Abstract
The locus alx, which encodes a putative transporter, was discovered previously in a screen for genes induced under extreme alkaline conditions. Here we show that the RNA region preceding the alx ORF acts as a pH-responsive element, which, in response to high pH, leads to an increase in alx expression. Under normal growth conditions this RNA region forms a translationally inactive structure, but when exposed to high pH, a translationally active structure is formed to produce Alx. Formation of the active structure occurs while transcription is in progress under alkaline conditions and involves pausing of RNA polymerase at two distinct sites. Alkali increases the longevity of pausing at these sites and thereby interferes with formation of the inactive structure and promotes folding of the active one. The alx locus represents the first example of a pH-responsive riboregulator of gene expression, introducing a novel regulatory mechanism that involves RNA folding dynamics driven by pH.
Keywords: RNA regulator, transcriptional pausing, alkaline conditions, translation control
RNA regulators of gene expression have become a focus of scientific attention, as it became evident that such molecules, widespread in both prokaryotes and eukaryotes, participate in regulation of almost all cellular processes. One class of regulatory RNAs comprises mRNA leaders that affect expression in cis by adopting different conformations in response to cellular and/or environmental signals, including stalled ribosomes, uncharged tRNAs, elevated temperatures, or small molecule ligands (Waters and Storz 2009). The first regulatory mRNA leaders to be described involved transcription attenuation. In these systems, stalled ribosomes lead to changes in mRNA structure, affecting transcription elongation through formation of terminator or anti-terminator structures. Later studies showed that specific sequences in transcripts encoding tRNA synthetases bind the corresponding uncharged tRNAs, and that mRNA leader structures of the so-called “RNA thermosensors” are sensitive to elevated temperatures (Narberhaus et al. 2006). More recently, riboswitches were defined: These leader sequences change their structure upon binding to small molecule ligands. Generally, riboswitches are composed of two domains: a highly conserved, natural aptamer that serves as a ligand-binding domain, and an expression platform that interfaces with RNA elements involved in gene expression, such as the Shine-Dalgarno sequence or transcription terminator structures. Hence, the control mechanisms employed by riboswitches are based largely on translation initiation and transcription termination (Winkler and Breaker 2005). Recent investigations have discovered different types of riboswitches, suggesting that the diversity of riboswitch function and/or control mechanisms could be greater than envisioned originally. For example, the glmS riboswitch is a metabolite-responsive ribozyme that undergoes self-cleavage activated by GlcN6P (Collins et al. 2007). Another example of an atypical riboswitch is metE of Bacillus calusii, which has two distinct elements: one that responds to S-adenosylmethionine, and another responding to coenzyme B12 (Sudarsan et al. 2006). Metal-sensing regulatory RNAs represent another unique group of riboregulators that control gene expression in response to changes in cellular magnesium levels (Cromie et al. 2006; Dann et al. 2007). Notably, outside the bacterial kingdom, so far only a single eukaryotic, thiamine–pyrophosphate-binding riboswitch has been characterized. It directs gene expression by regulating mRNA splicing in fungi, green algae, and land plants (Bocobza and Aharoni 2008).
Much of the regulation of gene expression of these regulatory mRNA leaders is accomplished during the elongation phase and involves pausing of RNA polymerase. Pausing, in these cases, allows for interaction with regulators that, later on, results in changes in RNA conformation. RNA polymerase pause sites have been identified in leaders of the his, trp, and gly operons of Escherichia coli and Bacillus subtilis (Winkler and Yanofsky 1981; Mooney et al. 1998; Grundy and Henkin 2004; Yakhnin et al. 2006). In B. subtilis, the expression of the trpEDCFBA operon is regulated in response to tryptophan availability by both transcription attenuation and translation control and involves pausing. It has been suggested that pausing at the trpEDCFBA leader provides the RNA-binding protein TRAP with a greater time frame to bind the nascent transcript and promote transcription termination. In addition, TRAP binding, assisted by pausing downstream from the site of termination, promotes the formation of an RNA structure in which translation of trpE is blocked (Yakhnin et al. 2006). Another instance in which transcriptional pausing at a specific location affects recruitment of regulators is illustrated by the flavin mononucleotide (FMN)-responsive riboswitch, where pausing provides the small metabolite FMN with more time to bind the RNA during its synthesis and thereby affects the kinetic parameters of this riboswitch (Wickiser et al. 2005). More generally, the importance of the sequential nature of transcription is highlighted by the adenine riboswitch, where it was shown that to achieve regulation the ligand must bind the RNA-binding domain before synthesis of the expression platform is completed (Lemay et al. 2006).
Previously, in a systematic search for small RNA-encoding genes in E. coli, we identified SraF, a small RNA of very low abundance (Argaman et al. 2001). Downstream from SraF is the locus ygjT, discovered independently in a genetic screen for genes induced under extreme alkaline conditions and thus denoted alx (Bingham et al. 1990). Subsequent studies confirmed that the expression of alx increases in response to high pH and identified alx as ygjT (Stancik et al. 2002; Hayes et al. 2006). Here we show that sraF is part of the 5′ untranslated region (UTR) of alx mRNA. Moreover, we show that this region serves as a pH-responsive RNA element (PRE) that brings about alx expression under alkaline conditions. The folding of PRE into a translationally active structure to produce Alx is kinetically controlled by RNA polymerase pausing in a pH-dependent manner.
Results
alx translation is increased in response to alkaline conditions
The proximity of SraF and alx prompted us to examine whether their expression is linked. By the use of sraF-alx′-lacZ transcription fusions with and without the SraF promoter, we found this promoter to direct the transcription of alx. In its absence, only background expression of lacZ could be detected, indicating that SraF is the 5′ UTR of alx mRNA (Table 1A, LB, lines 1,2).
Table 1.
alx expression regulation in response to alkaline conditions
Results are displayed as mean ± standard deviation.
aAll transcription fusions were transferred as a single copy onto the chromosome.
bThe 5′ UTR of alx is here denoted as PRE.
Previous studies showed that the expression of alx increased in response to alkali (Bingham et al. 1990; Stancik et al. 2002; Hayes et al. 2006). We examined whether this increase in alx expression is mediated by its 5′ UTR by comparing transcription and translation fusions under normal (LB medium at pH 6.8) and extreme alkaline (LBK medium at pH 8.4) conditions (Supplemental Material). We found that high pH led to a twofold to threefold increase in the levels of alx transcription (Table 1A, line 1). A similar increase of twofold to threefold was observed in fusions carrying the alx promoter (Palx) only, indicating that the increase in transcription fusion levels was due to an increase in promoter activity (Table 1A, line 3). In contrast, the translation fusion was induced sevenfold to eightfold in response to high pH stress (Table 1B, line 1). Moreover, in the absence of the 5′ UTR, transcription fusion levels remained unchanged (Table 1A, cf. lines 1 and 4), whereas alx translation levels were high and unresponsive to alkaline conditions (Table 1B, line 2). Taken together, the data suggest that the alx 5′ UTR serves as a PRE RNA that mediates the translation control of alx, leading to its induction under conditions of high pH.
Conserved sequence and structural elements in PRE
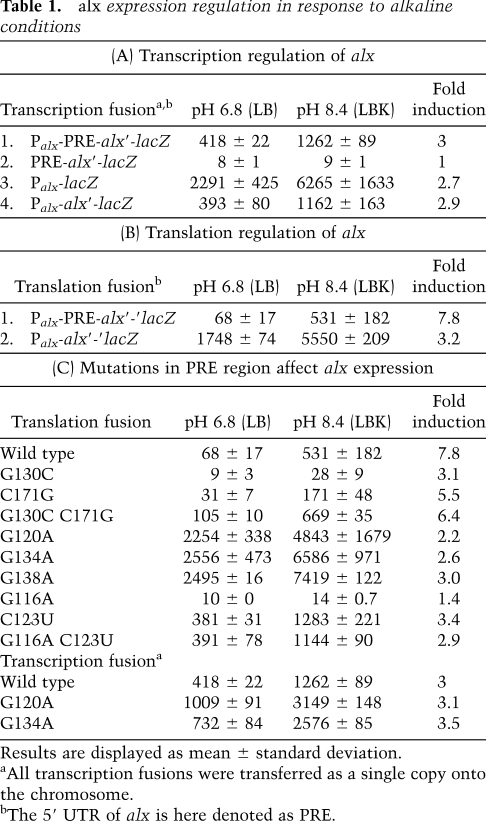
Various computer folding programs predict four stem–loop domains (A–D) in the 5′ UTR of alx mRNA (PRE) of E. coli (Hofacker 2003). By sequence gazing, we observed that 9 nucleotides (nt) in loop C were complementary to loop D (Fig. 1A). In addition, we noticed that PRE RNA harbors a putative loop E motif that overlaps the ribosome-binding site. This asymmetric motif belongs to the sarcin/ricin type of motifs. The motif is characterized by a consecutive row of non-Watson-Crick pairs and was identified in a number of RNA species, such as 5S RNA, 23S rRNA, 28S rRNA, Hairpin ribozyme, and more (Leontis and Westhof 1998, 2003; Leontis et al. 2002). Sequence comparison using BLAST of the PRE region to complete bacterial genomes revealed statistically significant alignment scores with enteric bacteria such as E. coli and species of Shigella, Salmonella, Serratia, Photorhabdus, Klebsiella, Erwinia, Sodalis, and Yersinia. By sequence and structural alignment, we found that the E. coli PRE secondary structure, the C/D complementary domains, and the loop E motif were preserved in these RNA orthologs (see Supplemental Figs. S1A, S2A). Although the loop E motif holding the 5′ and 3′ ends of the transcript is conserved in most sequences, it deviates from its consensus at the 3′ end in nine sequences.
Figure 1.
Proposed PRE-alx RNA conformations. Primary (A) and alternative (B) structures predicted to form under neutral pH (structure N) and high pH (structure H) conditions, respectively. The Shine-Dalgarno and the initiation codon of alx are in red. Complementary sequences in hairpins C and D are in blue. Arrows indicate specific nucleotide changes generated randomly or by site-directed mutagenesis. Brackets indicate multiple mutations or deletions. To form structure H, the lower part of hairpin C folds into the small hairpin S, while base-pairing between loops C and D forms hairpin C/D. The loop E motif in structure N is boxed (dashed line). The probing data obtained with the G116A mutant are displayed on structure N (A), while G134A data are shown on structure H (B). Circles indicate strong (filled circles) and weak (open circles) DMS modification sites. Triangles indicate strong (filled) and weak (open) nuclease T1 cleavage sites. A schematic representation of the genetic elements of PRE-alx RNA is shown at the bottom of the figure. Indicated are the promoter of alx (Palx); four hairpins of structure N, including two rho-independent transcription terminators at hairpins B and D (TB and TD, respectively); the Shine Dalgarno (SD); and the initiation codon of alx. Pausing sites identified and mapped in this study are indicated as PC and PD. The PRE region is in purple. A subset of PRE-alx transcripts terminates at the transcription termination signals at hairpins B and D (see also Supplemental Fig. S8). Arrows indicate stable RNA species. The two shorter species represent the previously characterized SraF.
Base-pairing between loops C and D facilitates translation
To examine the function of the putative interaction between loops C and D (C/D), we modified loops C and D to carry the corresponding complementary mutations that, when combined, would restore the formation of this putative duplex (mutations G130C and C171G) (Fig. 1A). LacZ assays of translation fusions of these mutants show that disrupting the possible interaction between loops C and D decreases both alx basal translation levels and the induction in response to high pH. Translation control is restored when these loops of PRE carry both mutations, indicating that base-pairing between these loops is essential for alx translation (Table 1C).
Mutations in PRE distinguish a translationally active structure from an inactive one
The occurrence of complementary domains within PRE strongly implies that this RNA could form an alternative structure, while the mutational data suggest that base-pairing between these domains results in formation of a translationally active structure. Taking into account computer prediction, experimental data, and structural considerations, we propose that PRE forms the alternative structure depicted in Figure 1B (for sequence alignments and conserved elements of the alternative structure, see also Supplemental Figs. S1B, S2B).
To unravel the changes in the primary structure of PRE that facilitate formation of the translationally active structure, we induced random mutations at this locus using Palx-PRE-alx′-′lacZ translation fusion plasmid and screened for nonregulated high-level expression mutants. We found these mutants to carry mutations disrupting the stem of hairpin C of PRE (G120A, G134A, G138A) (Fig. 1A). Functional studies of these translation fusions revealed a dramatic increase (35-fold to 40-fold) in alx basal level expression (Table 1C, LB). In contrast, the basal levels of the transcription fusions of these mutants increased by 1.5-fold to 2.5-fold compared with wild type, indicating that the increase in expression is due to an increase in translation. Together, the data show that PRE primary structure (denoted structure N for neutral pH) is translationally inactive and that its disruption leads to the formation of an alternative, translationally active structure (denoted structure H for high pH). Furthermore, the pH-dependent induction of these translation fusions was similar to that observed with their transcription fusions, indicating that the destabilization of hairpin C eliminated the response to alkaline conditions.
Formation of the main C/D hairpin in structure H is prompted by interference with the formation of hairpin C and requires that one strand of stem C is liberated to create a new structure (hairpin S) (see Fig. 1B). This small hairpin structure is stabilized by a terminal GNRA tetraloop that is closed by a non-Watson-Crick pair. To verify the existence of hairpin S and to investigate its function, we examined the phenotypes of strains carrying the G116A mutation and/or its corresponding compensatory mutation, C123U. The G116A mutation is predicted to have no effect on the stability of structure N, but is predicted to destabilize hairpin S (Fig. 1). LacZ assays demonstrate that the G116A mutant is totally inactive in translation (Table 1C). C123U on the other hand, slightly decreases the stability of structure N, replacing a C-G pair with U-G in stem C, hence increasing the translation of alx. The double mutation G116A C123U restores base-pairing and enables the formation of hairpin S of structure H. Functional assays of the G116A C123U mutant demonstrate that the combination of these two enables expression of alx at similar levels as those of C123U mutant alone, indicating that formation of hairpin S is important for the creation of the alternative active structure H.
Structure probing of PRE-alx RNA corroborates the existence of two conformations
LacZ assays of the low- and high-level expression mutants suggested that their RNAs each form one distinct structure; i.e., the translationally inactive structure (structure N) or the active one (structure H). Thus, we probed the structures of mutants G134A and G116A that are likely to maintain structures H and N, respectively, and compared them with wild type. We determined the secondary structures of these RNAs under conditions of neutral pH using dimethyl sulfate (DMS), which methylates adenosine and cytidine residues at N1 and N3 positions, respectively, and by subjecting the RNAs to partial cleavage by RNase T1 that is specific for single-stranded guanosine residues. The modified nucleotides and the cleavage sites were mapped by primer extension. The results displayed in Figure 2A and summarized in Figure 1, A and B, show that the positions of DMS modifications at the 5′-proximal region of these molecules are in agreement with the predicted hairpins A and B. Also, as expected, the loops of these structures were accessible to T1 cleavage. Downstream from hairpins A and B, DMS modifications of G116A and G134A RNAs are in agreement with the predictions carried out for these regions for structures N and H, respectively. It is worth noticing the differences in the positions and/or the strength of the modifications that are indicative of one structure or the other. For example, in the G116A mutant, the modification rates of residues A131 and A132, and C171 and C172, located in loops C and D of structure N, respectively, are higher than in the G134A mutant, where these nucleotides base-pair and form hairpin C/D. Conversely, in G134A RNA, residues A147–A153, located in loop C/D, and A137 and A164 of the interior loop of this stem are much more accessible to DMS modification than in mutant G116A. Similarly, loop S residues A119 and A121 are more accessible in G134A RNA, while nucleotides A114 and A115, positioned in the stem of this hairpin (in H), are inaccessible. These nucleotides are unpaired in G116A RNA, probably because most of its RNA is in structure N. Comparing the intensities of the modifications in wild type with those of the mutants indicates that G134A and G116A RNAs form, respectively, almost exclusively, structures H and N, while wild-type RNA is found in both conformations, predominantly in structure N. In vivo structure probing of wild-type, G134A, and G116A RNAs carried out under normal growth conditions further substantiated the in vitro probing data (Supplemental Fig. S3).
Figure 2.
Wild-type RNA forms two conformations, of which one is translationally active. (A) Structure probing using DMS of PRE-alx′ wild-type, G116A, and G134A RNAs synthesized in vitro with T7 RNA polymerase from PT7-PRE-alx′. Modification was carried out at pH 7.4. Reverse transcription (primer 1048) of untreated (−) and DMS-treated (+) RNA samples. The numbers on the right indicate sequence positions relative to the transcription start site. Arrows indicate increased (up arrows) and decreased (down arrows) modifications at G134A RNA compared with G116A and wild-type RNAs. Arrows of G116A RNA indicate increased or decreased modifications compared with G134A mutant and wild type. (In. loop) Interior loop. Nucleotides A119 and A121 are paired in structure N, located at the base of hairpin C (Paired). The summary of the structural mapping is presented in Figure 1. (B) 30S ribosomal subunit binding to PRE-alx′ wild-type, G134A, and G116A RNAs (toeprinting). Where indicated, RNA (as above) was incubated with fMet–tRNA and 30S subunits for the indicated times and then subjected to reverse transcription for another 10 min. The arrow indicates the toeprint termination site found 17 nt downstream from the AUG start codon. (C) Native gel analysis. (Top panel) RNA as above was separated on nondenaturing polyacrylamide gels and transferred to nylon membranes by electroblotting. To avoid detection of the short terminated transcripts, the membranes were probed with end-labeled primer complementary to sequences of alx (512). Arrows indicate the two conformations observed. (Bottom panel) To verify the integrity of this RNA, the samples were analyzed on denaturing 6% polyacrylamide and 7.8 M urea gel.
To visualize the different conformers and learn about their ratios in wild type and mutants G134A and G116A, we examined these RNAs on nondenaturing polyacrylamide gels. These native gels further demonstrated that G134A RNA is found almost exclusively in one structure, while the majority of wild-type and G116A RNAs are found in the other conformation (Fig. 2C). We also learned from these assays that a small fraction of G116A RNA forms a number of other conformations, of which a few appear as faint distinct bands and others appear as a smear (data not shown).
Differential ribosome binding of structures N and H
The functional and structural data indicated that structure N is translationally inactive, while structure H is active. We examined the accessibility of these structures to ribosomes in vitro by toeprinting assays. Incubation of in vitro synthesized RNA with 30S ribosome subunits and uncharged fMet–tRNA results in an mRNA–30S initiation complex, which blocks the elongation by reverse transcriptase in a primer extension assay. From this assay, we learned that G134A RNA was bound by 30S ribosome subunits, forming a translation initiation complex. The G116A mutant showed no binding at all, while wild type showed very little ribosome binding (Fig. 2B). These data indicate that the ribosome-binding site of alx is occluded in structure N and exposed in H, and that wild-type RNA is found mainly in the inactive structure N.
In vivo probing demonstrates that structure H is formed in response to high pH
The data indicating that wild-type alx mRNA is found mainly in the translationally inactive state has led us to suggest that alx expression increases in response to alkali through regulation of the ratio between the two structures. To examine changes in the ratio of the structures (inactive to active) in vivo, cells carrying plasmids expressing wild-type PRE-alx mRNA were exposed to DMS under normal growth and alkaline conditions. We discovered that, under alkaline conditions, the ratio of N to H changes in favor of structure H; thus, a greater fraction of wild-type RNA exists in the translationally active structure (Fig. 3A). No changes in the structure were detected in an unrelated RNA probed under the same conditions (Supplemental Fig. S9). Taken together, our data show that, under normal growth conditions, wild-type alx mRNA exists mainly in structure N, in which the alx ribosome-binding site is occluded. In response to alkali, the ratio of inactive to active (N to H) changes in favor of the translationally active structure H. In this structure, the alx ribosome-binding site is exposed, leading to its translation.
Figure 3.
Structure probing of wild-type RNA synthesized in vivo (A) or incubated after its synthesis in vitro (B) under neutral and high pH conditions. (A) Cells containing a plasmid carrying the locus Palx-PRE-alx′ were grown in LB (pH 6.8) or LBK (pH 8.4) medium and treated with DMS. Reverse transcription (primer 1048) of DMS-modified (+) and unmodified (−) RNA. The numbers on the right indicate sequence positions relative to the transcription start site. Arrows indicate increased (up arrows) and decreased (down arrows) modifications of wild-type RNA extracted from cells grown in high pH (LBK) compared with RNA from cells grown in neutral pH (LB). (In. loop) Interior loop. Fold increase of A137 and A152 intensity (indicative of the active structure H) in LBK versus LB is 3.8 and 3.0, respectively. Fold decrease of A131 and A155 intensity (indicative of the inactive structure N) in LBK versus LB is 2.7 and 2.4, respectively. (B) In vitro structure probing of wild-type RNA incubated at neutral or high pH buffer. RNA synthesized in vitro using T7 RNA polymerase was probed at 37°C under the indicated pH conditions (left) or subjected to heat denaturation (70°C) followed by probing in the indicated pH buffers at 37°C (right). Arrows indicate decreased modifications typical of structure H to structure N transition. Reverse transcription was done as above.
Structure H folds during transcription in response to high pH
The change in N to H ratio in response to high pH could suggest that wild-type RNA molecules can be converted from one structure to the other, or that structure H is formed under conditions of high pH during its synthesis. To examine whether the shift in N to H ratio occurs by refolding, in vitro synthesized wild-type RNA was incubated in high-pH buffer to allow refolding. Structure probing of this RNA showed no significant changes in the modification pattern (Fig. 3B, left panel). Subjecting the RNA to heat denaturation prior to its incubation at different pH conditions revealed a decrease in the modification of residues A151–A153 and A137, positioned in the stems of hairpins C and D, respectively, in structure N. This indicates that, upon heat denaturation, a transition from H to N takes place, suggesting that structure N is more stable and that refolding does not lead to formation of structure H (Fig. 3B, right panel).
To examine whether the shift in the N to H ratio detected in vivo occurs while transcription is in progress, the structure of wild-type RNA synthesized in vitro under different pH conditions by E. coli RNA polymerase was determined during its synthesis. Structure probing using DMS revealed that structure H forms when the RNA is being synthesized under conditions of high pH (Fig. 4A). With the increase in pH, the loop nucleotides of hairpin S (A119) and hairpin C/D (A147 and A149–A153), which are indicative of structure H, become more accessible to DMS modification. Likewise, nucleotides A137 and A164 of the interior loop of hairpin C/D become unpaired. In contrast, analysis of mutant G116A, which locks the RNA in structure N, shows that this RNA remains mostly in structure N.
Figure 4.
Cotranscriptional structure probing. (A) RNA (Palx-PRE-alx′) was transcribed in vitro by E. coli RNA polymerase for 5 min prior to the addition of DMS. Reverse transcription (primer 1048) of DMS-modified (+) and unmodified (−) RNA. The numbers on the right indicate sequence positions relative to the transcription start site. (In. loop) Interior loop. Ratios of intensities of A137 (indicative of the active structure H) to A131 (indicative of the inactive structure N) at pH 7.1, 7.8, and 8.1 were 10.5, 29.5, and 43 (wild-type RNA) and 0.9, 1.5, and 2 (G116A RNA), respectively. (B) Structure probing was done as above, except that, where indicated, NusA protein was added to the preincubation mix. (C) The same as above, except that, where indicated, Mg2+ (10 mM) was substituted for Mn2+ (2 mM). Ratios of intensities of A137 (indicative of the active structure) to A131 (indicative of the inactive structure) at pH 7.1 and 7.8 were 1 and 1.9, respectively.
Formation of the active structure involves transcriptional pausing
RNA folding during transcription elongation is influenced by properties such as RNA polymerase transcription speed and/or pausing (Landick 2006). The location and/or duration of pausing have been shown to alter folding of a newly transcribed RNA (Pan et al. 1999; Wickiser et al. 2005). As opposed to E. coli RNA polymerase, T7 RNA polymerase exhibits less pausing during elongation and its transcription speed is greater than that of E. coli (Sousa and Mukherjee 2003). Intriguingly, we found that the alkaline-dependent shift in the N to H ratio cannot be detected when T7 RNA polymerase is used instead of E. coli (Supplemental Fig. S4). Moreover, using T7 RNA polymerase, the ratio N to H leans toward structure N, as is evident from nucleotides such as A131–A132 and A137 that are indicative of structures N and H, respectively.
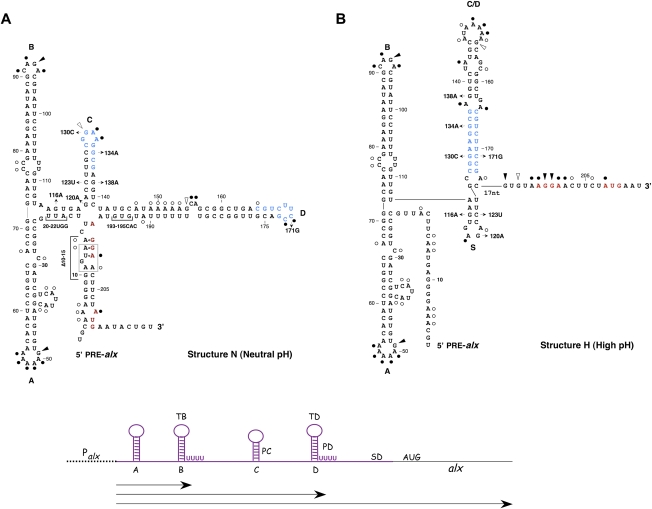
To learn whether transcription elongation of alx mRNA by E. coli RNA polymerase is indeed interrupted by pauses, we monitored synchronized, single-round in vitro transcription of wild-type RNA under neutral and high pH conditions. Two striking pause sites, denoted PC and PD, were identified and mapped to hairpins C (G138) and D (C182), respectively (Fig. 5A; Supplemental Fig. S5). The longevity of these incomplete transcripts is increased under alkaline conditions. The location of the pause sites prior to the completion of hairpins C and D strongly indicates that both concur with the formation of helix C/D and the prevention of hairpins C and D. By pausing at PC, RNA polymerase allows for formation of hairpin S, while pausing at PD indirectly promotes base-pairing between the complementary sequences in hairpins C and D (see the model in Fig. 6).
Figure 5.
Identification of transcriptional intermediates. (A) Synchronized single-round in vitro transcription of wild-type RNA (Palx-PRE-alx′) carried out using E. coli RNA polymerase under neutral and alkaline conditions (two left panels) or at neutral pH in the presence of NusA (right panel). After the formation of initiation complexes, transcription was allowed to proceed for the indicated times (in seconds) by adding all four unlabeled NTPs in excess and rifampicin. TB and TD indicate RNA products generated by the rho-independent transcription terminators at hairpins B and D, respectively. PC and PD indicate pausing sites at hairpins C (G138) and D (C182), respectively. Under neutral and high pH conditions, pause site C is present at more than half the maximal intensity at 4–6 and 2–8 sec of chase, respectively, while pause D is present at 8–10 and 4–8 sec, respectively. In the presence of NusA, pause sites C and D are present at more than half the maximal intensity at 6–10 and 8–12 sec of chase, respectively. (B) The same as above except that Mn2+ (2 mM) was used instead of Mg2+ (10 mM). Mn2+ eliminates pause D, delays and shortens pause C, and enhances pausing at a new site (open circle).
Figure 6.
A model for PRE-alx regulation by alkaline conditions. Under conditions of normal growth, PRE-alx forms a translationally inactive structure (structure N), whereas under extreme alkaline conditions when folding is kinetically controlled by RNA polymerase pausing, the newly synthesized RNA adopts an active structure (structure H) in which the ribosome-binding site (in red) is exposed for 30S ribosomal subunit binding. Formation of the active structure involves pausing of RNA polymerase at two sites located at hairpin C (PC) and hairpin D (PD). Upon pausing of RNA polymerase (shaded in light blue) at PC, the formation of hairpin C is prevented, and instead formation of hairpin S (indicated by a purple line) is promoted. Pausing at PD inhibits the formation of hairpin D, allowing base-pairing between the complementary sequences in hairpins C and D (in blue) and formation of the apical loop of hairpin C/D (in green).
The transcription elongation factor NusA is known to stabilize paused transcription complexes (Artsimovitch and Landick 2000). To test whether prolonged pausing is sufficient to alter the ratio of the structures, inactive to active, we examined whether NusA protein can replace the signal exerted by high pH. Structure probing of wild-type RNA synthesized by E. coli RNA polymerase under neutral pH conditions in the presence of NusA protein shows that NusA stimulates formation of the active structure (structure H), reproducing the effects of high pH under neutral conditions (Fig. 4B). These results indicate that alkaline conditions affect RNA polymerase pausing and confirm that pausing is an important step in the formation of the alternative structure H. In accordance with these results, single-round in vitro transcription of wild-type RNA carried out under neutral conditions in the presence of NusA demonstrates that NusA protein prolongs pausing at both PC and PD
Compared with Mg2+, Mn2+ is considered to have relaxed coordination requirements within RNA polymerase, thereby altering the characteristics of the active site of the enzyme. Mn2+ has been shown to promote transcription, possibly by suppressing pausing and/or by increasing bond formation rate (Kireeva and Kashlev 2009). We found that substituting Mg2+ for Mn2+ reduces the effects of high pH and the shift in the ratio of inactive to active is diminished, further confirming that alkaline conditions promote formation of the active structure through an effect on pausing longevity (Fig. 4C). Single-round in vitro transcription of wild-type RNA carried out under high pH conditions in the presence of Mn2+ reveals that Mn2+ shortens and delays the appearance of PC and abrogates pausing at PD (Fig. 5A,B). Notably, Mn2+ also enhances pausing at a new site that precedes PC.
Discussion
A limited number of RNA regulators have been characterized so far, and yet the diversity of their physiological roles and mechanisms of action suggests that these likely represent only a fraction of the contribution that RNA makes to gene regulation systems. Here we describe the first example of a pH-responsive RNA regulator. Moreover, this previously uncharacterized riboregulator exhibits a novel regulatory mechanism whereby pH influences RNA folding dynamics. Specifically, we show that the alx transcript forms a translationally inactive structure under neutral conditions, but when synthesized under high pH conditions, a translationally active structure is formed. Single point mutations freeze the RNA in one of the two structures, distinguishing the active conformation from the inactive one. In vitro transcription assays demonstrate that the active structure forms when transcription progresses under high pH conditions and involves pausing of RNA polymerase at two distinct sites. These assays also show that alkaline conditions prolong transcription pausing, extending the lifetimes of transcriptional intermediates formed at PC and PD. NusA protein, known to stabilize paused transcription complexes, prolongs pausing at both PC and PD, stimulating formation of the active structure under neutral conditions, thereby reproducing the effects of high pH. Conversely, the substitution of Mg2+ for Mn2+, known to affect the active site of RNA polymerase, inhibits the effects of high pH by changing the pattern of pausing at the PRE, and the shift in the ratio of inactive to active structure is diminished.
Over the years, several studies aimed at deciphering regulation of gene expression have discovered mechanisms that involve formation of alternative RNA structures. One classic example is the CIII protein of phage λ, which plays a role in the lysis–lysogeny decision of this phage. Studies of cIII expression regulation carried out almost two decades ago showed that the translation rate of the λ cIII gene is determined by two alternative mRNA structures that exist in equilibrium (Altuvia et al. 1989, 1991). The equilibrium between the structures shifts in a temperature-dependent manner. High temperatures (45°C) favor the energetically more stable structure in which the ribosome-binding site is occluded, while at 37°C the ratio shifts toward the active structure and CIII protein is produced. Similarly, the E. coli rpsO mRNA encoding ribosomal protein S15 exists in two conformations in equilibrium. The protein S15 controls its own levels by repressing its own translation. The conformation containing a pseudoknot is bound by 30S ribosomal subunits. In the absence of S15, this pseudoknot is rapidly unwound, leading to the formation of a productive initiation complex and rpsO translation. When S15 is in excess, it stabilizes the pseudoknot mRNA, trapping the ribosome in a preinitiation stage (Marzi et al. 2007).
While cIII and rpsO exhibit a dynamic equilibrium between two RNA structures, active versus inactive, some RNAs require de novo synthesis to form an alternative conformation. Indeed, cumulative data indicate that much of the regulation of gene expression in both prokaryotes and eukaryotes is accomplished during the elongation phase of transcription and involves pausing of RNA polymerase. Transcriptional pausing functions by halting the transcribing enzyme at positions or for times required for regulatory interactions. In some cases, it allows interaction with, or recruitment of, regulators. In other cases, pausing allows proper folding of the RNA (Landick 2006). The elementary steps of base-pairing are fast (microseconds) compared with transcription rates (milliseconds); however, folding into native states can be very slow, compared with transcription rates, with marked differences between in vitro and in vivo. Although base-pairing can initiate as soon as the emerging RNA strand reaches a sufficient length to allow folding, the formed base-pairing scheme does not necessarily correspond to the native folded state (Cruz and Westhof 2009). Therefore, sequence-specific pausing and the elongation speed of RNA polymerase can influence greatly the dynamics of folding. Indeed, the end result of stepwise folding is often entirely different from that displayed when a full transcript folds as a whole (as observed in most in vitro experiments). For example, pausing was shown to assist in the correct folding of certain E. coli noncoding RNAs, including SRP, RNase P, and tmRNA, specifically, by enabling temporary sequestration of nonnative helices (Wong et al. 2007).
The pH-dependent folding of PRE-alx RNA into an active structure requires de novo synthesis, during which RNA polymerase plays an active role in controlling which structure forms. When in vitro synthesized and heat-denatured PRE-alx RNA is incubated in high-pH buffer to allow refolding, only transition from the less stable active structure to the more stable inactive one can be detected, but not the reverse transition. The formation of the active structure occurs while transcription progresses under high pH conditions and involves pausing of the elongating complex at two distinct sites. We mapped these pausing sites to hairpin C and hairpin D. An intriguing question is how pausing at these sites aids in formation of the active structure. Biochemical studies indicate that, typically, 12 ± 1 nt of the nascent RNA are sequestered within the elongation transcription complex (Monforte et al. 1990). We suggest that pausing at nucleotide G138 results in sequestering of the complementary strand required to form the stem of hairpin C (see region G138–C127) (Fig. 6). Thus, the free transcript preceding position C127 is capable of forming hairpin S. Likewise, the pausing at C182 sequesters within RNA polymerase sequences required for the formation of hairpin D, but leaves exposed sequences complementary to loop C that consequently are available to anneal and form hairpin C/D. Interestingly, both mutants, G116A and G134A, exhibit differences in the pattern of pausing compared with wild-type; both G116A and G134A show weaker pausing at PD, mainly at high pH; G134A RNA also shows no pausing at PC (Supplemental Fig. S6). It is possible that, in addition to its effect on the stability of the inactive structure, the mutation G134A affects the specificity of the first pause site, causing RNA polymerase to bypass the site without pausing.
Another intriguing issue concerns how high pH affects the efficiency of pausing. Pause duration has been shown to be influenced by various parameters (Landick 2006), including the stability of the RNA–DNA hybrid (Palangat et al. 1998), interaction with regulatory proteins (Artsimovitch and Landick 2002), a nascent RNA hairpin positioned the right distance from an elemental pause site (Artsimovitch and Landick 2000), and dsDNA sequences that contact RNA polymerase downstream from the active site (Palangat et al. 2004). Alkaline conditions could affect the stability of the nucleic acid scaffold or RNA polymerase itself, and thus incur longer-lived pausing. Our results, showing that NusA protein stimulates formation of the active structure and reproducing the effects of high pH under neutral conditions, suggest that alkaline conditions, by affecting RNA polymerase, prolong transcriptional pausing, and thereby facilitate folding of the newly synthesized RNA into a translationally active structure. However, single-round in vitro transcription of an unrelated RNA shows that not all pause sites are affected by high pH (Supplemental Fig. S10). Because several mechanisms of pausing have been observed, it is plausible that a subset of pause sites that use a certain mechanism is sensitive to the effects of high pH, while others are not. Thus, it appears that high pH affects the interaction between RNA polymerase and pause sites only in a specific context. The mechanistic details of how high pH affects the longevity of pause complexes require elucidation.
The sequence and structural features of PRE-alx are highly conserved (Supplemental Figs. S1, S2). Sequence alignments performed on both structures, inactive (N) and active (H), reveal clear conservations and conspicuous variations among homologs. The most variable regions common to both structures are found at the 5′ and 3′ ends of the PRE, the apical region of hairpin A, and a major part of hairpin B. The rearrangement that reflects active versus inactive RNA structure involves mainly the four-way junction of structure N, which reorganizes into two consecutive hairpins in structure H. Notably, the basal region of hairpin D in structure N is rather variable and forms the apical part of hairpin C/D in structure H, while the very conserved hairpins C and D in structure N form the basal helix of hairpin C/D in structure H. Thus, in the active structure H, all of the conserved elements are in the core of the structure. Although the loop E motif holding the 5′ and 3′ ends of the transcript is conserved in most homologs, it deviates from consensus at the 3′ end in nine homologs. The observed pause sites (G138 and C182) are located at the 3′ ends of each of the conserved elements forming hairpins C and D in structure N and the basal helix C/D in structure H. Thus, those conserved segments are still occluded by the polymerase during pausing and cannot fold back on themselves and form the C and D hairpins as in structure N.
Barrick et al. (2004) presented previously an alignment of an element called yybP/ykoY, which is part of the RNA studied here. The alignment concerns only hairpins A and B. The domains on which we focus here, hairpins C and D, are neither aligned nor represented in their two-dimensional diagrams. However, an important point emerges: Gram-positive and Gram-negative bacteria present two distinct sets of aligned sequences. The sequences of the Bacillus family do not show evidence for the existence of hairpins C and D, while these occur in enteric bacteria; instead, they are mainly restricted to helices A and B. Furthermore, in the Bacillus family, the helix preceding the loop E-like motif seems to be an anti-terminator helix, since its 3′ strand can pair with a segment preceding a U-stretch, thereby forming a terminator helix. In enteric bacteria, however, new structured modules appeared that control ygjT expression. It is intriguing that the modules present in Gram-positive bacteria are predicted to control by transcription termination/anti-termination, while those in Gram-negative control translation, as often found for riboswitches in these groups.
The translationally active structure H has been shown genetically and biochemically to be bound efficiently by ribosomes. Detailed structure-probing data of the ribosome-binding region of wild-type RNA suggest that it is unavailable to ribosomes either because it is engaged in noncanonical base-pairing or because of steric hindrance (Supplemental Fig. S7). We examined whether the ribosome-binding site becomes available to ribosomes when unpaired by deleting the sequence opposite the ribosome-binding site (see Δ10–15 in Fig. 1A). LacZ assays and in vitro 30S ribosome binding show that although the ribosome-binding site of the Δ10–15 mutant is accessible to a short complementary oligonucleotide, as demonstrated in an RNase H-probing assay, it is unavailable to ribosomes, possibly because of steric hindrance. Indeed, site-directed mutagenesis to prevent base-pairing at the junction of the structure (AUU20–22UGG; GUG193–195CAC), rendered this mutant translationally active (Fig. 1A; Supplemental Table S3A). Together, these results indicate that structure N is translationally inactive also because of structural hindrance.
In addition to the structural rearrangement affecting translation, the formation of the active structure results in elimination of the second transcription terminator in hairpin D, which could account for some increase in alx expression in response to alkali. However, transcription fusions of mutants (e.g., G120A and G134A) in which the PRE-alx RNA forms mainly structure H exhibit only a modest increase (twofold) in LacZ activity, compared with wild-type PRE-alx. In contrast, translation fusions of these mutants display a dramatic increase (>30-fold) in LacZ activity. Therefore, the contribution of anti-termination to alx expression appears marginal. Also, Northern blot analysis of alx UTR short RNA species produced due to transcription termination at hairpins B and D shows that, under alkaline conditions, transcription termination at hairpin D does not decrease, excluding the possibility that regulation is achieved through transcription attenuation (Supplemental Fig. S8). Furthermore, transcription and translation fusions of Palx-PRE-alx′-lacZ carrying a mutation in the promoter of alx (A-68T) that prevents this promoter from being induced in response to alkali reveal that, although the transcription fusion shows no increase in expression in response to the stress, translation of alx′-′lacZ increases fourfold in response to alkali (Supplemental Table S3B,C). These results further confirm that high pH controls alx translation.
In vivo, the increase in alx expression in response to high pH is somewhat moderate. Several mechanistic aspects could account for this low increase in alx levels. It has been reported that only a fraction of transcribing RNA polymerase molecules are affected by a given pause signal; a subset of molecules bypasses the site without pausing (Landick 2006). In line with this, our in vivo structure-probing data as well as the functional data indicate that, under alkaline conditions, only a fraction of alx molecules form the active structure. In addition, the duration and the extent of pH change within the bacterial intracellular environment could account for the moderate change in the ratio of inactive to active observed under high pH. Bacterial cells encounter alkaline conditions in their external environment (e.g., alkaline soil) or within a host (e.g., mucosal tissue of the small intestines) (Padan et al. 2005). Studies of intracellular responses show that neutrophilic bacteria can maintain their steady-state cytoplasmic pH in a narrow range of 7.4–7.8 over an external pH range of 5.5–9. However, shifting cells from neutral to alkaline pH leads to rapid loss of this homeostasis, the rebuilding of which can take up to 30 min, while the cells acidify their cytoplasm. Under some circumstances, the new steady-state pH stabilizes at 8.1 and still allows growth (Zilberstein et al. 1984). Therefore, although extracellular pH is not a reliable indicator of the pH environment surrounding alx transcripts, it appears likely that induction of alx could occur before re-establishment of homeostasis. Finally, it remains to be seen whether, in vivo, the assembly of PRE-alx RNA into a functional structure is regulated also by the action of a host factor—either a repressor or an activator.
Prokaryotes deploy a variety of mechanisms to cope with alkalinization of their cytoplasm, including increased acid production through the use of amino acid deaminases and sugar fermentation, changes in the properties of the cell surface, and increased activity of transporters, such as monovalent cation/proton anti-porters (Padan et al. 2005). Considering that the pair PRE-alx is so remarkably conserved among enteric bacteria and that Alx itself shows sequence homology with membrane proteins with transport activity, it is intriguing how, under unique environmental niches, the function of this kinetically regulated pH sensor is exploited.
Materials and methods
In vitro structure probing of RNA generated by T7 RNA polymerase
RNA synthesized in vitro (1.75 pmol) using plasmid PT7-PRE-alx′ (pSA66; wild type and mutants) as template was preincubated for 15 min at 37°C in 14.5 μL of 50 mM Na-cacodylate (pH 7.4), 200 mM NaCl, and 10 mM MgCl2. Then, 0.5 μL of DMS (diluted 1:10 in ethanol) was added, and the reaction was allowed to proceed for 5 min. Reactions were stopped with phenol/chloroform and precipitated with ethanol in the presence of 0.3 M sodium acetate, 1 μL of Quick-Precip (Edge BioSystems), and 20 μg of yeast RNA. To follow possible refolding, the RNA was denatured at 70°C in water, incubated for 15 min at 37°C in the appropriate buffers (pH 7.2, 7.8, and 8.2), and probed as above. RNase T1 digestion of RNA samples (0.7 pmol) was carried out for 4 min at 37°C (after 15 min of preincubation) in 10-μL reactions containing 20 mM Tris-HCl (pH 7.2), 100 mM NaCl, 10 mM MgCl2, 10 μg of yeast RNA, and 0.05 U or 0.5 U of RNase T1 (Boehringer Mannheim). The reactions were stopped and treated as described above. To detect the modified and cleaved sites, the treated RNA (0.4 pmol and 0.1 pmol, respectively) was annealed with 0.6 pmol end-labeled primer (5 min at 70°C, followed by incubation for 5–10 min on ice), and then subjected to primer extension (60 min at 42°C) in 15-μL reactions containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, dNTPs (500 μM each), and 25 U MMLV reverse transcriptase (Promega). The extension products were separated on 6% polyacrylamide and 7.8 M urea gels, alongside sequencing reactions.
Cotranscriptional in vitro structure probing
RNA was synthesized using E. coli RNA polymerase (1 U; USB or Epicentre) at pH 7.1, 7.8, and 8.1 in 50-μL reactions containing 20 mM Tris-HCl, 150 mM KCl, 10 mM MgCl2, 5 mM DTT, NTPs (250 μM each), 20 U RNase inhibitor (CHIMERx), and 2.5 μg of plasmid DNA carrying Palx-PRE-alx′ (pSA64; wild-type and G116A mutant). The mixture was preincubated for 5 min at 37°C prior to the addition of the enzyme. After 5 min of synthesis at 37°C, 1.7 μL of DMS (diluted 1:10 in ethanol) was added. The reactions were stopped 5 min after the addition of DMS by phenol/chloroform extraction followed by ethanol precipitation in the presence of 0.3 M sodium acetate and 1 μL of Quick-Precip. To detect the modified sites, the treated RNA was annealed with 0.6 pmol end-labeled primer (5 min at 70°C, followed by incubation for 5–10 min on ice), and then subjected to primer extension (60 min at 43°C) in 15 μL reactions containing 0.5% (v/v) Tween 20, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 5 mM MgCl2, 10 mM DTT, dNTPs (1 mM each), and 25 U Expand reverse transcriptase (Roche). The extension products were separated on 6% polyacrylamide and 7.8 M urea gels, alongside sequencing reactions. Where indicated, NusA protein (250 or 500 nM final concentration) was added to the preincubation mix. To substitute Mg2+ for Mn2+, 2 mM MnCl2 was used instead of 10 mM MgCl2.
Single-round transcription
Single-round transcription reactions at pH 7.2 and 8.2 were performed as described in Landick et al. (1996) with slight modifications. To allow formation of a transcription complex followed by synthesis of the first 12 nt of PRE and the incorporation of label, 1.25 μg of plasmid DNA carrying Palx-PRE-alx′ (pSA64) was incubated for 10 min at 37°C in the presence of 10 μCi of α-32P-ATP, 20 mM Tris-HCl, 150 mM KCl, 10 mM MgCl2, 2.77 mM DTT, 1 μM ATP, 2.5 μM GTP, 20 μg/mL BSA, 150 μM GpC dinucleotide (Sigma), glycerol to 3.33% (v/v) (including glycerol of the enzyme storage buffer), and 0.5 U of E. coli RNA polymerase in a 45-μL volume. Thereafter, aliquots of 9 μL were transferred to prewarmed tubes and transcription was resumed by adding 1 μL of prewarmed solution of the appropriate pH containing NTPs (2.5 mM each), 20 mM Tris-HCl, 150 mM KCl, 10 mM MgCl2, and 150 μg/mL rifampicin (to block reinitiation). The reactions were stopped by freezing in liquid nitrogen. Samples were analyzed on 6% polyacrylamide and 7.8 M urea gels. To map pause sites, PCR-generated templates of Palx-PRE-alx′ ending at positions A137 and C182 of PRE were subjected to runoff transcription labeled as above for 10 min. Where indicated, NusA protein (500 nM final concentration) was added prior to the addition of RNA polymerase. To substitute Mg2+ for Mn2+, 2 mM MnCl2 was used instead of 10 mM MgCl2.
Acknowledgments
We are grateful to Knud Nierhaus for generously providing 30S subunits, and to Paul Roesch for NusA. This study was supported by The Israel Science Foundation, founded by The Israel Academy of Sciences and Humanities, grant number 499/06, and The Israel Science Foundation-Bikura Program, grant number 1342/05. S.A. and E.W. acknowledge financial support from BACRNAs, a Specific Targeted Research Project supported by the European Union's FP6 Life Science, Genomics and Biotechnology for Health, LSHM-CT-2005-018618. In memory of Amos Oppenheim.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.552209.
Supplemental material is available at http://www.genesdev.org.
References
- Altuvia S, Kornitzer D, Teff D, Oppenheim AB. Alternative mRNA structures of the cIII gene of bacteriophage λ determine the rate of its translation initiation. J Mol Biol. 1989;210:265–280. doi: 10.1016/0022-2836(89)90329-x. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Kornitzer D, Kobi S, Oppenheim AB. Functional and structural elements of the mRNA of the cIII gene of bacteriophage λ. J Mol Biol. 1991;218:723–733. doi: 10.1016/0022-2836(91)90261-4. [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham RJ, Hall KS, Slonczewski JL. Alkaline induction of a novel gene locus, alx, in Escherichia coli. J Bacteriol. 1990;172:2184–2186. doi: 10.1128/jb.172.4.2184-2186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocobza SE, Aharoni A. Switching the light on plant riboswitches. Trends Plant Sci. 2008;13:526–533. doi: 10.1016/j.tplants.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes & Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J Bacteriol. 2004;186:5392–5399. doi: 10.1128/JB.186.16.5392-5399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes ET, Wilks JC, Sanfilippo P, Yohannes E, Tate DP, Jones BD, Radmacher MD, BonDurant SS, Slonczewski JL. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 2006;6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan CL. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: His leader pause site as paradigm. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie. 2002;84:961–973. doi: 10.1016/s0300-9084(02)01463-3. [DOI] [PubMed] [Google Scholar]
- Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–1031. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Monforte JA, Kahn JD, Hearst JE. RNA folding during transcription by Escherichia coli RNA polymerase analyzed by RNA self-cleavage. Biochemistry. 1990;29:7882–7890. doi: 10.1021/bi00486a015. [DOI] [PubMed] [Google Scholar]
- Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: Recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol Rev. 2006;30:3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: New insights. Biochim Biophys Acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Meier TI, Keene RG, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- Palangat M, Hittinger CT, Landick R. Downstream DNA selectively affects a paused conformation of human RNA polymerase II. J Mol Biol. 2004;341:429–442. doi: 10.1016/j.jmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R, Mukherjee S. T7 RNA polymerase. Prog Nucleic Acid Res Mol Biol. 2003;73:1–41. doi: 10.1016/s0079-6603(03)01001-8. [DOI] [PubMed] [Google Scholar]
- Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Winkler ME, Yanofsky C. Pausing of RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry. 1981;20:3738–3744. doi: 10.1021/bi00516a011. [DOI] [PubMed] [Google Scholar]
- Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Yakhnin H, Babitzke P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP–RNA interaction. Mol Cell. 2006;24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Zilberstein D, Agmon V, Schuldiner S, Padan E. Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]