Abstract
Plants have developed their own defense strategies because they have no immune cells. A common plant defense strategy involves programmed cell death (PCD) at the infection site, but how the PCD-associated cell-autonomous immunity is executed in plants is not fully understood. Here we provide a novel mechanism underlying cell-autonomous immunity, which involves the fusion of membranes of a large central vacuole with the plasma membrane, resulting in the discharge of vacuolar antibacterial proteins to the outside of the cells, where bacteria proliferate. The extracellular fluid that was discharged from the vacuoles of infected leaves had both antibacterial activity and cell death-inducing activity. We found that a defect in proteasome function abolished the membrane fusion associated with both disease resistance and PCD in response to avirulent bacterial strains but not to a virulent strain. Furthermore, RNAi plants with a defective proteasome subunit PBA1 have reduced DEVDase activity, which is an activity associated with caspase-3, one of the executors of animal apoptosis. The plant counterpart of caspase-3 has not yet been identified. Our results suggest that PBA1 acts as a plant caspase-3-like enzyme. Thus, this novel defense strategy through proteasome-regulating membrane fusion of the vacuolar and plasma membranes provides plants with a mechanism for attacking intercellular bacterial pathogens.
Keywords: Membrane fusion, proteasome, cell-autonomous immunity, hypersensitive response, caspase activity, programmed cell death
Plants, unlike mammals, lack mobile immune cells. Instead, they have evolved a cell-autonomous immune system. Plants use different defense strategies to defend against different pathogens (Jones and Dangl 2006). One of these strategies, the hypersensitive response, confers broad-spectrum disease resistance in plants. The hypersensitive response is often accompanied by rapid and localized programmed cell death (PCD) at the infection site to prevent the growth and spread of pathogens into healthy tissues. This type of PCD is called hypersensitive cell death. Hypersensitive cell death is initiated by direct or indirect recognition of a pathogen avirulence (Avr) factor by a plant resistance (R) gene product and is controlled by multiple signal transduction pathways (Dangl and Jones 2001). Although the components of the signaling pathways that lead to cell death activation have been well-documented (Yang et al. 1997; Jones and Dangl 2006), little is known about how cell death is executed or whether plants share cell death mechanisms with animals.
Many studies have shown that activities similar to those of caspase-1 and caspase-3, the executors of animal apoptosis, are required for hypersensitive cell death of plants (for review, see Lam and del Pozo 2000; Woltering 2004; Bonneau et al. 2008). However, plants have no genes that are homologous to caspase-1 and caspase-3. Despite much effort, attempts to identify the proteinase with caspase-3-like activity have been unsuccessful. Here we provide evidence that the proteasome subunit PBA1 has DEVDase activity, an activity associated with caspase-3, and that this activity is responsible for bacterially induced hypersensitive cell death.
The proteasome system has a major role in the regulation of animal apoptosis (Orlowski 1999). However, there is not yet any direct evidence that the proteasome itself is involved in cell death and disease resistance in plants. In recent years, a combination of genetic screens and gene silencing techniques has identified several proteins with E3 ubiquitin ligase activity that play a role in defense signaling, indicating that ubiquitination is important for resistance of plants to pathogens (Dreher and Callis 2007; Craig et al. 2009). The importance of the ubiquitination pathway in plant defense signaling was revealed by the discovery that SGT1 protein regulates several R gene-mediated defense pathways (Austin et al. 2002; Azevedo et al. 2006). SGT1b can associate with SCF-type E3 ligase (Azevedo et al. 2002; Liu et al. 2002) and appears to promote SCF-mediated degradation of E3 ligase substrates in budding yeast (Kitagawa et al. 1999) and in the Arabidopsis auxin response pathway (Gray et al. 2003). Although polyubiquitination is primarily associated with proteasome-mediated function, it also has other functions not associated with proteasomes (Mukhopadhyay and Riezman 2007). However, it remains unclear whether the proteasome is involved in plant immunity.
We previously identified a vacuolar processing enzyme (VPE) as a “plant caspase-1” that is responsible for virus-induced hypersensitive cell death (Hatsugai et al. 2004; Kuroyanagi et al. 2005). VPE mediates a vacuolar collapse system, which necessarily releases vacuolar hydrolytic enzymes into the cytoplasm to prevent viral proliferation (Hatsugai et al. 2004, 2006; Hara-Nishimura et al. 2005). Although this system is effective at eliminating viruses within the cells, it could not be effective at preventing bacteria from proliferating outside the cells. Nevertheless, plant vacuoles accumulate a variety of antibacterial proteins. Until now, it has been a mystery how vacuolar antibacterial proteins reach the intercellular space where bacterial pathogens proliferate.
Here we show that fusion of a large central vacuole with the plasma membrane discharges vacuolar proteins to the outside of the cells, where they attack bacterial pathogens. The membrane fusion is normally suppressed and is triggered by a bacterial infection in a proteasome-dependent manner. This novel defense strategy provides plants with a mechanism for attacking intercellular bacterial pathogens.
Results
Fusion of a large central vacuole with the plasma membrane in response to bacterial infection
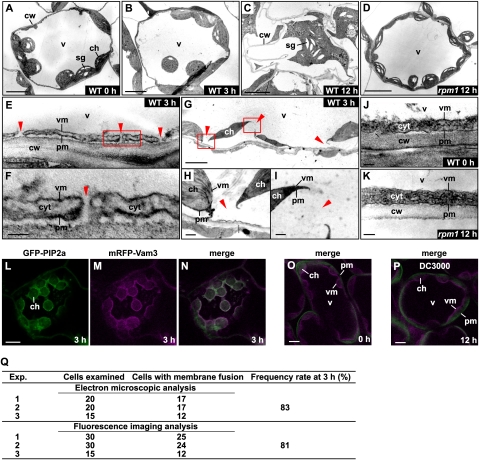
Arabidopsis thaliana (Col-0) are resistant to Pseudomonas syringae pv. tomato DC3000 having the Avr gene avrRpm1 (Pst DC3000/avrRpm1) (Mackey et al. 2002). We investigated ultrastructural changes in Col-0 leaf cells after inoculating them with Pst DC3000/avrRpm1. Figure 1A shows a typical cell of wild-type leaves with a vacuole (v) and many chloroplasts (ch). A magnified picture of a peripheral region of leaf cells shows an intact vacuolar membrane (Fig. 1J, vm) and a plasma membrane (Fig. 1J, pm). Twelve hours after the bacterial inoculation, we observed cell shrinkage and cytoplasmic aggregation in dead cells, which is a characteristic of hypersensitive cell death (Fig. 1C). At 3 h after the bacterial inoculation, the cells were alive (Supplemental Fig. 1) and looked intact in a low-power field of the cells (Fig. 1B). Surprisingly, however, we found that the vacuolar membrane was uniformly and frequently fused with the plasma membrane as shown in Figure 1E (indicated by red arrowheads). The membrane fusion occurred nearly simultaneously: We detected the fusion in 83% of the cells examined at 3 h (Fig. 1Q) and in most of the cells at 6 h (data not shown). The fusion resulted in the interconnection of vacuoles and the outside spaces of the plasma membrane in the leaf cells (Fig. 1F, indicated by red arrowheads), which makes it possible to discharge vacuolar contents into the outside of the cells (discussed below). Consequently, the loss of turgor of vacuoles caused plasmolysis, resulting in the separation of chloroplasts from the cell walls (Fig. 1G). The enlarged pictures show that the vacuolar and plasma membranes were disconnected by their fusion (Fig. 1H,I, indicated by red arrowheads).
Figure 1.
The vacuolar membrane fuses with the plasma membrane in leaves after infection of avirulent bacteria Pst DC3000/avrRpm1. (A–K) Electron micrographs of the wild-type leaves of Arabidopsis at 0 h (A,J), 3 h (B,E–I), and 12 h (C) after the inoculation of Pst DC3000/avrRpm1, and electron micrographs of the rpm1-1 mutant leaves at 12 h after the inoculation of the bacteria (D,K). The boxed areas in E and G are shown at higher magnifications in F, and H, and I, respectively. (E–I) Membrane fusions between plasma membranes and vacuolar membranes are indicated by red triangles. Bars: A–D,G, 5 μm; E,H,I, 0.5 μm; F,J,K, 0.1 μm. (L–P) Double-fluorescence images of the transgenic Arabidopsis leaves expressing both the plasma membrane-localized GFP-PIP2a and the vacuolar membrane-localized mRFP-VAM3 at 0 h (O) and 3 h (L–N) after the inoculation of Pst DC3000/avrRpm1 and at 12 h after the inoculation of Pst DC3000 (P). Bars, 10 μm. (cw) Cell wall; (pm) plasma membrane; (vm) vacuolar membrane; (v) vacuole; (ch) chloroplast; (cyt) cytosol; (sg) starch granule. (Q) Frequency rates of cells with the fused membranes at 3 h after the bacterial inoculation. Each of the electron microscopic and fluorescent imaging analyses was performed with three independent infected samples.
To observe membrane fusion in real time, we generated a transgenic plant that coexpressed a plasma membrane marker, GFP-PIP2a (Cutler et al. 2000), and a vacuolar membrane marker, mRFP-VAM3 (Fig. 1O; Ebine et al. 2008). An immunoblot analysis showed that leaves of the transgenic plant accumulated fusion proteins GFP-PIP2a and mRFP-VAM3 (Supplemental Fig. 2). The plasma membrane and vacuolar membrane were fluorescent in leaves of the transgenic plant (Fig. 1O), but not in leaves of the nontransformants (Supplemental Fig. 3). At 3 h after the Pst DC3000/avrRpm1 inoculation, the two fluorescences colocalized on both the plasma membrane and the vacuolar membrane and surrounded the chloroplasts (Fig. 1L–N). A statistical analysis showed that the colocalization of the fluorescences occurred in 81% of the cells examined at 3 h (Fig. 1Q), which corresponds to the value determined at 3 h by electron microscopy (described above). These results indicate that the vacuolar membrane fused with the plasma membrane in response to the bacterial infection. Membrane fusion then accelerated and was spread in most of the cells examined at 4 h (data not shown).
To see whether membrane fusion depends on the R gene, we used the rpm1 mutant and the virulent strain Pst DC3000 that does not have avrRpm1. Membrane fusion was not detected in cells of rpm1 after the inoculation of Pst DC3000/avrRpm1 (Fig. 1D,K). Similarly, membrane fusion was not caused by inoculation of Pst DC3000 (Fig. 1P). Taken together, these results indicate that an interaction between PRM1, a plant R gene product, and AvrRpm1, a pathogen Avr factor, is required for membrane fusion.
Discharge of vacuolar antibacterial proteins to the outside of the bacteria-infected cells
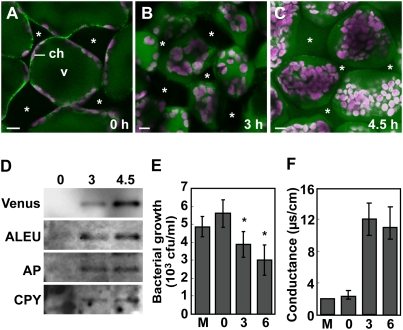
To see whether membrane fusion causes the discharge of vacuolar proteins to the outside of the cells, we infected the leaves of a transgenic plant that expressed a vacuolar-localized fluorescent protein (SP-Venus-2SC). The fluorescence became detectable outside the cells at 4.5 h after the inoculation of Pst DC3000/avrRpm1 (Fig. 2A–C). Chloroplasts, which were detected by their red autofluorescence, were drastically reorganized as a result of the loss of turgor of vacuoles. An immunoblot of the extracellular fluid from leaves detected the vacuolar-localized Venus and endogenous vacuolar proteolytic enzymes at 3 h after the inoculation of Pst DC3000/avrRpm1 and then detected increased levels at 4.5 h (Fig. 2D). The timing of the discharge of vacuolar proteins to the outside of the cells corresponded to that of membrane fusion in response to the bacterial infection. This suggests that fusion between the vacuolar and plasma membranes releases the vacuolar proteins outside the cells.
Figure 2.
Fusion between the vacuolar and plasma membranes causes the discharge of vacuole proteins to the intercellular space, which prevents proliferation of bacterial pathogens and activates PCD. (A–C) Fluorescence images of the transgenic Arabidopsis leaves with the vacuole-localized Venus at 0 h (A), 3 h (B), and 4.5 h (C) after the Pst DC3000/avrRpm1 inoculation. Bars, 10 μm. (Asterisks) Intercellular spaces; (v) vacuole; (ch) chloroplast with red autofluorescence. (D) An immunoblot of the extracellular fluid from the transgenic leaves at 0, 3, and 4.5 h after the bacterial infection to detect vacuole-localized Venus and proteinases. (ALEU) Aleurain; (AP) aspartyl protease; (CPY) carboxypeptidase Y. (E) Antibacterial activity of the extracellular fluid obtained before and at 3 and 6 h after bacterial inoculation of leaves. Bacterial growth for 12 h was determined in the presence of the extracellular fluid. Vertical bars indicate standard errors of three repeated experiments. Asterisks indicate statistically significant differences compared with that from 0 h (P < 0.05). (M) Mock inoculation. (F) Death of healthy leaf cells as measured by conductance after inoculation of extracellular fluid obtained before and at 3 and 6 h after bacterial inoculation of leaves. Conductance, an indicator of ion leakage, was measured at 2 h after application of the extracellular fluid. Vertical bars indicate standard errors of three repeated experiments. (M) Mock inoculation.
Interestingly, in the hours following bacterial inoculation, the extracellular fluid from the leaves increasingly suppressed the growth of Pst DC3000/avrRpm1 (Fig. 2E). Furthermore, the extracellular fluids became increasingly lethal to healthy leaves, as shown by the release of electrolytes from dead cells (Fig. 2F). These results indicate that vacuolar proteins with antibacterial activity and cell death induction activity are discharged to the outside of the cells.
Proteasome PBA1 subunit has DEVDase activity
Hypersensitive cell death is caused by proteinases with caspase-1-like and caspase-3-like activities. We reported previously that VPE has the activity of caspase-1 (YVADase activity) and mediates the collapse of the vacuolar membrane in response to viral infection, resulting in hypersensitive cell death (Hatsugai et al. 2004, 2006; Hara-Nishimura et al. 2005). However, the bacterially induced hypersensitive cell death was abolished by neither VPE deficiency (Supplemental Fig. 4) nor a caspase-1/VPE inhibitor, Ac-YVAD-CMK (Supplemental Fig. 4), indicating that VPE is not required for hypersensitive cell death in response to bacterial infection. This implied that the membrane fusion system for bacterially induced cell death is caused by proteinases other than VPE.
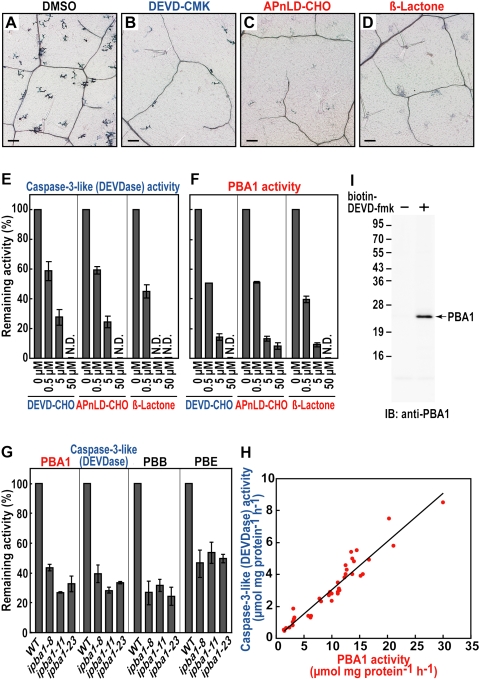
We found that a caspase-3 inhibitor (Ac-DEVD-CMK) and two proteasome inhibitors—Ac-APnLD-CHO, an inhibitor of the β1 subunit (Kisselev et al. 2003), and β-lactone, a general proteasome inhibitor (Fenteany et al. 1995)—effectively abolished the hypersensitive cell death (Fig. 3A–D). None of the inhibitors affected the bacterial growth themselves (Supplemental Fig. 5). The proteasome is responsible for the hydrolysis of intracellular proteins and is a large protein complex having three catalytic subunits (β1, β2, and β5) (for review, see Smalle and Vierstra 2004; Kurepa and Smalle 2008). In Arabidopsis, the genes for these subunits are called PBA1, PBB, and PBE, respectively (Yang et al. 2004).
Figure 3.
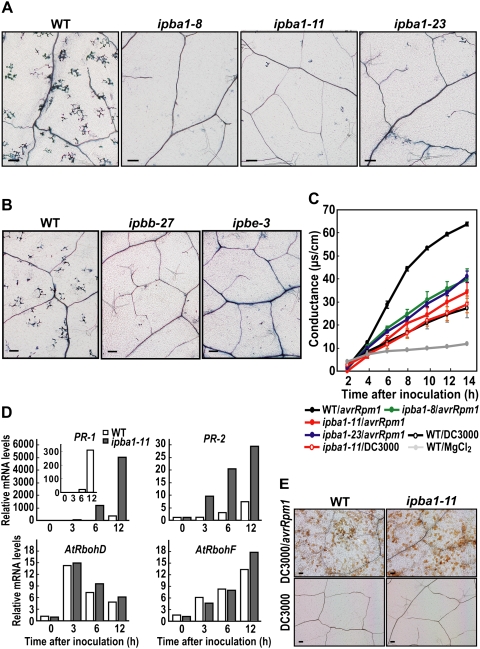
The plant proteasome, whose catalytic subunit (PBA1) exhibits a caspase-3-like (DEVDase) activity, is required for bacterially induced hypersensitive cell death in leaves. (A–D) Trypan-blue staining of dead cells of Arabidopsis leaves that were infected with Pst DC3000/avrRpm1 in the absence (A) or presence of caspase-3 inhibitor (B), an inhibitor to proteasome PBA1 subunit (C), and a general proteasome inhibitor (D). Bars, 100 μm. (E,F) Effects of proteinase inhibitors on DEVDase activity (E) and the PBA1 activity (F) in the infected leaves. Vertical bars indicate standard errors of three repeated experiments. (N.D.) Not detected. (G) The three ipba1 lines had reduced activities of not only the PBA1 subunit and DEVDase, but also the other proteasome subunits (PBB and PBE). Vertical bars indicate standard errors of three repeated experiments. (H) Correlation between the DEVDase and PBA1 activities in two or three plants of 17 ipba1 lines and wild type. (I) The extract of the infected leaves was pulled down with (+) or without (−) biotin-DEVD-fmk and were then subjected to immunoblot with anti-PBA1 antibody.
Arabidopsis leaves after an inoculation of bacteria had both an activity on a caspase-3 substrate (DEVD-MCA) (Fig. 3E) and PBA1 activity (Fig. 3F). The DEVDase activity was abolished by the two proteasome inhibitors in a dose-dependent manner as well as by Ac-DEVD-CHO (Fig. 3E). The PBA1 activity was completely abolished by Ac-DEVD-CHO as well as by Ac-APnLD-CHO and β-lactone (Fig. 3F), while the PBB and PBE activities were partially abolished by Ac-DEVD-CHO (Supplemental Fig. 6). For an in planta analysis, we established three independent RNAi lines of Arabidopsis (ipba1-8, ipba1-11, and ipba1-23) in which the PBA1 gene was specifically suppressed (Supplemental Fig. 7). Each of the three RNAi lines had a reduced PBA1 activity and a reduced DEVDase activity (Fig. 3G). The DEVDase activity and PBA1 activity were highly correlated (Fig. 3H). The three RNAi lines also had reduced PBB and PBE activities (Fig. 3G), possibly because of a reduced proteasome complex level in the absence of the PBA1 subunit.
To obtain more direct evidence that PBA1 interacts with DEVD, we performed a pull-down analysis with biotin-DEVD-fmk followed by an immunoblot with anti-PBA1 antibody. We found a positive signal on the blot (Fig. 3I), suggesting that PBA1 is responsible for the DEVDase activity. This result is consistent with the finding that the β1 subunits of animal and yeast proteasomes are caspase-like sites (Kisselev et al. 2003). Our findings unveil a plant enzyme with caspase-3-like activity that has long been reported to function in hypersensitive cell death.
Bacterially induced fusion of vacuolar membrane with plasma membrane is regulated by proteasome
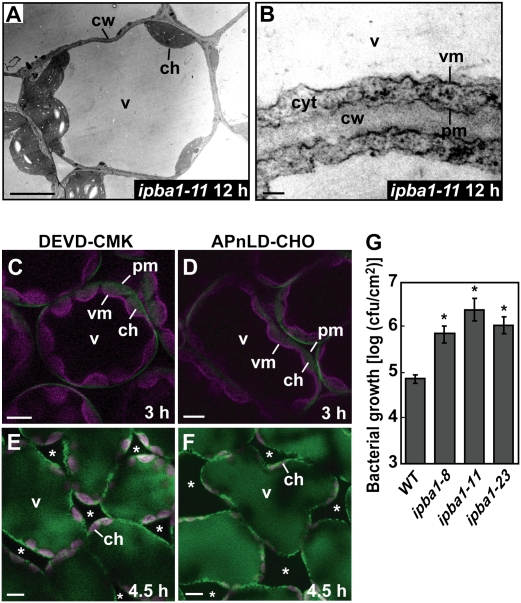
An in planta analysis provided evidence that proteasome is involved in cell-autonomous immunity based on the membrane fusion as follows. An ultrastructural analysis did not detect fusion of the vacuolar membrane and the plasma membrane in the ipba1-11 leaves even at 12 h after avirulent bacteria infection (Fig. 4A,B), although it detected fusion in wild-type leaves at 3 h (Fig. 1E–I). Furthermore, a fluorescence imaging analysis showed that treatment with a caspase-3 inhibitor (Ac-DEVD-CMK) and a PBA1 inhibitor (Ac-APnLD-CHO) suppressed not only membrane fusion (Fig. 4C,D) but also the discharge of vacuolar proteins outside the infected cells (Fig. 4E,F). To examine the effect of no discharge of the vacuolar proteins on the bacterial growth outside of the cells, we measured the number of bacteria in the three ipba1 lines. The numbers of bacteria in all of the ipba1 lines were much higher than the number in the wild type (Fig. 4G), but were two orders of magnitude lower than the number in the rpm1 mutant (discussed below; Grant et al. 1995). Together, these results indicate that the proteasome mediates the discharge of vacuolar contents for the purpose of attacking bacterial pathogens by regulating the membrane fusion between the vacuolar membrane and the plasma membrane in response to the bacterial infection.
Figure 4.
An in planta analysis showing that proteasome is involved in the cell-autonomous immunity based on the membrane fusion in association with the discharge of vacuolar proteins to the outside of the cells. (A,B) Electron micrographs of the ipba1-11 leaves at 12 h after the inoculation of Pst DC3000/avrRpm1. Bars: A, 5 μm; B, 0.1 μm. (C,D) Double-fluorescence images of the transgenic leaves expressing both the plasma membrane-localized GFP-PIP2a and the vacuolar membrane-localized mRFP-VAM3 at 3 h after the bacterial inoculation in the presence of a caspase-3 inhibitor (DEVD-CMK) (C) or a PBA1 inhibitor (APnLD-CHO) (D). Bar, 10 μm. (cw) Cell wall; (pm) plasma membrane; (vm) vacuolar membrane; (v) vacuole; (ch) chloroplast; (cyt) cytosol. (E,F) Fluorescence images of the transgenic leaves expressing the vacuole-localized Venus at 4.5 h after the bacteria inoculation in the presence of DEVD-CMK (E) or APnLD-CHO (F). Bar, 10 μm. (Asterisks) Intercellular spaces; (v) vacuole; (ch) chloroplast with red autofluorescence. (G) Numbers of the bacteria grown in the infected leaves of three ipba1 lines and wild type for 3 d were determined. Vertical bars indicate standard errors of five repeated experiments. Asterisks indicate statistically significant differences compared with wild-type leaves (P < 0.05).
Proteasome plays a role in the bacterially induced hypersensitive cell death
Hypersensitive cell death in the three ipba1 lines was monitored by trypan-blue staining. Deficiency of PBA1 suppressed the hypersensitive cell death in response to the infection of Pst DC3000/avrRpm1 (Fig. 5A). The PBA1 deficiency did not affect either susceptible cell death caused by the virulent strain Pst DC3000 (Supplemental Fig. 8A), or nonspecific cell death induced by methanol (Supplemental Fig. 9). These results suggest that the proteasome is required for R gene-mediated cell death. Growth of the virulent bacteria Pst DC3000 on ipba1-11 was slightly less than that on the wild type (Supplemental Fig. 8B), which is consistent with the report that defective cullin3-based E3 ligases were more resistant to the virulent bacteria P. syringae pv. maculicola ES4326 (Spoel et al. 2009). Deficiency of either PBB or PBE (Supplemental Fig. 10) also suppressed the hypersensitive cell death in response to the infection of Pst DC3000/avrRpm1 (Fig. 5B), suggesting that the lack of one subunit causes a defect in proteasome function.
Figure 5.
The plant proteasome is involved in the bacterially induced hypersensitive cell death. (A,B) Trypan-blue staining of the infected leaves showing that the three ipba1 lines (A) and the ipbb-27 and ipbe-3 lines (B) had much fewer dead cells than wild type. Bars, 100 μm. (C) The ipba1 and wild-type leaves were infected with either avirulent (DC3000/avrRpm1) or virulent (DC3000) bacteria. The ion leakages from dying and dead cells were measured by conductance. Vertical bars indicate standard errors of five repeated experiments. (D) Real-time RT–PCR showing the relative mRNA levels of four defense genes normalized to that of Actin in the ipba1-11 and wild-type leaves after the bacterial inoculation. (E) DAB staining showing production of hydrogen peroxide in the ipba1-11 and wild-type leaves after the inoculation of avirulent (DC3000/avrRpm1) and virulent (DC3000) bacteria. Bars, 100 μm.
Hypersensitive cell death was also quantitatively monitored by the release of electrolytes from dead cells. Ion leakage from the leaves of the three ipba1 lines was much less than that from the wild-type leaves (Fig. 5C), but was the same as the leakage in the wild-type leaves infected with the virulent bacteria Pst DC3000 (Fig. 5C). The quantitative analysis showed that PBA1 deficiency caused no hypersensitive cell death until 14 h after infection with the avirulent bacteria Pst DC3000/avrRpm1 (Fig. 5C). These results indicate that the proteasome confers immunity to bacterial pathogens through the use of hypersensitive cell death.
The plant defense system against pathogens is associated with the expression of defense genes and defense-related genes (van Loon et al. 2006). In the present study, after the bacterial inoculation, the expression levels of two defense genes (PR-1 and PR-2) increased in ipba1-11 as they did in the wild type (Fig. 5D), although their levels in ipba1-11 were higher than those in the wild type, possibly due to a defect of the proteasome-dependent degradation of some regulator for their gene expression (Spoel et al. 2009) in ipba1-11. Two defense-related genes (AtrbohD and AtrbohF) that encode NADPH oxidases are responsible for generating reactive oxygen intermediates (ROIs) that are involved in the hypersensitive response (Torres et al. 2002). The expressions of these genes increased in ipba1-11 as well as in the wild type (Fig. 5D). Consequently, ROI production was not significantly different between ipba1-11 and the wild type (Fig. 5E). The overall results indicate that a PBA1 deficiency does not suppress the induction of defense genes or ROI production, although it partially suppressed disease resistance through hypersensitive cell death.
Another avirulent bacterial strain, Pst DC3000/avrRpt2, also induces a proteasome-regulating defense system based on membrane fusion
To clarify whether the proteasome-regulating membrane fusion is specific to the strain Pst DC3000/avrRpm1 or not, we examined the defense response to another avirulent bacterial strain, Pst DC3000/avrRpt2. Double-fluorescence images of the transgenic leaves expressing both the plasma membrane-localized GFP-PIP2a and the vacuolar membrane-localized mRFP-VAM3 show that membrane fusion between the vacuolar and plasma membranes occurred at 7.5 h after inoculation (Fig. 6A–D). The membrane fusion also caused the discharge of vacuole-localized Venus to the outside of the cells at 10 h after the inoculation (Fig. 6F,G). A PBA1 inhibitor (Ac-APnLD-CHO) suppressed not only membrane fusion (Fig. 6E) but also the discharge of vacuolar proteins outside the infected cells (Fig. 6H). Pst DC3000/avrRpt2 induced membrane fusion and vacuolar protein discharge later than Pst DC3000/avrRpm1. This is consistent with a previous report that Pst DC3000/avrRpt2 induces hypersensitive cell death later than Pst DC3000/avrRpm1 (Mackey et al. 2002).
Figure 6.
Another avirulent strain, Pst DC3000/avrRpt2, also induces proteasome-regulating hypersensitive cell death through membrane fusion and discharge of vacuolar proteins to the outside of the cells. (A–E) Double-fluorescence images of transgenic leaves expressing both the plasma membrane-localized GFP-PIP2a and the vacuolar membrane-localized mRFP-VAM3 at 0 h (D) and 7.5 h (A–C,E) after the inoculation of avirulent bacteria Pst DC3000/avrRpt2 in the absence (A–D) and presence (E) of a PBA1 inhibitor (APnLD-CHO). Bars, 10 μm. (pm) Plasma membrane; (vm) vacuolar membrane; (v) vacuole; (ch) chloroplast. (F–H) Fluorescence images of the transgenic leaves with the vacuole-localized Venus at 0 h (F) and 10 h (G,H) after the inoculation of Pst DC3000/avrRpt2 in the absence (F,G) and presence (H) of APnLD-CHO. Bars, 10 μm. (Asterisks) Intercellular spaces; (v) vacuole; (ch) chloroplast with red autofluorescence. (I) Trypan-blue staining of the infected leaves with Pst DC3000/avrRpt2, showing that the three ipba1 lines had much fewer dead cells than wild type. Bars, 100 μm. (J) Ion leakage from dying and dead cells of the ipba1 and wild-type leaves that were infected with Pst DC3000/avrRpt2. Vertical bars indicate standard errors of three repeated experiments. (K) Numbers of the bacteria grown in the infected leaves of three ipba1 lines and wild type 3 d after inoculation were determined. Vertical bars indicate standard errors of three repeated experiments. Asterisks indicate statistically significant differences compared with wild-type leaves (P < 0.05).
Trypan-blue staining revealed that the Pst DC3000/avrRpt2-induced hypersensitive cell death was suppressed in all three ipba1 lines (Fig. 6I). Ion leakage from the Pst DC3000/avrRpt2-infected leaves of the ipba1 lines was much less than that from the wild-type leaves (Fig. 6J). These results indicate that Pst DC3000/avrRpt2 induces proteasome-mediated hypersensitive cell death, as does Pst DC3000/avrRpm1. The numbers of bacteria in all of the ipba1 lines were higher than the number in the wild type (Fig. 6K), indicating the reduction of disease resistance by a deficiency of PBA1. These results indicate that Pst DC3000/avrRpt2 induces a proteasome-dependent defense system based on membrane fusion, as does Pst DC3000/avrRpm1.
Discussion
A model for proteasome-dependent membrane fusion as a plant defense strategy against intercellular bacteria
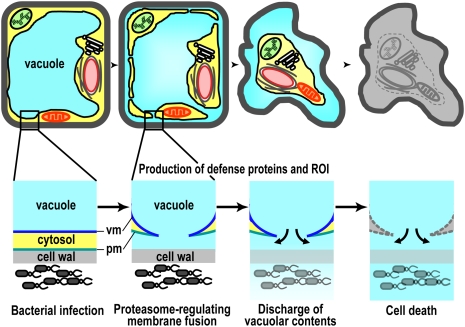
The proteasome-dependent defense strategy involves three processes as follows (Fig. 7). The first process concerns the fusion of a large central vacuole with the plasma membrane after bacterial infection. The fusion causes the discharge of vacuolar contents including antibacterial proteins and hydrolytic enzymes to the outside of the plasma membrane, resulting in wilted leaves (Supplemental Fig. 1). Vacuole-type defense proteins are up-regulated after the infection of bacterial pathogens (van Loon et al. 2006), but it has been a mystery how these proteins attack the bacteria in the intercellular space. Our new results solve this mystery. The second process is the production of defense proteins and ROI in the cells even after fusion of the membranes. In the third process, the cells induce hypersensitive cell death by the actions of the hydrolytic enzymes that accumulate on the outside of the cells. Unlike animal pathogens, phytopathogenic bacteria do not enter host cells but proliferate in the intracellular space of the leaves. Plants might have developed the unique characteristic “membrane fusion” in plant–pathogen interactions as a defense strategy against intercellular microbes.
Figure 7.
A model for a cell-autonomous immune system based on membrane fusion for attacking intercellular bacteria. Bacterial infection triggers the fusion of a large central vacuole with the plasma membrane in a proteasome-dependent manner. The fusion causes the discharge of vacuolar contents including antibacterial proteins and hydrolytic enzymes to the outside of the cells where bacteria proliferate. The defense proteins and ROI that have the ability to attack bacterial pathogens are produced even after fusion of the membranes for a while. Finally, the cells induce hypersensitive cell death by the actions of the hydrolytic enzymes that accumulate on the outside of the cells. (vm) Vacuolar membrane; (pm) plasma membrane.
This novel type of cell-autonomous immune system uses vacuoles, which exist in each cell of plants. Using vacuoles for defense makes sense for plants because plants do not have immune cells and so each cell has to provide its own defense against pathogens. The immune system based on membrane fusion differs markedly from the system associated with the membrane collapse of vacuoles, which we reported previously (Hatsugai et al. 2004, 2006; Hara-Nishimura et al. 2005), although both systems use vacuoles. Intriguingly, both systems use enzymes with caspase-like activities: The membrane fusion system uses PBA1 with caspase-3-like activity and the vacuolar collapse system uses VPE with caspase-1-like activity (Hatsugai et al. 2004, 2006; Hara-Nishimura et al. 2005). VPE-mediated vacuolar collapse necessarily releases vacuolar hydrolytic enzymes into the cytoplasm to prevent viral proliferation (Hatsugai et al. 2004, 2006; Hara-Nishimura et al. 2005). Although this system is effective at eliminating viruses within the cells, it could not be effective at preventing bacteria from proliferating outside the cells. Taken together, this study and our previous study suggest that plants have evolved a cell-autonomous immune system based on membrane fusion to inhibit proliferation of bacterial pathogens and a vacuolar collapse system to be unable to spread systemically viral pathogens.
Proteasome function is required for hypersensitive cell death
There has so far been no direct evidence linking the proteasome to cell death and disease resistance in plants. Our findings demonstrate that proteasome function is required for both efficient hypersensitive response development mediated by the R gene and full resistance against avirulent bacteria. On the other hand, basal host resistance was reported to require polyubiquitination but not a proteasome pathway in powdery mildew-inoculated barley (Dong et al. 2006). The proteasome-mediated defense system in this study might be responsible for the R gene-dependent resistance against bacterial pathogens, but not for the basal host defense against fungal pathogens.
Our finding that transcription of defense-related genes and the production of ROI were not inhibited in proteasome-defective plants (Fig. 5D,E) suggests that proteasome-mediated degradation is required for cell death activation but not for defense signaling. So far, no target proteins of proteasome-mediated degradation that are associated with disease resistance have been identified in plants. Identification of the proteasome target proteins would help to unravel the molecular mechanism underlying the membrane fusion-mediated defense system.
Proteasome PBA1 subunit might be responsible for plant caspase-3-like activity
Our finding that plants with defective PBA1 subunits have reduced caspase-3-like activity (Fig. 3G,H) suggests that the PBA1 subunit is the source of caspase-3-like activity that has long been reported to be present in plant PCD. Consistent with this finding, the amino acid sequence of the substrate pocket of the yeast β1 subunit (ScPRE3) (Fu et al. 1998), which cleaves a peptide bond at the C-terminal side of either glutamic acid or aspartic acid (Kisselev et al. 1999), is conserved in PBA1. Caspases also recognize aspartic acid residues (Earnshaw et al. 1999). PBA1 did not recognize the aspartic acid of YVAD that is recognized by caspase-1 (data not shown), although PBA1 recognized the aspartic acid at the P1 position of DEVD that is recognized by caspase-3.
On the other hand, PBB1 and PBB2 have a putative substrate pocket similar to the one in the yeast β2 subunit (ScPUP1) (Supplemental Fig. 11; Fu et al. 1998). The β2 subunit has trypsin-like activity in that it specifically recognizes basic amino acids (Kisselev et al. 1999). PBE1 and PBE2 have a putative substrate pocket that is similar to the one in the yeast β5 subunit (ScPRE2) (Supplemental Fig. 11; Fu et al. 1998). The β5 subunit has chymotrypsin-like activity in that it recognizes bulky hydrophobic amino acids (Kisselev et al. 1999). These results suggest that neither the PBB subunit nor the PBE subunit recognizes aspartic acid and that PBA1 is responsible for the caspase-3-like activity in Arabidopsis. Although the caspase-3 inhibitor partially abolished the activities of PBB and PBE (Supplemental Fig. 6), the inhibition was not dose-dependent, and thus was probably not competitive inhibition.
A possible role of proteasomes in membrane fusion
Our study demonstrates that membrane fusion, which is normally suppressed, is triggered by bacterial infection in a proteasome-dependent manner. How does the proteasome function in the fusion of the vacuolar and plasma membranes? One possibility is that some negative regulator suppresses membrane fusion in uninfected cells but is degraded by the proteasome in response to bacterial infection. In yeast, fusion of vacuolar membranes may require proteasomal degradation of ubiquitinated Ypt7, a yeast homolog of Rab7 GTPase (Kleijnen et al. 2007). A similar mechanism could function in the fusion of heterologous membranes. AtRab7, an Arabidopsis homolog, is induced during PCD after treatment with superoxide or infection with Pst DC3000/avrRpm1 (Mazel et al. 2004). Ubiquitination may also regulate membrane fusion events that reassemble fragmented organelles after mitosis in mammalian cells (Meyer and Popp 2008). These cellular membrane and organelle fusion events also involve SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors). SNAREs have been shown recently to have roles in plant immunity (Thordal-Christensen 2009), and thus might have a role in the fusion of the vacuolar and plasma membranes in response to bacterial infection.
Materials and methods
Plant materials and growth conditions
We used wild-type plants of A. thaliana (ecotype Columbia, Col-0), the rpm1-1 mutant that was kindly provided by Dr. Jeffery L. Dangl of University of North Carolina, and the transformants as follows. Plants were grown under 8 h of light in a day at 22°C.
Transformation of Arabidopsis
We generated the RNAi plants ipba1 for the PBA1 gene that encodes β1 subunit of proteasome, ipbb for the PBB genes that encode β2 subunits, and ipbe for the PBE genes that encode β5 subunits. Gateway Technology (Invitrogen) was used to produce the plasmids for RNAi. Partial cDNA fragments derived from the PBA1 gene (101–639 base pairs [bp]), the PBB1 gene (265–922 bp), and the PBE1 gene (112–776 bp) were amplified by RT–PCR using primer sets as follows: PBA1 set (5′-CACCCTAATTTATACTCGCAAAACTCA-3′ and 5′-TGTTATCTTTCCAAGCCTGGTCG-3′), PBB set (5′-CACCTGGTGTGATACTAGGGGCAGATA-3′ and 5′-TCACCAACTTCTGTGATTTCGAC-3′), and PBE set (5′-CACCATGAAGCTTGATACTAGTGGGTT-3′ and 5′-AATGTCGCATGGTAGATTGATCT-3′). Each of three amplified fragments was inserted into pENTER/D-TOPO (Invitrogen) by the TOPO reaction to produce the entry clone. The fragment of each entry clone was transferred into the destination vector pB7GWIWG2 (Plant System Biology). The recombinant pB7GWIWG2 contained two identical cDNA fragments in both sense and antisense orientations within the T-DNA region. We introduced the recombinant pB7GWIWG2 into Arabidopsis plants by using Agrobacterium tumefaciens (strain GV3101). We selected transformed lines (T1 transformants) on the medium containing appropriate antibiotics and then obtained T2 progeny for further analyses.
The transgenic plant that expresses GFP-PIP2a was obtained from ABRC (Arabidopsis Biological Resource Center) at Ohio State University according to information from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu). The transgenic plant that expresses mRFP-VAM3 was kindly provided by Dr. Takashi Ueda of University of Tokyo. By crossing these two transgenic plants, we generated the transgenic plants that coexpress both GFP-PIP2a and mRFP-VAM3.
Pathogen strains and pathology tests
We used Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and two avirulent strains (Pst DC3000/avrRpm1 and Pst DC3000/avrRpt2) that were provided by Dr. Jeffery L. Dangl. We cultured the bacteria in KB medium with kanamycin (30 μg/mL) and rifampicin (100 μg/mL) (Katagiri et al. 2002). After washing the cells twice in 10 mM MgCl2, we suspended them at 5 × 105 colony-forming units (CFUs) per milliliter for each experiment of trypan-blue-staining and in planta growth assay, and at 5 × 107 CFUs per milliliter for each experiment of trypan-blue staining, ion leakage, DAB staining, enzyme assay, RT–PCR analysis, and microscopic analysis. We inoculated the suspensions into the abaxial surfaces of leaves with needleless syringes. We monitored bacterial growth in the leaves as described (Katagiri et al. 2002).
Evaluation of cell death
We measured ion leakage from dying and dead cells principally as described (Mackey et al. 2002). We prepared four discs with a 4.5-mm diameter or two discs with a 7.5-mm diameter from leaves immediately after the inoculation of bacteria (5 × 107 CFUs per milliliter) or extracellular fluid that was prepared from the leaves as described below, respectively, and then floated them in 2 mL of distilled water for 30 min. We transferred the leaf discs into the 1.5 mL of water and incubated for 2 to 24 h. We measured conductance of the water with an electrical conductivity meter (B-173, Horiba). Alternatively, we visualized dying cells of the leaves at 12 h after inoculation of either Pst DC3000/avrRpm1 (5 × 105 CFUs per milliliter) or 40% methanol and at 24 h after the inoculation of Pst DC3000/avrRpt2 (1 × 106 CFUs per milliliter) by trypan-blue staining (Koch and Slusarenko 1990).
Detection of H2O2
We used 3,3′-diaminobenzidine (DAB) to visualize the production of H2O2 in leaves as described previously (Thordal-Christensen et al. 1997). We harvested the leaves at 2 h after the bacteria inoculation (5 × 107 CFUs per milliliter) and then infiltrated them with DAB solution. The leaves were placed in a plastic box under high humidity for 10 h. The leaves were fixed with a solution (60% [v/v] ethanol, 20% [v/v] lactic acid, 20% [v/v] glycerol) for 5 h, were decolorized in a chloral hydrate solution (2.5 g/mL) for 20 h, and were then visualized with a light microscope.
Treatment with proteinase inhibitors
We used proteinase inhibitors Ac-YVAD-CMK (Peptide Institute, Osaka, Japan), Ac-YVAD-CHO (Peptide Institute), Ac-DEVD-CMK (Calbiochem), Ac-DEVD-CHO (Peptide Institute), Ac-APnLD-CHO (BIOMOL), and clasto-lactacystin β-lactone (BIOMOL). To determine the effects of proteinase inhibitors on hypersensitive cell death, we mixed each proteinase inhibitor, whose final concentration was 1 mM in 2% (v/v) dimethylsulfoxide (DMSO), with the bacterial suspension and 0.02% Silwet L-77 just before the inoculation.
Pull-down assay
The extract from the leaves at 1–2 h after the inoculation of Pst DC3000/avrRpm1 (5 × 107 CFUs per milliliter) was incubated with or without biotin-DEVD-fmk (Calbiochem) for 1 h at room temperature and then incubated with streptavidin-conjugated agarose (Vector Laboratories, Inc.). After extensive washing, the agarose beads were boiled in SDS sample buffer (100 mM Tris-HCl at pH 6.8, 4% [v/v] SDS, 5% [v/v] 2-mercaptoethanol). The pulled-down samples were subjected to SDS-PAGE followed by immunoblot with anti-PBA1 antibody (diluted, 5000-fold; BIOMOL) (Yang et al. 2004).
Enzyme assay
Leaves were harvested at 1–2 h after the bacteria inoculation (5 × 107 CFUs per milliliter) and homogenized in 20S Proteasome Assay Buffer (BIOMOL) with 5 mM ATP. The leaf extracts were preincubated with 0.5–50 μM each proteinase inhibitor (Ac-DEVD-CHO, Ac-APnLD-CHO, and clasto-lactacystin β-lactone) for 1 h at 30°C. Fluorogenic substrates conjugated with α-(4-methyl-coumary-7-amide) (MCA) were added to the extracts. The caspase-3 substrate was Ac-DEVD-MCA (Peptide Institute) (100 and 500 μM) and the proteasome substrates were Ac-nLPnLD-MCA (BIOMOL) (100 and 500 μM), Ac-RLR-MCA (BIOMOL) (10 and 50 μM), and Suc-LLVY-MCA (Peptide Institute) (10 and 50 μM). Proteinase activities were expressed as the fluorescence at 465 nm.
Quantitative RT–PCR
We infected the leaves with Pst DC3000/avrRpm1 (5 × 107 CFUs per milliliter) and prepared total RNA from the leaves at 0, 3, 6, and 12 h after inoculation. cDNA was synthesized from 1 μg of total RNA with Ready-to-Go RT–PCR beads (GE Healthcare). RT–PCR was performed with the RNA and gene-specific primer sets (Supplemental Table 1). We used a primer set for Actin1 as a control (Supplemental Table 1). For quantitative RT–PCR, the cDNA was quantified with a gene-specific-primer set (Supplemental Table 2) and the TaqMan Gene expression Assay Kit (Applied Biosystems) in a 7500 Fast Real-Time PCR System (Applied Biosystems). The experiments were repeated two times. The relative quantity of target mRNA was calculated using Actin as a standard.
Preparation of extracellular fluids
Extracellular fluids were prepared as described (Sanmartin et al. 2007). Arabidopsis leaves at the indicated time after inoculation with Pst DC3000/avrRpm1 (5 × 107 CFUs per milliliter) were vacuum-infiltrated in 20 mM Tris-HCl (pH 7.5) and 50 mM NaCl for 15 min. After drying with a paper towel, the leaves were subjected to centrifugation at 900g to obtain extracellular fluid. The extracellular fluid was subjected to immunoblot with each antibody against GFP, aleurain, aspartyl protease, and carboxypeptidase Y. The extracellular fluid was used for an ion leakage assay to evaluate the PCD-inducing activity as described above. For evaluation of antibacterial activity of the extracellular fluid, we cultured the bacteria with the extracellular fluid in KB medium for 12 h and then counted the CFUs of the bacteria on KB plates.
Electron microscopic analysis
Arabidopsis leaves that were infected with Pst DC3000/avrRpm1 (5 × 107 CFUs per milliliter) were vacuum-infiltrated for 1 h with a fixative, dehydrated, and embedded in Epon, as described previously (Hatsugai et al. 2004). Ultrathin sections were examined with a transmission electron microscope (model 1200EX; JEOL) at 80 kV.
Confocal laser-scanning microscopy
The fluorescent images were inspected with a confocal laser-scanning microscope (model LSM510 META; Carl Zeiss) using either the 488-nm line of a 40-mW Ar/Kr laser for both GFP and Venus or the 543-nm line of a 1-mW He/Ne laser for mRFP and chlorophyll autofluorescence. The images were analyzed with LSM image examiner software (Carl Zeiss). The data were exported as 12-bit TIFF files and processed using Adobe Photoshop CS (Adobe Systems).
Acknowledgments
We thank Drs. J. Dangl, University of North Carolina, for donating the Pst DC3000 strains and Arabidopsis rpm1-1 mutant; C. Somerville, Carnegie Institution, for providing the CS84725 seeds; T. Kunieda, Kyoto University, for Venus-transformed Arabidopsis; and T. Kawasaki, H. Yoshioka, and K. Shirasu for technical advice. This work was supported by PREST and CREST of the Japan Science and Technology Corporation; by Grants-in-Aid for Scientific Research (nos. 16085203 and 17107002) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; and by the Global Center of Excellence Program “Formation of a Strategic Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem” of MEXT.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1825209.
Supplemental material is available at http://www.genesdev.org.
References
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau L, Ge Y, Drury GE, Gallois P. What happened to plant caspases? J Exp Bot. 2008;59:491–499. doi: 10.1093/jxb/erm352. [DOI] [PubMed] [Google Scholar]
- Craig A, Ewan R, Mesmar J, Gudipati V, Sadanandom A. E3 ubiquitin ligases and plant innate immunity. J Exp Bot. 2009;60:1123–1132. doi: 10.1093/jxb/erp059. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Dong W, Nowara D, Schweizer P. Protein polyubiquitination plays a role in basal host resistance of barley. Plant Cell. 2006;18:3321–3331. doi: 10.1105/tpc.106.046326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell. 2008;20:3006–3021. doi: 10.1105/tpc.107.057711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics. 1998;149:677–692. doi: 10.1093/genetics/149.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Hatsugai N, Kuroyanagi M, Nakaune S, Nishimura M. Vacuolar processing enzyme: An executor of plant cell death. Curr Opin Plant Biol. 2005;8:404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305:855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Nishimura M, Hara-Nishimura I. A cellular suicide strategy of plants: Vacuole-mediated cell death. Apoptosis. 2006;11:905–911. doi: 10.1007/s10495-006-6601-1. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana–Pseudomonas syringae interaction: March 27, 2002. In: Somerville C, Meyerowitz E, editors. The Arabidopsis book. American Society of Plant Biologists; Rockville, MD: 2002. http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell. 1999;4:395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Garcia-Calvo M, Overkleeft HS, Peterson E, Pennington MW, Ploegh HL, Thornberry NA, Goldberg AL. The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J Biol Chem. 2003;278:35869–35877. doi: 10.1074/jbc.M303725200. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Kirkpatrick DS, Gygi SP. The ubiquitin–proteasome system regulates membrane fusion of yeast vacuoles. EMBO J. 2007;26:275–287. doi: 10.1038/sj.emboj.7601486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Smalle JA. Structure, function and regulation of plant. proteasomes. Biochimie. 2008;90:324–335. doi: 10.1016/j.biochi.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J Biol Chem. 2005;280:32914–32920. doi: 10.1074/jbc.M504476200. [DOI] [PubMed] [Google Scholar]
- Lam E, del Pozo O. Caspase-like protease involvement in the control of plant cell death. Plant Mol Biol. 2000;44:417–428. doi: 10.1023/a:1026509012695. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. Role of SCF ubiquitin–ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell. 2002;14:1483–1496. doi: 10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF, III, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Mazel A, Leshem Y, Tiwari BS, Levine A. Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e) Plant Physiol. 2004;134:118–128. doi: 10.1104/pp.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Popp O. Role(s) of Cdc48/p97 in mitosis. Biochem Soc Trans. 2008;36:126–130. doi: 10.1042/BST0360126. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Orlowski RZ. The role of the ubiquitin–proteasome pathway in apoptosis. Cell Death Differ. 1999;6:303–313. doi: 10.1038/sj.cdd.4400505. [DOI] [PubMed] [Google Scholar]
- Sanmartin M, Ordonez A, Sohn EJ, Robert S, Sanchez-Serrano JJ, Surpin MA, Raikhel NV, Rojo E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc Natl Acad Sci. 2007;104:3645–3650. doi: 10.1073/pnas.0611147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H. Vesicle trafficking in plant pathogen defence. In: Baluska F, Mancuso S, editors. Signaling in plants. Springer-Verlag; Berlin, Heidelberg: 2009. pp. 287–301. [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Woltering EJ. Death proteases come alive. Trends Plant Sci. 2004;9:469–472. doi: 10.1016/j.tplants.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Yang Y, Shah J, Klessig DF. Signal perception and transduction in plant defense responses. Genes & Dev. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- Yang P, Fu H, Walker J, Papa CM, Smalle J, Ju YM, Vierstra RD. Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem. 2004;279:6401–6413. doi: 10.1074/jbc.M311977200. [DOI] [PubMed] [Google Scholar]