Summary
Changes in gene expression contribute to the long-lasting regulation of the brain’s reward circuitry seen in drug addiction, however, the specific genes regulated and the transcriptional mechanisms underlying such regulation remain poorly understood. Here, we used chromatin immunoprecipitation coupled with promoter microarray analysis to characterize genome-wide chromatin changes in the mouse nucleus accumbens, a crucial brain reward region, after repeated cocaine administration. Our findings reveal several interesting principles of gene regulation by cocaine and of the role of ΔFosB and CREB, two prominent cocaine-induced transcription factors, in this brain region. The findings also provide novel and comprehensive insight into the molecular pathways regulated by cocaine – including a new role for sirtuins (Sirt1 and Sirt2) –which are induced in the nucleus accumbens by cocaine and, in turn, dramatically enhance the behavioral effects of the drug.
Introduction
Repeated use of addictive drugs such as cocaine causes long-lasting changes in the brain’s reward circuitry, a key component of which is the nucleus accumbens (NAc). Accordingly, a major goal in the field has been to uncover the molecular mechanisms underlying addiction-associated neuroadaptations in this brain region. It has been hypothesized that one such mechanism is the regulation of gene expression (Hyman et al., 2006), and there have been numerous studies that have documented altered expression of genes, through candidate gene approaches or through gene expression arrays, in the NAc (Freeman et al., 2001; McClung and Nestler, 2003; Yao et al., 2004). As well, several transcription factors have been shown to be altered in the NAc after chronic cocaine exposure, including ΔFosB (a Fos family protein) (Hiroi et al., 1997; Nestler, 2008) and CREB (cAMP response element binding protein) (Carlezon et al., 2005), which have both been related directly to the behavioral abnormalities that characterize an addicted state.
ΔFosB is induced uniquely by chronic cocaine exposure and persists for several weeks after drug cessation due to the unusual stability of the protein (Nestler, 2008). Increasing evidence supports the view that this induction of ΔFosB increases rewarding responses to, and incentive motivation for, cocaine (Colby et al., 2003; Hiroi et al., 1997; Kelz et al., 1999; McClung and Nestler, 2003; Nestler, 2008). In contrast, CREB activity is induced in response to acute or chronic cocaine administration and serves a homeostatic role by reducing sensitivity to drug reward and mediating negative emotional symptoms during drug withdrawal (Carlezon et al., 2005).
However, all investigations of cocaine-induced changes in gene expression to date have focused by necessity on measures of steady-state mRNA levels, which may not reflect the transcriptional regulation of the encoding genes. Recent advances in chromatin biology have made it possible for the first time to extend this level of knowledge to direct examination of transcriptional mechanisms. Thus, we now know, largely from studies of non-neural tissue, that the state of activation or repression of a gene is typically reflected in the covalent modifications of histone proteins in the gene’s vicinity (Kouzarides, 2007). The vast majority of reports indicate that increased acetylation of histone H3 or H4 is highly predictive of gene activation, while increased methylation of H3 at K9 or K27 (Lys9 or 27) is predictive of gene repression. Chromatin immunoprecipitation (ChIP), where tissue is lightly fixed to crosslink DNA with histones and other DNA-binding proteins and then immunoprecipitated for a protein of interest (e.g., acetylated H3), can be used to assess the extent to which a given gene is associated with these markers of activation or repression. For example, we recently demonstrated by use of ChIP that chronic administration of cocaine or related psychostimulants increases or decreases histone acetylation in the NAc at the promoters of several genes whose mRNA levels are known to be increased or decreased, respectively, by the drug. Moreover, we directly implicated ΔFosB in mediating some of these effects by carrying out ChIP for this transcription factor under these conditions (Kumar et al., 2005; Renthal et al., 2008).
ChIP represents a powerful advance in establishing regulation of gene transcription by cocaine, but traditional methods are still limited by the analysis of individual genes of interest one at a time. Moreover, regulation of a gene may be mediated by many types of coincident alterations of histones and related modifications (Kouzarides, 2007). These considerations argue for the importance of analyzing the immunoprecipitated DNA, not for an individual gene, but genome-wide using ChIP-chip assays, to gain a global view of genes that show markers of activation or repression after cocaine. In the present study, we mapped the genomic effects of chronic cocaine in the NAc by performing ChIP-chip for acetylated and methylated histones and for ΔFosB and phospho-CREB (the activated form of this transcription factor). We then demonstrate that one family of novel target genes, the sirtuins, discovered by ChIP-chip to be regulated in the NAc by chronic cocaine, contributes directly to addiction-related behaviors. These findings provide fundamentally new insight into cocaine’s regulation of gene transcription in this brain reward region.
Results
Cocaine regulation of histone acetylation and methylation in the NAc
To extend our previous ChIP studies (see Renthal and Nestler, 2008), which identified that cocaine or amphetamine administration alters histone acetylation and methylation at specific genes in the NAc, we mapped the genome-wide promoter binding of these histone modifications in the NAc from mice treated chronically with cocaine (20 mg/kg/day for 7 days, i.p.) or saline. To do this, we performed ChIP with antibodies directed against polyacetylated H3 (K9 and K14), polyacetylated H4 (K5, K8, K12, and K16), or dimethylated H3 (K9 and K27) on independent pools of NAc lysates, with each pooled sample representing bilateral NAc dissections from 8 mice. These histone modifications have been widely shown to reflect the state of gene activation or repression (Kouzarides, 2007). The immunoprecipitated DNA, as well as input (total) DNA, was sheared and then repaired, amplified, and labeled with a fluorescent dye with the use of ligation-mediated PCR (Sikder et al., 2006). Cy5-labeled immunoprecipitated DNA and Cy3-labeled total DNA were mixed and hybridized to MM8 NimbleGen promoter microarrays (Madison, WI), which span the promoters of ~20,000 genes. After pre-processing, normalization, and identification of binding sites using Mpeak, statistical analysis of cocaine-induced changes in promoter occupancy was performed. Since the distribution of fold changes was slightly short-tailed, we performed a non-parametric analysis using 3.1 standard deviations as a cutoff, which would roughly correspond to p < 0.001 under normal assumptions (see Supplemental Methods). These unique datasets provided a novel view of cocaine-regulated gene targets and the regulatory mechanisms in this critical brain reward region.
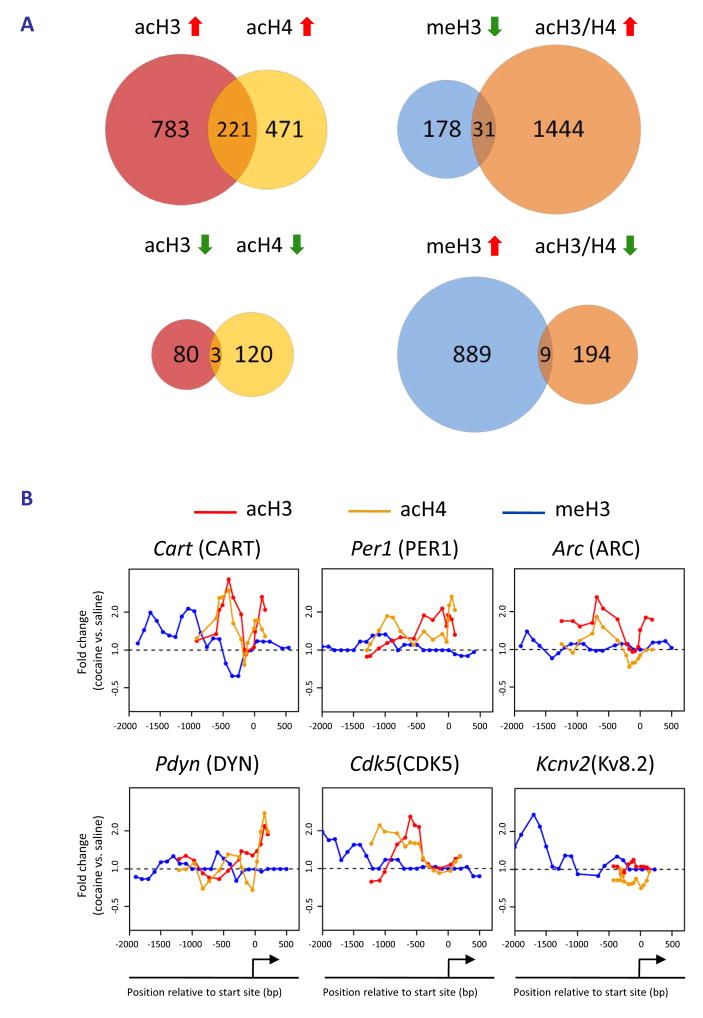
Repeated cocaine administration induced hyperacetylation of histone H3 at 959 gene promoters in NAc and hypoacetylation at 83 gene promoters compared to saline-treated controls (Fig. 1A). Similarly, chronic cocaine caused hyperacetylation of histone H4 at 692 gene promoters and hyopacetylation at 123. It is interesting to note that several fold more genes showed H3 or H4 hyperacetylation after chronic cocaine compared to hypoacetylation. This is consistent with our earlier findings, from DNA expression arrays, that the predominant effect of cocaine is gene activation (McClung and Nestler, 2003; Renthal et al., 2007), and with more recent findings that reductions in histone acetylation block cocaine’s behavioral effects while increases in acetylation augment cocaine action (Kumar et al., 2005; Renthal et al., 2007). A striking finding from the present study is that only about 15% of regulated gene promoters showed increased acetylation of both H3 and H4 (see Fig. 1A). Even less overlap was observed with genes hypoacetylated at H3 and H4 (1%). While these overlaps are statistically significant, the very small amount of overlap suggests that acetylation of one histone is sufficient to induce transcription in the NAc in vivo, consistent with cell culture data (Kurdistani et al., 2004; McCool et al., 2007). Moreover, it suggests that cocaine-induced signaling in the NAc converges on at least two mechanistically distinct pathways for histone acetylation of H3 vs. H4, which overlap sparsely. Cocaine-induced changes in H3 acetylation occurred at a larger number of genes than H4 acetylation, consistent with our earlier demonstration that chronic cocaine induces selective H3 acetylation at several candidate gene targets (FosB, Cdk5 [cyclin-dependent kinase 5], Bdnf [brain-derived neurotrophic factor]), with no induction of H4 acetylation observed (Kumar et al., 2005). However, we did observe increased H4 acetylation at several hundred gene promoters after chronic cocaine in the present study, which underscores the importance of this genome-wide investigation.
Fig. 1. Regulation of histone acetylation and methylation at gene promoters in the NAc by chronic cocaine.
A. Venn diagrams of genes that show altered levels of H3 or H4 acetylation and H3 methylation (dimethyl-K9/K27) binding 24 hrs after chronic (7 days) cocaine administration. B. Patterns of cocaine-induced changes in H3 and H4 acetylation and H3 methylation at 6 representative gene promoters previously implicated in cocaine action.
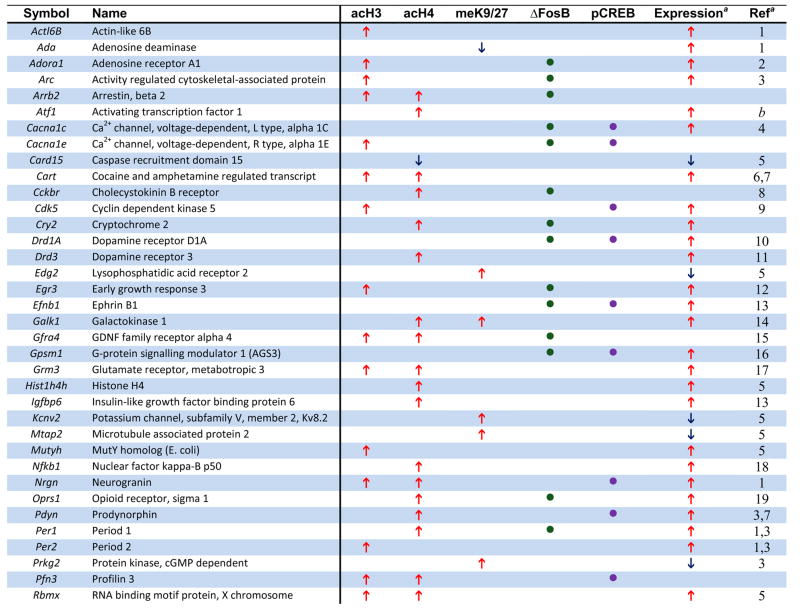
Numerous genes, shown in previous studies to be upregulated in NAc after chronic cocaine, were found in our ChIP-chip data to be associated with hyperacetylation of H3 or H4. Examples of such genes are listed in Table 1 (see Supplemental Tables S1-S3 for complete gene lists). Importantly, many of these individual genes have been directly related to aspects of neural and behavioral plasticity associated with cocaine exposure (see below). These observations provide important validation for our ChIP-chip approach and highlight the potential significance of the >1000 other genes that show similar markers of activation in this study. To experimentally determine the quality of our new ChIP-chip data sets, we performed quantitative ChIP on independent cohorts of saline and cocaine-treated mice. Analysis of 20 randomly selected target genes revealed that our ChIP-chip data have a false-positive rate of 25% – 15 out of 20 genes demonstrated statistically significant cocaine-induced changes in histone acetylation and 3 additional genes showed non-significant trends (see Supplemental Information). These findings demonstrate that our new gene lists are of excellent quality, especially given that they are derived from in vivo tissue samples of adult brain.
Table 1.
Examples of Genes that Show Chromatin Regulation by Chronic Cocaine.
 |
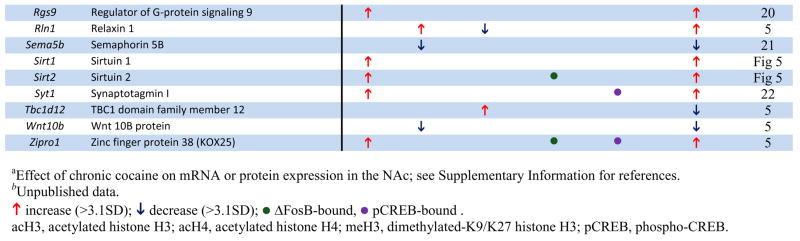 |
The genome-wide acetylation patterns of H3 and H4 show interesting features similar to previous reports in cell culture (Li et al., 2007) (Supplemental Fig. S1). Levels of acetylated H3 and H4 were maximal between −500 to +200 bp, with acetylated H3 forming a bimodal peak and acetylated H4 forming a single peak over transcription start sites. Notably, these overall patterns were not significantly altered by chronic cocaine administration, indicating that, on average, cocaine increases the amount of acetylation on specific promoters (Fig. 1A) rather than altering the spatial distribution of acetylation within promoters (Supplemental Fig. S1). Representative examples of cocaine-regulated gene promoters illustrate both prototypical and less common patterns of H3 and H4 acetylation (Fig. 1B). The gene encoding CART (cocaine- and amphetamine-regulated transcript) showed significant hyperacetylation of both H3 and H4 at an overlapping region of its promoter, each of which was very similar to the genome-wide average shown in Supplemental Fig. S1. Two other genes that are upregulated by cocaine (Fosnaugh et al., 1995; Sivam, 1989) and display acetylation patterns similar to the genome-wide average are Arc (activity regulated cytoskeletal-associated protein) and Pdyn (prodynorphin). Distinct from CART, however, the upregulation of ARC and PDYN by cocaine are associated with significant acetylation of a single histone only, H3 or H4, respectively. Per1 (period 1, a circadian gene) is also upregulated by cocaine (McClung and Nestler, 2003) and is only hyperacetylated significantly on H4, but it displays a pattern of acetylation distinct from the genome-wide average. While the acetylation peak closest to the transcription start site overlaps with the region of ΔFosB binding (Table 1 and data not shown), the significance of the more upstream peak is unknown and may represent a region of novel regulatory elements. However, most of the genes regulated by cocaine show only one peak of altered acetylation, suggesting a single regulatory region important for its cocaine-induced activity. Another illustrative gene is Cdk5, which shows selective hyperacetylation of H3 that correlates with cocaine-induced upregulation of its transcription (Bibb et al., 2001). Moreover, the selective increase in acetylated H3 at the Cdk5 promoter after chronic cocaine is consistent with our previous findings using quantitative ChIP (Kumar et al., 2005) and further supports the quality of our genome-wide study. In addition to the genes highlighted here, we identified numerous other cocaine-regulated genes that display significant alterations in histone acetylation on their promoters (see Table 1). Examples include Adcy3 (adenylyl cyclase 3), Adora1 (adenosine receptor A1), Drd3 (dopamine receptor D3), and Rgs9 (regulator of G protein signaling 9). Each of these genes has been implicated in cocaine action in the NAc (see Supplemental References for Table 1 in Supplemental Information). Taken together, a clear pattern emerges from these findings (Fig. 1, Table 1, and Supplemental Tables S1–3): cocaine-induced increases in acetylation of either H3 or H4 correlates strongly with elevated gene expression in the NAc in vivo and reveals numerous novel genes implicated in cocaine action.
Methylation of H3 at K9 and K27 in promoter regions is involved not only in silencing heterochromatin, but has also been shown to repress genes in transcriptionally active euchromatin (Kouzarides, 2007). It was, therefore, of interest to characterize this marker of gene repression in NAc after chronic cocaine. We focused on dimethyl-K9/K27, because preliminary ChIP experiments revealed that these modifications were the most dynamically regulated in the NAc by cocaine. A key advantage of this analysis is that it provides a highly novel look at genes that are largely repressed or silenced by cocaine, a relatively underexplored adaptation.
We found that chronic cocaine reduced levels of H3 dimethyl-K9/K27 at the promoter regions of 209 genes while increasing it at 898 genes in the NAc compared to saline-treated controls (Fig. 1A; see Supplemental Table S4). Interestingly, the genome-wide distribution of this methylation mark occurred on a much broader promoter span (from −1600 to +500 bp) compared to that observed for histone acetylation (Supplemental Fig. S1), similar to the pattern of histone methylation observed in cultured cell lines and yeast (Li et al., 2007). Although the genome-wide distribution did not change after chronic cocaine exposure, cocaine increased histone methylation levels at several-fold more genes than it decreased. This finding suggests that cocaine represses the expression or dampens the induction of many genes, despite the predominant effect of cocaine being gene activation (McClung and Nestler, 2003). Moreover, just as few genes showed significant changes in acetylation of both H3 and H4, there was minimal (~2%) overlap between genes that showed reduced histone acetylation and those that showed increased H3 methylation after chronic cocaine (see Fig. 1A and Supplemental Table S5). These novel findings suggest that, for the vast majority of cocaine-regulated gene promoters in the NAc, activation or repression involves independent alterations in histone acetylation or methylation.
Moreover, relatively few cocaine-regulated genes were associated with reductions of histone acetylation or methylation, suggesting that the most common mechanisms of cocaine-induced gene regulation in the NAc involve increases in histone acetylation for gene activation (rather than demethylation) or histone methylation for gene repression (rather than deacetylation). For example, the gene encoding the K+ channel subunit, Kcnv2, which is downregulated by cocaine (Renthal et al., 2007), showed increased H3 dimethyl-K9/K27 at its promoter without a change in histone acetylation, while PDYN and ARC, which are upregulated by cocaine (Fosnaugh et al., 1995; McClung and Nestler, 2003; Sivam, 1989), showed increased acetylation on their promoters without any changes in methylation (see Fig. 1B). Although increases in acetylation or methylation predominate, a subset of genes show reduced H3 acetylation or methylation after chronic cocaine exposure (e.g., the genes encoding semaphorin 5b and adenosine deaminase). Thus, hypoacetylation and hypomethylation may play an important role in gene regulation at a smaller subset of genes in response to chronic cocaine exposure.
Role of ΔFosB in the genomic effects of cocaine in the NAc
ΔFosB has been shown to play an important role in addiction: its overexpression in the NAc increases an animal’s responses to the rewarding effects of cocaine and of morphine, while overexpression of a dominant negative antagonist, termed ΔcJun, causes the opposite effects (Nestler, 2008). A previous gene expression array study identified genes in the NAc whose expression is altered upon overexpression of ΔFosB or ΔcJun (McClung and Nestler, 2003). However, this study could not provide information as to which of these genes are direct targets for ΔFosB or regulated indirectly via other mechanisms. To address this question, we carried out ChIP for ΔFosB in mice treated with chronic cocaine or saline. Since a change in the absolute level of a transcription factor on a given promoter is not necessarily an indication of its activity, we simply identified the gene promoters significantly occupied by ΔFosB under saline and cocaine-treated conditions independently. After pre-processing and normalization, we generated a high-confidence gene list at a false discovery rate of 5% using established methodology (see Supplemental Methods). In chronic cocaine-treated mice, ΔFosB was bound to nearly 40% more promoters (1553) than it was in saline-treated mice (1113) (see Supplemental Tables S6 and S7), which is consistent with the several fold induction of ΔFosB in the NAc by cocaine (Nestler, 2008).
To determine how ΔFosB binding affects the expression of its target genes in the NAc after chronic cocaine exposure, we identified which ΔFosB gene targets also showed significant cocaine-induced changes in chromatin markers of gene activation or repression (see Supplemental Table S8). Roughly 13% of the significant ΔFosB gene targets in cocaine-treated mice exhibited coincident increases in histone acetylation, which suggests that ΔFosB may be acting as a transcriptional activator at these genes (Fig. 2A, Supplemental Table S8). ΔFosB is also known to act as a transcriptional repressor at certain genes, and while very few ΔFosB-bound genes showed hypoacetylation 24 hrs after chronic cocaine (Supplemental Fig. S2), roughly 8% of ΔFosB targets showed increased histone H3K9/27 methylation (Fig. 2A). These genes may represent targets where ΔFosB acts as a repressor. While statistical analysis did not reveal a significant interaction between ΔFosB binding and histone acetylation or methylation on a global scale, the overlapping gene lists may represent the specific targets where ΔFosB affects gene expression 24 hrs after the last dose of cocaine. Since we only analyzed ΔFosB binding 24 hrs after the final cocaine dose, ΔFosB may promote chromatin and transcriptional changes at many additional gene promoters with different kinetics.
Fig. 2. Regulation of ΔFosB and phospho-CREB binding at gene promoters in the NAc by chronic cocaine.
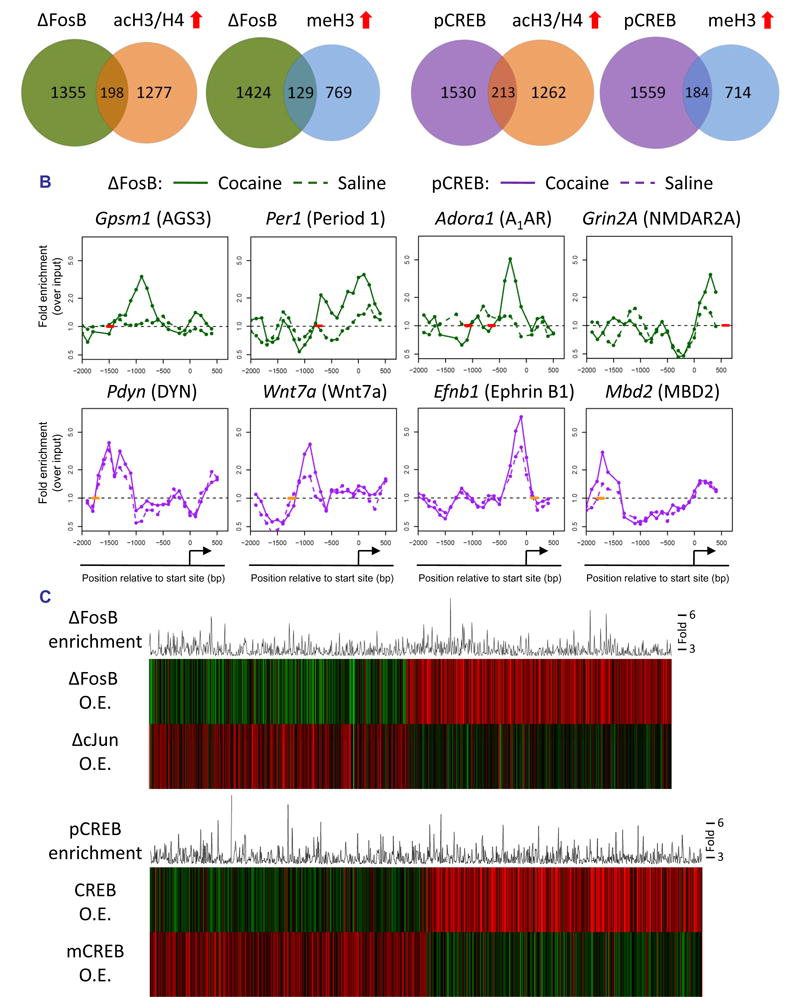
A. Venn diagrams of genes that show significant levels of ΔFosB or phospho-CREB binding, or of H3/H4 acetylation or H3 methylation, after chronic (7 days) cocaine. B. Patterns of ΔFosB (green) and phospho-CREB (purple) binding at representative gene promoters after chronic cocaine (solid line) or saline (dotted line) treatment. Short bold lines on the x-axes indicate positions of consensus or near-consensus AP1 (red) or CRE (orange) sites. C. The top panel illustrates significant ΔFosB target genes from ChIP-chip (histogram) after chronic cocaine exposure and how expression of the encoded mRNAs are regulated upon inducible overexpression of either ΔFosB or its dominant negative antagonist ΔcJun in the NAc (heatmaps) (ρ = −0.09, p = 0.005). The bottom panel illustrates significant phospho-CREB target genes from ChIP-chip (histogram) after chronic cocaine exposure and how expression of the encoded mRNAs are regulated upon inducible overexpression of either CREB or its dominant negative antagonist mCREB (heatmaps) in the NAc (ρ = −0.3, p < 1E-16).
The large majority of ΔFosB-bound genes identified here appear to be bona fide targets, since levels of their mRNA’s are significantly regulated after inducible overexpression of ΔFosB or its dominant-negative ΔcJun in the NAc (Fig. 2C), which provides important validation of our ChIP-chip data. Interestingly, increased binding of ΔFosB to gene promoters is associated with either increased or decreased mRNA expression, consistent with its complex role as both a transcriptional activator and repressor depending on the gene in question (McClung and Nestler, 2003; Renthal et al., 2008).
ΔFosB displayed significant binding to a broad region of promoters genome-wide (−1500 to +250 bp) (Supplemental Fig. S1), similar to that of H3 dimethyl-K9/K27. As with histone acetylation and methylation, chronic cocaine did not significantly affect the average position of the genome-wide pattern of ΔFosB binding. ΔFosB was found to be strongly enriched on many genes previously implicated in behavioral responses to cocaine, as well as others that now warrant further investigation. Fig. 2B illustrates ΔFosB binding across the promoters of four representative genes after chronic cocaine. For each promoter, the figure shows where ΔFosB binding is enriched relative to the gene’s transcription start site and the location of the nearest consensus AP1 (activator protein 1) site (TGA[C/G]TCA) or near-consensus site (1 bp different). One ΔFosB target gene, Gpsm1 (AGS3, activator of G-protein signaling 3) is upregulated for weeks after cocaine exposure in the NAc and has been shown to be critically involved in behavioral responses to cocaine (Bowers et al., 2004). The stability and persistence of ΔFosB during weeks of drug withdrawal further support its role as a key regulator of AGS3 transcription during this period. Examples of genes where ΔFosB is associated with promoter hyperacetylation are Grin2a (NDMAR2A subunit) (Hemby et al., 2005), Per1 (Period 1), and Adora1 (Adenosine receptor A1) (Toda et al., 2002), each of which is known to be upregulated after chronic cocaine exposure and implicated in behavioral responses to cocaine (see Table 1). Together, these data indicate that the induction of ΔFosB in the NAc by cocaine regulates numerous gene targets, which now offer a much more complete understanding of the complex mechanisms by which this transcription factor mediates sensitized drug reward.
Role of CREB in the genomic effects of cocaine in the NAc
As stated earlier, repeated cocaine administration induces CREB activity in the NAc, which then feeds back and attenuates the rewarding effects of cocaine (Carlezon et al., 2005). Cocaine activates CREB by increasing its phosphorylation at Ser133. Phosphorylation of CREB, which is often bound to responsive genes at CRE (cAMP response element) sites even in the absence of its phosphorylation, contributes to the recruitment of transcriptional co-activators such as the histone acetyltransferase, CBP (CREB binding protein), to promote gene transcription (Mayr and Montminy, 2001). Therefore, to gain insight into the transcriptional actions of CREB in the NAc after chronic cocaine, we carried out ChIP for phospho-CREB followed by promoter array assays. These data were then analyzed as described above for ΔFosB, by generating a high-confidence list of gene targets with a false discovery rate of 5%. After chronic cocaine exposure, we found that phospho-CREB was bound significantly to the promoters of 1743 genes, approximately 38% more genes than were occupied in saline-treated mice (1259) (Fig. 2A; Supplemental Tables S9 and S10). Roughly 12% of the phospho-CREB-bound genes after chronic cocaine exhibited cocaine-induced increases in histone acetylation, a marker of gene activation (Fig. 2A; Supplemental Table S11), suggesting that these genes are targets where phospho-CREB promotes transcription 24 hrs after chronic cocaine exposure. Interestingly, 10% of genes to which phospho-CREB is bound exhibit coincident increases in H3 dimethyl-K9/K27, a marker of repression, which suggests that phospho-CREB acts as a transcriptional repressor at certain genes (Mayr and Montminy, 2001). The overlapping gene lists of significant phospho-CREB binding and cocaine-induced changes in histone acetylation or methylation may represent the most likely targets where phospho-CREB alters gene activity 24 hrs after the last dose of cocaine.
Among the genes that show significant phospho-CREB binding after chronic cocaine are many that have previously been implicated in cocaine and CREB action, such as Pdyn, Fos (c-Fos), Nrgn (neurogranin), and Grin2A (Carlezon et al., 1998; Hemby et al., 2005; McClung and Nestler, 2003; Zhang et al., 2006), among other examples given in Fig. 2B, Table 1, and Supplemental Table S9. For each gene promoter illustrated in Fig. 2B, the figure shows where phospho-CREB binding is enriched relative to the gene’s transcription start site and the location of the nearest consensus CRE site (TGACGTCA) or near-consensus site (1 bp different).
As observed for ΔFosB, many gene promoters (e.g., Ephb1, Wnt7a, Mbd2) showed greater enrichment of phospho-CREB binding in cocaine-treated mice vs. saline-treated mice, which is likely related to the increased levels of phospho-CREB present in the NAc at this time point. Interestingly, however, this pattern was not observed for all phospho-CREB targets (e.g., Pdyn, c-Fos): these genes showed equivalent levels of phospho-CREB binding under cocaine-and saline-treated conditions despite higher levels of expression of these genes after cocaine. These findings suggest that changes in gene activity can be coordinated by recruitment of other cofactors, such as CBP (Levine et al., 2005; Zhang et al., 2005), to a constant level of phospho-CREB on the promoter. Moreover, on average, the genome-wide spatial pattern of phospho-CREB binding relative to gene transcription start sites, which exhibits a relatively sharp peak between −500 and +200 bp and a weaker peak beyond −500 bp up to −1500 bp (Supplemental Fig. S1), is not altered by chronic cocaine exposure.
To gain insight into how phospho-CREB binding after chronic cocaine may alter gene activity, we compared our phospho-CREB ChIP-chip data to gene expression data from mice overexpressing CREB or its dominant negative, mCREB, in the NAc (McClung and Nestler, 2003). Importantly, inducible overexpression of mCREB in the NAc oppositely regulates most of the gene expression changes observed after CREB overexpression (Fig. 2C). The high degree of overlap between phospho-CREB binding and CREB-induced changes in gene expression provides further confidence in our phospho-CREB ChIP-chip data and suggests that most genes to which phospho-CREB is bound after chronic cocaine are indeed bona fide CREB targets.
A notable feature of our prior gene expression analysis of ΔFosB and CREB overexpression in the NAc (McClung and Nestler, 2003) was the identification of a subset of genes regulated by both transcription factors. Using ChIP-chip, we found a significant 20% overlap between the cocaine-induced ΔFosB- and phospho-CREB-bound genes (Supplemental Fig. S2; Supplemental Table S12). The 20% overlap observed here is not as strong as that found in our earlier overexpression study, but this may reflect a temporal disconnect between transcription factor binding and changes in steady-state mRNA levels (e.g., co-activators are not immediately recruited) or indirect effects of CREB and ΔFosB overexpression on gene regulation. Nevertheless, many of the 323 overlapping genes, including the genes encoding AGS3, the D1 receptor, the CCKA receptor (cholecystokinin A receptor), IkBε (inhibitor of kappaB), 5-HT2AR (serotonin 2A receptor), and the NMDA2A receptor subunit (see Supplemental Table S12 for complete list) have been implicated previously in cocaine responses and likely contribute to the potent action of these two transcription factors on cocaine-induced behaviors (Carlezon et al., 2005; Nestler, 2008). The list of overlapping ΔFosB and CREB target genes generated here offers a rich array of genes that influence cocaine action at the level of the NAc.
Discovery of a novel pathway involved in cocaine action: Studies of SIRT1 and SIRT2
Thus far, our discussion has highlighted examples of known cocaine targets that show altered histone modifications or ΔFosB or phospho-CREB binding in our ChIP-chip studies. However, the arrays also provide rich lists of many novel genes, not heretofore implicated in cocaine action, which show robust and highly significant markers of activation or repression in response to chronic cocaine. It was, therefore, important to evaluate the predictive power of these gene lists for revealing fundamentally new insight into the molecular pathophysiology of cocaine action in the NAc. One gene family was of particular interest: the sirtuins. Sirtuins–also referred to as SIRT’s (silent information regulator of transcription)–are categorized as Class III NAD-dependent histone deacetylases, which in addition to deacetylating histones also deacetylate other cellular proteins, such as tubulin, p53 (a tumor suppressor transcription factor), and NFκB (nuclear factor kappa B) (Denu, 2005). Sirtuins are highly conserved from bacteria to mammals–7 forms have been identified in mouse and human–and have been implicated in diverse processes, including cell morphology, growth, apoptosis, general metabolism, and aging (Michan and Sinclair, 2007), although very little is known about their function in the nervous system.
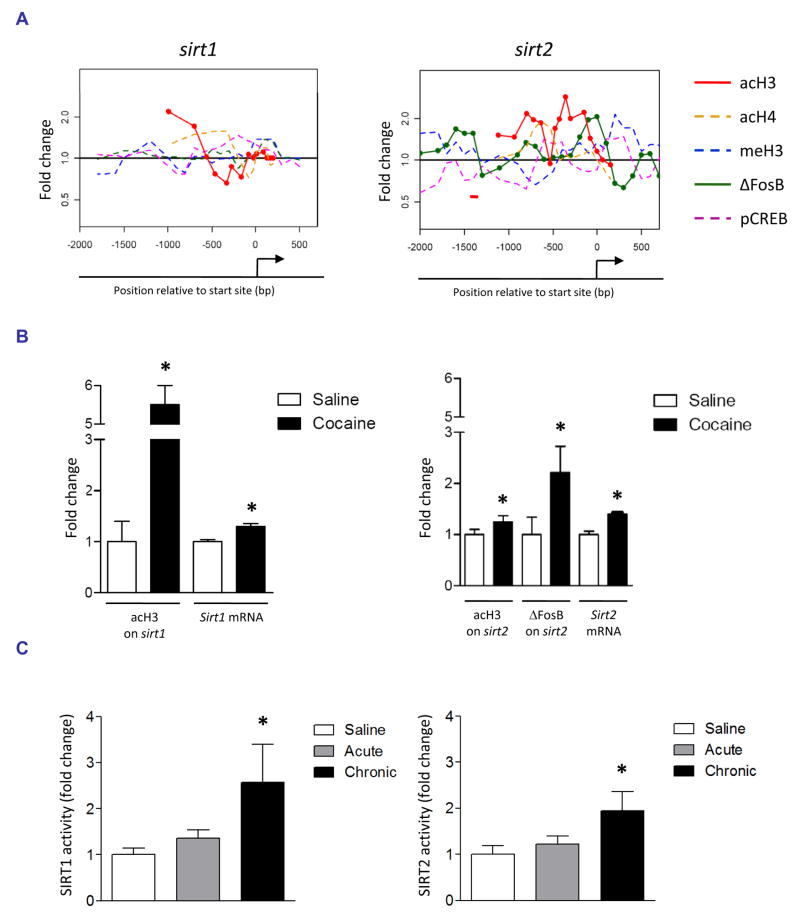
Our ChIP-chip studies identified significant enrichment of ΔFosB on the Sirt2 promoter after chronic cocaine exposure, indicating a potential role in cocaine responses (Fig. 3A, Table 1). After analyzing the promoter of Sirt2 in more detail, we noticed that there was also a strong increase in acetylated H3 downstream of the AP1/ΔFosB-binding site that we had overlooked because it fell just below the 3.1 SD cutoff. In an independent cohort of mice, we confirmed that chronic cocaine significantly increases levels of ΔFosB and acetylated H3 on the Sirt2 promoter (Fig. 3B) and that this is associated with increased Sirt2 mRNA expression in the NAc (Fig. 3B). Moreover, we found significant cocaine-induced H3 acetylation of a related sirtuin, Sirt1, in this brain region, a finding which we validated in an independent ChIP experiment and showed is associated with a significant increase in Sirt1 mRNA levels (Fig. 3A, 3B). To determine whether the increases observed in histone acetylation, ΔFosB-binding, and transcription of Sirt1 and Sirt2 are associated with changes in functional protein, we incubated cocaine-treated NAc lysates with a fluorescent substrate of SIRT1 or SIRT2. Consistent with the ChIP-chip, qChIP, and mRNA data, we found that chronic cocaine significantly increases both SIRT1 and SIRT2 catalytic activity in the NAc (Fig. 3C). Acute exposure to cocaine does not alter SIRT1 or SIRT2 activity, suggesting that the upregulation of sirtuins in NAc may contribute to the chronic neuroadaptations involved in drug addiction. Together, these data illustrate the predictive quality of our ChIP-chip analyses and suggest that our stringent cutoff of 3.1 SD may even underestimate the number of cocaine-regulated promoters.
Fig. 3. Validation of sirtuins as a novel target for cocaine in the NAc.
A. Changes in histone H3 and H4 acetylation, H3 K9/K27 methylation, and ΔFosB and phospho-CREB binding at the Sirt1 and Sirt2 gene promoters in the NAc after chronic (7 days) cocaine. A short red bold line along the x-axis indicates the position of an AP1 site. Significant changes are shown as solid lines. B. Quantitative ChIP confirmed cocaine-induced increases in H3 acetylation at the Sirt1 (left) and Sirt2 (right) gene promoters in an independent cohort of mice (p < 0.05, n = 3–6). Cocaine-induced ΔFosB binding was also confirmed for the Sirt2 promoter (p < 0.05, n = 3–6). This chromatin regulation is associated with significant increases in Sirt1 and Sirt2 (p < 0.05, n = 7–8) mRNA levels in the NAc. C. As well, SIRT1 and SIRT2 catalytic activity was significantly increased in NAc after chronic cocaine administration (p < 0.05, n = 7–8).
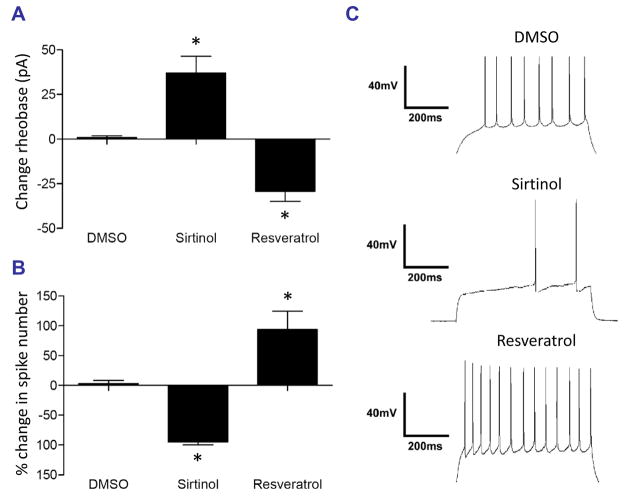
The cocaine-induced upregulation of SIRT1 and SIRT2 in the NAc prompted us to examine how these enzymes affect the physiological activity of NAc neurons. To do this, we performed whole-cell current-clamp recordings of medium spiny neurons (MSNs) in acute NAc slices incubated with a pharmacological inhibitor (sirtinol) or activator (resveratrol) of sirtuins. We found that 30 μM sirtinol nearly silenced NAc neurons, reducing the number of spikes elicited by 100 pA current injection by ~90% compared to vehicle (0.1% DMSO). Moreover, the minimum amount of current needed to elicit an action potential (rheobase) was significantly higher in sirtinol-treated slices (Fig. 4A), consistent with a decrease in NAc MSN excitability (Fig. 4B). Conversely, the sirtuin activator resveratrol (50 μM) potently increased NAc MSN excitability while reducing rheobase, suggesting that cocaine-induced increases in SIRT1 and SIRT2 activity may be associated with increased NAc excitability. Individual traces are shown in Fig. 4C, which illustrate the dramatic effects of inhibiting or activating sirtuins on the functional activity of NAc MSN neurons.
Fig. 4. Sirtuin regulates the electrical excitability of NAc neurons.
A. Incubation (20 min) of acute NAc slices from adult mice with the sirtuin inhibitor, sirtinol (30 μM), caused a significantly higher rheobase compared to control (DMSO-treated) slices (ANOVA: F (2,13) = 24.64, p < 0.0001, Tukey’s post-hoc compared to DMSO, *p < 0.05). Conversely, slices incubated with 50 μM resveratrol, a sirtuin activator, exhibited a significant reduction in rheobase (*p < 0.05). B. A 100 pA injection into NAc neurons incubated with sirtinol (30 μM) elicits significantly fewer action potentials compared to control, while incubation with resveratrol (50 μM) results in significantly more firing than controls (ANOVA: F (2, 13) = 25.38, p< 0.0001, Tukey’s post-hoc compared to DMSO, *p < 0.05). C. Example traces from DMSO-, sirtinol-, and resveratrol-treated slices illustrate the robust physiological effects of manipulating sirtuins on NAc neurons.
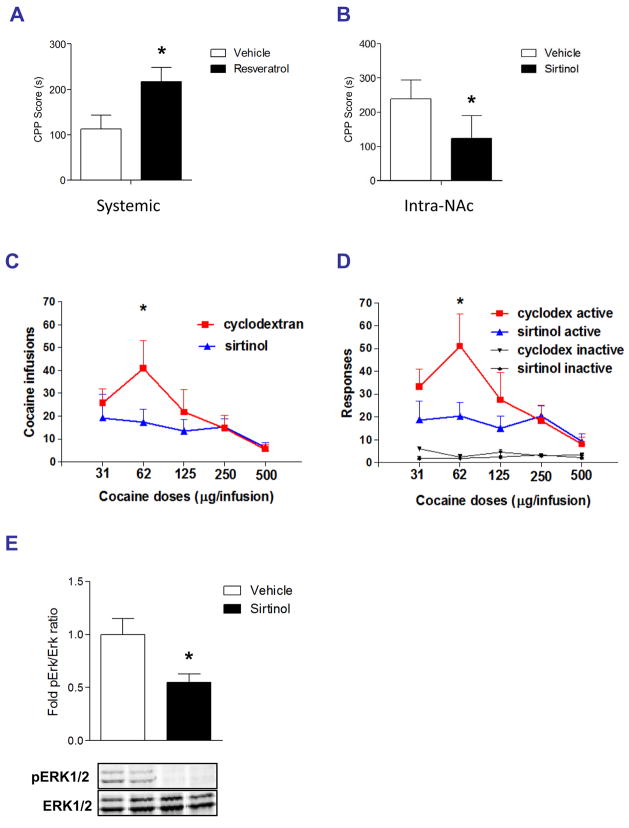
We next tested directly whether elevated sirtuin activity in the NAc is involved in behavioral responses to cocaine. To do this, we used resveratrol and sirtinol in the conditioned place preference assay, where animals learn to prefer a cocaine-paired environment. We administered resveratrol systemically since this drug is known to penetrate the brain after systemic administration (Baur and Sinclair, 2006), and sirtinol directly into the NAc using an osmotic minipump connected to guide cannulae. Similar to our electrophysiological observations, resveratrol and sirtinol exerted opposite effects on cocaine reward, with resveratrol significantly enhancing cocaine place conditioning and sirtinol significantly attenuating it (Figs. 5A, 5B).
Fig. 5. Sirtuins regulate behavioral responses to cocaine.
A. Systemic administration of the sirtuin agonist, resveratrol (20 mg/kg ip, dissolved in 5% hydroxypropyl β-cyclodextrin vehicle), increases the rewarding effects of cocaine (5 mg/kg) in the conditioned place preference paradigm (p < 0.05, n = 9–12). B. Intra-NAc delivery of the sirtuin antagonist, sirtinol (50 μM in 5% hydroxypropyl β-cyclodextrin), decreases the rewarding effects of 10 mg/kg cocaine (right). Data are expressed as mean ± s.e.m. (n = 9–12 in each group), *p < 0.05 by t-test. C. Intra-NAc delivery of sirtinol (100 μM) in rats that were trained to self-administer cocaine significantly reduced the number of cocaine infusions at the threshold dose of 62 μg/infusion (*p < 0.05, n = 5–7). D. The sirtinol-induced decrease in cocaine self-administration was specific to the active (cocaine-associated) nose poke apertures, as they behaved normally at the inactive apertures. E. Sirtinol significantly reduces ERK1/2 phosphorylation under depolarizing conditions in acute NAc slices ex vivo (*p < 0.05, n = 4).
While these data directly implicate sirtuins in regulating the rewarding effects of cocaine, cocaine addiction is marked by motivation to take drug, which is best assayed in the self-administration paradigm. Thus, we delivered sirtinol or vehicle via osmotic minipump directly into the NAc of rats, which were subsequently trained to self-administer cocaine. Sirtinol had no effect on the acquisition phase of cocaine self-administration (data not shown). However, while vehicle-infused rats displayed normal cocaine-seeking behavior throughout the ascending and descending limbs of the cocaine dose-response curve, sirtinol-infused rats displayed a dramatic reduction in cocaine self-administration (Fig. 5C). Sirtinol did not affect responding on the inactive nose poke aperture, which provides an important control for the lack of effect of sirtinol on general locomotor responses (Fig. 5D). These findings demonstrate that sirtuin activity in the NAc is an essential regulator of cocaine self-administration behavior, and further highlight the role played by this novel biochemical pathway in cocaine action.
Inhibition of Class I and II HDAC’s in the NAc has been shown to increase behavioral sensitivity to cocaine (Kumar et al., 2005; Renthal et al., 2007), while we show here that inhibition of Class III HDAC’s (sirtuins) reduces it. Given the role of sirtuins in regulating numerous cytosolic substrates apart from histone proteins, it is possible that these downstream effectors are more important contributors than histone deacetylation to sirtuin regulation of cocaine action. Indeed, it was recently shown in primary neuronal cultures that SIRT1 is required for normal activation of ERK, a protein well known to exert potent control over behavioral responses to cocaine (Li et al., 2008). To determine if ERK activation requires SIRT activity in the adult NAc, we used ex vivo slice pharmacology to bathe NAc slices in a depolarizing buffer (to induce ERK activation) plus either sirtinol or vehicle. We observed a significant reduction in ERK1/2 phosphorylation in the sirtinol-treated NAc while total ERK1/2 levels remained unchanged (Fig. 5E). These findings demonstrate that sirtinol inhibits ERK activation in the NAc, which may be one mechanism by which sirtinol antagonizes NAc excitability as well as cocaine reward and self-administration.
Together, these several lines of evidence demonstrate the ability to translate ChIP-chip data of chromatin and transcriptional regulation by cocaine to identify new molecular pathways involved in cocaine addiction.
Molecular pathway analysis of the “cocaine transcriptome”
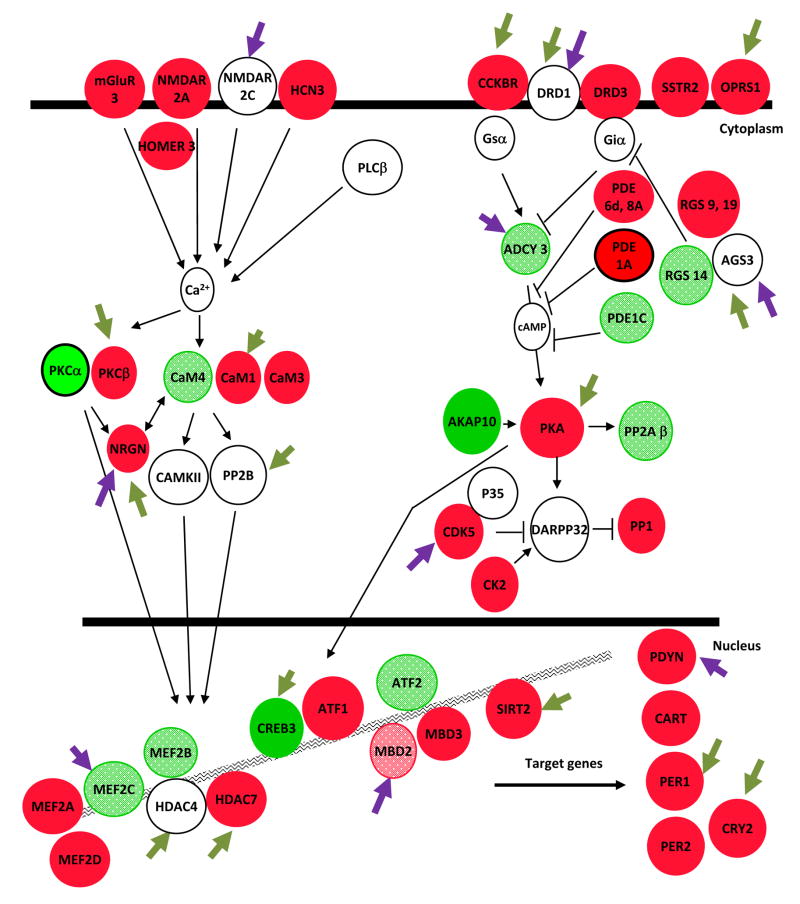
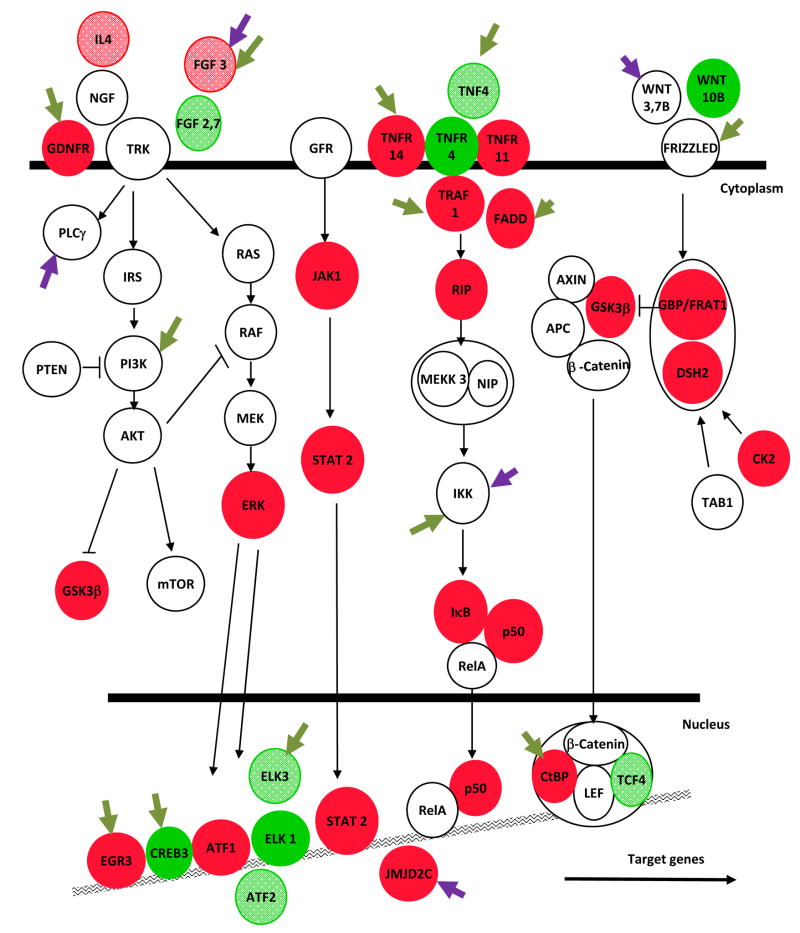
To gain a more global view of the genes regulated by cocaine in the NAc, we subjected our gene sets to Ingenuity molecular pathway analysis software (see Supplemental Fig. S3). The most significantly regulated molecular pathways based on altered histone acetylation were cAMP signaling, long term potentiation or depression, RXR signaling, and neurotrophin signaling (e.g., ERK/MAP kinase and PTEN/PI-3-kinase/Akt) pathways. Each of these pathways is known to play integral roles in the cellular and behavioral responses to chronic cocaine, and their enrichment in our analyses supports the predictive quality of this study. Notably, chronic cocaine also regulated several glutamate receptors and downstream calcium signaling molecules, including subunits of NMDA and AMPA receptors, metabotropic receptors, and HOMER3. The upregulation of glutamate signaling is consistent with previous reports, which demonstrate that chronic cocaine increases the sensitivity of NAc neurons to glutamate (Kalivas, 2004). Examples of these highly regulated pathways are shown in Figs. 6A and 6B. The panels illustrate the profound and complex effects of cocaine on second messenger- and growth factor-regulated molecular pathways in the NAc, and how the individual genes in these pathways are influenced by alterations in histone acetylation or methylation. The panels also identify which proteins in these pathways show significant ΔFosB or phospho-CREB binding at their gene promoters, again illustrating the important influence of these transcription factors in cocaine action.
Fig. 6. Molecular pathway analysis of the genomic effects of cocaine in the NAc.
Chronic cocaine-induced molecular changes in the NAc were identified by ChIP-chip for changes in acetylated H3 and H4, H3 dimethyl-K9/K27, ΔFosB, and phospho-CREB binding followed by rigorous statistical analysis (>3.1SD) and Ingenuity molecular pathway examination. The Key defines the different types of regulation shown in the figure. Alterations in second messenger (A) and growth factor (B) regulated pathways are shown. Red indicates modifications associated with gene activation (increased histone acetylation or decreased methylation); bright green, gene repression (decreased histone acetylation or increased methylation). Dark green arrows indicate genes that show significant alterations in ΔFosB binding, purple arrows phospho-CREB binding. See Supplemental Information for definitions of the gene abbreviations used in the figure.
Among the molecular pathways regulated by cocaine are several not heretofore appreciated in cocaine action. In addition to the sirtuins discussed above, examples include protein ubiquitination, Toll-like receptor signaling, and FGF signaling. Indeed, protein ubiquitination signaling regulates dendritic spine morphology and neuronal function (Pak and Sheng, 2003), and NAC-1, a cocaine-induced gene, can shuttle the proteasome and a ubiquitin ligase into spines in response to neural activity (Shen et al., 2007). While NAC-1 was not regulated at the time point used in our ChIP-chip analyses, many ubiquitin signaling molecules were regulated in response to cocaine and, like NAC-1, now warrant further investigation.
In addition to identifying novel pathways involved in cocaine action, we have gained new insight into the layers of complex regulation that control these pathways. For most of the pathways we investigated, chronic cocaine promotes the expression of an intricate network of activating and inhibiting molecules. This is surprising because we know that many of these same pathways are functionally upregulated several fold after chronic cocaine. For example, in the NFκB pathway, there is increased acetylation on the genes coding both the transcriptional activator, p50, and its inhibitor, IκB (see Fig. 6B). However, we know that NFκB activity is upregulated in the NAc after chronic cocaine (Ang et al., 2001; Russo et al., 2009), suggesting that the increased acetylation on IκB may serve to limit the primary activation of p50 and its upstream activators, TNFR and TRAF1. Similar complex regulation is seen for the dopamine-cAMP pathway. Cocaine functionally upregulates cAMP signaling in NAc (Carlezon et al., 2005), but simultaneously increases histone acetylation on the genes for the Gi-coupled D3 receptor and an inhibitor of Gi-signaling, RGS9. The NFκB and cAMP pathways exemplify the intricate cocaine-induced regulation of intracellular signaling in the NAc, and thereby underscore the importance of genome-wide studies which permit the parallel assessment of multiple molecular components of each pathway.
Discussion
This study provides a genome-wide assessment of chromatin and transcriptional alterations in the NAc in response to repeated cocaine administration. Among the many genes that show regulation by cocaine are a large number reported in previous studies to show altered mRNA or protein expression in the NAc after chronic cocaine (see examples in Table 1). For instance, the cocaine-induced genes that encode ARC, CART, CDK5, NFκB, PDYN, σ-opioid receptor, and Period 1 and 2 (Freeman et al., 2001; McClung and Nestler, 2003; Yao et al., 2004) (see also Supplemental References in Supplemental Information) were shown in the present study to exhibit increased binding of acetylated H3 or H4, with either attenuated or unchanged methylation of H3 at K9/K27. Genes that are known to be downregulated by cocaine, such as the voltage-gated potassium channel Kv8.2 and the microtubule associated protein MTAP2 (Renthal et al., 2007), were often associated with increased methylation of H3 at K9/K27 (Table 1). For each type of histone modification, cocaine induced increases in acetylation or methylation at many more genes than it induced decreases (Fig. 1A). Although there are a few examples where reductions in acetylation or methylation are associated with respective changes in gene expression, our data suggest that chronic cocaine more commonly regulates transcription by either increasing histone H3 or H4 acetylation (to elevate mRNA levels), or by increasing histone H3 dimethyl-K9/27 (to reduce mRNA expression). However, these are observations of genome-wide data and exceptions likely exist. There is also a subset of genes which is highly regulated at the chromatin level but show no detectible change in steady-state mRNA expression. For example, mRNA’s for HDAC4 and myocyte-enhancer factors 2A and 2D (MEF2A and MEF2D) are not altered by cocaine (data not shown), but their promoters are dramatically altered after cocaine treatment by histone modifications and/or transcription factor binding. These are particularly interesting examples given the potent influence of HDAC4 (Kumar et al., 2005) and of MEF2 (Pulipparacharuvil et al., 2008) in the NAc on cocaine responses, and may illustrate a new layer of regulation not previously appreciated. Similar disconnects between gene activity and gene or protein expression have been observed during cocaine withdrawal (Self et al., 2004). Bdnf is a good example where hyperacetylation of its promoter (Kumar et al., 2005) does not correlate with an immediate increase in steady state BDNF expression, however, during cocaine withdrawal, levels of BDNF protein become significantly elevated (Grimm et al., 2003). Similarly, gene expression arrays found increased MEF2D expression in the NAc after extinction from cocaine self-administration (personal communication, D.W. Self). Thus, histone acetylation at certain genes may represent a priming mechanism to facilitate subsequent gene induction. Taken together, our study corroborates numerous established molecular targets of cocaine action in the NAc, and demonstrates the power of ChIP-chip assays to uncover in a comprehensive manner the genomic targets through which cocaine induces neural and behavioral plasticity in this critical brain reward region. A still further advance would be to determine whether these various cocaine-induced changes in NAc occur in neurons vs. glia (Bowers and Kalivas, 2003).
Likewise, results of the present study provide novel insight into the target genes through which ΔFosB and CREB contribute to the genomic effects of cocaine. Several genes previously identified as targets of ΔFosB and CREB, inferred from gene expression array studies of mice overexpressing the transcription factors or their dominant negative antagonists in the NAc (McClung and Nestler, 2003), were identified in this study. Examples include neurogranin, Period 1, GABAA receptor subunits, and MEF2C, to name a few. The present findings thereby indicate that many of the ΔFosB and CREB target genes determined through overexpression studies are indeed direct, physiological targets for these transcription factors in the NAc in vivo. Although to the best of our knowledge there have been no prior genome-wide ChIP-chip studies for FosB or ΔFosB, we should note that many of the phospho-CREB-bound genes observed here after chronic cocaine exposure (e.g., Pdyn, c-Fos, Mef2c, Cdk5) were also identified as CREB targets by previous CREB ChIP-chip studies (Impey et al., 2004; Tanis et al., 2007; Zhang et al., 2005). While it is known that CREB targets can differ between cell types (Cha-Molstad et al., 2004; Zhang et al., 2005), observing many of the same genes regulated by CREB from cultured cells to brain gives us high confidence in the predictive quality of our data.
Of all the genes in the NAc that show markers of activation or repression after chronic cocaine, roughly 14% exhibit altered levels of ΔFosB binding and roughly 16% exhibit altered levels of phospho-CREB binding (Fig. 2A; Supplemental Fig. S2; Supplemental Tables S8, S11). This is interesting in that our previous DNA expression array study found that similar fractions of genes regulated in the NAc after 5 days of cocaine were also regulated upon ΔFosB overexpression or CREB overexpression, respectively (McClung and Nestler, 2003). The number of cocaine-regulated genes influenced by ΔFosB increased to >25%, whereas that for CREB decreased to 5%, after 4 weeks of cocaine administration, which illustrates the importance of performing genome-wide ChIP-chip and gene expression studies after longer periods of cocaine exposure and different time points of cocaine withdrawal.
As stated, the dramatic cell type differences that have been reported for the genomic targets of a given transcription factor between even two types of cultured cells means that an absolutely crucial next step in the field is to define modes of chromatin regulation that occur in the brain in vivo. Indeed, well beyond identifying lists of genes that show interesting patterns of chromatin regulation by cocaine, results of the present study reveal several novel principles by which cocaine regulates gene expression in the NAc of behaving animals. Among the lessons revealed are that most cocaine-regulated genes show altered acetylation either of histone H3 or of H4, with changes at H3 predominating, and alterations either in histone acetylation or in histone methylation, but only rarely both modifications together on the same gene. Another striking lesson is that phospho-CREB exerts complex transcriptional effects and can act as a transcriptional activator or repressor in the brain in vivo. This is also true for ΔFosB, which has been shown previously (Kumar et al., 2005; McClung and Nestler, 2003; Renthal et al., 2008) and observed in our ChIP-chip data to act as either a transcriptional activator or repressor depending on the gene promoter and conditions involved.
There have been numerous studies of cocaine regulation of gene expression in the NAc and other brain regions by gene expression arrays. This research has revealed large numbers of transcripts that are altered in response to cocaine administration. The ChIP-chip studies reported here enable the coordinated use of both approaches to identify a smaller set of genes in which the field can place greater confidence as being bona fide targets of cocaine and the key transcription factors which mediate cocaine’s effects. Moreover, this work begins to describe the specific mechanisms underlying these cocaine-induced transcriptional changes and reveals fundamentally new insight into the genome-wide patterns of chromatin regulation by cocaine in the NAc. Together, this new insight has led to the identification of a novel family of genes involved in cocaine action in the NAc, the sirtuins, which, as we have shown, play an essential role in addiction-like behavior.
We identified Sirt1 and Sirt2 from our ChIP-chip analyses of ΔFosB target genes that also were regulated by histone acetylation. We then identified significant increases in both Sirt1 and Sirt2 mRNA and protein activity in the NAc after chronic cocaine administration. We showed further that elevated sirtuin activity in the NAc increases the electrical excitability of NAc MSNs and potentiates the rewarding effects of cocaine. Finally, we demonstrated that pharmacological inhibition of sirtuins specifically in the NAc reduced both cocaine’s rewarding effects as well as the motivation to self-administer the drug. Thus, sirtuins appear to act downstream of ΔFosB and may contribute to a positive-feedback loop in which repeated drug exposure increases levels of ΔFosB and sirtuins, which in turn enhances the motivation to take additional drug. These findings raise the possibility of using SIRT1/2 inhibitors as potential treatment agents for cocaine addiction.
Taken together, our ChIP-chip analyses have revealed a novel family of proteins involved in cocaine responses and underscore the vast clinical potential of the many other new gene targets identified in this study for the development of more effective treatments of cocaine and potentially other drug addictions.
Methods
Animals and reagents
Male C57/BL/6 mice 10–12 weeks old were obtained from Jackson Laboratory and housed on a 12-hour light-dark cycle with access to food and water ad libitum. Mice were injected i.p. with cocaine (20 mg/kg, Sigma) or saline once per day for 7 days. 24 hrs after the last dose, NAc was punch dissected from mice as described previously (Kumar et al., 2005).
Chromatin immunoprecipitation and microarray analysis
ChIP was performed for acetylated histone H3 and H4, dimethyl-K9/K27 H3, ΔFosB, and phospho-CREB as described previously (Kumar et al., 2005) with minor modifications. Immunoprecipitated DNA was amplified via ligation-mediated PCR and hybridized to NimbleGen mouse MM8 promoter arrays. See Supplemental Information for detailed ChIP-chip methods. Quantitative ChIP was performed as described above except that the immunoprecitated DNA was directly quantified by quantitative real-time PCR.
RT-PCR
NAc punches were homogenized in Trizol (Invitrogen, Carlsbad, CA) and processed according to the manufacturer’s instructions. Purified RNA was reverse transcribed using Bio-Rad iScript Kit (Hercules, CA). cDNA was quantified with real-time PCR using SYBR Green (Applied Biosystems, Foster City, CA). Each reaction was run in triplicate and analyzed using the ΔΔCt method as described previously (Tsankova et al., 2006).
Electrophysiological and behavioral analyses
Detailed methods for characterizing the effect of a sirtuin inhibitor and activator on the electrical excitability of NAc neurons and on behavioral responses to cocaine are provided in Supplemental Methods in Supplemental Information.
Supplementary Material
Acknowledgments
We would like to thank Thomas Green, Shibani Mukherjee, Kwang-Ho Choi, and Colleen McClung for thoughtful discussions, Hoang-Trang Truong and Geetha Kalahasti for technical assistance, and Shaday Michan and David Sinclair for reagents and discussions. This work was supported by grants from NIDA.
Footnotes
Conflict of interest statement: The authors report no conflicting interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ang E, Chen J, Zagouras P, Magna H, Holland J, Schaeffer E, Nestler EJ. Induction of nuclear factor-kappaB in nucleus accumbens by chronic cocaine administration. J Neurochem. 2001;79:221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, Yan Z, Sagawa ZK, Ouimet CC, Nairn AC, et al. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- Bowers MS, Kalivas PW. Forebrain astroglial plasticity is induced following withdrawal from repeated cocaine administration. Eur J Neurosci. 2003;17:1273–1278. doi: 10.1046/j.1460-9568.2003.02537.x. [DOI] [PubMed] [Google Scholar]
- Bowers MS, McFarland K, Lake RW, Peterson YK, Lapish CC, Gregory ML, Lanier SM, Kalivas PW. Activator of G protein signaling 3: a gatekeeper of cocaine sensitization and drug seeking. Neuron. 2004;42:269–281. doi: 10.1016/s0896-6273(04)00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, Duman RS, Neve RL, Nestler EJ. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc Natl Acad Sci U S A. 2004;101:13572–13577. doi: 10.1073/pnas.0405587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby CR, Whisler K, Steffen C, Nestler EJ, Self DW. Striatal cell type-specific overexpression of DeltaFosB enhances incentive for cocaine. J Neurosci. 2003;23:2488–2493. doi: 10.1523/JNEUROSCI.23-06-02488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Fosnaugh JS, Bhat RV, Yamagata K, Worley PF, Baraban JM. Activation of arc, a putative “effector” immediate early gene, by cocaine in rat brain. J Neurochem. 1995;64:2377–2380. doi: 10.1046/j.1471-4159.1995.64052377.x. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Nader MA, Nader SH, Robertson DJ, Gioia L, Mitchell SM, Daunais JB, Porrino LJ, Friedman DP, Vrana KE. Chronic cocaine-mediated changes in non-human primate nucleus accumbens gene expression. J Neurochem. 2001;77:542–549. doi: 10.1046/j.1471-4159.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J Neurosci. 2003;23:742–747. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemby SE, Horman B, Tang W. Differential regulation of ionotropic glutamate receptor subunits following cocaine self-administration. Brain Res. 2005;1064:75–82. doi: 10.1016/j.brainres.2005.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Brown JR, Haile CN, Ye H, Greenberg ME, Nestler EJ. FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine’s psychomotor and rewarding effects. Proc Natl Acad Sci U S A. 1997;94:10397–10402. doi: 10.1073/pnas.94.19.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Kalivas PW. Glutamate systems in cocaine addiction. Curr Opin Pharmacol. 2004;4:23–29. doi: 10.1016/j.coph.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Jr, Whisler K, Gilden L, Beckmann AM, Steffen C, Zhang YJ, Marotti L, Self DW, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci U S A. 2005;102:19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS- 2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation- dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- McCool KW, Xu X, Singer DB, Murdoch FE, Fritsch MK. The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J Biol Chem. 2007;282:6696–6706. doi: 10.1074/jbc.M609519200. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Transcriptional mechanisms of addiction: role of deltaFosB. Philos Trans R Soc London, B Biol Sci. 2008;363:3245–3255. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, 3rd, Truong HT, Alibhai I, Kumar A, Montgomery RL, Olson EN, Nestler EJ. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Wilkinson MB, Mazei-Robison M, Dietz DM, Maze I, Krishnan V, Renthal W, Graham A, Birnbaum S, Green TA, et al. NF-kB signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29:3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self DW, Choi KH, Simmons D, Walker JR, Smagula CS. Extinction training regulates neuroadaptive responses to withdrawal from chronic cocaine self-administration. Learning and Memory. 2004;11:648–657. doi: 10.1101/lm.81404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Korutla L, Champtiaux N, Toda S, LaLumiere R, Vallone J, Klugmann M, Blendy JA, Mackler SA, Kalivas PW. NAC1 regulates the recruitment of the proteasome complex into dendritic spines. J Neurosci. 2007;27:8903–8913. doi: 10.1523/JNEUROSCI.1571-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder D, Johnston SA, Kodadek T. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem. 2006;281:27346–27355. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- Sivam SP. Cocaine selectively increases striatonigral dynorphin levels by a dopaminergic mechanism. J Pharmacol Exp Ther. 1989;250:818–824. [PubMed] [Google Scholar]
- Tanis KQ, Duman RS, Newton SS. CREB Binding and Activity in Brain: Regional Specificity and Induction by Electroconvulsive Seizure. Biol Psychiatry. 2007 doi: 10.1016/j.biopsych.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S, McGinty JF, Kalivas PW. Repeated cocaine administration alters the expression of genes in corticolimbic circuitry after a 3-week withdrawal: a DNA macroarray study. J Neurochem. 2002;82:1290–1299. doi: 10.1046/j.1471-4159.2002.01083.x. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–638. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang L, Jiao H, Zhang Q, Zhang D, Lou D, Katz JL, Xu M. c- Fos facilitates the acquisition and extinction of cocaine-induced persistent changes. J Neurosci. 2006;26:13287–13296. doi: 10.1523/JNEUROSCI.3795-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.