriptional control. Philos Trans R Soc Lond B Biol Sci. 2003;358:181–188. doi: 10.1098/rstb.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker FJH, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA. 2005;11:14–28. doi: 10.1261/rna.7181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- Brangeon J, Sabar M, Gutierres S, Combettes B, Bove J, Gendy C, Chetrit P, des Francs-Small CC, Pla M, Vedel F, et al. Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotiana sylvestris NMS1 nuclear mutant. Plant J. 2000;21:269–280. doi: 10.1046/j.1365-313x.2000.00679.x. [DOI] [PubMed] [Google Scholar]
- Brennicke A, Leaver CJ. Encyclopedia of life sciences. Wiley; Chichester, UK: 2007. Mitochondrial genome organization and expression in plants. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–1021. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell. 2007;19:3256–3265. doi: 10.1105/tpc.107.054841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J, Scott R, Hamil J. Blackwell Scientific; Oxford, UK: 1988. Transformation of dicotiledonous plant cells using the Ti plasmid of A. tumefaciens and the Ri plasmid of A. rhizogenes. [Google Scholar]
- Fromm M, Taylor LP, Walbot V. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc Natl Acad Sci. 1985;82:5824–5828. doi: 10.1073/pnas.82.17.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D, Binder S. Expression of the plant mitochondrial genome. In: Logan D, editor. Plant mitochondria. Blackwell; Ames, IA: 2007. pp. 50–95. [Google Scholar]
- Garmier M, Carroll AJ, Delannoy E, Vallet C, Day DA, Small ID, Millar AH. Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 2008;148:1324–1341. doi: 10.1104/pp.108.125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Girard-Bascou J, Choquet Y, Rochaix JD. Trans-splicing mutants of Chlamydomonas reinhardtii. Mol Gen Genet. 1990;223:417–425. doi: 10.1007/BF00264448. [DOI] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R. Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci. 1997;94:3436–3441. doi: 10.1073/pnas.94.7.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH. Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 2005;139:598–609. doi: 10.1104/pp.105.065532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: The Arabidopsis subcellular database. Nucleic Acids Res. 2007;35:213–218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747–420751. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BD, Barkan A. Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 2001;20:872–879. doi: 10.1093/emboj/20.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BD, Kulhanek DJ, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardestrom P. Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant. 2005;124:403–409. [Google Scholar]
- Keren I, Klipcan L, Bezawork-Geleta A, Kolton M, Shaya F, Ostersetzer-Biran O. Characterization of the molecular basis of group II intron RNA recognition by CRS1-CRM domains. J Biol Chem. 2008;283:23333–23342. doi: 10.1074/jbc.M710488200. [DOI] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S. Proteome dynamics during plastid differentiation in rice. Plant Physiol. 2007;143:912–923. doi: 10.1104/pp.106.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. The mitochondrial DNA of land plants: Peculiarities in phylogenetic perspective. Curr Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci. 2009;106:4537–4542. doi: 10.1073/pnas.0812503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger J-M. Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur J Biochem. 1993;217:831–838. doi: 10.1111/j.1432-1033.1993.tb18311.x. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Landan G, Graur D. Heads or tails: A simple reliability check for multiple sequence alignments. Mol Biol Evol. 2007;24:1380–1383. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 2006;34:2782–2790. doi: 10.1093/nar/gkl328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maayan I, Shaya F, Ratner K, Mani Y, Lavee S, Avidan B, Shahak Y, Ostersetzer-Biran O. Photosynthetic activity during olive (Olea europaea) leaf development correlates with plastid biogenesis and Rubisco levels. Physiol Plant. 2008;134:547–558. doi: 10.1111/j.1399-3054.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L. Higher plant mitochondria. Plant Cell. 1999;11:571–586. doi: 10.1105/tpc.11.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Wang Y, Liu XQ. An intron-encoded protein assists RNA splicing of multiple similar introns of different bacterial genes. J Biol Chem. 2005;280:35085–35088. doi: 10.1074/jbc.C500328200. [DOI] [PubMed] [Google Scholar]
- Merendino L, Perron K, Rahire M, Howald I, Rochaix JD, Goldschmidt-Clermont M. A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 2006;34:262–274. doi: 10.1093/nar/gkj429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Kazuhiko U, Haruo O. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Mohr G, Lambowitz AM. Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res. 2003;31:647–652. doi: 10.1093/nar/gkg153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Sakurai N. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial nad4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:772–783. doi: 10.1093/pcp/pcj051. [DOI] [PubMed] [Google Scholar]
- Neuwirt J, Takenaka M, Van Der Merwe JA, Brennicke A. An in vitro RNA editing system from cauliflower mitochondria: Editing site recognition parameters can vary in different plant species. RNA. 2005;11:1563–1570. doi: 10.1261/rna.2740905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Noah JW, Lambowitz AM. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003;42:12466–12480. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters NM, Chapron A, Giritch A, Grandjean O, Lancelin D, Lhomme T, Vivrel A, Small I. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J Mol Evol. 2000;50:413–423. doi: 10.1007/s002390010044. [DOI] [PubMed] [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ. Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell. 2000;12:319–341. doi: 10.1105/tpc.12.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J-B, Ytterberg AJ, Sun Q, van Wijk KJ. New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J Biol Chem. 2004;279:49367–49383. doi: 10.1074/jbc.M406763200. [DOI] [PubMed] [Google Scholar]
- Perron K, Goldschmidt-Clermont M, Rochaix J-D. A multiprotein complex involved in chloroplast group II intron splicing. RNA. 2004;10:704–711. doi: 10.1261/rna.5237804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney BS, Thelen JJ. Proteomic characterization of a triton-insoluble fraction from chloroplasts defines a novel group of proteins associated with macromolecular structures. J Proteome Res. 2005;4:497–506. doi: 10.1021/pr049791k. [DOI] [PubMed] [Google Scholar]
- Pineau B, Mathieu C, Gerard-Hirne C, De Paepe R, Chetrit P. Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMSII mutant lacking nad7. J Biol Chem. 2005;280:25994–26001. doi: 10.1074/jbc.M500508200. [DOI] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan L, Vogel C, Srivastava A. Analysis of context sequence surrounding translation initiation site from complete genome of model plants. Mol Biotechnol. 2008;39:207–213. doi: 10.1007/s12033-008-9036-9. [DOI] [PubMed] [Google Scholar]
- Robart A, Zimmerly S. Group II intron retroelements: Function and diversity. Cytogenet Genome Res. 2005;110:589–597. doi: 10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, de Kouchkovsky Y. Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 2000;124:1239–1250. doi: 10.1104/pp.124.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 Pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN, Saldanha RJ, D'Souza LM, Lambowitz AM. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol. 2002;318:287–303. doi: 10.1016/S0022-2836(02)00054-2. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ. PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 2009;37:969–974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Brennicke A, Jonatha MG. RNA editing in plant mitochondria: Assays and biochemical approaches. In Methods Enzymol. 2007;424:437–439. doi: 10.1016/S0076-6879(07)24020-0. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- Uyttewaal M, Mireau H, Rurek M, Hammani K, Arnal N, Quadrado M, Giegé P. PPR336 is associated with polysomes in plant mitochondria. J Mol Biol. 2008;375:626–636. doi: 10.1016/j.jmb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Walker NS, Stiffler N, Barkan A. POGs/PlantRBP: A resource for comparative genomics in plants. Nucleic Acids Res. 2007;35:852–856. doi: 10.1093/nar/gkl795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell. 2007;19:2606–2623. doi: 10.1105/tpc.107.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Zsigmond L, Rigo G, Szarka A, Szekely G, Otvos K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 2008;146:1721–1737. doi: 10.1104/pp.107.111260. [DOI] [PMC free article] [PubMed] [Google Scholar]